Abstract
Thermal proteome profiling (TPP) is an invaluable tool for functional proteomics studies that has been shown to discover changes associated with protein–ligand, protein–protein, and protein–RNA interaction dynamics along with changes in protein stability resulting from cellular signaling. The increasing number of reports employing this assay has not been met concomitantly with new approaches leading to advancements in the quality and sensitivity of the corresponding data analysis. The gap between data acquisition and data analysis tools is important to fill as TPP findings have reported subtle melt shift changes related to signaling events such as protein posttranslational modifications. In this study, we have improved the Inflect data analysis pipeline (now referred to as InflectSSP, available at https://CRAN.R-project.org/package=InflectSSP) to increase the sensitivity of detection for both large and subtle changes in the proteome as measured by TPP. Specifically, InflectSSP now has integrated statistical and bioinformatic functions to improve objective functional proteomics findings from the quantitative results obtained from TPP studies through increasing both the sensitivity and specificity of the data analysis pipeline. InflectSSP incorporates calculation of a “melt coefficient” into the pipeline with production of average melt curves for biological replicate studies to aid in identification of proteins with significant melts. To benchmark InflectSSP, we have reanalyzed two previously reported datasets to demonstrate the performance of our publicly available R-based program for TPP data analysis. We report new findings following temporal treatment of human cells with the small molecule thapsigargin that induces the unfolded protein response as a consequence of inhibition of sarcoplasmic/endoplasmic reticulum calcium ATPase 2A. InflectSSP analysis of our unfolded protein response study revealed highly reproducible and statistically significant target engagement over a time course of treatment while simultaneously providing new insights into the possible mechanisms of action of the small molecule thapsigargin.
Keywords: thermal proteome profiling, functional proteomics, InflectSSP, unfolded protein response, Thapsigargin
Graphical Abstract
Highlights
-
•
InflectSSP for computational and statistical analysis of thermal proteome profiling.
-
•
Novel integrative replicate analyses for calculation of p-values for melt shifts.
-
•
Melt shift coefficient provides new metric for TPP hit prioritization.
-
•
InflectSSP provides highly reproducible detection of temporal target engagement.
-
•
Identification of candidate downstream effectors of the inhibitor thapsigargin.
In Brief
In this work, we describe our computational workflow for statistical analysis of thermal proteome profiling data known as InflectSSP. InflectSSP has been optimized for sensitive and reproducible detection and prioritization of protein melt shift changes in biological thermal proteome profiling datasets. Analysis of induction of the unfolded protein response with the small molecule inhibitor thapsigargin using InflectSSP revealed significant melt shift changes that shed new light on the potential mechanisms of action of inhibition of the ER calcium channel SERCA2.
The biophysical based cellular thermal shift assay and thermal proteome profiling (TPP) have been used for nearly a decade to study biochemical phenomena in the cellular context (1, 2). Since the initial TPP report (1), research groups have leveraged this workflow to identify targets of small molecules and have offered an approach for target and/or off-target identification for drug discovery. Additionally, it has been clearly shown that TPP studies can be used to understand the functional proteome of different cellular states. Recent studies have used TPP to observe protein stability differences across: cell cycle (3, 4), protein complex stability (5), thermal stability of proteins across evolution (6), RNA–protein interactions (7), viral infection (8), and posttranslational modifications (9, 10, 11, 12). Altogether, these studies illustrate the far-reaching potential for TPP to acquire functional proteomics data giving insights into biochemical and biophysical changes that occur in cells and tissues under different experimental conditions. Investigation of protein stability in its native environment has the potential to gain deeper understanding of signaling, protein modifications, and relationships within the cell that have not been observed with other methods.
Data analysis in a TPP experiment is a multistep process that can be automated through calculation algorithms. Algorithms that have been reported by the scientific community include TPP-TR, Inflect, MSstatsTMT, and Rtpca (13, 14, 15, 16). MSstatsTMT is a more general workflow for protein quantification of proteins with TMT labeling, while Rtpca is used in thermal proximity coaggregation analysis of TPP datasets. TPP-TR and Inflect are used for analysis of melt curve data generated from TPP and related experiments.
TPP-TR (17) uses input data from any number of experiments with filters such as peptide spectrum matches (i.e. PSMs) to calculate melt temperatures (called at 50% of max abundance) and melt shifts individually for each replicate experiment (control versus condition). While the original iteration of the TPP-TR program did not report use of significance calculation (18), the output of the current version in bioconductor includes p-value calculation for each protein and experiment (condition versus control). The related Inflect program calculates melts much like “TPP” but uses the inflection point in the melt curve to determine the melt temperature. Melt curve fitting in Inflect uses four parameter log fit (4PL) which we have shown improves curve fitting and inflection point calculation as reported in our prior study (13). The use of inflection point rather than 50% of max abundance of protein signal also provides more accurate melt temperature calculations for various protein populations including heat resistant proteins and other proteins with unique melt curve characteristics (13).
In the work described herein, we have added multiple improvements to the Inflect program to optimize computational analysis and maximize relevant biological insights obtained from TPP datasets. A primary goal of this work was the addition of functionality for calculation of single melt curves for all biological replicates in an experiment. Incorporation of this functionality allows for assessment of experiment variability such as biological replicate dynamics to be accounted for in the melt shift calculation. This is an important aspect of TPP analysis that has yet to be reported in other programs. Second, we wanted to not only add filters and p-value–based outputs but also provide key information to the user on how the filters impact the melt shifts and how they are calculated. To improve the sensitivity and selectivity of the computational workflow for biologically relevant proteins, we have incorporated a z-score and p-value–based assessment. We have also included an output with calculation of a new melt coefficient score to aid in prioritization of hits from TPP experiments. Finally, we wanted to add bioinformatic outputs and quality indicators for the user to improve ease of use of InflectSSP. The output of a TPP experiment can consist of many proteins with unknown relationships, and relative importance of proteins melts can be unclear. A bioinformatic plug-in to the InflectSSP workflow allows for the user to make hypotheses and design follow-up experiments in a manner guided by functional annotations.
In this work, our new program InflectSSP was developed to address these gaps in TPP data analysis while building upon TPP analysis improvements made in our prior work (13). We used InflectSSP to reanalyze two publicly available data sets from Kalxdorf et al. (19) and Sridharan et al. (20) and validate its utility and performance relative to these prior studies. Additionally, we designed a temporal experiment through cell treatment with a small molecule inhibitor of the endoplasmic reticulum (ER) calcium pump (SERCA2A) and inducer of the unfolded protein response (UPR) that we reasoned should cause multiple types of simultaneous changes in protein homeostasis. This dataset from our group was used to develop an additional novel functionality of our program; the calculation of a “melt coefficient” that can be used to rank melt shifts based on quality of the respective melts. Overall, our findings show that InflectSSP has increased sensitivity and selectivity for identification of likely changes in the functional proteome while providing multiple statistical metrics including p-value with and without post hoc correction that can be used for cutoff-based analysis of any individual dataset.
Experimental Procedures
Data Analysis
TPP experiments were analyzed using InflectSSP as described in the main text. LC-MS/MS data was analyzed using Proteome Discoverer. Bioinformatics analysis was done using annotations from STRING and DAVID databases (21, 22). For DAVID analysis, a background set of proteins was used that consisted of the list of proteins observed in the respective mass spectrometry experiments.
Publicly Available Data Sets
Two independent data sets were used in our case study. Data from Sridharan et al. (20) and Kalxdorf et al. (19) consist of normalized abundance values at each temperature in the thermal gradients along with search outputs including number of peptide spectrum matches (PSMs) and unique peptides (UPs). In the case of the dataset from Kalxdorf et al., data was collected in an experiment where K562 cells were treated with 1 mM dasatinib (protein tyrosine kinase inhibitor) for 60 min with the goal of identifying cell surface proteins. Three replicate data sets were used for this analysis. In the case of the Sridharan experiment, the data sets from Jurkat cell crude lysates were treated with 2 mM Na-ATP for 10 min. Two replicate experiments were used for this analysis from the Sridharan data set. The data from the source publication including abundance for each protein along with their PSM and UP attributes were used for analysis. In the case of each data set, the qupm columns were used to indicate the number of UPs while the qusm column was used for the number of spectral matches. Details regarding the file fields used for data are summarized in supplemental Table S1.
Statistical DOE Analysis
In silico experiment design and data analysis were conducted using JMP version 16 (SAS Institute). The JMP design of experiment (DOE) design tool was used to select the combination of PSM, UP, R2, and p-value limits for the in silico experiment that would provide a full understanding of “main effects” and “interactions”. Data from the Kalxdorf and Sridharan data sets were each analyzed in series in an automated fashion using the various combinations of filter settings specified in the DOE. The results from the analysis (i.e. number of proteins of interest) were analyzed with respect to each of the inputs using the Fit Model tool in the JMP program. Scaled estimates for each term were calculated by the JMP program and compared to each other to understand the relative impact of each term (i.e. PSM) on the output (i.e. number of proteins).
R Analysis
InflectSSP was coded in R programming language and uses the following functions: readxl, data.table, plotrix, tidyr, ggplot2, xlsx, httr, jsonlite, GGally, network, stats, RColorBrewer, svglite.
Cell Culture and Treatment
HEK293A cells (Invitrogen) transduced with a lentivirus encoding an ATF4-firefly luciferase transcriptional reporter gene were used for the thapsigargin treatment experiments. Cells were cultured at 37 °C with CO2 at 5% and water for humidification. Medium consisted of Corning Dulbecco’s Modified Eagle Medium (10-013CV) supplemented with 10% fetal bovine serum. Cells were grown adherently using 10 cm and 15 cm diameter tissue culture plates. Cultures were passaged every few days to maintain viability and were also periodically checked for absence of mycoplasma contamination. Treatment experiments consisted of removing growth medium and replacement with fresh media supplemented with thapsigargin or dimethyl sulfoxide (DMSO). Thapsigargin experiments used a 1 mM stock of the drug (Sigma T9033-1 MG) dissolved in DMSO. Post treatment for 1 h, the media was aspirated from the plates and cells were washed with 1X PBS prior to either lysis or removal from the plates. Lysis was used for Western blot experiments, while removal of cells from the plates with rubber scraper was used for TPP experiments; details are described in respective sections.
