During Drosophila border cell migration, the B-type lamin, Lam, maintains nuclear envelope integrity, stabilizes the lead cell protrusion, and promotes cluster invasion between nurse cells. The nucleus may function as a wedge to promote this collective, confined in vivo movement.
Abstract
Cells migrate collectively through confined environments during development and cancer metastasis. The nucleus, a stiff organelle, impedes single cells from squeezing into narrow channels within artificial environments. However, how nuclei affect collective migration into compact tissues is unknown. Here, we use border cells in the fly ovary to study nuclear dynamics in collective, confined in vivo migration. Border cells delaminate from the follicular epithelium and squeeze into tiny spaces between cells called nurse cells. The lead cell nucleus transiently deforms within the lead cell protrusion, which then widens. The nuclei of follower cells deform less. Depletion of the Drosophila B-type lamin, Lam, compromises nuclear integrity, hinders expansion of leading protrusions, and impedes border cell movement. In wildtype, cortical myosin II accumulates behind the nucleus and pushes it into the protrusion, whereas in Lam-depleted cells, myosin accumulates but does not move the nucleus. These data suggest that the nucleus stabilizes lead cell protrusions, helping to wedge open spaces between nurse cells.
Introduction
Collective cell migration is critical for development and wound healing, and promotes cancer metastasis (Rørth, 2009; Friedl and Gilmour, 2009). One challenge for collective groups moving in vivo is the difficulty of squeezing into small spaces between cells, for example, during intravasation and extravasation (Stoletov et al., 2010; Wyckoff et al., 2000; Reymond et al., 2013), or through dense extracellular matrix (ECM). How groups of cells physically crawl into narrow paths and navigate complex tissue environments is not well understood.
Nuclear deformations have been widely observed in cells migrating through tight spaces in vivo (Yamauchi et al., 2005; Stoletov et al., 2010; Denais et al., 2016; Raab et al., 2016; Kalukula et al., 2022). Since the nucleus can pose a physical barrier to movement (Calero-Cuenca et al., 2018; Friedl et al., 2011), a major research focus is to determine how nuclei contribute to, obstruct, or adapt to confined migrations through natural environments. Many studies have focused on the nucleus in single-cell migration (Yamada and Sixt, 2019; McGregor et al., 2016), while how nuclei affect collective, confined in vivo migration is less understood.
The nucleus is typically the largest and stiffest organelle and is mechanically supported by networks of lamin filaments. A- and B-type lamins are proteins that assemble into distinct intermediate filament networks underneath the nuclear envelope and are primary contributors to nuclear mechanics (Hetzer, 2010; Davidson and Lammerding, 2014; Wintner et al., 2020; Lammerding et al., 2004). Higher expression of lamins, particularly A-type lamins, increases nuclear stiffness (Swift et al., 2013; Ferrera et al., 2014). A-type lamin tends to be expressed at higher levels in stiff tissues while B-type expression is higher during development and in soft tissues such as the brain (Swift et al., 2013; Hutchison, 2014). In vitro, mouse embryonic fibroblasts, cancer cell lines, mesenchymal stem cells, and hematopoietic cells depleted of A-type lamins have more deformable nuclei and migrate faster through narrow artificial channels or dense ECM (Rowat et al., 2013; Davidson et al., 2014; Harada et al., 2014; Shin et al., 2013), so nuclear stiffness can impede confined migration. In support of this idea, fast migratory cells, such as circulating white blood cells, downregulate A-type lamins (Shin et al., 2013) and are able to move rapidly through narrow channels in silicone devices (Rowat et al., 2013; Raab et al., 2016; Thiam et al., 2016). In contrast to the notion that nuclear rigidity is a barrier to migration, several studies report that lamin A/C depletion reduces the ability of multiple cell types, including mesenchymal stem cells, melanoma cells, and dendritic cells, to move in confinement (Lee et al., 2021; Lomakin et al., 2020). Thus, lamins may have distinct effects on migration depending on the cell type, migration environment, and migration mode.
There are multiple, not necessarily mutually exclusive, models for how nuclear mechanical properties contribute to confined migration. When the nuclei of a variety of cell types including HeLa or primary zebrafish progenitor cells are compressed, they respond by unfolding indentations in the nuclear envelope and triggering cytosolic phospholipase A2 (PLA2)–stimulated cortical actomyosin contractions, which promote extensive blebbing of the plasma membrane and escape from the confinement (Lomakin et al., 2020; Venturini et al., 2020). This mechanism, which is activated when the cell is confined to a space smaller than the nuclear diameter, is referred to as the nuclear ruler (Lomakin et al., 2020; Venturini et al., 2020). The nuclear piston model, initially reported in primary human fibroblasts (Petrie et al., 2014) and later observed in other cell types including confined, mesenchymal stem cells (Lee et al., 2021), proposes that hydrostatic pressure builds up in confined cells, activating mechanosensitive ion channels that open and cause swelling of large, “lobopodial” protrusions (Lee et al., 2021). Again, blebbing accompanies the response to confinement. In leukocytes, the nucleus is positioned near the front of the cell and acts as a mechanical gauge, allowing the cell to identify the path of least resistance (Renkawitz et al., 2019). A key open question is how nuclei sense, respond, and contribute to confined cell migration in vivo, particularly in cells that migrate collectively in between other cells.
Here, we use the border cell cluster in the Drosophila ovary as a model to study how nuclei change shape during and contribute to collective, confined, in vivo cell migration. We show that the nucleus of the lead cell rapidly deforms as the cluster moves in between tightly apposed nurse cells. Lead cell nuclei undergo the most significant shape changes, elongating as they move into forward-directed protrusions and then recovering a more circular shape. Nuclear movement correlates with protrusion expansion. We further show that reduced lamin expression delays migration. While both A- and B-type lamins are expressed, only the B-type lamin, Lam, is required to maintain nuclear integrity and promote border cell movement in between nurse cells. Lam-depleted cells extend transient forward protrusions that do not enlarge properly, as well as ectopic protrusions, ultimately resulting in undirected movement and failed invasion between nurse cells. Cortical nonmuscle myosin II flashes push the nucleus into protrusions in control cells, whereas in Lam-depleted cells, the flashes initially accumulate behind the nucleus but then move past the nuclei and ultimately constrict the protrusion. We did not detect some hallmarks of the nuclear piston or nuclear ruler, such as blebbing, in border cells. The data suggest that the nucleus promotes invasion of the border cell cluster into a space that is initially much smaller than even a single nucleus, possibly by functioning as a wedge.
Results
The lead cell nucleus transiently deforms as the lead cell protrusion widens and border cells move between nurse cells
To study the role of nuclei in collective cell migration, we used the well-established model of border cell migration in the Drosophila egg chamber. At stage 9, the border cell cluster delaminates from a layer of epithelial cells and squeezes in between germline cells, termed nurse cells, to reach the oocyte by stage 10 (Fig. 1, A–C; Montell et al., 2012). Border cells consist of a pair of inner, nonmotile polar cells and four to six outer motile cells (Fig. 1 B). Typically, one outer border cell extends a large protrusion toward the oocyte and leads the cluster, though the leader can change over time. Lead border cells extend and retract protrusions, probing for chemoattractants and available space. The border cells select the central path between the nurse cells because they contain slightly larger spaces (Dai et al., 2020; Fig. 1 D); however, even the largest spaces are much smaller than the cluster.
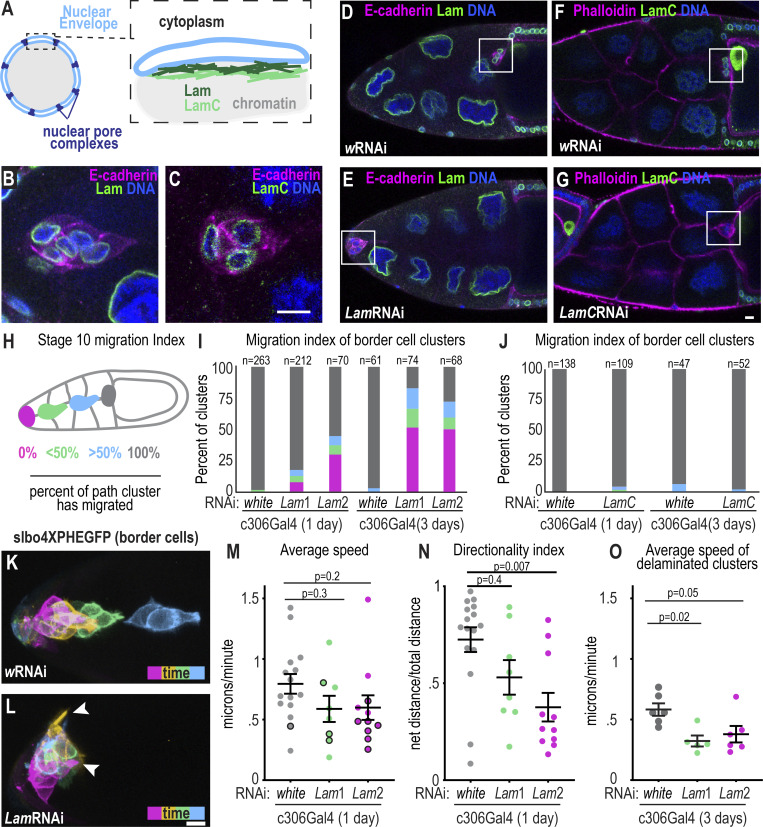
Figure 1.
