Abstract
To determine the types of Enterocytozoon bieneusi strains associated with intestinal microsporidiosis, we developed a rapid and efficient approach for typing parasites obtained from stool specimens by PCR-restriction fragment length polymorphism (PCR-RFLP). Typing was based on DNA polymorphism of the ribosomal DNA internal transcribed spacer (ITS) region of E. bieneusi. RFLPs generated with two restriction enzymes (NlaIII and Fnu4HI) in PCR-amplified ITS products were used to classify strains into different lineages. This approach was successfully used to differentiate 78 strains that had been obtained from the stools of 65 patients with intestinal microsporidiosis. Among the 78 strains, we found four genetically unrelated lineages, showing the genetic diversity of E. bieneusi. Type I strains of E. bieneusi were found in a majority of the samples, accounting for 51 (78%) of the 65 microsporidiosis cases. In contrast, type II, III, and IV strains were found in only 8 (12%), 3 (5%), and 3 (5%) cases, respectively. All strains of E. bieneusi we have tested so far fall into one of four different lineages, and this study shows that human intestinal microsporidiosis is most often associated with type I strains. PCR-RFLP analysis of the ITS region of E. bieneusi should be useful for epidemiological studies.
Microsporidia are obligate intracellular protozoan parasites that can infect vertebrates as well as invertebrates. Over 1,000 species of microsporidia have been described, but reports of human infections were rare before the AIDS epidemic. Since then, microsporidia have been recognized as opportunistic pathogens in patients with AIDS. Different species of microsporidia have been shown to infect humans, and among these species, Enterocytozoon bieneusi (7) is by far the most frequently identified. E. bieneusi infection is responsible for chronic diarrhea and wasting in immunocompromised patients, such as patients with AIDS and organ transplant recipients (2, 15, 17, 23). Substantial variation in the progression and severity of disease is observed among cases of intestinal microsporidiosis, and these differences are presumably due to several factors, including host immune defenses and phenotypic differences in parasite strains (23).
However, modes of transmission and sources of human infection by E. bieneusi are largely unknown (11, 23). Epidemiologic investigations of a number of infectious diseases have shown that amplification methods could be extremely valuable tools for laboratory-based investigations (20). This approach has proven successful in defining different strains of Encephalitozoon cuniculi, a zoonotic microsporidian that can infect humans and a wide variety of other mammals (6, 8, 10, 14). At least three strains of this species have been defined, on the basis of the number of 5′-GTTTT-3′ repeats present in the internal transcribed spacer (ITS) region of the ribosomal DNA. We wished to study the genetic diversity of E. bieneusi strains by a similar approach. In this report, we describe the development of an amplification-based assay for genetic analysis of the ITS region of E. bieneusi that should be useful for epidemiological studies.
MATERIALS AND METHODS
Patient specimens.
Seventy-eight stool specimens were obtained over a 4-year peiod (1994 to 1997) from 65 patients with intestinal microsporidiosis seen at the Department of Infectious Diseases, Hôpital Saint-Louis, Paris, and the Hôtel-Dieu, Lyon, France (15–17). All patients were immunocompromised. Of these patients, 63 were infected with human immunodeficiency virus (HIV) and 2 were not HIV infected but had undergone heart-lung and kidney transplantation. The diagnosis of microsporidial infection in these patients was made by the detection of typical spores in stools by two different techniques as previously described (16). The species-level identification of E. bieneusi was made by transmission electron microscopy analysis of duodenal biopsy samples (38 patients) and/or by PCR with DNA extracted from duodenal biopsy samples or stools (44 patients) with primer sets specific for the small-subunit rRNA gene of E. bieneusi as previously described (4, 12).
DNA extraction.
Fresh stools were inactivated for 2 h at 65°C and stored at −20°C until DNA extraction. Microsporidian DNA was extracted from 100 μl of filtrated stool suspension by the guanidium thiocyanate-based method described by Boom et al. (1), as adapted for stool samples by van der Hoek et al. (21). A 100-bp DNA size marker was used as a control for DNA extraction.