TPP Experiments
Prior to execution of the TPP workflow, cell pellets were removed from the freezer and resuspended in lysis buffer (40 mM Hepes pH 7.5, 200 mM NaCl, 5 mM beta glycerophosphate (anhydrous basis), 0.1 mM sodium orthovanadate, 2 mM TCEP, 10 mM MgCl2, 0.4% NP40, Roche EDTA free mini complete protease inhibitor). Cells were lysed in 1.5 ml micro tubes (Diagenode) by sonication using a Bioruptor sonication system (Diagenode) with cycles of 30 s/30 s off for 60 min. Total protein concentration of each sample was determined by a protein assay with lysates diluted to a protein concentration of ∼5 mg/ml for subsequent temperature treatment. Note samples in the same experiment replicate were adjusted to the same concentration. Aliquots of the adjusted supernatants (50 μl) were transferred to PCR tubes after which the samples were heated and cooled using a gradient procedure. The heat treatment consisted of 2 min at 25 °C, 3 min at given temperature per gradient, 2 min at 25 °C, followed by 4 °C. Gradient temperatures for TPP experiments consisted of 25.0, 35.0, 39.3, 50.1, 55.2, 60.7, 74.9, and 90.0 °C. In the case of the single replicate experiment, the gradient consisted of 25.0, 35.0, 40.9, 51.2, 55.2, 60.7, 74.9, and 90.0 °C. Heat-treated samples were centrifuged to pellet insoluble protein, while the supernatant was reserved and precipitated in 20% TCA.
Sample Preparation for LC-MS/MS and Mass Spectrometry
Dried pellets were resuspended in 8M urea in 100 mM Tris pH 8.5. Samples were reduced with TCEP and alkylated with chloroacetamide as previously reported (23). Reduced and alkylated samples were digested with LysC/trypsin (Promega) followed by quenching with formic acid. Quenched samples were desalted with Waters C18 columns and then isobarically labeled with Thermo Scientific TMTPro labels as previously reported (24). The labeling scheme for the first two biological replicates is shown in supplemental Fig. S1. The labeling scheme for the third biological replicate is shown in supplemental Fig. S2. Samples were fractionated into eight fractions using Waters C18 columns and then analyzed on the LC-MS instruments.
Search Parameters and Acceptance Criteria (MS/MS and/or Peptide Mass Fingerprint data)
In the case of the first two biological replicate experiments, there were two total technical replicates of the thapsigargin datasets using two different LC-MS instruments. In the case of one technical replicate, Nano-LC-MS/MS analyses were performed on an Exploris 480 mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nLC HPLC system (Thermo Fisher Scientific). The peptides were eluted using a mobile phase gradient for 180 min at 400 nl/min to ensure elution of all peptides. The heated capillary temperature was kept at 275 °C and ion spray voltage was kept at 2.5 kV using a FAIMS source with a compensation voltage of −50 V. During peptide elution, the mass spectrometer method was operated in positive ion mode, programmed to select the most intense ions from the full MS scan using a top speed method. Exploris MS1 parameters include the following: microscans 1; MS1 resolution 60 k; automatic gain control (AGC) target 3E6; and scan range 375 to 1600 m/z. Exploris data-dependent MS/MS parameters include the following: microscans 1; resolution 45 k; AGC target 2E5; maximum IT 87 ms; isolation window 0.7 m/z; fixed first mass 110 m/z; and HCD normalized collision energy 35. The respective data-dependent settings were set with parameters: apex trigger as “-”; charge exclusion as “1,7,8, >8“; multiple Charge. States as “all”; peptide match as “preferred”; exclude isotopes as “on”; dynamic exclusion of 30 s; if idle “pick others” using Xcalibur software (available from Thermo Fisher Scientific).
A technical replicate analysis of the first two biological replicate samples was also performed on a Lumos mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nLC HPLC system (Thermo Fisher Scientific) for 180 min. The mass spectrometer method was operated in positive ion mode during a 170 min gradient, programmed to select the most intense ions from the full MS scan using a top speed method. Lumos MS1 parameters include the following: microscans 1; MS1 resolution 120 k; standard AGC; and scan range 400 to 1600 m/z. Lumos data-dependent MS/MS parameters include the following: microscans 1; resolution 50 k; normalized AGC target of 250%; isolation window 0.7 m/z; fixed first mass 100 m/z; and HCD normalized collision energy 34. The respective data-dependent settings were set with parameters: exclude isotopes as “on”; dynamic exclusion of 60 s using Xcalibur software (Thermo Fisher Scientific).
In the case of the third biological replicate, samples were analyzed twice (technical replicates) on an Exploris 480 mass spectrometer (Thermo Fisher Scientific) coupled to an EASY-nLC HPLC system (Thermo Fisher Scientific). The peptides were eluted using a mobile phase gradient for 175 min at 300 nl/min. During peptide elution, the mass spectrometer method was used as above. Exploris MS1 parameters include the following: one microscans; MS1 resolution 120 k; custom AGC; and scan range 400 to 1750 m/z. Exploris data-dependent MS/MS parameters include the following: one microscans; resolution 45 k; normalized AGC target of 200%; isolation window 0.7 m/z; fixed first mass 100 m/z; and HCD normalized collision energy 32.0. The respective data-dependent settings were set with parameters: exclude isotopes as “on”; dynamic exclusion of 30.0 s. The data were recorded using Xcalibur software (Thermo Fisher Scientific).
The resulting RAW files from the time course experiments were subjected to protein FASTA database search using Proteome Discoverer 2.4.0.305 (Thermo Fisher Scientific). The SEQUEST HT search engine was used to search against a human protein database from the UniProt repository containing 20,350 human proteins (2019) and common contaminant sequences such as proteolytic enzymes (FASTA file used available on MassIVE under MSV000090867 and in ProteomeXchange under PXD038752). Specific search parameters used were trypsin as the full proteolytic enzyme, peptides with a max of two missed cleavages, precursor mass tolerance of 20 ppm, and a fragment mass tolerance of 0.5 Da. Minimum and maximum peptide length were set to 6 and 144, respectively, with max number of peptides reported at 10. Spectrum matching parameters in the search were set to True for “Use Neutral Loss” for all ions, and weight of b and y ions were set to 1 with all others at 0. Max equal and dynamic modifications per peptide were set to 3 and 4, respectively. Static modifications were TMTPro label on lysine (K) and the N termini of peptides (+304.207 Da). Percolator false discovery rate (FDR) cutoff filtering was set to a strict setting of 0.01 (1% FDR). Total ion abundance values at the protein level were summed from UPs and used for quantification for melt shift calculation at the protein level. The output from this database search is included as supplemental Table S2.
The resulting RAW files from 1 h experiment (three replicates) were subjected to protein FASTA database search using Proteome Discoverer 2.5.0.400 (Thermo Fisher Scientific). The SEQUEST HT search engine was used to search against a human protein database from the UniProt repository containing 20,290 human proteins and common contaminant sequences such as proteolytic enzymes (FASTA file used available on MassIVE under MSV000090867 and in ProteomeXchange under PXD038752). Specific search parameters used were trypsin as the full proteolytic enzyme, peptides with a max of two missed cleavages, precursor mass tolerance of 10 ppm, and a fragment mass tolerance of 0.02 Da. Minimum and maximum peptide length were set to 6 and 144, respectively, with max number of peptides reported at 10. Spectrum matching parameters in the search were set to True for “Use Neutral Loss” for all ions, and weight of b and y ions were set to 1 with all others at 0. Max equal and dynamic modifications per peptide were set to 3 and 4, respectively. Static modifications were TMTPro label on lysine (K) and the N termini of peptides (+304.207 Da) and carbamidomethyl (+57.021 Da). Dynamic Modifications included acetyl (+42.011), Met-loss (−131.040), and Met-loss+Acetyl (−89.030). Percolator FDR cutoff filtering was set to a strict setting of 0.01 (1% FDR). Total ion abundance values at the protein level were summed from UPs and used for quantification for melt shift calculation at the protein level. The output from this database search is included as supplemental Table S3.
The mass spectrometry proteomic data (including .pdResult, .mzTab, .mzML, and .raw files) have been deposited to the ProteomeXchange Consortium via the MassIVE partner repository with the data set identifier and doi:10.25345/C5VM4325J. A combined search was also completed on all of the 1-h experiment data (including the third biological replicate). RAW files and search results from this analysis can be found in MassIVE repository: MSV000090932, ftp://massive.ucsd.edu/MSV000090932/.
InflectSSP Analysis
Settings used in melt shift analysis for main figures in this manuscript are described in supplemental Table S4.
TPP Analysis
TPP program version 3.22.1 was used for comparative analysis of the thapsigargin datasets. User-specified filters for the program were set to maximum range. Filtering was only done on the output of the program by selecting proteins with p-values that were <0.05 across all replicates.
Experimental Design and Statistical Rationale
The Sridharan experiment analysis used two publicly available biological replicate data sets, while the Kalxdorf experiment analysis used three publicly available biological replicate data sets (total number available from each respective source). The thapsigargin data sets from our group consisted of two biological replicates, and this number of experiments was chosen based on the number of temperatures that could be successfully multiplexed using available TMT labels. Each of the three data set used both a condition (treatment) and control (vehicle) to calculate melt shifts. In silico experiment design and statistical data analysis were conducted using JMP version 16 (SAS Institute). TPP experiments were analyzed using InflectSSP version 1.5 (described herein). The InflectSSP program has an optional FDR adjustment which can be used by users to increase the specificity of the data set being analyzed. To account for potential biological and workflow variability in data sets analyzed, an FDR calculation was not used in the analysis described herein. The z-score–based p-value calculated by the InflectSSP program was deemed acceptable for the sigmoidal data analysis that was being analyzed. Traditional coefficient of determination (R2) is also available for use in the InflectSSP program as it is used in the field for describing the adequacy of sigmoidal fit.