Leader cell nuclei transiently deform during the onset of border cell migration. (A) Overview of the Drosophila ovary, ovariole, and egg chamber stages. (B) Illustration of the border cells (green) and polar cells (yellow). (C) A max projection of images from a time lapse of a stage 9 egg chamber to show the border cell cluster movement over time. The egg chamber is shown with differential interference contrast and border cells expressing GFP-moesin are labeled with a different color for each time point. (D) A stage 9 egg chamber expressing fruitlessGal4;UAS-GFP-moesin (border cells);UASdsRED.NLS (nuclei) and incubated with 10 kDa dextran-Alexa647 to label junctures between cells. (D′and D″) The inverted fluorescent image of the 10 kDa dextrans in both XY and YZ views to show possible paths for border cells to migrate through. Arrowheads mark central extracellular spaces where the border cells migrate. Scale bars: 10 μm. (E) Individual and mean ± SEM values for diameters of the indicated features. n = 10 (clusters), 58 (cells and nuclei), 63 (>3 nurse cell [NC] junctures), 10 (initial juncture). One-way ANOVA followed by a post-hoc Tukey was performed, all diameters are significantly different from one another, P < 0.0001 for all comparisons except initial juncture versus >3 NC juncture, P = 0.004. Measurements are from N = 10 egg chambers. (F and G) XY (F) and YZ (G) images of border cells and junctures from a time-lapse series labeled with the indicated markers. Yellow box marks the same juncture shown in F and G. White box in F shows the leading cell’s nucleus in grayscale. (H) Plot showing individual juncture widths relative to time of protrusion entry (green) and nuclear entry (magenta) normalized to the size of the initial juncture. A mixed effects model (REML) followed by Tukey’s multiple comparison test was performed for statistics. N = 10 egg chambers. (I and J) Example images from time-lapse series of border cell nuclei (I) or polar cell nuclei (J) labeled with UAS-dsRED.NLS expressed by c306Gal4 (I) or UpdGaL4 (J). Magenta arrowheads label the lead cell’s nucleus. (K and L) Plots showing individual and average ± SEM nuclear aspect ratios (K) and changes in nuclear aspect ratio (L) for each individual nucleus. n = 20 (leader), 43 (follower), and 24 (polar) cell nuclei. One-way ANOVA followed by Tukey post-hoc was performed, data were collected from N = 10 movies for border cells and N = 12 movies for polar cells. Scale bars: 10 μm. Genotypes and experimental replicates are reported in Table S2.
To assess the relative sizes and dynamics of the clusters, cells, nuclei, and available spaces, we acquired time series images at delamination. We labeled extracellular spaces (junctures) with fluorescent 10 kiloDalton (kDa) dextrans (Fig. 1 D), nuclei with a dsRED-tagged nuclear localization signal (NLS), and border cells with a GFP-tagged actin-binding domain of moesin (Fig. 1 D). Border cell clusters were ∼15–20 μm in diameter, whereas individual border cells averaged ∼7.5 μm and each nucleus measured ∼5 μm (Fig. 1 E). The junctures that clusters initially moved into during delamination averaged ~1.5 μm in diameter, which was approximately threefold narrower than even a single border cell nucleus (Fig. 1, E and F). As border cells delaminated, the leading protrusion widened, significantly expanding the juncture (Fig. 1, F–H; Fig. S1 A; and Video 1). These data suggest that the leading protrusion might pry nurse cells apart before nuclear translocation.
Figure S1.
Changes in nuclear shape occur rapidly in migrating border cell clusters. (A) Plots of the average ± SEM of each indicated parameter relative to the time of delamination. n = 10 movies. (B) Nuclear aspect ratio of each leading cell’s nucleus aligned to the protrusion axis, n = 10. (C) Representative plot showing an example where nuclear width and protrusion width simultaneously oscillate while the protrusion tip width changes variably. (D) Individual (dots) and average ± SEM correlation between nuclear width and protrusion base width or between protrusion tip and base width for the first border cell protrusion of each movie. A two-sided, unpaired t test yields P = 0.02. (E) Average difference between the protrusion base and protrusion tip. (E and F) n = 10 leading cells. (F) Images from a time-lapse series with the leading cell nucleus marked with GFP.NLS. Images were acquired every 20 s. Bottom right: projection of time points. (G) ZY view of border cell cluster leading cell and following cell over time to show the expansion and shrinking of the juncture as the cluster moves through it. (H–J) Example images of nuclei from time-lapse series of control leader, follower, or RacN17 dominant negative expressing border cells. Right: projection of time points. (K) Plot of the average change in nuclear aspect ratio; bars: mean ± SEM; dots: value for one nucleus; n = 51 (control), n = 29 (RacN17). A Mann–Whitney test was used to test for statistical significance. Scale bars: 10 µm. Genotypes and experimental replicates reported in Table S2.
Video 1.
Border cell cluster undergoing delamination. Single z-slices from a stage 9 egg chamber expressing fruitlessGal4; UAS-GFP-moesin actin-binding domain (border cells, green) UAS-dsRED.NLS (nuclei, magenta), and incubated with 10 kDa dextran-Alexa 647 (dextrans, cyan). The time interval is 3 min. Scale bar: 10 µm. Frame rate: 3 frames/s. Playback speed is 540× real time.
The lead cell nucleus elongated as it moved into the protrusion (Fig. 1 F and Fig. S1 A) and then recovered a rounder shape as the protrusion widened (Fig. 1 F and Fig. S1, A and B), so nuclear shape changed as nuclei transiently elongated into protrusions (Fig. 1 F and Fig. S1 B; and Video 1). The width of the juncture expanded by approximately fivefold as first the protrusion and then the nucleus moved in (Fig. 1, F–H). Nuclear diameter and protrusion base width tended to oscillate simultaneously when nuclei were in the protrusion (Fig. S1, C and D). In contrast, the tip of the spear-shaped protrusion was typically several microns narrower than the protrusion base and its size fluctuations did not correlate consistently with the base diameter (Fig. S1, C–E). Thus, nuclear translocation into the protrusion correlates with further expansion of the space between substrate nurse cells.
Lead cell nuclei tended to deform when entering the base of the protrusion (Fig. 1, F and I; Fig. S1 F; and Videos 2 and 3). This deformation occurred in <1 min (Fig. S1 F and Video 2), unlike the hours-long process of nuclear deformation observed in breast cancer cells and fibroblasts migrating in silicone channels (Davidson et al., 2014; Denais et al., 2016). The nuclei of follower border cells also elongated and deformed, though not as much as lead cell nuclei (Fig. 1, I, K, and L), and entry of follower cells further widened the path (Fig. S1 G). The inner pair of nonmotile polar cells had the lowest and least variable nuclear aspect ratio (Fig. 1, J–L; and Video 3). These data indicate nuclear dynamics vary based on position and/or cell type, and the lead cell nucleus experiences the most nuclear deformation during collective border cell migration.
Video 2.
The nucleus in the leading cell undergoes rapid changes in shape. A max projection of a border cell cluster expressing Sqh-mCherry to and GFP.NLS at 20-s time intervals. Right: inverted grayscale of GFP.NLS. Scale bar: 10 µm. Frame rate: 10 frames/s, playback speed: 200× real time.
Video 3.
Border cell nuclei undergo dynamic shape changes while polar cells maintain nuclear shape. Top: border cell nuclei labeled with UAS-dsRED.NLS and clusters labeled with UAS.GFP-Moesin in lines expressing c306Gal4 (polar cells did not highly express dsRED.NLS). Bottom: border cell clusters expressing UpdGal4;UAS-dsRED.NLS to label polar cell nuclei and UAS-GFP-moesin to label cells. The time interval is 3 min. Scale bar: 10 µm. Frame rate: 2 frames/s. Playback speed: 360× real time.
To test if transient changes in nuclear shape occurred spontaneously or due to forces resulting from migration, we measured nuclear shapes in immobile border cells expressing a dominant negative form of the small GTPase Rac (RacN17). RacN17-expressing cells lack lead protrusions and completely fail to move (Murphy and Montell, 1996). RacN17-expressing border cells had significantly lower variation in nuclear shape compared with control border cell nuclei (Fig. S1, H–K). Thus, outer border cell nuclei, particularly when leading the cluster, undergo transient shape changes as they migrate to a greater extent than inner polar cells, which may be insulated from compressive forces by the surrounding cells.
B-type lamin promotes border cell movement between nurse cells
We next asked how nuclear lamins affect the ability of border cells to migrate into confined space. A- and B-type lamins form separate intermediate filament networks that confer mechanical support to the nuclear envelope (Wintner et al., 2020; Davidson and Lammerding, 2014; Xie et al., 2016). The Drosophila genome encodes one A-type lamin called LamC and one B-type lamin named Lam (Bossie and Sanders, 1993; Lenz-Böhme et al., 1997; Fig. 2 A). We found that both Lam (Fig. 2 B) and LamC (Fig. 2 C) antibodies stained the periphery of border cell nuclei, and staining at the nuclear rim was diminished in cells expressing the corresponding RNAi (Fig. 2, D–G; and Fig. S2, A–G). Expressing two different UAS-RNAi lines, referred to as Lam1 RNAi and Lam2 RNAi, which target separate regions of the Lam gene, with a Gal4 that drives expression in border and polar cells (c306Gal4), caused migration defects (Fig. 2, E, H, and I). In contrast, neither a control RNAi line (white (w) RNAi) nor LamC RNAi impaired migration (Fig. 2, D, F–J). Gal4 is more active at 29°C than at lower temperatures. When we incubated Lam RNAi-expressing flies at 29°C for 1 d, we observed variable knockdown and clusters with the most effective knockdown exhibited more severe migration defects (Fig. S2, E and H). When we incubated flies for 3 d at 29°C, the lamin depletion was stronger (Fig. S2 F). Migration defects were also stronger: 82% of clusters expressing Lam1 RNAi and 72% of clusters expressing Lam2 RNAi showed incomplete migration at stage 10. In both lines, half of border cell clusters failed to move in between nurse cells (Fig. 2 I).
Figure 2.
