The nutrient sensor O-GlcNAc transferase modifies proteins with the O-GlcNAc moiety. In this issue, Capotosti et al. (2011) reveal that O-GlcNAc transferase not only glycosylates the cell-cycle regulator host cell factor 1 but activates it through proteolytic cleavage, providing a surprising link between metabolism and epigenetic regulation of the cell cycle.
Host cell factor 1 (HCF-1) is an evolutionarily conserved epigenetic regulator that undergoes an unusual proteolytic processing event to generate stably associated fragments that regulate different stages of the cell cycle (Kristie et al., 2010). In vertebrates, the molecular identity of the factor responsible for HCF-1 cleavage has been cloaked in mystery. With a plot twist worthy of a best-selling detective novel, Capotosti et al. now unmask an unexpected culprit—the nutrient-sensing epigenetic regulator O-linked GlcNAc transferase (OGT) (Figure 1). Their findings provide compelling evidence that OGT has a dual enzymatic role, catalyzing both O-GlcNA-cylation and cleavage of HCF-1. This partnering of two epigenetic regulators to control cell-cycle progression has important implications for fields as diverse as viral pathogenesis, stem cell biology, and diseases of aging including cancer, diabetes, and neurodegeneration.
Figure 1. OGT GlcNAcylates and Cleaves HCF-1 for Cell-Cycle Progression.
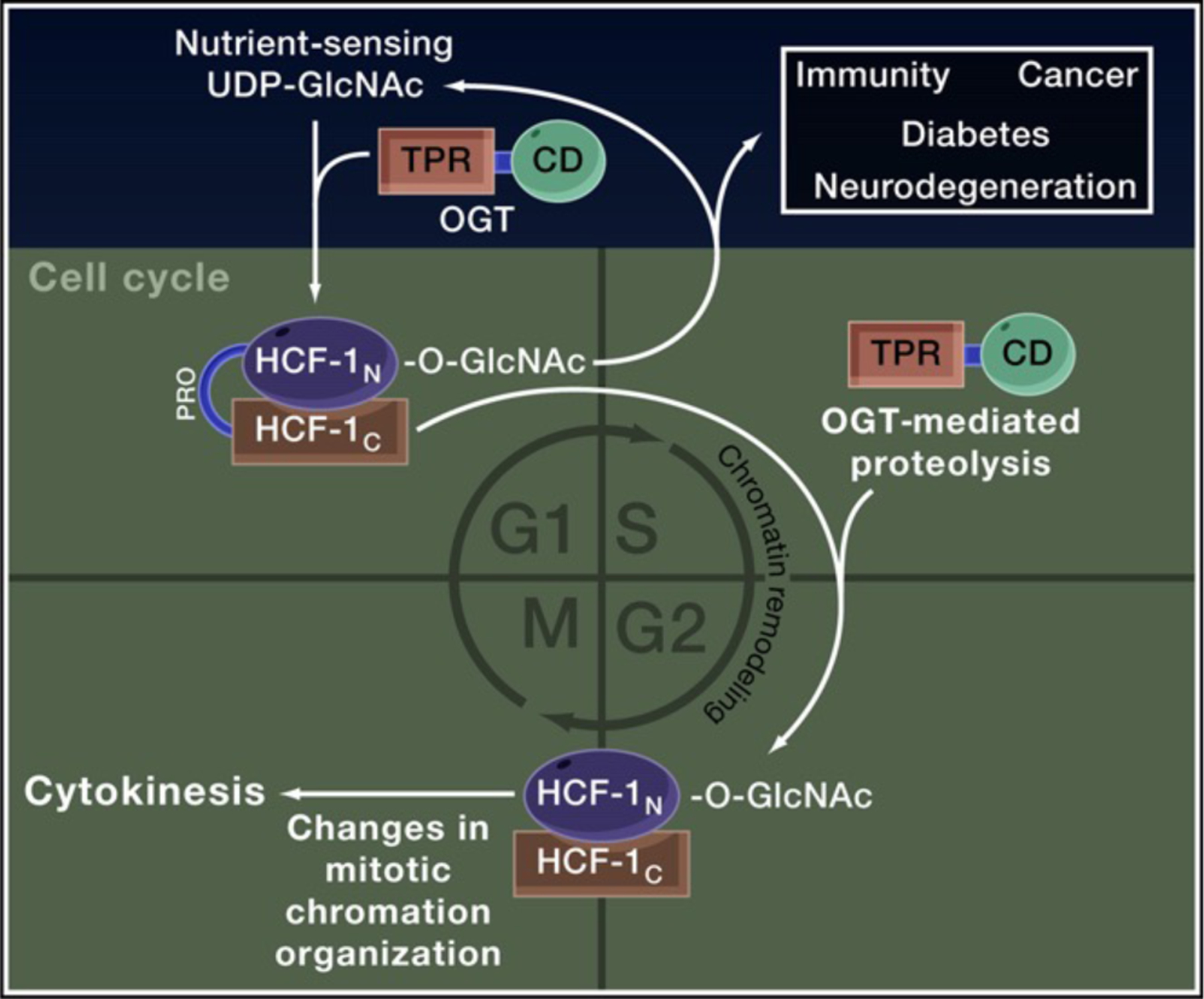
O-GlcNAc transferase (OGT) couples the nutrient-dependent synthesis of UDP-GlcNAc to the O-GlcNAcylation of numerous targets including the cell-cycle regulator host cell factor 1 (HCF-1). OGT is also necessary and sufficient for proteolysis of the HCF-1PRO domain; together O-GlcNAcylation and cleavage of HCF-1 promote changes in chromatin organization that are required for progression through G1-S and M phases of the cell cycle. OGT and HCF-1 are tethered together in a tight complex and share a number of known binding partners and targets, participating in chromatin remodeling and cell-cycle regulation to influence stem cell fate, development, and disease-related processes including immunity, cancer, diabetes, and neurodegeneration.