Results
Initial Assessment of InflectSSP Workflow
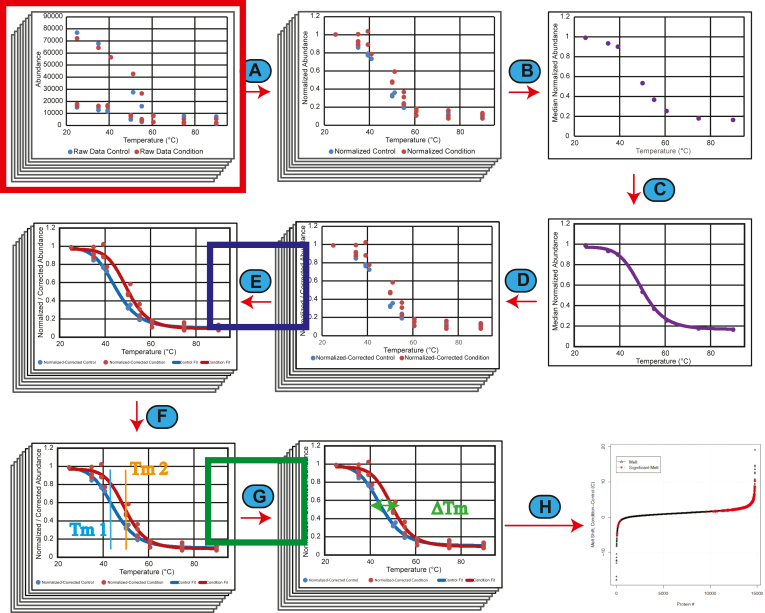
Inflect was developed as an R package for the analysis of TPP experiments. A goal of our ongoing development of the Inflect workflow was to increase the sensitivity and selectivity for changes in protein thermal stability when analyzing TPP (and similar assay) results. Using the output from a TPP experiment, normalized ion abundance values at different temperature treatments are used as input for the melt curve analysis in Inflect to determine the inflection point of the curve as the melt temperature (Tm) of the protein of interest. For the development of InflectSSP, additional parameters were developed to consider their respective impact on the output of the melt shift calculation workflow. The InflectSSP data analysis workflow that was used for our assessment is described pictorially in Figure 1. Step A in the workflow imports data from the source directory. The current version of InflectSSP allows for the import of multiple experiments which can consist of an unbalanced number of experiment files for condition and control. The normalization step divides each abundance value by the abundance observed at the lowest temperature so that results from separate experiments can be analyzed together. This data normalization is completed at the “protein level” for each protein and for each experiment factor (vehicle or drug). If for instance there is a vehicle treatment and a drug treatment (2 factors) in the 8-temperature heat treatment experiment and a total of 5000 proteins have been identified, there will be 80,000 abundance values at the end of this “Normalization” step. Step B in the algorithm converts the normalized abundance data from the protein level to proteome level. Specifically, the median abundance is measured at each temperature across all factors. In our example, if there are 80,000 abundance values at the end of step A with eight heat treatments, this “Quantitation” will yield eight total values. Step C or “Curve Fit 1” determines the three or four parameter log fit coefficients that best describe the variability observed in step B. The purpose of steps B and C are to describe how well the heat treatment step compared to ideal melt behavior. If, for example, there is a subtle increase or decrease for one temperature in the heat gradient (e.g. in the heating block), the curve would depart from sigmoidal shape. A 4PL is used to describe the curve but changes to a three-parameter log fit (3PL) if there are challenges with curve fit convergence by the program. The equations that are used for the log fits are shown in supplemental Fig. S3. Step D is the “Correction” step that adjusts the normalized abundance value at each temperature for each protein based on how well actual values meet predicted values in step C (“Curve Fit 1”). While step C is done at the proteome level, step D is done at the protein level. In this step, a correction factor is first calculated for each temperature based on how much the actual values depart from predicted values in step C. The correction factor at each temperature is then used for each protein in the experiment. For example, if the actual values are 1% greater than those predicted at 35 °C, the normalized abundance values for each individual protein at 35 °C are decreased by 1% percent to allow for normalized abundance values to fit with ideal melt behavior. If there are 5000 proteins going into step B and there are 100 proteins excluded due to low PSM or UP, there would be a total of 4900 proteins going into step D and therefore 78,400 normalized abundance values being corrected. “Curve Fit 2” in step E is executed for each individual protein in the experiment at the protein level. In our example, 4900 proteins across two factors would be used to fit 9800 curves.
Fig. 1.
Graphical overview of the computational steps in InflectSSP.A, abundance data from each protein normalized by dividing each abundance value at each temperature by the abundance at the lowest temperature. B, median of all abundance values across proteome at each temperature and for each condition and control across all replicates. C, curve fitting of proteome data sets. D, correction of individual normalized abundance values for each protein based on difference between actual and predicted in previous step. E, curve fitting for each protein melt curve based on corrected normalized abundance values. R2 for curve fits are also calculated. F, calculation of melt temperature for each condition and control across all proteins. G, calculation of melt shift along with melt shift p-value. H, result reporting that describes the melt shifts calculated in the experiment with outputs in various formats (plots and tables). Potential locations in the data analysis workflow where quality of the data analysis can be evaluated. The red box indicates the number of peptide spectrum matches (PSMs) and number of unique peptides (UPs) that are reported with the protein/abundance data from the MS experiment and proteomic search. The blue box represents where the coefficient of determination (R2) is calculated based on the fit of the melt curves. The green box represents where the melt shift p-values are calculated in the workflow with the melt shifts.
Like “Curve Fit 1” in step C, a 4PL is first used before using a 3PL fitting. If neither set of equations converges in the program, the protein is excluded from further analysis. The melt temperature for each protein is calculated in step E using the inflection point of the melt curves. The inflection point is defined as the temperature where the second derivative of the fit equation equals 0. In the process of generating the curve fits and the associated inflection point, it is possible that the curve fitting algorithm can converge on a set of optimal parameters. The fit curve, however, may not represent what would be biologically likely. To address this challenge, the 3PL fit is used if a calculated melt temperature is less than or greater than the temperature range used during the heat treatment. An example of how this operation allows for more biologically representative results is shown in supplemental Fig. S4. In this example where a 4PL fit was initially used, the melt is calculated to be 71.3 °C, but when the 3PL fit was used (to better reflect biological conditions), a more realistic melt of 57.0 °C is observed. Step F or “Melt Calculation” is completed using the fit curves for each protein. Specifically, the melt temperature for each protein is calculated as the inflection point in the sigmoidal curves that are calculated in the previous step. This process is also linked with the previous “Curve Fit 2.” If the calculated melt temperature is less than the lowest temperature or greater than the highest temperature in the heat treatment, a 3PL will be used. This process has been implemented in InflectSSP to avoid artificially large melt shifts that result from melt curve shapes that are not anticipated to reflect biological conditions. The “Melt Calculation” is completed in step G where the control melt temperature is subtracted from the condition melt temperature to determine the magnitude of shifts for each protein in the experiment. In our example to this point, the output of this step would be 4900 melt shifts. Step H consists of generating summary outputs from the program. These outputs include a rank order or waterfall plot that describes the melt shifts of each rank-ordered protein in the experiment, tables that summarize all melt shifts from the experiment along with their calculated “melt coefficient”, STRING-based network diagrams, and tables that summarize nodes of interest in the STRING diagram. A plot is also generated that summarizes the “Melt Shift Coefficient” across p-value ranges for the analysis.
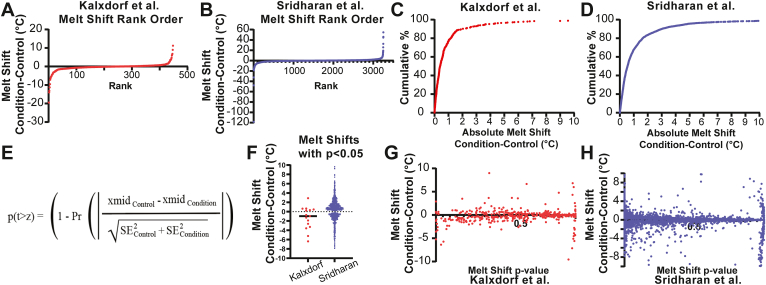
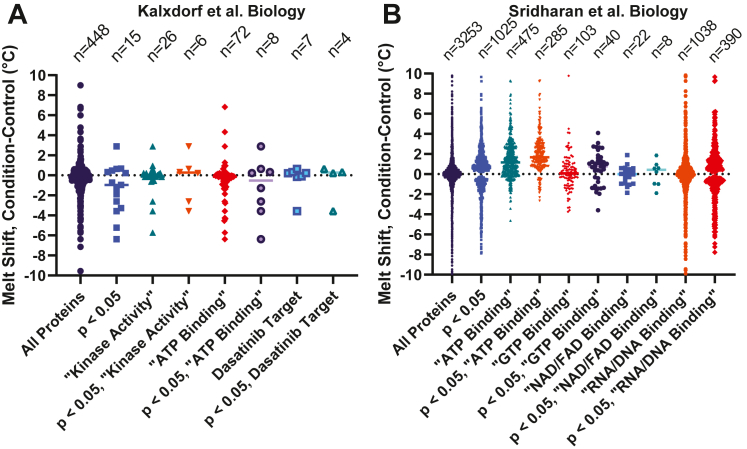
The workflow used to analyze data from the Kalxdorf et al. and Sridharan et al. data sets is described in the Experimental Procedures section of this report. In the case of the dataset from Kalxdorf et al., data was collected in an experiment where K562 cells were treated with 1 mM dasatinib (protein tyrosine kinase inhibitor) for 60 min. Three replicate data sets were used for this analysis. In the case of the Sridharan experiment, the data sets from Jurkat cell crude lysates were treated with 2 mM Na-ATP for 10 min. Two replicate experiments were used for this analysis from the Sridharan data set. The rank order plots of the calculated melt shifts for each protein in these data sets are shown in Figure 2, A and B, and these panels reflect the wide range of melt shifts in each set. While 10 to 20% of the protein melt shifts are greater than 2 °C and around 4% of the proteins are greater than 5 °C (Fig. 2, C and D), it is not easily discernible from melt shifts alone which proteins have a significant melt shift and which are within the variability of the experiment. One possible method for determining significance would be to report proteins with melt shifts that are greater than an absolute limit based on the mean and SD. At the same time, this approach may not allow for selection of proteins with subtle changes. This fact guided our work to provide objective quality control criteria based on statistical metrics to apply cutoffs for selecting proteins of interest in these data sets.
Fig. 2.
InflectSSP outputs from reanalysis of public datasets. Melt shift rank order plots for (A) Sridharan and (B) Kalxdorf data sets. Pareto chart for absolute melt shifts for (C) Sridharan and (D) Kalxdorf data sets. X-axis max for (C) and (D) have been set at 10 °C for ease of viewing. E, formula describing the derivation of the melt shift p-value. F, melt shifts for proteins with melt shift p-values <0.050 in each of the two data sets. G, melt shift versus melt shift p-value for Sridharan data set. H, melt shift versus melt shift p-value for Kalxdorf data set.