PCR amplification.
Primers for PCR were chosen to amplify the ITS region of E. bieneusi. The forward primer Eb.gc (5′-TCAGTTTTGGGTGTGGTATCGG-3′), complementary to positions 1 to 22, was designed by using the GenBank sequence of E. bieneusi (22, 24) (accession no. L20290). The reverse primer Eb.gt (5′-GCTACCCATACACACATCATTC-3′) was designed to be complementary to positions 189 to 210 of the GenBank sequence of E. bieneusi (22, 24) (accession no. L20290). The PCR was performed in 50 μl of reaction mixtures that contained the following: 2.5 μg of each primer/ml, 200 μmol of each deoxynucleoside triphosphate/liter, 75 mmol of Tris-HCl (pH 9.0)/liter, 20 mmol of (NH4)2SO4/liter, 0.01% Tween 20, 2 mmol of MgCl2/liter, and 2 U of heat-stable DNA polymerase (Goldstar; Eurogentec, Seraing, Belgium). DNA amplification was performed on a Hybaid (Teddington, Middlesex, United Kingdom) touch down. The amplification procedure included 3 min of initial denaturation at 94°C (1 cycle), followed by 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 60 s of extension at 72°C for 40 cycles. A 10-min extension at 72°C was used after the 40 cycles. Each set of reactions included a negative control (a tube containing all the reagents but not the template DNA), to ensure the absence of contamination of samples during analysis, and a positive control. Culture spores of Encephalitozoon intestinalis, E. cuniculi, and Encephalitozoon hellem were also tested to assess the specificity of the primer set Eb.gc-Eb.gt. A 10-μl aliquot from each reaction mixture was run on a 1.2% agarose gel (agarose standard; Bioprobe, Montreuil, France) and was stained with ethidium bromide to visualize the amplified-PCR products under UV illumination.
DNA sequencing of PCR products.
To assess genetic diversity among stool isolates, amplification products of the ITS regions from six independent samples were gel purified from 1.2% agarose gels run in 1× Tris-borate-EDTA with a Geneclean II DNA purification kit (Bio 101, Vista, Calif.) by following the manufacturer’s protocol. For each specimen, the nucleotide sequence of the amplified product was determined by a commercial company (Euro Sequence Genes Services, Evry, France) on an ABI 377 sequencer using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase FS (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). Both strands were sequenced with the primers used for the PCR. The nucleotide sequence of each specimen was compared to the others and with the E. bieneusi ITS GenBank sequence (accession no. L20290) with Sequence Navigator software (Perkin-Elmer, Applied Biosystems Division).
Restriction endonuclease digestion of PCR products.
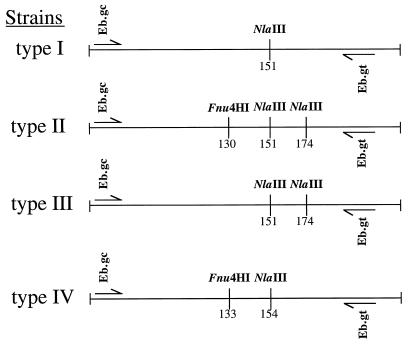
Two restriction endonucleases, NlaIII and Fnu4HI (New England Biolabs, Beverly, Mass.), were selected from the DNA sequencing data with DNA Strider 1.2 software (Commisariat Energie Atomique, Gif-sur-Yvette, France) to digest PCR products obtained from amplification of stool DNA extracts (Fig. 1). Ten microliters of PCR product was digested in two separate tubes with 10 U of each restriction enzyme in a final volume of 20 μl, as described previously (18). The restriction digests were electrophoresed through polyacrylamide gels (Gibco BRL, Paisley, Scotland), stained with ethidium bromide, and photographed under UV illumination. All restriction fragment length polymorphism (RFLP) patterns were compared visually and classified as matching if the numbers and molecular weights of the bands were identical.
FIG. 1.