B-type lamin promotes border cell delamination. (A) Schematic of the nuclear envelope and the two Drosophila lamin filament networks made of Lam and LamC proteins. (B and C) Images of fixed border cell clusters stained with Lam (B) and LamC (C). Scale bar: 10 μm. (D–G) Images of fixed stage 10 egg chambers for the indicated RNAi and antibody stains. Flies were incubated for 1 d at 29°C. The oocyte is always shown on the right and the white box marks the border cell cluster. (H) Schematic of migration index used to score stage 10 egg chambers. (I and J) Plots showing migration indexes for indicated conditions. n = number of egg chambers. Migration index: magenta: 0%, green: <50%, blue: >50%, and gray: 100%. (I) Statistical test, 1 d at 29°C: Fisher’s exact test with Bonferroni correction yields significant difference in percentage with complete migration, wRNAi versus Lam1RNAi, P < 0.0002, and wRNAi versus Lam2RNAi, P < 0.0002. Statistical test, 3 d at 29°C: Fisher’s exact test with Bonferroni correction for difference in percentage with complete migration, wRNAi versus Lam1RNAi, P < 0.0002, and wRNAi versus Lam2RNAi, P < 0.0002. (J) Statistical test 1 d at 29°C: Fisher’s exact test for difference in percentage with complete migration in wRNAi versus LamCRNAi 1 d, P = 0.08, and Fisher’s exact test wRNAi versus LamCRNAi 3 d yields P = 0.3. (K and L) Maximum projections of time frames from time-lapse series of stage 9 egg chambers expressing slbo4xPHEGFP to mark border cell membranes in control (wRNAi, K) and Lam-depleted border cells (Lam1RNAi, L) after 1 d at 29°C. Time points relative to the start of imaging: magenta = 0 min, yellow = 33 min, green = 54 min, blue = 108 min. Arrowheads mark protrusions. (M) Cluster speed of individual (dots) and mean ± SEM (bars) in control and Lam-depleted clusters. Black outlines: border cell clusters that did not move away from the anterior during the imaging session. Kruskal–Wallis Test. (N) Individual (dots) and mean ± SEM directionality index of clusters measured as total path traveled/net distance. Kruskal–Wallis Test. (M and N) n = 16 (w), 8 (Lam1), and 11 (Lam2) egg chambers. (O) Individual and mean ± SEM speed of delaminated clusters after 3 d incubation at 29°C; n = 6 (w), 5 (Lam1), and 6 (Lam2) egg chambers. One-way ANOVA followed by Tukey post-hoc was performed for statistical testing. Genotypes and experimental replicates are reported in Table S2.
Figure S2.
Stronger depletion of lamins correlates with higher frequency of migration defects. (A–D) Images of Lam (A and B) and LamC (C and D) for the indicated conditions. (E–G) Plots showing the mean ± SEM percentage of border cells with the indicated depletion (full, partial, or none) of Lam or LamC at the nuclear periphery for the indicated conditions. (E) A Kruskal–Wallis Test was performed to compare means of cells with a full knockdown (KD): wRNAi versus Lam1, P = 0.006 and w versus Lam2 P < 0.0001. (F) Kruskal–Wallis Test was performed to compare those with a full KD: w versus Lam1, P < 0.0001 and w versus Lam2, P < 0.0001. (G) A Kruskal–Wallis Test was performed to compare the mean of cells with a full KD; wRNAi versus LamC RNAi, P < 0.0001. (H) Plot showing the individual and average ± SEM percent of cells with an indicated knockdown in Lam-depleted clusters with complete (n = 36) or incomplete (n = 31) migration. A Kruskal–Wallis test was performed. Data shown are from line Lam2 RNAi after 1 d at 29°C. (I) Migration index of stage 9 egg chambers for the indicated conditions to quantify the percentage that moved away from the anterior (gray). n = number of egg chambers, after 1 d at 29°C. A Fisher’s exact test with a Bonferroni correction to compare percentage with delamination yields P = 0.0003 for wRNAi versus Lam1RNAi and P < 0.0002 for wRNAi versus Lam2RNAi. (J and K) Migration index for the indicated conditions; n = number of egg chambers. A Fisher’s exact test with a Bonferroni correction to compare percentage with complete migration. P values for J: wRNAi versus Lam1RNAi, P < 0.0002; wRNAi versus Lam2RNAi, P < 0.0002. P values for K: wRNAi versus Lam1 RNAi, P = 0.4; wRNAi versus Lam2RNAi, P > 0.9. KD condition: 1 d at 29°C. (L and M) Images of fixed border cells from stage 9 egg chambers stained for LBR with LacZ overexpressed (OE; L) or LBR overexpressed (M) with c306Gal4 for 1 d at 29°C. (L′ and M′) Grayscale images of LBR staining. (N and O) Images of Hoechst (DNA) staining of stage 9 egg chambers overexpressing LacZ (N) or LBR (O) kept at 29°C for 1 d. Scale bars: 10 μm. Arrowhead: elongated nucleus. (P) Plots measuring nuclear shape in nuclear aspect ratio from Hoechst channel of fixed border cell. Each dot represents an individual nucleus, and the middle and error bars represent the mean ± SEM. A Mann–Whitney test was used for statistical testing. N = 3 experimental replicates, n = 112 (Lac), and n = 123 (LBR) nuclei. (Q and R) Plots of migration indexes of stage 10 egg chambers in the noted conditions. n = number of egg chambers counted. A Fisher’s exact test was used to compare percentage with complete migration and yields P < 0.0001 for Q and P = 0.5 for R. Genotypes and experimental replicates reported in Table S2.
To assess how lamins affect border cell motility and behavior, we performed live imaging of stage 9 egg chambers. During delamination, lead border cells from controls rounded up and extended one main protrusion in between nurse cells before moving toward the oocyte (Fig. 2 K and Video 4). In contrast, Lam-depleted clusters extended short-lived and ectopic protrusions (Fig. 2 L and Video 4). Clusters were mobile and sometimes moved between anterior follicle cells and germ cells instead of taking their normal path between nurse cells (Fig. 2, L and M). Lam-depleted clusters exhibited less directional persistence (Fig. 2 N). In the 1-d RNAi-treated clusters, movement away from the anterior end of the egg chamber was significantly delayed (Fig. S2 I), but clusters that delaminated exhibited similar migration speeds to controls (Fig. 2 M), likely due to incomplete knockdown, though it was not possible to assess knockdown efficiency in living samples. With a 3-d incubation, the delamination defect was more penetrant, and those lamin-depleted clusters that did delaminate migrated slower (Fig. 2 O). Together, we conclude that a partial Lam knockdown delays delamination, whereas a more severe Lam-depletion hinders delamination and slows migration.
Video 4.
Control border cells maintain a leading protrusion as they delaminate and migrate while Lam-depleted clusters form ectopic protrusions and have undirected movement. Movies of egg chambers with a control border cells (top, c306Gal4; UAS-wRNAi) or a Lam-depleted cluster (bottom, c306Gal4;UAS-Lam2RNAi) expressing slbo4XPH-EGFP to mark the border cell membranes (green) and UbiHisRFP to mark all nuclei (magenta). Right panel shows slbo4XPH-EGFP channel in grayscale. Time interval is 3 min. Scale bar: 10 µm. Frame rate: 5 frames/s. Playback speed: 900× real time.
To test if Lam is required in the outer border cells or polar cells, we crossed UAS-Lam RNAi lines to fruitlessGal4, which is expressed in border but not polar cells, and UpdGal4, which is expressed in polar but not border cells. Lam RNAi expressed with fruitlessGal4 but not with UpdGal4 caused incomplete migration at stage 10 (Fig. S2, J and K). We conclude the Drosophila B-type lamin is required in the outer, motile border cells.
B-type lamins are required to maintain nuclear shape and integrity
Depletion of either A-type or B-type lamin disrupts the nuclear envelope permeability barrier in cultured cells (Vargas et al., 2012). To test how Lam and LamC depletion affect border cell nuclei, we performed live imaging of dsRED.NLS, which is retained in the nucleus when the nuclear permeability barrier is intact. In control (Fig. 3, A and A′) and LamC-depleted (Fig. 3, B and B′) nuclei, dsRED.NLS remained inside the nucleus in all cells observed. By contrast, some Lam-depleted cells exhibited dsRED.NLS throughout the cell (Fig. 3, C, C′, and D). Live imaging also revealed that a subset of Lam-depleted leading cell nuclei in protrusions herniated backward, whereas neither LamC-depleted nor control nuclei exhibited such herniations (Fig. 3, E–G). Lam-depleted nuclei within protrusions were even more elongated than controls (Fig. 3 H), consistent with the proposed role of B-type lamins in nuclear elasticity (Wintner et al., 2020; Harada et al., 2014). A subset of leaders had reduced circularity in Lam-depleted but not LamC-depleted nuclei (Fig. 3 I). Lam-depleted nuclei were also slightly smaller than controls (Fig. 3, J–L) though cell area was not affected (Fig. 3 M). We conclude that Lam is required to maintain border cell nuclear integrity, shape, and size, while LamC is less critical.
Figure 3.