The absolute magnitude of melt shifts in a TPP experiment may not be sufficient for determining which proteins are significantly stabilized or destabilized for functional proteomics interrogation. The reason for this limitation is that the calculated melts may not account for experimental variability. The melt curve inflection point or melt temperature may be distinct, but the variability of the abundance values around the calculated melt from the curve fit may be large enough that the melt temperatures between condition and control are not significantly different from each other. Selection of a protein based on the magnitude of the shift alone would potentially cause the investigator to focus on proteins and pathways that are not actually affected by the experimental conditions. To address this deficiency in the calculation pipeline, we have updated our existing version of Inflect to allow for analysis of biological replicate experiments to determine statistical significance of melt shifts between conditions. Other filters or quality control steps were also added to the analysis pipeline to remove proteins that increase the “noise” of the calculated melt shift (i.e. outliers). Three steps in the described analysis workflow (Fig. 1) were identified as possible opportunities for inserting control criteria. The mass spectrometry and proteomics search (designated by the red box in Fig. 1) is one place where criteria can be set. The number of UPs for each protein is one output that is reported by proteomic search algorithms such as Proteome Discoverer.† This value indicates the number of reported peptide fragments that are unique to a protein in the source proteomics database. Since the sequence of peptides determined from a “bottom-up” mass spectrometry experiment is based on experimental spectra, the number of PSMs are also reported by Proteome Discoverer.† The PSM are the number of spectra from the experiment that match with spectra from the search database within the set cutoff criteria. These two variables offer quality control criteria for screening the performance of the mass spectrometry and associated data search experiment. In our analysis, we used the UP and PSM reported in step A (Fig. 1) to conduct the exclusion of proteins at step D after the proteome curve fit is conducted. These filters allow for exclusion of proteins with low numbers of total identifications independent of their summed ion abundance value. These two filters were inserted prior to the curve fitting steps in step E since a low UP or PSM value could result in lower confidence in the identity of the protein associated with the peptide(s). The exclusion was set after the overall proteome curve fitting (step C) to ensure that the abundance values of the peptides still affect the total proteome abundance. The distribution of PSM and UP across the two data sets is shown in supplemental Fig. S5, A and B. Both sets of distributions for both data sets are skewed to low number of total PSMs and UPs as is commonly observed for data dependent acquisition-based bottom-up proteomics experiments. Considering that the multiplexing allowed for by TMT-based isobaric labeling facilitates high-dimensional TPP dataset generation with multiple temperature datapoints, this challenge cannot be fully addressed using alternative acquisition strategies such as data-independent acquisition. However, recent work has reported acquisition strategies for performing TPP using data-independent acquisition (25). Normalized melt curves generated from such an approach would also be compatible with Inflect-SSP.
A second step in the analysis workflow where filtering was identified is step E where the curve fitting is conducted (blue box in Fig. 1). One statistical tool that can be used to describe the quality of fit for the melt curve is the coefficient of determination (R2). While the coefficient of determination is not necessarily a strong measure of nonlinear fit (26, 27), it is a widely used parameter in the scientific community with practical utility. This step in the process was identified as a point where variability in the cell culture, heat treatment, and MS portions of the experiment could be characterized. This was also a way of incorporating variability from replicate experiments. The distributions of R2 for each of the two data sets are shown in supplemental Fig. S5C and reflect the fact that the quality of fit is tends towards high-quality values of 1.
A third point in the overall workflow where possible data filtering was identified is step G where the melt shifts are calculated (green box in Fig. 1). The difference in melt temperature between condition (i.e. treatment) and control (i.e. vehicle) in an experiment is defined as the melt shift. A positive shift is generally interpreted as a protein with increased thermal stability, while a negative shift is a decrease in protein thermal stability from experiment conditions. The magnitude and direction of each shift do not provide sufficient information to determine whether a shift is significantly greater than experimental variability. Experiment variability can be captured through the execution of biological and technical replicates that are input into the InflectSSP workflow. The melt curves are fit for each protein in step D (Fig. 1) using either 4PL or 3PL, respectively, depending on the success with analysis convergence. To quantify the signal-to-noise ratio in these melt shifts, we have established a z-score accompanied by a p-value calculation. The equation in Figure 2E is used by the current version of InflectSSP to calculate a p-value for each protein melt shift. The calculation is based on the difference in melt temperature normalized by the standard error calculated by the “nls” function (R “stats” package). Our z-score uses a 1 SD criteria for the evaluation of the p-value. The definition of this p-value is the likelihood that you would reject the null hypothesis (no difference in melt temperature) when the null hypothesis is true. supplemental Fig. S5D shows melt shift p-value distributions for each of the two data sets and indicates that while the Sridharan data set has more statistically significant melts (lower p-value), the Kalxdorf data set has a more even distribution of melt p-values with less statistically significant shifts. Figure 2F shows the distribution of melt shifts that have p-values less than 0.05 and reinforces the fact that melt shifts that meet the p-value of <0.05 are not necessarily large in magnitude, clearly showing the utility of a statistical method that considers the reproducibility of biological replicates. The plots in Figure 2, G and H show the relationship between the calculated melt shift p-value and the melt shifts across both data sets. These results indicate that the magnitude of the melt shift is not necessarily correlated with melt shift p-value. These results suggest that the magnitude of the melt shift is insufficient for determining the significance of the shift as many of the proteins with low p-values also have very small shifts.
The p-value calculated by our workflow describes the confidence that the melt shift is greater than or less than 0 °C. Multiple correction testing is also included in InflectSSP, incorporated as an optional FDR correction into p-value calculation. The FDR calculation is done using the p.adjust function in R along using “fdr”. We used noncorrected p-value in the assessments described herein but both options are available to users of the InflectSSP package to fit user needs.
Impact of Quality Control Criteria on Program Performance
Once we identified potential quality control criteria, we wanted to understand the relative impact of each filter on the final output from the workflow. To evaluate the relative impact of these criteria on the performance of the algorithm, we established an objective approach using numeric outputs along with associated limits. “Significant Proteins” were defined as those that met all specified criteria (protein PSM, protein UP, curve R2, and melt shift p-value), while “Biologically Relevant” proteins were those relevant to the experiment. In the case of the Sridharan et al. data set, “ATP Binding” Gene Ontology (GO) Molecular Function (MF) term was used to identify the number of proteins that have previously been described and annotated as likely to be biologically relevant following treatment with 2 mM Na-ATP. In the case of the Kalxdorf et al. data set, “Kinase Activity” GO MF term was used to determine those proteins that have previously been described and annotated as likely to be relevant to this dataset which uses treatment with 1 mM of dasatinib, a kinase inhibitor. The first output calculated for the evaluation was the “percent of biologically relevant” proteins. This value was determined by dividing the number of “significant” proteins that were biologically relevant (as defined by the criteria above) by the total number of biologically relevant proteins (given the defined criteria) in the overall data set and multiplying by 100. The number of “significant” proteins that were not “biologically relevant” were also calculated for each of these data sets to give insights into the sensitivity and specificity of the InflectSSP analysis.
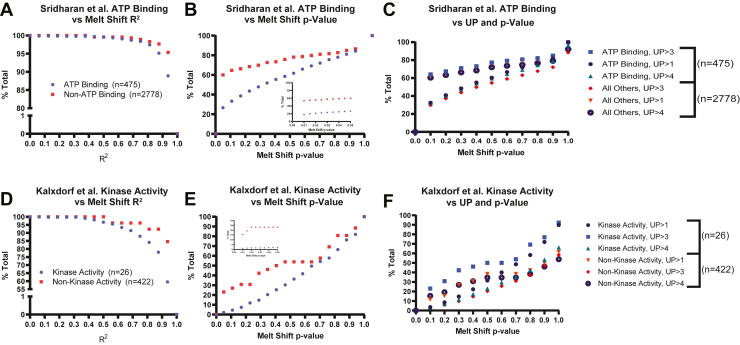
Our in silico experiment was designed using JMP with the goal of understanding main effects (i.e., limit on PSM alone) and interactions (i.e., limit on PSM being affected by the number of UPs) on the two outputs. supplemental Table S5 shows the ranges for each quality control variable that were used in our assessment. The InflectSSP program was run serially using each of the settings in the experiment design. Outputs from the in silico experiment (i.e., percentage of biologically relevant proteins) were further analyzed using JMP statistical program version 16. Specifically, the results were modeled using the inputs of the experiment using nonlinear systems. The results from this statistical analysis are shown in supplemental Fig. S6. As shown in supplemental Fig. S6, A and B, the models developed in the program described 97 to 98% of the variability in the outputs. The scaled estimates for each term in the models are shown in supplemental Fig. S6, C and D and quantify the relative leverage that each term has on the output of the model (i.e., percent of biologically relevant proteins). These results indicate that the melt shift p-value has the largest impact of all quality control parameters examined. The number of PSMs, number of UPs, and the curve R2 each have a relative impact that is 5 to 15% that of the melt shift p-value term. To graphically illustrate the relative impact of these variables, the percent of biologically relevant and nonbiologically relevant proteins was plotted versus R2 (Fig. 3, A and D) and melt shift p-value (Fig. 3, B and E). The impact of the p-value coupled with the number of UPs are shown in Figure 3, C and F and reflect how the melt shift p-value has a larger impact on the percent of relevant and nonrelevant proteins in comparison to the other four quality control inputs. The wide separation of proteins that are annotated as “ATP binding” or “Kinase Activity” versus all other proteins in Figure 3, B and E indicates that the use of p-value–based cutoffs for TPP dataset analysis will have a large impact on the specificity and selectivity of the findings. The benefits of p-value–based selectivity in the datasets were observed with p-value cutoffs ≤0.5, but the largest separation in proteins for each term group was observed at cutoffs ≤0.1.
Fig. 3.
Analysis of2mM Na-ATP treatmentdata from Sridharan etal.using design of experiment (DOE). (A), percent of total “All Others” and ATP-binding proteins as a function of the R2 used as criteria for selection of proteins of interest. B, percent of total “All Others” and ATP-binding proteins as a function of melt shift p-value criteria with inset showing 0 to 0.05. C, percent of total “All Others” and ATP-binding proteins as a function of melt shift p-value and the number of unique peptides. Analysis of 1 mM dasatinib treatment data from Kalxdorf et.al. D, percent of total nonkinase and kinase activity proteins as a function of the R2 used as criteria for selection of proteins of interest. E, percent of total nonkinase and kinase activity proteins as a function of melt shift p-value criteria with inset showing 0 to 0.05. F, percent of total nonkinase and kinase activity proteins as a function of melt shift p-value and the number of unique peptides.