Diagram of the E. bieneusi ITS region showing the locations of the primers used for PCR amplification of the 210-bp fragment and the polymorphic restriction sites used for identification of types.
RESULTS
PCR amplification of the E. bieneusi ITS region.
DNA was easily extracted from all stool specimens. Use of the Eb.gc-Eb.gt primer set resulted in efficient amplification of all specimens tested. For three (5%) stool specimens, however, a 10-fold dilution of released DNA was necessary to remove PCR inhibitors. No other modification of the protocol was made for these three samples, and the results of amplification were as unambiguous as those obtained with the other samples. The positions of the primer set Eb.gc-Eb.gt within the ITS sequence predicted that a 210-bp fragment would be generated. Fragments of the appropriate size were visualized on agarose gels following PCR of released DNA obtained from all stool specimens, but not from culture spores of E. intestinalis, E. cuniculi, or E. hellem (data not shown).
Nucleotide sequence of the ITS regions of six E. bieneusi isolates.
The sequences of four amplified DNA fragments obtained from stool samples matched the sequence of the GenBank database (accession no. L20290) for the ITS region of E. bieneusi, confirming that we successfully amplified E. bieneusi from the stools of our patients. The nucleotide sequences of the two other amplified DNA fragments, however, showed one and seven mismatches (97% identity) when compared with the E. bieneusi ITS GenBank sequence. The E. bieneusi ITS GenBank sequence was classified as type I, as were the four amplified fragments that matched the ITS E. bieneusi GenBank sequence. The amplified fragment that showed one mismatch was still classified as type I (no restriction site), and the amplified fragment that showed seven mismatches was classified as type II.
Typing analysis.
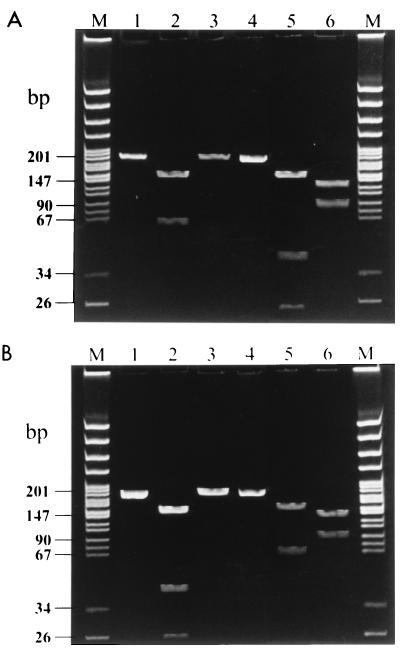
RFLP analysis of the amplification products of the ITS region was then performed on the 78 E. bieneusi stool isolates. For all samples, digestion of amplicons with NlaIII and Fnu4HI produced distinctive fragments detectable in ethidium bromide-stained polyacrylamide gels (Fig. 2). The sizes of the DNA fragments in the various RFLP profiles were all in accordance with the positions of the restriction enzyme cleavage sites on the ITS region as deduced from their sequences (Table 1). Among the 78 stool specimens we found four genetically unrelated lineages. Type I strains of E. bieneusi were found in a majority of the samples, accounting for 51 (78%) of the 65 microsporidiosis cases (Table 2). In contrast, type II, III, and IV strains were found in only 8 (12%), 3 (5%), and 3 (5%) of the 65 cases, respectively (Table 2). No association was seen between the types of the parasite strains and clinical presentation, and no mixed infection was noted. However, stool specimens from 2 non-HIV-infected patients with intestinal microsporidiosis contained a type II strain, while only 6 of 63 (9.5%) HIV-infected patients were found to shed type II strains in their stools (Table 2).
FIG. 2.