B-type lamins are required to maintain nuclear shape and integrity. (A–C) Images of border cells from time-lapse series in stage 9 egg chambers with the indicated markers and conditions. Yellow arrowheads point to cells with dsRED.NLS throughout the cell. (A–C′) Grayscale images of nuclei (dsRED.NLS). (D) Plot measuring the percentage of cells with dsRED.NLS throughout the cell versus in the nucleus. A Fisher’s exact test with a Bonferroni correction for multiple testing was performed: (w versus Lam1 P = 0.01; w versus Lam2, P = 0.009; w versus LamC, P > 0.9). n = number of nuclei. (E and F) Images of border cells from time-lapse series with the indicated markers and conditions. Yellow arrowhead marks backward nuclear herniation. (E′ and F′) Grayscale images of nuclei (HisRFP). (G) Plot with the percent of nuclei with herniations in each condition. A Fisher’s exact test with a Bonferroni correction for multiple testing was performed: (w versus Lam1 P = 0.001; w versus Lam2,P = 0.0009; w versus LamC, P > 0.9). n = number of nuclei. (H) Plot of individual and mean ± SEM nuclear aspect ratios when the nucleus extended into the protrusion (defined as >1 SD above average nucleus distance to polar cell boundary). n = 8 (w), 3 (Lam1), and 5 (Lam2) leader nuclei. A one-way ANOVA with post-hoc Tukey was performed. (I) Plot of individual and average ± SEM values for nuclear circularity. A Kruskal–Wallis test was performed. n = 15 (w), 13 (LamC), 12(Lam1), and 16 (Lam2) leader cell nuclei. (J and K) Images of fixed border cell clusters stained with indicated markers for wRNAi (J) and Lam2RNAi (K) border cells. (J′ and K′) Grayscale images of DNA (Hoechst). (L and M) Measurements of nuclear area and cell area. Each individual area value shown as dots and bars representing mean ± SEM. Nuclei number (L); n = 290 (w), 201 (Lam1), and 221 (Lam2). Cell number (M), n = 80 (w), 152(Lam1), and 104 (Lam2). Statistical test for L and M: Kruskal–Wallis test. Scale bars: 10 μm. KD conditions: 1 d at 29°C except the cell area (M), which was 3 d at 29°C. Genotypes and experimental replicates reported in Table S2.
Effects of Lam and LamC overexpression on border cell migration
In some contexts, A-type lamins impede migration (Davidson et al., 2014; Rowat et al., 2013); however, overexpression of LamC did not disrupt border cell movement (Fig. 4, A and B). Overexpressing Lam using a re-encoded, RNAi-resistant version (LamRE) also did not impede migration; however, stronger Lam overexpression with a different construct impaired migration (Fig. 4, C–F). Strong Lam overexpression perturbed nuclear shape (Fig. 4 D), consistent with the proposed role of B-type lamins in regulating nuclear membrane abundance (Prüfert et al., 2004). Expression of LamRE fully rescued Lam RNAi migration defects (Fig. 4, G and H), whereas LamC overexpression only partially rescued Lam RNAi migration defects (Fig. 4, I–K). We conclude that optimal Lam levels are critical for border cell migration.
Figure 4.
Lamin overexpression compensates for Lam RNAi, but strong Lam overexpression impedes border cell migration. (A–D) Representative images of stage 10 egg chambers with overexpression (OE) of the indicated UAS constructs in border cells by c306Gal4. The white box marks border cell clusters. (E) Migration indices for the indicated conditions. n = number of egg chambers. LacZ is the control. A Fisher’s exact test with a Bonferroni correction to test for proportion with complete migration yields a significant result between LacZ versus Lam OE, P <0.0003. All other comparisons to LacZ are not significant. (F) Plot of cluster speed with individual movies (dots) and mean ± SEM plotted. n = 7 (LacZ), 6 (LamRE), 7 (Lam), and 4 (LamC) egg chambers. Statistical test: one-way ANOVA followed by post-hoc Tukey. (G–J) Representative images of stage 10 egg chambers for the indicated conditions. White box surrounds border cell cluster. (K) Plot of the mean ± SEM percentages of egg chambers with completed migration, each dot represents one experimental replicate (N). n (number of egg chambers): wRNAi/LacZ: 47; LacZ/Lam1RNAi: 290; Lam1RNAi/LamRE: 59; Lam1RNAi/LamOE: 73; LamC/Lam1RNAi: 149. Statistical test: Brown–Forsythe and Welch followed by Dunnett’s Multiple Comparison test. Genotypes and experimental replicates reported in Table S2. Scale bars: 10 μm.
To further test the impact of perturbing nuclear structure on border cell migration, we investigated the effects of overexpressing the Lamin B receptor (LBR). LBR is an inner nuclear membrane protein that is responsible for the multilobed nuclei that are proposed to allow neutrophils to squeeze into and out of blood vessels and tissues (Hoffmann et al., 2002). LBR overexpression causes irregular nuclear morphology in Drosophila embryos (Hampoelz et al., 2016) and interferes with the nuclear ruler mechanism in HeLa and melanoma cells (Lomakin et al., 2020). In control (Fig. S2 L) and LBR-overexpressing border cells (Fig. S2 M), LBR localized at the nuclear periphery. Nuclei overexpressing LBR were more elongated than control nuclei (Fig. S2, N–P). To test if LBR overexpression disrupted migration in border cells or polar cells, we crossed UAS-LBR and a control (UAS-LacZ) to fruitlessGal4 and UpdGal4. Overexpression of LBR with fruitlessGal4, but not UpdGal4, caused significant migration defects, indicating LBR overexpression affects the outer, motile border cells (Fig. S2, Q and R). Thus, LBR overexpression disrupts nuclear shape and impedes border cell movement between nurse cells, supporting that border cells require specific nuclear properties and biochemical composition to move within their naturally confined environment.
B-type lamin promotes expansion of the lead protrusion
To assess how Lam depletion might affect border cell invasion between nurse cells, we first determined if Lam RNAi-expressing border cells are specified normally. Border cell fate specification, cluster formation, and movement require the activity of multiple transcription factors and their downstream targets (Montell et al., 2012). In addition to their mechanical roles, lamins are required for chromatin organization, gene expression, and cell survival (Davidson and Lammerding, 2014; Chen et al., 2019; Harada et al., 2014). So, we tested the effect of Lam RNAi on border cell fate, cluster formation, and expression of genes required for initiation of migration. Lam-depleted border cells still formed clusters (Fig. S3, A–C) with similar circularity (Fig. S3 D) and cell numbers (Fig. S3 E) as controls. Further, Lam-depleted clusters had similar F-actin (Fig. S3 F) and E-cadherin (Fig. S3, G and H) levels to controls. STAT activation, which is essential for border cell specification (Silver and Montell, 2001), was actually increased approximately twofold in Lam-depleted cells at stage 8 (Fig. S3, I–K), which is consistent with reports that Lam and the nuclear-membrane-associated protein, Dysfusion, limit STAT signaling (Wu et al., 2022; Petrovsky and Großhans, 2018). Increased STAT in border cells does not impede their migration (Silver and Montell, 2001; Silver et al., 2005). Ectopic STAT can cause additional border cell clusters to form; however, we did not observe extra border cells in Lam-depleted clusters, suggesting that the elevated STAT signaling was insufficient to induce extra border cells. Notch activity, which is required for border cell delamination (Wang et al., 2007), was similar to controls (Fig. S3, L and M). We conclude that lamin is not required for border cell specification.
Figure S3.
Lam-depleted border cells specify and express key border cell genes. (A–C) Representative images of stage 9 border cells stained with the indicated markers in control (A) and Lam1RNAi (B) and Lam2RNAi (C). Left: merged images. Right: grayscale images of F-actin, E-cadherin, and singed staining. (D) Plot of individual (dots) and average ± SEM (bars) cluster circularity, 1 d at 29°C. One-way ANOVA with Tukey post-hoc was performed. n = 27 (w), 63 (Lam1), and 49 (Lam2) clusters. (E) Plot of average ± SEM numbers of border cells in a cluster, 1 d at 29°C. A Krusal–Wallis test was performed. (F and G) Mean F-actin (F) and E-cadherin (G) levels for each cluster (dots) and the average ± SEM (bars). KD condition: 1 d at 29°C (see also Fig. S5 for 3-d F-actin analysis). n = 27 (w), 63 (Lam1), and 49 (Lam2) clusters. One-way ANOVA with Tukey post-hoc was performed for each plot. (H) Clonal analysis of the ratio of mean peripheral E-cadherin in a LamRNAi clone divided by the mean of a wildtype clone. KD condition: 3 d at 29°C. The middle bars show the mean ± SEM. A Wilcoxon test was performed. n = 61 (Lam1) and 36 (Lam2) clusters. (I and J) Confocal images of STAT activity reporter (10XSTATGFP) surrounding polar cells (asterisks) for the indicated conditions. LamRNAi line shown: Lam1RNAi. KD condition: 1 d at 29°C. (K) Plot showing individual (dots) and mean ± SEM (bars) measures of STAT fluorescence normalized to the mean of the control. n = 27 (w), 45 (Lam1), and 13 (Lam2) clusters. A Kruskal–Wallis test was used for statistical testing. (L) A border cell cluster expressing hsFlpAyGal4 UAS-GFP UAS-LamRNAi clones (GFP+ cells marked with arrowheads) and wildtype clones with the indicated markers a merge of channels. (L′) Grayscale image of notch responsive element RFP. (M) Plot showing the fluorescence intensity (F.I.) of the nuclear notch responsive element relative to wildtype clones in the cluster. KD condition: 3 d at 29°C. Middle bars show the mean ± SEM. n = 43 clusters. A Wilcoxon test was used to test for statistical upregulation, P = 0.06, ns. Scale bars: 10 µm. Genotypes and experimental replicates reported in Table S2.