As a result of the multivariate analysis, the melt shift p-value alone was used to further reanalyze these two data sets for additional comparative analyses with PSM and UP set to 0 and R2 set to 1. The proteins found to have “significant” melt shifts (p < 0.05) were 15 of 448 for the Kalxdorf data set (Fig. 4A) and 1025 of 3253 for the Sridharan data set (Fig. 4B). The large difference in the number of proteins filtered by the p-value reflects the variability in the values from each data set. This result also reflects the ability of the quality control limit to decrease the large number of protein thermal stability changes. Proteins that met these criteria for each of the two data sets were then further analyzed from a biological perspective using MF GO terms for the proteins identified in the data sets. Since the Kalxdorf experiment treated cells with 1 mM dasatinib (a kinase inhibitor), proteins with “Kinase Activity” or “ATP Binding” terms were identified. Targets that have been reported for dasatinib (28) were also used to group melt shifts. The InflectSSP workflow identified approximately 10 to 50% of the proteins from each of these biological categories (“Kinase Activity”, “ATP Binding”, “Dasatinib Target”). This same analysis was used for the Sridharan data set which was collected where cells were treated with 2 mM Na-ATP. MF terms used to classify sets of proteins in the original Sridharan report were used in this assessment (Fig. 4B) including “ATP Binding”, “GTP Binding”, “NAD Binding”, “FAD Binding”, “RNA Binding,” and “DNA Binding.” As shown in Figure 4B, 40 to 50% of proteins from each of these terms also had melt shift p-values <0.05. The number of proteins that fit these criteria in our analysis were then compared with the number of proteins that were reported in the Sridharan set as significantly changed in stability by ATP addition. Note that Sridharan et. al used a 2D-TPP dataset with multiple concentrations of ATP, whereas we analyzed changes at a single concentration point. Count of proteins with significant stability changes in categories annotated for ATP Binding, GTP Binding, NAD/FAD Binding, and RNA/DNA Binding were 285/315, 40/55, 8/28, 390/82 proteins, respectively, between our report and by Sridharan et al (20). Generally, these results indicate that the melt shift p-value of 0.05 provides a good filter for identifying proteins of interest from a biological perspective with similar findings in the ATP Binding category being of particular interest from this study (again 285 detected as significant in our report compared to 315 in Sridharan et. al). Of note, InflectSSP had increased sensitivity in the categories of “RNA/DNA Binding” with 390 proteins with melt shift p-values <0.05 relative to 82 significant changes in Sridharan et. al. Since ATP is a nucleotide component of both RNA and DNA, some proteins which interact with those macromolecules can make contacts with free nucleotide as well. Indeed, ATP has been shown to function as a hydrotrope to maintain solubility of RNA-binding proteins such as FUS by preventing fibrillization, which for FUS is associated with a cytotoxic form of the protein found in amyotrophic lateral sclerosis (29). Therefore, the increased sensitivity provided by InflectSSP could be important for identifying additional RNA/DNA-binding proteins whose stability is altered because of ATP or other nucleotide-binding events.
Fig. 4.
Melt shiftdistribution of quantitatedproteinsfrom reanalysis of public datasets. Melt shifts shown are calculated from Kalxdorf et al. (A) and Sridharan et al. (B) data sets. Melt shift data point groups are based on their melt shift statistical significance along with the biological significance as indicated by Gene Ontology (GO) Molecular Function (MF) terms. Note that y-axis ranges have been abbreviated for ease of data viewing. The total number of data points (including those outside of the axis range) is shown above each group.
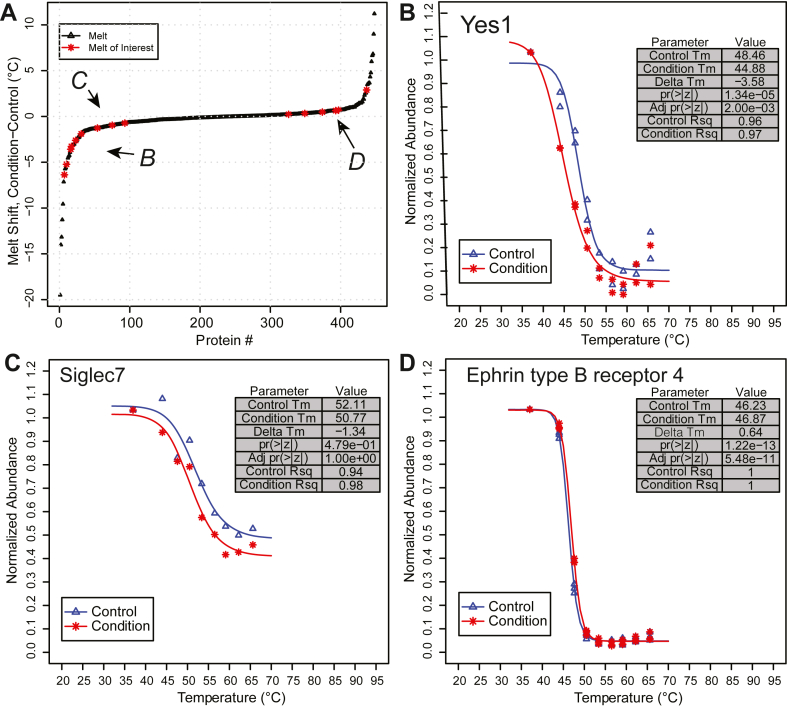
To better understand the result of the data filtering process, individual melt curves were examined further. The rank order plot of melt shifts from the Kalxdorf data set (from InflectSSP analysis) is shown in Figure 5A. Proteins with various magnitude and significance of melt shifts are highlighted in Figure 5, B–D. Figure 5B shows the example where the melt shift magnitude is large coupled with a significant p-value. Yes1, a SRC family kinase, has been investigated in dasatinib therapy (30). Figure 5C, melt curves for Siglec7, shows an example of a large magnitude shift with a p-value that does not meet criteria of 0.05. This example in Figure 5C shows why proteins with large melt shifts (that would normally be considered as significant) are removed from consideration as a program output. Finally in the case of Figure 5D, the magnitude of shift for Ephrin type-B receptor 4 is small, while the p-value criterium is met. This result in Figure 5D, is even more relevant when it is considered that this protein has ATP binding and kinase MF according to Uniprot (31). This receptor has also been reported to be a secondary target of dasatinib (32) and thus, further provides biological relevance of the findings and validation of the workflow. Overall, these curves show how the p-value can assist in the differentiation of melt shifts based on the associated experiment variability.
Fig. 5.
Selected InflectSSP outputsfromtheKalxdorf1mM dasatinibtreatmentdata set.A, rank order plot of protein melt shifts. Proteins with select characteristics have been highlighted by letters based on their location on the plot, (B) where the absolute melt shift is large and the p-value for the melt shift is <0.05, (C) where the absolute melt shift is still high but the p-value is >0.05, (D) where the melt shift is negative and the p-value for the melt shift is <0.05 thus being considered significant regardless of the shift magnitude.
Bioinformatic Reporting: STRING Analysis
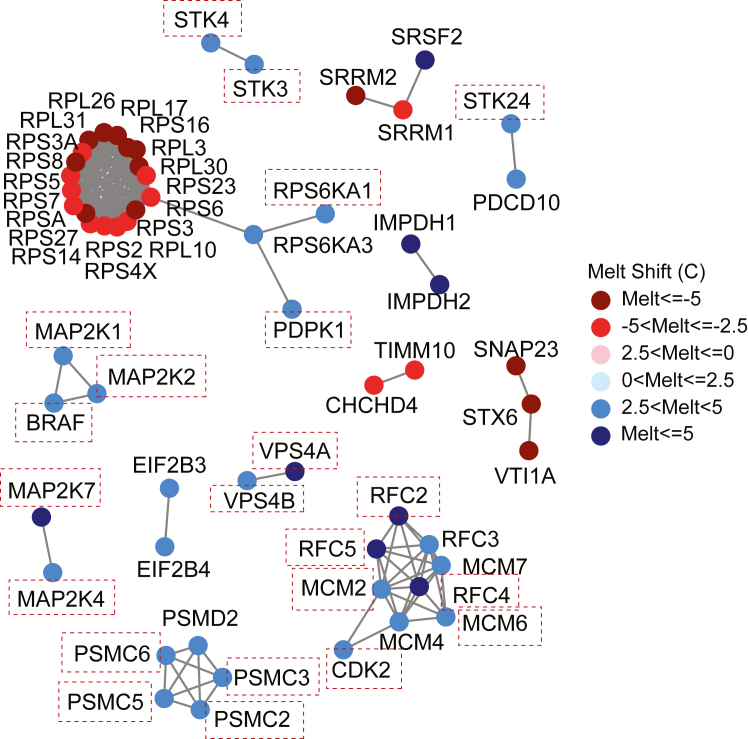
Since TPP experiments could have 5000 to 10,000 melt shifts depending on biological system, it may be advantageous in early studies to analyze data sets using bioinformatic approaches. One bioinformatic tool that has been integrated into the InflectSSP program is STRING (33). STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) is an online database supported by the STRING Consortium (Swiss Institute of Bioinformatics, EMBL and others) that reports protein relationships and interactions that have been reported or assumed based on other homologous proteins (22). Interactions between queried proteins are reported using network diagrams where the connections between nodes are determined based on confidence specified by the user. The confidence in the interactions is based on the number and type of reports where an interaction is reported. The STRING database is accessed through InflectSSP using an application program interface and integrates the melt shifts for significant proteins using a network. An example of the network generated from the Sridharan et al. data is shown in Figure 6. Protein nodes for all significant proteins in the results along with associated interactions and relationships reported through STRING are shown. Colors of the nodes vary depending on the melt shift for each protein where deep red and dark blue are the destabilized and stabilized melt shifts (respectively) as shown in the legend to the right of Figure 6. ATP-binding proteins in this figure are shown by the orange squares and further demonstrate the biological relevance of the workflow output. This output also suggests that one possible explanation for some of the melt shifts is an interaction between some of the proteins. In the case of MAP2K1, MAP2K2, and BRAF, the three are all stabilized and have all been reported to interact with each other based on documentation in STRING.
Fig. 6.
Bioinformatics analysis conducted using Sridharan data set where Jurkat cells were treated with2mM Na-ATP. Proteins with melt shifts having p-values <0.05 and melt shift magnitudes greater than 3 °C are shown as nodes in the network. Melt shift temperature is designated by the color of the nodes with legend on the right. The STRING network analysis for significant proteins based on specified STRING significance of 0.95. Proteins related to ATP binding are highlighted in orange dashed squares.