RFLP analysis of PCR products of strains of the four E. bieneusi lineages generated with primer set Eb.gc-Eb.gt and digested by either NlaIII or Fnu4HI. (A) Lanes: M, molecular size markers corresponding to pBR322 DNA digested by MspI (New England Biolabs); 1 to 3, PCR products from type I strains without digestion and following digestion by NlaIII and Fnu4HI, respectively; 4 to 6, PCR products from type II strains without digestion and following digestion by NlaIII and Fnu4HI, respectively. (B) Lanes: M, molecular size markers; 1 to 3, PCR products from type III strains without digestion and following digestion by NlaIII and Fnu4HI, respectively; 4 to 6, PCR products from type IV strains without digestion and following digestion by NlaIII and Fnu4HI, respectively. The different strains can be distinguished by comparing the numbers and sizes of bands.
TABLE 1.
Sizes of restriction enzyme digests
| E. bieneusi strain type | Fragment size(s) (bp)
|
|
|---|---|---|
| NlaIII | Fnu4HI | |
| I | 151, 59 | 210 |
| II | 151, 36, 23 | 130, 80 |
| III | 151, 36, 23 | 210 |
| IV | 154, 56 | 133, 77 |
TABLE 2.
Prevalence of E. bieneusi strain types in stool specimens
| Clinical status of patients (n) (no. of patients) | No. (%) of isolates
|
|||
|---|---|---|---|---|
| Type I | Type II | Type III | Type IV | |
| HIV infected (63) | 51 (81) | 6 (9) | 3 (5) | 3 (5) |
| Not HIV infected (2) | 0 (0) | 2 (100) | 0 (0) | 0 (0) |
| Total | 51 (78) | 8 (12) | 3 (5) | 3 (5) |
For nine HIV-infected patients, we further tested whether the same strain of E. bieneusi was found in stools over time by analyzing sequential stool specimens. For each of the nine patients, all stool specimens displayed the same RFLP profile over time (data not shown).
DISCUSSION
In the present report, we describe a rapid and efficient method for determining the types of unrelated human E. bieneusi strains isolated from stool specimens of patients with intestinal microsporidiosis.
Different PCR assays have been used previously for detection of E. bieneusi in clinical samples from patients with intestinal microsporidiosis, using the single-stranded rRNA gene as the target (3, 4, 9, 12). This locus, however, is not sufficiently polymorphic to allow strain typing (9, 22, 24), and E. bieneusi was thought therefore to be a relatively homogeneous entity. Consequently, we chose to develop a PCR-RFLP assay based on the putative polymorphism of the ITS region of E. bieneusi to study the genetic diversity of E. bieneusi. This approach has proven successful for the typing of E. cuniculi strains (6, 8, 10, 14).
In our study, the presence of restriction sites in the ITS region of E. bieneusi permitted the identification of individual strains by DNA fingerprinting with RFLP analysis. Indeed, PCR amplification of the ITS region of E. bieneusi, followed by RFLP analysis, allowed us to assign all 78 isolates tested to one of four different lineages. Also, the same strains were found in sequential stool specimens from the same patients, suggesting the clonal nature of E. bieneusi infection in the gastrointestinal tract. Among the 65 patients we studied, type I strains of E. bieneusi were much more prevalent than type II, III, or IV strains. It is, however, not clear yet whether this high prevalence of type I strains in humans simply reflects a common source of strains that lead to human infection. Alternatively, this high prevalence could be related to the pathogenicity of this strain in a particular type of host, namely, HIV-infected patients. Indeed, stool specimens of the two immunocompromised patients without HIV infections contained only type II strains, and this type occurred in only 9% of HIV-infected patients (Table 2). However, no evident association between clinical symptoms and strain type was seen in our study, as all of our patients were symptomatic.
Interestingly, E. bieneusi has recently been found in stool specimens from a pig and from simian immunodeficiency virus-infected macaques (5, 13). Only the sequence of E. bieneusi obtained from the pig is available (GenBank accession no. U61180) (5). This strain belongs to the type IV lineage, which was found in only 5% of our patients, but in none of these cases was contact with pigs documented. Furthermore, preliminary results of a recent study seemed to indicate the presence of E. bieneusi in surface water (19), a likely source of human contamination. Undoubtedly, the comparison of the types of E. bieneusi isolates from human, animal, and environmental sources will provide a better understanding of the epidemiology of E. bieneusi infections.