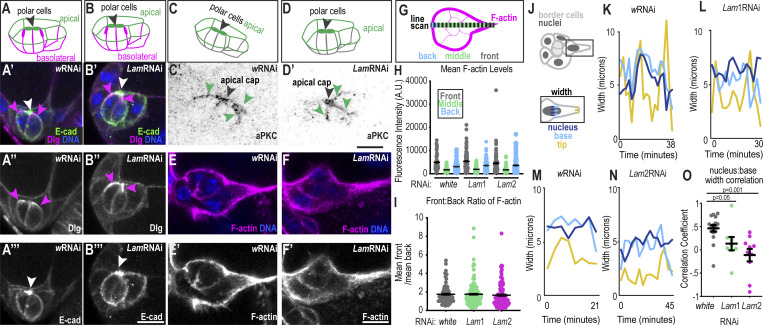
Next, we investigated how lamins affect cluster polarity given some reports that the nucleus is required for cell polarization while others show that polarity can develop even in enucleated cells (Graham et al., 2018; Lee et al., 2007). Border cells maintain three types of polarity that are important for migration: (1) apical–basolateral polarity, (2) inside–outside polarity, and (3) front–back polarity (Montell et al., 2012; Pinheiro and Montell, 2004; Wang et al., 2018; Duchek et al., 2001; McDonald et al., 2008; Assaker et al., 2010; Luo et al., 2019). Polar cells had an apical enrichment of E-cadherin and were found on the inside of control (Fig. S4 A) and Lam-depleted clusters (Fig. S4 B). E-cadherin was also enriched at cell–cell junctions in both conditions (Fig. S3, A–C; and Fig. S4, A and B). Similar to controls (Fig. S4, A and C), Lam RNAi clusters (Fig. S4, B and D) had lateral localization of Discs large (Dlg) and apical enrichment of atypical Protein Kinase C (aPKC). Further, control (Fig. S4 E) and Lam RNAi clusters (Fig. S4 F) displayed enrichment of F-actin on the outside of the cluster compared with inside and in forward-directed protrusions (Fig. S4, G–I), although Lam-depleted clusters also formed ectopic side protrusions (Fig. 2 L). These data indicate border cells retain apicobasal and inside/outside cluster polarity upon Lam-depletion, although abnormal protrusion suggested abnormal leading/lagging cluster polarization.
Figure S4.
Effects of lamins on cluster polarity and protrusion dynamics. (A–B′) Schematics of polarity orientation (A and B) and merged images of control (A′) and LamRNAi (B′) clusters marked with E-cadherin to mark the apical surfaces, Dlg to mark basolateral surfaces, and Hoecsht to mark DNA. (A″–B‴) Dlg grayscale image. (A‴ and B‴) E-cadherin grayscale image. White arrowhead: polar cell apical cap; magenta arrowheads: lateral Dlg. (C–D′) Schematic (C and D) and inverted grayscale images (C′ and D′) of clusters stained for aPKC as an apical marker. Black arrowhead: polar cell apical cap; green arrowheads: apical border cell surface. (E and F) Images of border cells stained with Phalloidin to mark F-actin and Hoechst to mark DNA. (E′ and F′) Grayscale images of F-actin. (G) Schematic of line scale acquired across cluster. The binning of regions were for front: 0–10% length, middle: 11–89% length, and back: 90–100% length. (H) Individual and mean ± SEM F-actin levels in indicated clusters and position. Statistical test: Brown Forsythe and Welch followed by Dunnett’s Multiple Comparison; means for front, back, or middle F-actin are all not significantly different from LamRNAis for relative position, P > 0.9. (I) Individual and mean ± SEM ratios of the front:back intensity for each cluster. Statistical test: Kruskal–Wallis test, wRNAi versus Lam1RNAi: P > 0.9; wRNAi versus Lam2RNAi: P = 0.24. Number of clusters analyzed (n) in H and I: n = 78 (w), n = 134 (Lam1), and n = 95 (Lam2). (J) Schematic showing measurement of width in the leading cell. (K–N) Example plots showing the width of the protrusion tip, base, and nucleus for the indicated conditions for an individual cluster over time. (O) Plot with dots of individual and mean ± SEM correlations between nuclear width and base width for each cluster’s first protrusion. A Kruskal–Wallis Test was performed. N = 16 (wRNAi), N = 8 (Lam1RNAi), and N = 10 (Lam2RNAi) movies for each condition shown in C–E and J. Scale bars: 10 μm. Genotypes and experimental replicates reported in Table S2.
The observations that nuclei normally move into leading protrusions, which then widen, and that Lam depletion impedes delamination and migration without affecting cluster specification or polarity suggested that border cell nuclei, in particular the lead cell nucleus, might function as a wedge to stabilize and enable enlargement of the lead protrusion. To explore this possibility further, we compared nuclear shape changes to protrusion shape changes over time. In controls, the movement of leading cell nuclei into protrusions corresponded with a widening of the protrusion neck (Fig. 5 A and Video 5). In lamin-depleted clusters, nuclei extended forward but did not remain in the protrusion neck, and protrusions narrowed (Fig. 5 B and Video 5). On average, protrusions were shorter and thinner in Lam RNAi-expressing cells compared with controls (Fig. 5, C and D). In control leading cells, nuclear movement forward into the protrusion was associated with wider protrusions, while backward movement correlated with narrow protrusions (Fig. 5 E). In Lam-depleted cells, protrusion width was not significantly changed by nuclear movement (Fig. 5 E). Further, nuclear width and protrusion base width did not correlate as well upon Lam depletion compared with control clusters (Fig. S4, J–O). We conclude that Lam is required to maintain and facilitate expansion of the leading protrusion.
Figure 5.
B-type lamins promote the expansion of a single protrusion. (A and B) Single z-slice images from time-lapse series of migrating control border cells (wRNAi, A) or Lam-depleted border cells that delaminate but fail to expand the protrusion (B). White bars show protrusion width. Inset in B shows the lead cell nucleus with a backward herniation (arrowhead). Scale bars: 10 μm. (C–E) Plots showing average ± SEM (bars) and individual movie values (dots) of the average length (C) and width of protrusions (D and E) for the indicated conditions. n = 16 (w), 8 (Lam1), and 10 (Lam2) border cell clusters. (E) Average protrusion width relative to nuclear movement for each RNAi line; a number of leading cells with indicated condition left to right, n = 12, 16, 12, 6, 8, 6, 8, 10, 8. One-way ANOVAs with Tukey post-hocs were performed for each plot.
Video 5.
Nuclear movement corresponds with protrusion widening while Lam-depleted cells do not maintain protrusions. Time-lapse movies of controlRNAi border cells (top, c306Gal4; UAS-wRNAi) or Lam-depleted border cells (bottoms, c306Gal4; UAS-Lam2RNAi) expressing slbo4XPHEGFP to mark the border cells (green) and UbiHisRFP to mark all nuclei (magenta). The time interval is 3 min. Right panels show grayscale of UbiHisRFP. Scale bar: 10 µm. Frame rate: 2 frames/s. Playback speed: 360× real time.
The result that Lam promotes protrusion expansion is, in principle, consistent with the nuclear piston mechanism (Lee et al., 2021). One feature of the piston effect is that there is a pressure gradient and diffusion barrier between the front and back of the confined cell (Petrie et al., 2014). So we tested for a diffusion barrier in the leading cell by illuminating a photoactivatable GFP-ɑtubulin in leading border cell protrusions in front of the nucleus and then measuring its diffusion to the opposite side of the nucleus at the back of that cell (Fig. 6 A and Video 6). GFP diffused rapidly (Fig. 6 A). For comparison, we photoactivated at the basal side of epithelial follicle cells, which should not have a diffusion barrier, and assessed diffusion to the apical side of the nucleus (Fig. 6 B and Video 6). The ratio of GFP fluorescence between activated:unactivated regions were similar between the follicle cell and border cell over time (Fig. 6 C), and diffusion time constants in unactivated regions were not significantly different between protruding border cells and epithelial follicle cells (Fig. 6 D). We conclude that border cells lack a key hallmark of the nuclear piston.
Figure 6.
There is no detectable diffusion barrier in the leading border cell. (A and B) Images from-time lapses series where a photoactivatable (PA) GFP-ɑtubulin was activated in a border cell (A) or a posterior follicle cell (B). (A′ and B′) Images with regions of interest (ROIs) that were measured. ROIA: activated region (blue box); ROIB: unactivated region (yellow box). Scale bar: 10 μm. Time is relative to the start of the movie and photoactivation occurred at 6 s. (C) Plot showing the mean ± SEM of the ratio of the F.I. of GFP in front:back ROIs over time. Experiments were performed on three different experimental days and n = number of movies is displayed for each condition. (D) Time constants are calculated from the slope of the Boltzmann sigmoidal fitted curve (see Materials and methods). A Kruskal–Wallis test was performed. Genotypes and experimental replicates are reported in Table S2.
Video 6.
Photoactivation of GFP-αtubulin. Time-lapse movie of border cells expressing slboLifeAct RFP and UAS-PA-GFPαtubulin and photoactivated at a region of interest in a follicle cell (A) or border cell (B). Time interval is 2 s. Scale bar: 10 µm. Frame rate: 10 frames/s. Playback speed: 20× real time.
Myosin II cortical flashes correspond with nuclear movement and shape changes
An additional mechanism by which lamins may promote border cell delamination is through the nuclear ruler. The nuclear ruler model proposes that when cells are confined to a space narrower than the nuclear diameter, the nuclear envelope stretches out, leading to calcium release and recruitment of cytosolic PLA2 (cPLA2) to the nuclear membrane. This calcium initiates a signaling cascade that recruits myosin to the cell cortex in many cell types including HeLa and Zebrafish cells (Lomakin et al., 2020; Venturini et al., 2020) and stimulates massive blebbing, which results in cells escaping the confinement. Border cells do not normally exhibit blebs, and there is no identifiable cPLA2 encoded in the fly genome (Ben-David et al., 2015). There is a calcium-independent PLA2 (iPLA2), so we tested its effect on migration. Neither of the two independent null mutants (Lin et al., 2018) exhibited any border cell migration defect (Fig. S5 A). We also used a genetically encoded calcium indicator, GCaMP, to evaluate calcium dynamics in border cells. While earlier-stage follicle cells show rapid pulses of calcium with the GCaMP sensor as reported previously (Sahu et al., 2017), migrating border cells did not exhibit spatial or temporal changes in GCaMP fluorescence (Fig. S5, B–E and Video 7). Therefore, border cells also lack key features of the nuclear ruler mechanism.