Analysis of a Novel Dataset Using InflectSSP
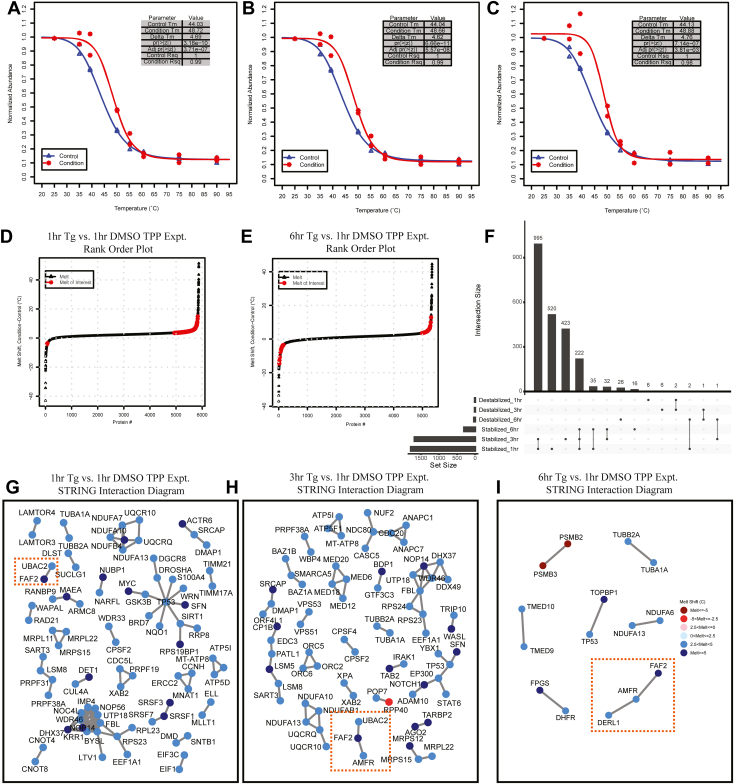
Once we confirmed the suitability of the InflectSSP program in analyzing publicly available data sets, we applied InflectSSP to a complex temporal TPP study with ± small molecule inhibitor (SMI) treatment. The use of a temporal dataset allows us to assess reproducibility of SMI target identification over a larger number of biological replicate conditions while allowing for identification of temporal changes that occur directly/indirectly because of SMI response. Our novel TPP dataset interrogates cellular response to 1 μM thapsigargin, a SERCA2A inhibitor (34) and inducer of the UPR (35). The UPR is one of the mechanisms by which eukaryotic cells can address the presence of unfolded protein(s) in the ER through slowing of global translation, increasing abundance of select proteins and if necessary, driving apoptosis (36). The goal of this experiment was to induce the UPR with an SMI and quantify changes in protein stability over time. In this experiment, adherent cultures were treated with 1μM thapsigargin (in DMSO) over 1, 3, or 6 h along with a DMSO control. The treatments were conducted in biological duplicate after which the cells were harvested, lysed, and supernatant heat treated over an eight temperature gradient. The post heat treatment samples were processed through reduction, alkylation, and LysC digestion followed by multiplexing using TMTPro within a 16-plex and analysis by LC-MS/MS. Protein abundance across the treatments and melt temperatures are summarized in supplemental Fig. S7. Following database search and calculation of total ion abundances, normalized abundance values from our experiment were then processed through the InflectSSP pipeline. The DMSO 1 h dataset (n = 2) was used as the control in the pipeline with each of the three time point datasets obtained following thapsigargin treatment. The melt shifts at each time point for the thapsigargin target, SERCA2A, are shown in Figure 7, A–C. Each set of melt curves for this protein show reproducible increased stability relative to DMSO. Rank order plots from InflectSSP for the 1-h and 6-h data sets are shown in Figure 7, D and E. These plots show the change in melt shift of significant proteins from the 1 to 6-h time frame. One explanation for this observation is that post treatment, cellular proteins are initially engaged by either chaperones or degradation receptors during the initial stressed state following 1 μM thapsigargin treatment. Over the 6-h period, the UPR allows proteins to be either degraded or folded resulting in a change in overall protein stability across the proteome. An upset plot with the number of overlapping proteins over the 1-, 3-, and 6-h experiments is shown in Figure 7F. The large number of overlapping stabilized proteins common to the three time points (222, ∼10% of all stabilized proteins) reflect a set of proteins with a consistent response to the treatment over the 6-h time period. Also of note, 44% of all stabilized proteins were reproducibly detected in the one- and 3-h time points. These data suggest that some of the intial protein-level responses to 1 μM thapsigargin treatment is resolved by 6 h (Fig. 7F). This resolution or change in levels of stress induction are reinforced by results from another lab where HEK293 cells were treated with thapsigargin over a time course of 0 to 30 h with intervals of ∼4 h (37). In Lin et al., there was reduced activation of UPR sensor (ATF6) with prolonged ER stress and reduced abundance of other downstream UPR targets such as CHOP, BiP, and ATF4. STRING networks were also generated using InflectSSP (Fig. 7, G–I) for proteins with melt shifts >3.5 C along with reported or predicted interactions that were also changing (using interaction score cutoff in STRING of 0.99). As expected, based on the rank order plots, the number of nodes in these diagrams decreases over the 6-h period. It can also be seen that there are some nodes that consistently appear in all three outputs. One group of proteins that is present in the 1-, 3-, and 6-h reports are FAF2, UBAC2, and AMFR (highlighted in the orange squares). FAF2 (or UBXD8) has been reported to play an important role in the ER-associated degradation process (ERAD) (38). FAF2 also has been reported to interact with UBAC2 to affect trafficking of FAF2 from the ER (39). The E3-ligase AMFR has then also been shown to interact with UBAC2 in the degradation of particular targets (40). Altogether, the consistent trend towards stabilization of proteins in our data set and the resolution of cellular responses over the time course likely reflects the biological responses to the SMI including reduction of translation and degradative type responses that occur during treatment with the UPR inducer Tg (37, 41). Overall, the thapsigargin TPP time course experiments clearly illustrate the utility and reproducibility of the InflectSSP workflow in detecting protein stability changes in a complex temporal dataset following SMI treatment.
Fig. 7.
InflectSSP output from analysis of TPP time course experiments where HEK293A cells were treated with thapsigargin for 1, 3, or 6 h or DMSO for 1 h.A–C, melt curves for SERCA2A at (G) 1 h, (H) 3 h, and (I) 6 h relative to DMSO at 1 h. D and E, rank order plots for all protein melt shifts in the time course experiments where proteins with melt shift p-value <0.05, melt shifts >3.5 °C or <−3.5 °C are highlighted in red. Plots are of melt shifts calculated between (D) DMSO at 1 h versus thapsigargin at 1 h, (E) DMSO at 1 h versus thapsigargin at 6 h. F, upset plot describing overlap of stabilized and destabilized proteins in the three time points in the thapsigargin time course experiment. G–I, STRING interaction diagrams for proteins with reported interactions, melt shifts >3.5 °C or <−3.5 °C, and STRING confidence score = 0.99. Diagrams are for (G) DMSO at 1 h versus thapsigargin at 1 h, (H) DMSO at 1 h versus thapsigargin at 3 h, (I) DMSO at 1 h versus thapsigargin at 6 h. Orange boxes in all three interaction diagrams highlight ERAD protein interactions that are present in all three data sets. ERAD, ER-associated degradation process; TPP, thermal proteome profiling.
FDR Correction
In the work described herein, we employed FDR correction in the calculation of melt shift p-values but did not always use adjusted p-values for target prioritization or designation of a data filtering cutoff. As has been discussed by others, the use of multiple correction testing is not always an easy choice in proteomic experiments as it can greatly affect the sensitivity of analysis (42). The FDR correction is also useful when simultaneously conducting a large number of hypothesis tests (i.e GWAS experiments) (43). In our hypothesis testing, we are only comparing the melt shift of single proteins between two conditions (and not relative shifts between all proteins in the dataset). Consequently, we were confident in our choice to limit use of FDR in p-value correction. To further explore the utility of FDR in our studies, we investigated the type of proteins that were observed as significant with and without FDR correction. We used proteins of interest along with STRING to determine the impact of correction. This analysis is summarized in supplemental Fig. S8A. When FDR correction was used, we observed that DNA-binding protein RFXANK had a statistically significant melt shift (supplemental Fig. S8B). While this protein has no reports of directly being associated with thapsigargin treatment, the gene has been reported to be a predicted target of the well-characterized UPR transcription factor ATF4 (44). When the FDR correction was then turned off for our analysis, however, we were able to observe several other proteins that have reported interactions with RFXANK including SLC35E1 (supplemental Fig. S8C) and MEF2BNB (supplemental Fig. S8D). In the case of MEF2BNB, this protein has been associated with the UPR sensor Ire1 (45). SLC35E1 is a transporter that aids in nucleotide-sugar movement across the ER during glycoprotein formation (46). Observation of these melt shifts both with and without FDR correction helped to confirm that we were able to observe biologically relevant proteins both with and without FDR correction. Users are able to use these cutoffs within InflectSSP in a customized manner to provide a high degree of flexibility for analysis of TPP datasets.
Comparison of InflectSSP with TPP Analysis Program and Calculation of Melt Coefficient
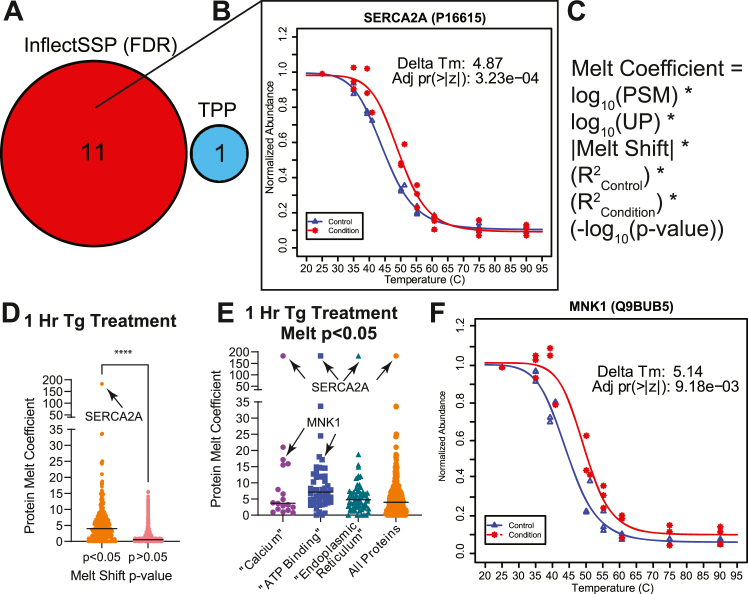
We were next interested in comparing the results from InflectSSP with those that are reported using the previously reported open-source R package, “TPP” (18). We analyzed our 1-h data set using both InflectSSP along with TPP version 3.22.1. To increase the number of replicates for analysis, we generated an additional biological replicate for the 1 μM thapsigargin treatment at the 1 h time point (n = 3). Since the “TPP” program uses multiple correction testing in its calculation of melt shift p-values, we also used the FDR optional function in execution of InflectSSP to keep the comparison equivalent. As shown in Figure 8A, the number of proteins identified as significant (adjusted p-value <0.05) was 1 for the “TPP” workflow, while InflectSSP observed a total of 11 significant proteins with no overlap in significantly changing protein melt shifts observed between the computational workflows. One of the proteins observed to be significantly stabilized by InflectSSP (no significance in “TPP”) was the well-described target of thapsigargin, SERCA2A. The melt curves for this protein are shown in Figure 8B (with full output in supplemental Fig. S9). The melt curves generated by the “TPP” program for SERCA2A are shown in supplemental Fig. S10 and demonstrate that change in stability of this protein is not observed when using the “TPP” program. One potential reason for the disparity in results (at least for SERCA2A) is that each workflow determines the melt temperature differently. In the case of InflectSSP, the melt temperature is the inflection point in the curve while in the case of TPP, it is temperature at 0.5 abundance. As we have described in previous work (13), the definition of melt can contribute significantly to the identification of proteins in a TPP experiment and thereby alter the determination of melt shift significance.