ACKNOWLEDGMENTS
This work was supported in part by grants from SIDACTION (Fondation pour la Recherche Médicale), Association des Professeurs de Pathologie Infectieuse et Tropicale (APPIT), Agence Nationale de Recherche sur le SIDA (ANRS 034, 035, and 054), and the Centre d’Etudes et de Recherches en Infectiologie.
We thank M. Rabodonirina for providing us with stool specimens from organ transplant recipients.
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canning E U. Microsporidia. In: Kreier J P, Baker J R, editors. Parasitic protozoa. 2nd ed. Vol. 6. New York, N.Y: Academic Press, Inc.; 1993. pp. 299–385. [Google Scholar]
- 3.da Silva A J, Schwartz D A, Visvesvara G S, de Moura H, Slemenda S B, Pieniazek N J. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J Clin Microbiol. 1996;34:986–987. doi: 10.1128/jcm.34.4.986-987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David F, Schuitema A R J, Sarfati C, Liguory O, Hartskeerl R A, Derouin F, Molina J M. Detection and species identification of intestinal microsporidia by polymerase chain reaction in duodenal biopsies from human immunodeficiency virus-infected patients. J Infect Dis. 1996;174:874–877. doi: 10.1093/infdis/174.4.874. [DOI] [PubMed] [Google Scholar]
- 5.Deplazes P, Mathis A, Müller C, Weber R. Molecular epidemiology of Encephalitozoon cuniculi and first detection of Enterocytozoon bieneusi in faecal samples of pigs. J Eukaryot Microbiol. 1996;43:93S. doi: 10.1111/j.1550-7408.1996.tb05018.x. [DOI] [PubMed] [Google Scholar]
- 6.Deplazes P, Mathis A, Baumgartner R, Tanner I, Weber R. Immunologic and molecular characteristics of Encephalitozoon-like microsporidia isolated from humans and rabbits indicate that Encephalitozoon cuniculi is a zoonotic parasite. Clin Infect Dis. 1996;22:557–559. doi: 10.1093/clinids/22.3.557. [DOI] [PubMed] [Google Scholar]
- 7.Desportes I, Le Charpentier Y, Galian Y, Bernard F, Cochand-Priollet B, Lavergne A, Ravisse P, Modigliani R. Occurrence of a new microsporidian: Enterocytozoon bieneusi n.g., n.sp., in the enterocytes of a human patient with AIDS. J Protozool. 1985;32:250–254. doi: 10.1111/j.1550-7408.1985.tb03046.x. [DOI] [PubMed] [Google Scholar]
- 8.Didier E S, Vossbrinck C R, Baker M D, Rogers L B, Bertucci D C, Shadduck J A. Identification and characterization of three Encephalitozoon cuniculi strains. Parasitology. 1995;111:411–421. doi: 10.1017/s0031182000065914. [DOI] [PubMed] [Google Scholar]
- 9.Fedorko D P, Nelson N A, Cartwright C P. Identification of microsporidia in stool specimens by using PCR and restriction endonucleases. J Clin Microbiol. 1995;33:1739–1741. doi: 10.1128/jcm.33.7.1739-1741.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollister W S, Canning E U, Anderson C L. Identification of microsporidia causing human disease. J Eukaryot Microbiol. 1996;43:104S–105S. doi: 10.1111/j.1550-7408.1996.tb05026.x. [DOI] [PubMed] [Google Scholar]
- 11.Hutin Y, Sombardier M N, Sarfati C, Decazes J-M, Modai J, Molina J M. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Risk factors for intestinal microsporidiosis in patients infected with human immunodeficiency virus (HIV), abstr. I-150; p. 271. [Google Scholar]