Figure S5.
Border cells do not require the LINC complex for migration and lack calcium dynamics but have rapid flashes of myosin around nuclei. (A) Migration indices for the indicated conditions. iPLA2 mutants are iPLA2-VIA[Delta174] and iPLA2-VIA[Delta192], which are both predicted to excise the start codon and have been validated as null alleles by antibody staining (Lin et al., 2018). N = 3 experimental replicates. (B and C) Select images of border cells (B) or follicle cells (C) expressing UAS-GCaMP6S driven by c306Gal4. Arrowhead: calcium pulse. (D) Plot showing the individual and mean +/ SEM fluorescent intensity of GCAMP6s. n = 7 (follicle) and n = 6 (border) cells from independent clusters. A two-sided unpaired t test was performed. (E) Plot of the maximum change in border cells and follicle cells with flashes. Individual (dots) and mean ± SEM (bars) are shown. A Welch’s t test was performed. (F and G) Example images of Sqh-mCherry in control (F) and Lam-depleted clusters (G) after 3 d incubation at 29°C. Arrowheads: myosin flashes. (H and I) Example images of border cell clusters stained for Klar and indicated markers in control (H) and klarRNAi (I). (J) Migration index for indicated conditions. Fisher’s exact test with Bonferroni correction to compare percentage with complete migration (wRNAi versus klarRNAi: P = 0.003, wRNAi versus koiRNAi: P > 0.9, and wRNAi versus Msp300RNAi, P > 0.9). Genotypes and experimental replicates reported in Table S2.
Video 7.
Calcium dynamics in border cells. Time-lapse series of egg chambers expressing c306Gal4; UAS-GCaMP6s. Time interval is 5 s. Scale bar: 10 µm. Frame rate: 3 frames/s. Playback speed: 15× real time.
We then tested the effects of lamins on myosin II dynamics because myosin II is essential for border cell migration. Myosin II accumulates at the cortex of border cell clusters in transient foci or flashes (Majumder et al., 2012). Myosin II flashes at the back help clusters delaminate (Majumder et al., 2012) while flashes on the side and front retract protrusions (Mishra et al., 2019b) and counteract nurse cell compression (Aranjuez et al., 2016). We used a fluorescently tagged myosin light chain (Sqh-mCherry) to observe the effect of Lam RNAi on myosin. In both control (Fig. 7 A and Fig. S5 F) and Lam-depleted (Fig. 7 B and Fig. S5 G) clusters, Sqh-mCherry appears transiently in “cortical flashes.” Myosin flashes occurred at similar frequencies in control and Lam-depleted clusters and throughout the cortex (Fig. 7, A–C; Fig. S5, F and G; and Video 8), indicating that Lam is not required for myosin recruitment to the cortex.
Figure 7.
Myosin II cortical flashes correspond with nuclear movement and shape changes. (A and B) Example images of Sqh-mCherry in control (A) and Lam-depleted clusters (B) after 1 d at 29°C (see also Fig. S5, F and G, for 3 d at 29°C). Sqh-mCherry aggregates at apical surfaces in polar cells (pc) as previously reported (Mishra et al., 2019b) and has been covered to focus on cortical flashes. Yellow arrowheads point to Sqh flashes. (C) Average number of myosin flashes observed divided by the total number of time frames acquired. RNAi condition: 3 d at 29°C. n = 9 (w), 8 (Lam1), and 8 (Lam2) egg chambers. Statistical test: one-way ANOVA with post-hoc Tukey. (D) Images from a time-lapse series showing myosin flashes around the nucleus. Yellow arrowheads point to flashes. (E and F) Average change in nuclear position along the anterior–posterior axis of the egg chamber (E), and average nuclear aspect ratio change (F) relative to presence and position of myosin flashes. N = 16 movies; each dot represents one nucleus; n = number of nuclei: 22 (no flash), 12 (front), 21 (side), and 16 (back). Bars: mean ± SEM. Statistical tests E and F: one-way ANOVA followed by Tukey post-hoc. (G and H) Images of control and Sqh-depleted clusters showing that nuclei stay behind protrusion necks upon sqhRNAi, 1 d at 29°C. (I) Plot of the change in nuclear aspect ratio of leader cells. Dots: individual nuclei, n = 6 (control) and 13 (sqh). Middle bar and error bars: mean ± SEM. 1 d at 29°C. Each dot represents one nucleus. Statistical test: Mann–Whitney Test. (J and K) Working model for how nuclei and myosin coordinate delamination and invasion into confined space. Genotypes and experimental replicates reported in Table S2. Scale bars: 10 μm.
Video 8.
Myosin II dynamics in control and Lam-depleted clusters. Time-lapse movies of border cells expressing Sqh-mCherry for control (left) or Lam-depleted (right) clusters. Polar cells are obscured with a black circle noted at the start of the movie to focus on the border cell cortical flashes. The time interval is 20 s. Scale bar: 10 µm. Frame rate: 2 frames/s. Playback speed: 40× real time.
In control clusters, cortical myosin flashes correlated with nuclear movement. When myosin accumulated behind the nucleus, it moved forward into the protrusion and the nucleus changed shape; in contrast, flashes in front of the nucleus correlated with backward nuclear movement (Fig. 7, D–F; and Videos 2 and 9). Myosin flashes accumulated behind the lead cell nucleus in both control and Lam-depleted clusters (Fig. 7, A–C; Fig. S5, F and G; and Videos 8 and 9). In Lam-depleted clusters with failed delamination, rather than pushing the nucleus forward, the flashes behind the nucleus moved ahead of the nucleus and the protrusion retracted (Fig. 7 B and Fig. S5 G; and Video 8). These data suggest myosin flashes accumulate behind Lam-depleted nuclei but fail to push them forward.
Video 9.
Myosin II flashes correspond with nuclear movement. Time-lapse series of border cells expressing Sqh-mCherry and UAS-GFP.NLS driven by c306Gal4. The time interval is 20 s. Scale bar: 10 µm. Frame rate: 3 frames/s. Playback speed: 60× real time.
To test the functional significance of myosin, we expressed sqh RNAi and evaluated the effect on nuclear shape and movement. A partial Sqh depletion reduced leader cell nuclear shape changes during migration (Fig. 7, G–I). A more penetrant Sqh depletion resulted in long and long-lived protrusions and failed delamination (Video 10) as reported previously (Majumder et al., 2012; Mishra et al., 2019b). In these clusters, nuclei failed to enter the protrusion (Fig. 7 H and Video 10). We conclude myosin forces deform and position nuclei.
Video 10.
Myosin II depletion results in long-lived protrusions and little nuclear movement. Time-lapse series of an egg chamber expressing UbiHisRFP, slbo4XPH-EGFP, c306Gal4;UAS-sqhRNAi after incubation at 29°C for 3 d acquired at 3-min time intervals. Scale bar: 10 µm. Frame rate: 5 frames/s. Playback speed: 900× real time.
Nuclei are connected to the cytoskeleton through the linker of the nucleoskeleton and cytoskeleton (LINC) complex (Crisp et al., 2006). However, prior work suggests nuclei in migrating dendritic cells and mesenchymal stem cells can be deformed and moved independently of the LINC complex, specifically when moving in 3D through pores smaller than the nuclear diameter (Jahed and Mofrad, 2019; Lee et al., 2021; Thiam et al., 2016). So, we tested if LINC complex components were involved in border cell migration. Depletion of LINC complex components, Klar, Koi, or Msp300 with previously tested RNAi lines (Collins et al., 2017; Perillo and Folker, 2018), did not cause delamination or major migration defects (Fig. S5, H–J). These results suggest myosin may have a more direct mechanical effect on nuclei. Together, the data indicate that cortical myosin flashes push nuclei into protrusions in a Lam-dependent manner. Further, Lam is required for the nucleus to stay in the protrusion and for the protrusion to enlarge, which facilitates cluster movement into the tiny spaces between nurse cells (Fig. 7 J).
Discussion
In vivo, cells migrate through diverse terrains: they can move on basement membranes or through ECM gels, but they can also squeeze between cells or in spaces between ECM and cells (Mishra et al., 2019a; Rørth, 2009; Yamada and Sixt, 2019). An important difference between cell movement through matrix versus between cells is that migrating cells can secrete proteases to enlarge spaces in matrix (Wolf et al., 2007, 2013; Yamada and Sixt, 2019). However, cells migrating between other cells, for example, during transendothelial migration or during embryonic development, do not have the option to degrade the substrate. Instead, they must either deform themselves sufficiently to squeeze through or push the substrate cells apart. How cells, especially cell collectives, achieve this is not well understood. Here, using the border cells as a model, we report the dynamics of nuclei and discover a novel role for the nucleus in this collective, confined, in vivo cell migration.
We propose a nuclear wedge model (Fig. 7 J), whereby F-actin–rich protrusions begin to separate nurse cells by breaking nurse cell–nurse cell adhesions. Then, the nucleus moves into the protrusion, preventing its collapse and allowing it to enlarge. Myosin flashes at the cortex appear to push the lead cell nucleus into the protrusion during successful delamination. However, when these flashes move past the nucleus, they retract the protrusion, impeding forward invasion (Fig. 7 K).
The B-type lamin, Lam, is essential for expansion of the leading border cell protrusion and movement into confined space. When Lam-depleted nuclei enter protrusions, they become more elongated than wildtype nuclei and sometimes herniate backward, indicating that they encounter rearward forces, likely generated by myosin-mediated contractions and/or nurse cell compression (Fig. 7 K). Despite transient accumulation of myosin behind Lam-depleted nuclei, they do not always progress into the protrusion. These observations suggest that the normal mechanical properties of the nucleus are important for myosin-mediated forces to push it into the protrusion. Alternatively or in addition, Lam may have other, less direct roles that affect the cell cortex to stabilize protrusions.