Fig. 8.
Comparison of InflectSSP outputs with “TPP” program and integration of “melt coefficient”. 1 h thapsigargin experiment used (n = 3). A, number of significant proteins identified through InflectSSP and “TPP”. Melt curves for SERCA2A (target of thapsigargin) by InflectSSP (B). C, melt coefficient formula that incorporates attributes of melt shift to quantify significance of a melt shift. D, distribution of melt coefficients for proteins with p-value <0.05 and ≥ 0.05. E, melt coefficient distribution with associated relevant GO terms for proteins with melt p-value <0.05. F, melt curves for protein of interest, MNK1, identified in (E). TPP, thermal proteome profiling.
The 1-h thapsigargin dataset was also analyzed without FDR correction to reduce the potential for loss of biologically significant changes (i.e., false negatives). When this analysis was done, there were a significant number of proteins of interest, and we wanted to determine whether we could rank order the proteins based on the “quality” of their melts. To answer this question, we incorporated a melt coefficient calculation into the InflectSSP program (calculation shown as Fig. 8C) that accounts for mass spec data quality considerations, melt shift magnitude, curve correlation, and melt shift p-value. We used the calculated coefficient from InflectSSP to distinguish which shifts had the strongest data supporting statistical significance. A comparison of the “melt coefficients” for proteins with melt shift p-values <0.05 and ≥ 0.05 (non-adjusted) is shown in Figure 8D. As shown in this panel, the average melt coefficient for the melts with p-value <0.05 are indeed higher than proteins with nonsignificant melts as expected. We also observed that the protein with the highest melt coefficient was SERCA2A, the target of thapsigargin. Since 1 μM thapsigargin treatment affects calcium through impact on an ATP binder in the ER, we were curious whether related GO terms could be observed in the proteins with p < 0.05. As seen in Figure 8E, many proteins with high melt coefficients and GO terms associated with “Calcium”, “ATP Binding,” and “Endoplasmic Reticulum” were observed consistent with the known function of the thapsigargin target SERCA2. One of the proteins with a high coefficient in both the “Calcium” and “ATP Binding” categories was MNK1, Figure 8F (with full output in supplemental Fig. S11). MNK1 is a MAP kinase that plays a role in protein translation through its interaction with cap-binding protein eIF4E (47). This finding affords a biological linkage with both calcium and the UPR through a drug that affects both by its inhibition of SERCA2. MNK1 has been implicated in Ca++ signaling and translational control including phosphorylation of mRNA cap-binding protein, EIF4E (48). It has been previously reported that MNK1 phosphorylation levels increase following thapsigargin treatment (49). Phosphorylation can impact protein stability in some cases (9), but changes in MNK1 phosphorylation have not previously been associated with a change in its thermal stability. Our findings using InflectSSP provide new insights into SMI mechanism of action with MNK1 showing biophysical state changes as a consequence of thapsigargin treatment. These results helped to support the utility of the melt coefficient in designating proteins of interest through prioritization of the InflectSSP output.
Discussion
TPP experiments and their associated results offer great potential for identification of intracellular changes to proteins and complexes; however, the data remains challenging to analyze and interpret because of the large number of potential variables that could have an impact on the final list of significant hits. In this work, we have sought to focus our analysis on the cutoff metrics that show clear impact on the sensitivity and selectivity of likely functional hits within the proteome. Our InflectSSP workflow is one methodology that can be used to calculate melt shifts from an experiment and then determine which shifts are significant from a statistical perspective. Our use of z-score with associated p-value calculation for biological replicate analysis provides an objective method for ascertaining the significance of a melt shift. This p-value is a valuable quality control criteria that we have shown improves the selectivity of the data analysis pipeline. We have also added peptide, PSM, and curve fit correlation limits to our workflow so that a user can vary the quality control filters in the analysis of experiments if desired. Unique bioinformatic tools have also been integrated into our workflow that allow for potential groups of targets and/or downstream effectors from TPP and related experiments to be rapidly identified. Finally, a “melt coefficient” calculation as incorporated into our program to further aid in identification of proteins of interest based on the quality of their melts. Three data sets have been used in our assessment of this R-based program, and results using these data sets help to validate the approach as an essential tool for TPP data analysis. Our results show changes in stability of proteins that are biologically relevant to the respective data set experiments. In the case of the novel thapsigargin dataset from our group, we have shown benefits of our analysis workflow as compared to the publicly available “TPP” program. Specifically, we observe the stabilization of MNK1 in the thapsigargin experiment. The melt coefficient calculations developed for InflectSSP show remarkable specificity for identification of this small molecule target SERCA2 in our UPR studies. Additional changes were also detected using the melt coefficient calculation and an FDR adjusted p-value cutoff that includes MNK1. MNK1 has not been previously implicated in SERCA2-dependent signaling or in the mechanism of action for thapsigargin. However, knockout of MNK1 has been shown to upregulate SERCA2 mRNA levels in adipose tissue (50), suggesting that they may have a functional connection. MNK1 is also the primary kinase responsible for eIF4E phosphorylation at Ser209 (51). Thapsigargin is known to induce the UPR through alteration of Ca++ homeostasis in the ER; however, our findings could suggest that MNK1 is a downstream effector of thapsigargin. This is intriguing since MNK1-dependent phosphorylation of eIF4E Ser209 has been shown to be required for preferential translation of ATF4 (52), a transcription factor that is canonically translated during the UPR.
Future work that could improve on this existing data program would include incorporation of multiple treatments in the analysis. The current version of InflectSSP allows for comparison of a single treatment with a single vehicle condition. The use of multiple treatments is a common strategy in TPP experiments and would therefore be a useful feature of an analysis pipeline. Incorporation of other melt determination strategies (i.e. nonparametric) would also be valuable for the InflectSSP pipeline.
InflectSSP is available at https://CRAN.R-project.org/package=InflectSSP with instructions on how to use the program in supplemental Fig. S12. Example outputs for the program are highlighted in supplemental Fig. S13.
Data Availability
RAW files, proteomics analysis results along with supplemental LC-MS/MS experiment information from the Thapsigargin data set (including .pdResult, .mzTab, .mzML, and .raw files) have been deposited into the MassIVE archive under accession number MSV000090867, doi:10.25345/C5VM4325J and in ProteomeXchange under PXD038752. The abundance values for this dataset are also included as Supplement_PDResults_Timecourse.xlsx.
A search was also completed on all of the 1-h experiment data (including the third biological replicate). RAW files and search results from this analysis can be found in MassIVE repository: MSV000090932, ftp://massive.ucsd.edu/MSV000090932/. The abundance values for this dataset are also included as Supplement_PDResults_1Hr.xlsx.
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We would like to thank the Mosley and Wek labs and Dr Ron Wek for multiple discussions regarding the project and manuscript. Dr Wek also provided partial support for Kirk Staschke and Neil McCracken for this project through National Institutes of Health, National Institute of General Medical Sciences R35GM136331. The IU Center for Proteome Analysis performed the mass spectrometry acquisition for this project and is partially supported by the Indiana Clinical and Translational Sciences Institute which is funded by Award Number UL1TR002529 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. The Indiana University Precision Health Initiative and the IU Simon Comprehensive Cancer Center also supported the acquisition of instrumentation in the Center for Proteome Analysis used for this project. We would like to thank Dr Emma Doud for her insights and discussions regarding public dataset reporting.
Funding and additional information
A portion of this work was supported by the National Institutes of Health, National Institute of Neurological Disorders and Stroke grant R01NS121550 (to A. L. M.), the Indiana University Diabetes and Obesity Research Training Program, DeVault Fellowship (to N. A. M.), the Showalter Research Trust (to A. B. W. and A. L. M.), and by the Indiana University Melvin and Bren Simon Cancer Center Support Grant in support of A. L. M. and A. B. W. (P30CA082709). The content is solely the responsibility of the authors and does not necessarily represent the official views of funders.
Author contributions
N. A. M., A. B. W., K. A. S., and A. L. M. conceptualization; N. A. M., H. L., A. M. R., H. R. S. W., A. B. W., K. A. S., and A. L. M. methodology; N. A. M., A. B. W., and K. A. S. investigation; N. A. M., H. L., A. M. R., H. R. S. W., and A. L. M. formal analysis; N. A. M., H. L., A. M. R., H. R. S. W., and A. L. M. data curation; N. A. M. and A. L. M. writing–original draft; N. A. M., H. L., A. M. R., H. R. S. W., A. B. W., K. A. S., and A. L. M. writing–review and editing.
Footnotes
Proteome Discoverer is a product of Thermo Fisher Scientific.