Although the HCF-1 protein was originally identified as a target for the Herpes simplex virus VP16 transcriptional activator, subsequent work has shown that HCF-1 normally coordinates passage through the cell cycle (Goto et al., 1997). The N-terminal subunit, HCF-1N, promotes progression through G1, whereas the C-terminal subunit, HCF-1C, regulates mitosis and cytokinesis. In vertebrates, these domains are separated by six conserved HCF-1PRO repeats, which contain many proline and threonine residues and an invariant glutamate that marks the site of proteolysis. Cleavage at this site is required for HCF-1 regulation of M phase events. Although HCF-1 activity in all animals is regulated by proteolysis, the HCF-1 sequence differs greatly between invertebrates and vertebrates such that different animal lineages employ distinct mechanisms for HCF-1 processing. Lower metazoans employ a threonine-directed endopeptidase (taspase) to sever the two HCF-1 subunits, but vertebrates use a different strategy (Capotosti et al., 2011).
Multiple pieces of evidence hint that posttranslational modification of HCF-1 might be important for cleavage. Capotosti et al. show that inhibitors of phosphorylation do not alter processing, but a presumptive OGT inhibitor and depletion of OGT by RNA interference prevent HCF-1 maturation. Although O-GlcNAcylation of HCF-1 depends on the presence of the threonine-rich HCF-1PRO repeats, it is several threonines in the HCF-1N N-terminal subunit that are glycosylated. OGT does more than simply glycosylate HCF-1 before it is cleaved. Capotosti et al. show that OGT glycosylates and then remains bound to a cleavage-resistant HCF-1 mutant. Furthermore, they find that purified recombinant OGT cleaves the HCF-1PRO repeat in vitro. The authors suggest that O-GlcNAc-catalyzed autoproteolysis of HCF-1 does not efficiently cleave HCF-1PRO repeats in vitro. Providing an in vivo validation of this conclusion, the authors replace the vertebrate HCF-1PRO domain with a taspase cleavage site. Although this variant is efficiently processed by taspase, it is neither recognized nor cleaved by OGT, and this heterologous HCF-1 does not rescue HCF-1 function. Together, these data strongly suggest that OGT-dependent glycosylation and cleavage are critical for the M phase functions of HCF-1, building a compelling case that OGT is the long-sought HCF-1 protease (Figure 1).