Lead cell protrusions serve multiple functions in migrating border cells. They are sensory structures that probe for chemoattractants and available space (Dai et al., 2020). Additionally, mechanical feedback from the dominant lead protrusion inhibits side and rear protrusion (Cai et al., 2014; Mishra et al., 2019b). The data presented here suggest an additional function for the lead cell protrusion—physical enlargement of the available space in front of the cluster, which has also been observed in cells migrating through hydrogels in vitro (Lee et al., 2021).
The thin, short, and unstable protrusions observed in Lam-depleted border cells suggest that the mechanical and/or biochemical properties of the nucleus normally facilitate enlargement of the lead cell protrusion and prevent its collapse, which is essential for the cluster to advance into the tiny spaces between nurse cells. Unable to maintain a single large protrusion toward the center of the egg chamber, Lam-depleted cells extend side protrusions, and clusters either fail to migrate or attempt abnormal trajectories, similar to the phenotype described for E-cadherin knockdown (Cai et al., 2014; Mishra et al., 2019b).
The leading border cell shares features with a variety of single cells migrating in confined environments in vitro and in vivo. In cells migrating in confined hydrogels, in neuronal progenitor cell-on-cell migrations, and in glioma cells invading the neural cortex, nuclei are pushed from behind by myosin similar to what we observe in the leading border cell (Beadle et al., 2008; Martini and Valdeolmillos, 2010; Lee et al., 2021)). In the original description of the nuclear piston (Petrie et al., 2014), myosin activity in front of the nucleus pulled the nucleus into the protrusion in a LINC-complex-dependent manner, which caused a diffusion barrier to form and pressure to build, followed by blebbing and enlargement of a “lobopodial” protrusion, which is equally wide at the tip and the base (closer to the nucleus). In contrast, in mesenchymal stem cells migrating in hydrogels, myosin activity at the rear moves the nucleus, independently of the LINC complex, to generate a piston effect that facilitates protrusion expansion (Lee et al., 2021). While myosin activity similarly pushes the lead border cell nucleus from the rear to facilitate protrusion expansion in a LINC-complex-independent manner, several hallmarks of the nuclear piston model are absent from border cells. These include blebbing, which is indicative of increased pressure, the diffusion barrier, and lobopodial protrusions. Although we sometimes observe transient broadening of the protrusion ahead of the lead border cell nucleus, somewhat reminiscent of a piston effect, the absence of other hallmarks suggests an alternative mechanism. Further, the tip of the border cell lead protrusion tends to be narrower than the base, supporting the idea that the nucleus stabilizes the protrusion base, and protrusion growth in front of the nucleus is likely due to F-actin polymerization since Rac activity and F-actin are elevated there (Murphy and Montell, 1996; Wang et al., 2010; Fulga and Rørth, 2002). Protrusions vary in size and shape during border cell migration, so it is also possible that more than one mechanism contributes to moving border cells through the extremely confined egg chamber environment.
Most hallmarks of the nuclear ruler were also absent from border cells, including calcium fluctuations, blebbing, and a requirement for cytosolic PLA2. One similarity between the mechanism operating in border cells and the nuclear ruler is the accumulation of cortical myosin. The experiments that uncovered the nuclear ruler severely compressed cells using cantilevers that were themselves not deformable, whereas border cells push nurse cells aside. This difference in the source of compression and material properties of the microenvironment may contribute to differences in cellular responses. The relatively mild deformations of lead border cell nuclei at least superficially resemble those of proteolytically active cells migrating through dense matrix (Friedl et al., 2011), which can relieve compressive forces by matrix degradation. Perhaps the ability to deform nurse cells similarly reduces the mechanical load on border cells.
Some cells, like border cells, use the nucleus to counteract environmental confinement, while other cells, such as neutrophils, reduce lamin expression instead to squeeze through tight spaces. Still other cells overcome the barrier of confinement, not by modulating the properties of the nucleus, but rather those of the cortex. For example, the transcription factor, Dfos, controls cortical actin levels to counteract tissue compression and permit nucleus movement during macrophage invasion into spaces between the ectoderm and mesoderm in Drosophila embryos (Belyaeva et al., 2022). In these cells, lamin levels do not normally affect migration, but reduced lamin can rescue invasion when cortical tension is impaired, illustrating how cells can use multiple mechanisms to move through confined environments. Whether lamins promote migration in confinement, impede it, or are irrelevant seems to depend on a combination of the mechanical properties of the nuclei and cortex of the migrating cells as well as the microenvironment. For example, egg chambers are estimated to be 100-fold softer than the silicone (Polydimethylsiloxane) used to fabricate microchannels (Johnston et al., 2014; Lamb et al., 2021). We speculate that the ability of the nucleus to wedge open spaces may be limited to relatively soft microenvironments and would likely be insufficient to overcome a barrier 100-fold stiffer than an egg chamber.
We found a role for B-type but not A-type lamin in border cell migration. A- and B-type lamins contribute to nuclear stiffness (Wintner et al., 2020) and have been reported to affect cell migration in prior studies (McGregor et al., 2016). However, we find that the sole A-type lamin in Drosophila, LamC, is not essential in border cells. It is possible that this is because LamC is normally expressed at a lower level than Lam, which would explain how overexpression of LamC can partially rescue Lam RNAi even though LamC RNAi does not cause a phenotype. Expression levels of A-type lamins tend to scale with tissue stiffness and are important in stiff environments, while B-type lamins dominate in softer environments (Swift et al., 2013). The egg chamber is a relatively soft environment, consistent with the pattern that B-type lamins might be more critical, similar to its role in neural tissues, where A-type lamins are downregulated (Jung et al., 2012). An alternative, but not mutually exclusive, hypothesis is that B-type lamin might have a unique function compared to A-type lamin. This might explain why A-type lamin can only partially compensate for the loss of the B-type lamin.
Follower border cells undergo less severe nuclear deformations compared with leader cells. Follower cells contribute to overall cluster motility (Campanale et al., 2022) and further expand the opening between nurse cells as the cluster advances. Polar cells undergo very little deformation. Since severe deformations can result in nuclear rupture and DNA damage (Irianto et al., 2017; Denais et al., 2016; Raab et al., 2016), absorption of the brunt of nuclear deformation by the lead cell could in principle serve to protect the rest of the cells in a moving collective and thereby provide a selective advantage to collective movement, which might be particularly relevant in the context of tumor metastasis.
Materials and methods
Drosophila genetics
Drosophila stocks used in this study are described in Table S1. Flies were maintained in cornmeal-yeast food. To combine Gal4s with different UAS-lines, five to seven virgin females were crossed to two to three males in the presence or absence of the temperature-sensitive repressor Gal80TS and flies were transferred into new vials every 2–3 days; all progeny used were heterozygous for constructs. For lines without Gal80TS, crosses were maintained at 25°C. For Gal4 lines with Gal80TS, crosses were kept at 18°C and incubated at 29°C for 3 d prior to dissection. For FlpOUT experiments to generate mosaic clones, flies were heat-shocked for 1 h at 37°C, restored to 25°C for 4 h, heat-shocked again for 1 h, and fattened for 3 d at 29°C. Crosses were fattened for both 1 d and 3 d at 29°C prior to dissection for stage 10 as specified in Table S2, figures, and figure legends. 2–5-d-old fly progeny were supplemented with yeast for each experiment, and control and RNAi or overexpression flies in each experiment were matched in age. Table S2 reports genotypes and conditions for each analysis.
LamRE construct
The re-encoded, RNAi-resistant LamRE construct was generated to re-encode the DNA of the regions targeted by Lam1 and Lam2 RNAi lines in the coding sequence while keeping the same codon sequence. The re-encoded Lam coding sequence was ordered as a gene block (IDT, see coding sequence below). Then, it was introduced into a pUAST-attB vector (RRID:DGRC_1419) with in-fusion cloning (#638947; takara bio). The UAS-LamRE construct was sent to Bestgene to generate a transgene on chromosome 3 (stock RRID: BDSC_8622 [y1 w67c23; P{CaryP}attP2]) using phiCI integrase-mediated site-specific transgenesis.