- 12.Liguory O, David F, Sarfati C, Schuitema A R J, Hartskeerl R A, Derouin F, Modai J, Molina J M. Diagnosis of infections caused by Enterocytozoon bieneusi and Encephalitozoon intestinalis using polymerase chain reaction in stool specimens. AIDS. 1997;11:723–726. doi: 10.1097/00002030-199706000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield K G, Carville A, Shvetz D, MacKey J, Tzipori S, Lackner A A. Identification of an Enterocytozoon bieneusi-like microsporidian parasite in simian-immunodeficiency-virus-inoculated macaques with hepatobiliary disease. Am J Pathol. 1997;150:1395–1405. [PMC free article] [PubMed] [Google Scholar]
- 14.Mathis A, Michel M, Kuster H, Müller C, Weber R. Two Encephalitozoon cuniculi strains of human origin are infectious to rabbits. Parasitology. 1997;114:29–35. doi: 10.1017/s0031182096008177. [DOI] [PubMed] [Google Scholar]
- 15.Molina J M, Sarfati C, Beauvais B, Lémann M, Lesourd A, Ferchal F, Casin I, Lagrange P, Modigliani R, Derouin F, Modai J. Intestinal microsporidiosis in human immunodeficiency virus-infected patients with chronic unexplained diarrhea: prevalence and clinical and biologic features. J Infect Dis. 1993;167:217–221. doi: 10.1093/infdis/167.1.217. [DOI] [PubMed] [Google Scholar]
- 16.Molina J M, Goguel J, Sarfati C, Chastang C, Desportes-Livage I, Michiels J-F, Maslo C, Katlama C, Cotte L, Leport C, Raffi F, Derouin F, Modai J. Potential efficacy of fumagillin in intestinal microsporidiosis due to Enterocytozoon bieneusi in patients with HIV-infection: results of a drug screening study. AIDS. 1997;11:1603–1610. doi: 10.1097/00002030-199713000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Rabodonirina M, Bertocchi M, Desportes-Livage I, Cotte L, Levrey H, Piens M A, Monneret G, Celard M, Mornex J F, Mojon M. Enterocytozoon bieneusi as a cause of chronic diarrhea in a heart-lung transplant recipient who was seronegative for human immunodeficiency virus. Clin Infect Dis. 1996;23:114–117. doi: 10.1093/clinids/23.1.114. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Sparfel J M, Sarfati C, Liguory O, Caroff B, Dumoutier N, Gueglio B, Billaud E, Raffi F, Molina J M, Miegeville M, Derouin F. Detection of microsporidia and identification of Enterocytozoon bieneusi in surface water by filtration followed by specific PCR. J Eukaryot Microbiol. 1997;44:78S. doi: 10.1111/j.1550-7408.1997.tb05792.x. [DOI] [PubMed] [Google Scholar]
- 20.Swaminathan B, Barrett T J. Amplification methods for epidemiologic investigations of infectious diseases. J Microbiol Methods. 1995;23:129–139. [Google Scholar]
- 21.van der Hoek L, Boom R, Goudsmit J, Snijders F, Sol C J A. Isolation of human immunodeficiency virus type 1 (HIV-1) RNA from feces by a simple method and difference between HIV-1 subpopulations in feces and serum. J Clin Microbiol. 1995;33:581–588. doi: 10.1128/jcm.33.3.581-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velásquez J N, Carnevale S, Guarnera E A, Labbé J H, Chertcoff A, Cabrera M G, Rodríguez M I. Detection of the microsporidian parasite Enterocytozoon bieneusi in specimens from patients with AIDS by PCR. J Clin Microbiol. 1996;34:3230–3232. doi: 10.1128/jcm.34.12.3230-3232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber R, Bryan R T, Schwartz D A, Owen R L. Human microsporidial infections. Clin Microbiol Rev. 1994;7:426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Wittner M, Tanowitz H B, Cali A, Weiss L M. Ribosomal RNA sequences of Enterocytozoon bieneusi, Septata intestinalis and Ameson michaelis: phylogenetic construction and structural correspondence. J Eukaryot Microbiol. 1994;41:204–209. doi: 10.1111/j.1550-7408.1994.tb01498.x. [DOI] [PubMed] [Google Scholar]