OGT contains no obvious protease-like domains, so how does it catalyze HCF-1 proteolysis? The authors examine two possibilities. OGT is part of an enzyme superfamily with known hydrolases as members, and as such, it could retain cryptic protease activity. Based on similarities between the OGT-mediated cleavage of HCF-1PRO and the proteolysis of E. coli LexA by its coprotease, RexA, the authors favor a model in which OGT promotes a conformational change in the HCF-1PRO domain, repositioning the cleavage site and the Nterminal O-GlcNAc moieties to stimulate hydrolysis (Capotosti et al., 2011).
it under the control of stress-triggered, nutrient-responsive O-GlcNAcylation and thus has profound implications for fields ranging from viral pathogenesis and stem cell biology to diseases of aging. Viruses, like Herpes simplex, notoriously subvert the host cell-cycle machinery for maintaining dormancy and viral production (Kristie et al., 2010). Integrating both stress and nutrient availability, host O-GlcNAcylation is a likely regulator of viral latency. Stem cell pluripotency and self-renewal may also be regulated by OGT and HCF-1 as both are components of the Oct4-centered transcription factor network implicated in stem cell pluripotency (Love et al., 2010b). In adulthood, obesity and nutrient excess are associated with an increased risk for diabetes, cardiovascular disease, and cancer; genome-wide association studies have linked
The recent finding that OGT modulates cell-cycle dynamics through its modification and processing of HCF-1 provides definitive support for a growing body of evidence linking O-GlcNAc to epigenetics and disease (Hanover et al., 2010 and Love et al., 2010b). O-GlcNAc is added to and removed from proteins by separate enzymes in a manner reminiscent of dynamic phosphorylation. OGT couples the nutrient-dependent synthesis of UDP-GlcNAc to the O-GlcNAcylation of Ser/Thr residues of a variety of targets. This link between nutrient availability and O-GlcNAcylation of proteins through the hexosamine signaling pathway provides cells with a mechanism to sense and respond to a variety of environmental conditions. Some of the known O-GlcNAcylated proteins include signaling kinases, nuclear pores components, various chromatin-remodeling enzymes, and RNA polymerase II. In addition, one isoform of OGT is targeted to mitochondria where it coordinates apoptotic and metabolic functions by adding O-GlcNAc to key mitochondrial proteins (Hanover et al., 2010). The O-GlcNAc moiety is removed by the O-GlcNAcase, MGEA5. Together the enzymes of O-GlcNAc cycling balance the activity of numerous kinases, influencing the robustness of cell proliferation and tissue homeostasis decisions (Butkinaree et al., 2010; Hanover et al., 2010). Both OGT and MGEA5 have been previously implicated in cell-cycle progression (Wang et al., 2010) and epigenetic regulation (Hanover et al., 2010). In Drosophila, OGT is encoded by the homeotic gene ‘‘Super sex combs,’’ a gene previously linked to the epigenetic regulation of the polycomb and trithorax groups (Love et al., 2010b). In C. elegans, mutations that affect O-GlcNAc cycling provide insight into the signaling cascades triggered by stress, infection, and aging (Love et al., 2010a). In vertebrates, OGT and HCF-1 coexist in deacetylase and methyltransferase chromatin-remodeling complexes, providing yet another link to epigenetic regulatory events (Wysocka et al., 2003).
The pairing of OGT with HCF-1 regulates cell-cycle progression by placing the OGT and MGEA5 loci to these same diseases (Hanover et al., 2010; Butkinaree et al., 2010). The recently uncovered connection between OGT and HCF-1 provides an important clue for understanding how the environmentally responsive hexosamine-signaling pathway intersects with human development and disease, highlighting the importance of the O-GlcNAc modification in epigenetic regulation of cell proliferation and signaling during development, adulthood, and senescence.
REFERENCES
- Butkinaree C, Park K, and Hart GW (2010). Biochim. Biophys. Acta 1800, 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, and Herr W (2011). Cell 144, this issue, 376–388. [DOI] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, and Nishimoto T (1997). Genes Dev 11, 726–737. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, and Love DC (2010). Biochim. Biophys. Acta 1800, 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie TM, Liang Y, and Vogel JL (2010). Biochim. Biophys. Acta 1799, 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Ghosh S, Mondoux MA, Fukushige T, Wang P, Wilson MA, Iser WB, Wolkow CA, Krause MW, and Hanover JA (2010a). Proc. Natl. Acad. Sci. USA 107, 7413–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Krause MW, and Hanover JA (2010b). Semin. Cell Dev. Biol 21, 646–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, and Hart GW (2010). Sci. Signal 3, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Myers MP, Laherty CD, Eisenman RN, and Herr W (2003). Genes Dev 17, 896–911. [DOI] [PMC free article] [PubMed] [Google Scholar]