Supplementary Data
References
- 1.Savitski M.M., Reinhard F.B., Franken H., Werner T., Savitski M.F., Eberhard D., et al. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346 doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- 2.Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 3.Dai L., Zhao T., Bisteau X., Sun W., Prabhu N., Lim Y.T., et al. Modulation of protein-interaction states through the cell cycle. Cell. 2018;173:1481–1494.e13. doi: 10.1016/j.cell.2018.03.065. [DOI] [PubMed] [Google Scholar]
- 4.Becher I., Andres-Pons A., Romanov N., Stein F., Schramm M., Baudin F., et al. Pervasive protein thermal stability variation during the cell cycle. Cell. 2018;173:1495–1507.e18. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peck Justice S.A., Barron M.P., Qi G.D., Wijeratne H.R.S., Victorino J.F., Simpson E.R., et al. Mutant thermal proteome profiling for characterization of missense protein variants and their associated phenotypes within the proteome. J. Biol. Chem. 2020;295:16219–16238. doi: 10.1074/jbc.RA120.014576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarzab A., Kurzawa N., Hopf T., Moerch M., Zecha J., Leijten N., et al. Meltome atlas-thermal proteome stability across the tree of life. Nat. Methods. 2020;17:495–503. doi: 10.1038/s41592-020-0801-4. [DOI] [PubMed] [Google Scholar]
- 7.Liang Y.Y., Bacanu S., Sreekumar L., Ramos A.D., Dai L., Michaelis M., et al. CETSA interaction proteomics define specific RNA-modification pathways as key components of fluorouracil-based cancer drug cytotoxicity. Cell Chem. Biol. 2022;29:572–585.e8. doi: 10.1016/j.chembiol.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 8.Hashimoto Y., Sheng X., Murray-Nerger L.A., Cristea I.M. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat. Commun. 2020;11:806. doi: 10.1038/s41467-020-14586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith I.R., Hess K.N., Bakhtina A.A., Valente A.S., Rodriguez-Mias R.A., Villen J. Identification of phosphosites that alter protein thermal stability. Nat. Methods. 2021;18:760–762. doi: 10.1038/s41592-021-01178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potel C.M., Kurzawa N., Becher I., Typas A., Mateus A., Savitski M.M. Impact of phosphorylation on thermal stability of proteins. Nat. Methods. 2021;18:757–759. doi: 10.1038/s41592-021-01177-5. [DOI] [PubMed] [Google Scholar]
- 11.Huang J.X., Lee G., Cavanaugh K.E., Chang J.W., Gardel M.L., Moellering R.E. High throughput discovery of functional protein modifications by hotspot thermal profiling. Nat. Methods. 2019;16:894–901. doi: 10.1038/s41592-019-0499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieitez C., Busby B.P., Ochoa D., Mateus A., Memon D., Galardini M., et al. High-throughput functional characterization of protein phosphorylation sites in yeast. Nat. Biotechnol. 2021;40:382–390. doi: 10.1038/s41587-021-01051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCracken N.A., Peck Justice S.A., Wijeratne A.B., Mosley A.L. Inflect: optimizing computational workflows for thermal proteome profiling data analysis. J. Proteome Res. 2021;20:1874–1888. doi: 10.1021/acs.jproteome.0c00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Childs D., Kurzawa N., Franken H., Doce C., Savitski M., Huber W. 2019. TPP: Analyze Thermal Proteome Profiling (TPP) Experiments. R Package Version 3.14.0. Bioconductor version: Release (3.17) [Google Scholar]
- 15.Huang T., Choi M., Tzouros M., Golling S., Pandya N.J., Banfai B., et al. MSstatsTMT: statistical detection of differentially abundant proteins in experiments with isobaric labeling and multiple mixtures. Mol. Cell. Proteomics. 2020;19:1706–1723. doi: 10.1074/mcp.RA120.002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurzawa N., Mateus A., Savitski M.M. Rtpca: an R package for differential thermal proximity coaggregation analysis. Bioinformatics. 2021;37:431–433. doi: 10.1093/bioinformatics/btaa682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Childs D., Kurzawa N., Franken H., Doce C., Savitski M., Huber W. 2022. TPP: Analyze Thermal Proteome Profiling (TPP) Experiments, Version 3.22.1. Bioconductor version: Release (3.17) [Google Scholar]
- 18.Franken H., Mathieson T., Childs D., Sweetman G.M., Werner T., Togel I., et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 19.Kalxdorf M., Gunthner I., Becher I., Kurzawa N., Knecht S., Savitski M.M., et al. Cell surface thermal proteome profiling tracks perturbations and drug targets on the plasma membrane. Nat. Methods. 2021;18:84–91. doi: 10.1038/s41592-020-01022-1. [DOI] [PubMed] [Google Scholar]
- 20.Sridharan S., Kurzawa N., Werner T., Gunthner I., Helm D., Huber W., et al. Proteome-wide solubility and thermal stability profiling reveals distinct regulatory roles for ATP. Nat. Commun. 2019;10:1155. doi: 10.1038/s41467-019-09107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H., Van der Jeught K., Zhou Z., Zhang L., Yu T., Sun Y., et al. Atractylenolide I enhances responsiveness to immune checkpoint blockade therapy by activating tumor antigen presentation. J. Clin. Invest. 2021;131 doi: 10.1172/JCI146832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Justice S.A.P., McCracken N.A., Victorino J.F., Wijeratne A.B., Mosley A.L. Boosting detection of low abundance proteins in thermal proteome profiling experiments by addition of an isobaric trigger channel to TMT multiplexes. bioRxiv. 2020 doi: 10.1101/2020.12.30.424894. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.George A.L., Sidgwick F.R., Watt J.E., Martin M.P., Trost M., Marín-Rubio J.L., et al. A comparison of quantitative mass spectrometric methods for drug target identification by thermal proteome profiling. bioRxiv. 2023 doi: 10.1101/2023.02.15.528618. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiess A.-N., Neumeyer N. An evaluation of R2 as an inadequate measure for nonlinear models in pharmacological and biochemical research: a Monte Carlo approach. BMC Pharmacol. 2010;10:6. doi: 10.1186/1471-2210-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colin Cameron A., Windmeijer F.A.G. An R-squared measure of goodness of fit for some common nonlinear regression models. J. Econom. 1997;77:329–342. [Google Scholar]
- 28.Li J., Rix U., Fang B., Bai Y., Edwards A., Colinge J., et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang J., Lim L., Song J. ATP binds and inhibits the neurodegeneration-associated fibrillization of the FUS RRM domain. Commun. Biol. 2019;2:223. doi: 10.1038/s42003-019-0463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redin E., Garmendia I., Lozano T., Serrano D., Senent Y., Redrado M., et al. SRC family kinase (SFK) inhibitor dasatinib improves the antitumor activity of anti-PD-1 in NSCLC models by inhibiting Treg cell conversion and proliferation. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2020-001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium T.U. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melnick J.S., Janes J., Kim S., Chang J.Y., Sipes D.G., Gunderson D., et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lytton J., Westlin M., Hanley M.R. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 1991;266:17067–17071. [PubMed] [Google Scholar]
- 35.Sehgal P., Szalai P., Olesen C., Praetorius H.A., Nissen P., Christensen S.B., et al. Inhibition of the sarco/endoplasmic reticulum (ER) Ca(2+)-ATPase by thapsigargin analogs induces cell death via ER Ca(2+) depletion and the unfolded protein response. J. Biol. Chem. 2017;292:19656–19673. doi: 10.1074/jbc.M117.796920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walter P., Ron D. The unfolded protein response: from stress pathway to Homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 37.Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller B., Klemm E.J., Spooner E., Claessen J.H., Ploegh H.L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olzmann J.A., Richter C.M., Kopito R.R. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi J.H., Zhong X., McAlpine W., Liao T.C., Zhang D., Fang B., et al. LMBR1L regulates lymphopoiesis through Wnt/β-catenin signaling. Science. 2019;364 doi: 10.1126/science.aau0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Márton M., Bánhegyi G., Gyöngyösi N., Kálmán E., Pettkó-Szandtner A., Káldi K., et al. A systems biological analysis of the ATF4-GADD34-CHOP regulatory triangle upon endoplasmic reticulum stress. FEBS Open Bio. 2022;12:2065–2082. doi: 10.1002/2211-5463.13484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuken S.R., McNerney M.W. Costs and benefits of popular P-value correction methods in three models of quantitative omic experiments. Anal. Chem. 2023;95:2732–2740. doi: 10.1021/acs.analchem.2c03719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noble W.S. How does multiple testing correction work? Nat. Biotechnol. 2009;27:1135–1137. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plaisier C.L., O’Brien S., Bernard B., Reynolds S., Simon Z., Toledo C.M., et al. Causal mechanistic regulatory network for glioblastoma deciphered using systems genetics network analysis. Cell Syst. 2016;3:172–186. doi: 10.1016/j.cels.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bae D., Moore K.A., Mella J.M., Hayashi S.Y., Hollien J. Degradation of Blos1 mRNA by IRE1 repositions lysosomes and protects cells from stress. J. Cell Biol. 2019;218:1118–1127. doi: 10.1083/jcb.201809027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander S.P., Benson H.E., Faccenda E., Pawson A.J., Sharman J.L., Spedding M., et al. The concise guide to pharmacology 2013/14: transporters. Br. J. Pharmacol. 2013;170:1706–1796. doi: 10.1111/bph.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waskiewicz A.J., Johnson J.C., Penn B., Mahalingam M., Kimball S.R., Cooper J.A. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 1999;19:1871–1880. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyronnet S., Imataka H., Gingras A.-C., Fukunaga R., Hunter T., Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi Y., Yang Y., Hoang B., Bardeleben C., Holmes B., Gera J., et al. Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene. 2016;35:1015–1024. doi: 10.1038/onc.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Sandeman L.Y., Kang W.X., Wang X., Jensen K.B., Wong D., Bo T., et al. Disabling MNK protein kinases promotes oxidative metabolism and protects against diet-induced obesity. Mol. Metab. 2020;42 doi: 10.1016/j.molmet.2020.101054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 2004;24:6539–6549. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan H., Li X., Xu X., Leibowitz B.J., Tong J., Chen L., et al. eIF4E S209 phosphorylation licenses myc- and stress-driven oncogenesis. Elife. 2020;9 doi: 10.7554/eLife.60151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RAW files, proteomics analysis results along with supplemental LC-MS/MS experiment information from the Thapsigargin data set (including .pdResult, .mzTab, .mzML, and .raw files) have been deposited into the MassIVE archive under accession number MSV000090867, doi:10.25345/C5VM4325J and in ProteomeXchange under PXD038752. The abundance values for this dataset are also included as Supplement_PDResults_Timecourse.xlsx.
A search was also completed on all of the 1-h experiment data (including the third biological replicate). RAW files and search results from this analysis can be found in MassIVE repository: MSV000090932, ftp://massive.ucsd.edu/MSV000090932/. The abundance values for this dataset are also included as Supplement_PDResults_1Hr.xlsx.