LamRE sequence: 5′-ATGTCGAGCAAATCCCGACGTGCTGGCACCGCCACGCCGCAGCCCGGCAACACCTCCACCCCCCGGCCGCCATCGGCGGGTCCGCAGCCGCCGCCCCCCAGCACCCATAGCCAAACCGCTAGCTCCCCGCTGTCCCCGACGCGCCATAGCCGTGTCGCTGAAAAAGTCGAATTGCAAAATTTGAATGACCGTTTGGCTACGTATATCGATCGCGTCCGTAATTTGGAAACCGAAAATAGCCGTCTGACGATTGAAGTCCAAACGACGCGCGATACCGTGACCCGTGAAACGACGAATATTAAAAATATTTTTGAAGCTGAATTGTTGGAAACCCGTCGCTTGCTGGACGATACCGCCCGCGACCGTGCCCGCGCTGAAATTGACATTAAACGCCTGTGGGAAGAAAATGAAGAACTGAAAAATAAATTGGATAAAAAAACGAAAGAATGCACCACTGCTGAGGGCAATGTCCGCATGTACGAGTCGCGCGCCAACGAGCTGAACAACAAATACAACCAGGCCAACGCCGATCGGAAGAAGCTTAACGAAGACCTGAATGAGGCGCTAAAGGAGCTGGAGAGACTGCGTAAGCAGTTCGAGGAAACGCGGAAGAACCTGGAACAGGAGACACTGTCGCGCGTTGACCTGGAGAACACCATTCAGAGTCTGCGCGAGGAGCTCTCGTTCAAGGATCAGATCCATTCGCAGGAGATCAATGAGTCGCGCCGCATCAAACAGACAGAGTATAGCGAGATCGACGGTCGCCTCAGCTCCGAGTACGATGCCAAGTTGAAGCAGTCGCTGCAGGAGCTGCGCGCCCAGTACGAGGAGCAGATGCAGATTAATCGCGATGAAATCCAGTCCCTCTACGAGGACAAGATCCAACGACTGCAAGAGGCCGCCGCACGCACATCCAATTCCACGCACAAGTCCATCGAGGAGCTGCGCTCCACTCGTGTGCGTATCGATGCGCTCAACGCCAATATCAACGAACTGGAGCAAGCCAATGCCGACCTCAATGCGCGGATCCGTGATCTGGAGCGCCAGCTGGACAACGATCGCGAACGCCACGGTCAAGAGATAGACCTTCTCGAGAAGGAGCTCATTCGGCTGCGCGAAGAGATGACGCAACAGCTCAAGGAGTACCAGGACCTTATGGACATCAAGGTCTCCCTGGATTTGGAAATCGCCGCATACGACAAGCTGCTGGTGGGCGAGGAGGCTCGTTTGAACATCACCCCAGCCACCAACACGGCCACAGTGCAGTCCTTTAGCCAGTCGCTGCGCAACTCCACGCGAGCCACGCCATCGCGTCGCACTCCCTCTGCTGCCGTGAAGCGCAAACGCGCCGTGGTCGACGAGTCGGAGGATCACAGCGTCGCCGATTACTATGTGTCCGCCAGTGCCAAGGGCAACGTGGAGATCAAGGAGATCGATCCCGAGGGCAAGTTCGTAAGGCTGTTCAACAAGGGCAGCGAGGAGGTGGCCATCGGTGGCTGGCAGCTGCAACGCCTGATTAATGAAAAGGGCCCCAGCACGACCTATAAATTTCACCGCAGCGTCCGCATTGAACCCAACGGAGTCATTACGGTGTGGAGCGCCGATACGAAAGCTAGCCATGAACCCCCCAGCTCCCTGGTCATGAAAAGCCAAAAATGGGTGAGCGCTGATAATACCCGCACCATCCTGTTGAATAGCGAAGGAGAAGCTGTCGCTAACTTGGACCGTATTAAACGTATCGTCAGCCAGCATACCAGCAGCAGCCGCTTGAGCCGCCGCCGTTCCGTCACGGCTGTCGATGGAAACGAACAACTGTATCATCAACAAGGAGACCCCCAACAAAGCAATGAAAAATGTGCTATCATGTAG-3′.
Immunostaining and fixed imaging
Ovaries were dissected in Schneider’s media (Thermo Fisher Scientific) supplemented with 20% heat-inactivated fetal bovine serum (Sigma-Aldrich) and 1× antimycotic/antibiotic (VWR) and adjusted to a pH of 6.85–6.95. Ovaries were then fixed in 4% paraformaldehyde in 1X phosphate-buffered saline (PBS) for 15 min. Ovarioles were washed 3 × 10 min with 1X PBS + 0.4% triton (PBST) and incubated in primary antibodies overnight at 4°C. Antibodies used include Lam (1:10, #ADL84.12; Developmental Studies Hybridoma Bank [DSHB]), E-cadherin (1:15, #DCAD2; DSHB), LamC (1:20, #LC28.26; DSHB), singed (1:20, #sn 7c; DSHB), Klar (1:10, #KLARC-9C10; DSHB), Lamin B receptor (1:200 gift from Dr. Georg Krohne), aPKC (1:500; Santa Cruz Technologies), and Dlg (1:20, #4F3; DSHB). The following day, ovaries were washed with 1X PBST followed by 3 × 10 min washes in 1X PBST. Then, ovaries were incubated with secondary antibodies (Goat anti-Rat IgG [H+L] Cross-Adsorbed Secondary Antibody, Alexa Fluor 647, 488, or 568, Invitrogen, Goat anti-Mouse IgG [H+L] Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 and 568; Invitrogen), phalloidin-Atto-647N (1:500; Sigma-Aldrich), and Hoechst (1:500; Invitrogen) for 1 h at room temperature. Ovaries were washed with 1X PBST followed by 3 × 10 min washes in 1X PBST. After the last wash, ovaries were suspended in vectashield (H-1000; Vector Laboratories) and incubated overnight at 4°C before mounting onto slides. Details for reagents and sources are listed in Table S3.
Slides were imaged on a Zeiss LSM780 confocal microscope with a 40× 1.1 N.A. 0.62-mm long working distance water objective. ZEN software was used to acquire images, and Image J (FIJI) was used to process images.
Live imaging
Ovarioles were dissected from ovaries in live imaging media (Schneider’s media [Thermo Fisher Scientific] supplemented with 20% heat-inactivated fetal bovine serum [Sigma-Aldrich] and 1× antimycotic/antibiotic [VWR] with a pH of 6.85–6.95) and resuspended in live imaging media with 0.4 mg/ml bovine insulin (I1882; Sigma-Aldrich) and 1% agarose and immediately mounted for imaging. Stage 9 egg chambers were imaged on an inverted Zeiss LSM800 confocal microscope fitted with a Plan-Apochromat 40×, 1.2 NA multi-immersion objective with the collar set for water immersion at time intervals specified in figures and movies. ZEN software was used to acquire images, and Image J (FIJI) was used to process images.
For imaging with dextrans to mark extracellular spaces, fluorescent dextrans were added to the media prior to mounting (100 μg/ml; Dextran Alexa 647 10,000 MW; D22914; Thermo Fisher Scientific).
Image analysis
Diameter measurements
For each movie, the diameters of clusters, cells, nuclei, and junctures in the central path were measured prior to delamination for the first time frame (Fig. 1 E). The outline of each cell, nucleus, and the entire cluster were manually traced from fluorescent markers in ImageJ (GFP-moesin for border cells, and dsRED.NLS for nuclei and 10 kDa dextran-Alexa647 junctures).
Protrusion shape analysis
The time of delamination (Fig. 1 F and Fig. S1, A and B) was scored as the full detachment of all border cells from the anterior of the egg chamber and set as time = 0 for the measurements and all time points were set relative to delamination = 0 min. UAS-GFP-moesin actin-binding domain was expressed in border cells by fruitlessGal4 or slbo4XPHEGFP and was used to mark the border cells’ boundaries. To measure protrusion length, a line that was parallel to the long axis of the egg chamber was drawn from the polar cell boundary to the protrusion tip. To measure protrusion width, lines were manually drawn across the middle of the protrusion neck parallel to the long axis of the egg chamber (for Fig. S1 A and Fig. 5, C–E) or at the tip or base of the protrusion (Fig. S1, C–E; and Fig. S4, J–O) and measured in Image J for each time point.
Nuclear movement and shape analysis
To measure nuclear position in the protruding cells, a line was manually drawn from the back of the nucleus to the nearest polar cell membrane marked with slbo4XPHE-GFP for each time point. The change in nuclear position was measured as the change in length of the line between two time points and divided by the time interval. Only the largest protrusion was scored for each time point if there were multiple protrusions, and analysis was only performed in protruding cells. To set a threshold for forward, neutral, or backward nuclear movement, the average and standard deviation (SD) were calculated for the data set, and forward movement was defined as >1 SD greater than the mean, neutral was defined as within 1 SD from the mean, and backward was defined as >1 SD lower than the mean.
To measure the lead cell’s nuclear aspect ratio relative to the migration axis lines (Fig. S1, A and B; and Fig. 3 H), lines were manually drawn across the center of the nucleus marked with dsRED.NLS parallel and perpendicular to the long axis of the egg chamber. The aspect ratio was calculated by dividing the parallel nuclear diameter by the perpendicular nuclear diameter.
To measure the nuclear aspect ratio and circularity in a more automated manner (and regardless of egg chamber orientation) from time-lapse series for comparison of leader, follower, and polar cell nuclei, the central z-slice of the channel with UAS.dsRED.NLS in each nucleus was cropped from each time point to prevent overlap of nuclei during semiautomated analysis. Nuclei were segmented into binary images using a custom Matlab code, and shape parameters were measured over time using the analyze particle tools in ImageJ. We manually verified if the segmentation worked properly and whether the nucleus was in view or not. If the automated analysis did not properly segment the nucleus at a time point, a manual trace was performed. Samples were blinded throughout the analysis. All time frames were included unless the nucleus went out of focus during the imaging session. The average nuclear aspect ratio was automatically generated in ImageJ measurements. The change in nuclear aspect ratio was calculated by taking the absolute value between two aspect ratios of adjacent time points.
For fixed imaging analysis of nuclear shape, the Hoecsht signal in fixed border cells proved difficult to segment automatically. Therefore, each nucleus in a border cell cluster was traced manually in Image J by using the Hoechst signal to mark the nucleus in stage 9 egg chambers. All border cells’ nuclei were included unless Hoechst staining did not allow distinguishing of a nucleus from the background or if a nucleus was out of focus. ImageJ measurements were used to calculate the area and aspect ratio of traces of nuclei in fixed images.
Myosin II flash analysis
Each movie was aligned so that the x axis was the long axis of the egg chamber and the anterior was on the left. The egg chamber was aligned over time points to correct for drift using the StackReg plugin in ImageJ. Myosin II flashes were manually scored for each time point relative to their position of the nucleus aligned to the migration axis (front: further from the anterior than nucleus, side: parallel in x axis to nucleus, back: closer to anterior than nucleus). It was also noted that there was no flash near the nucleus at the time point. Nuclear movement defined as x axis displacement was measured by tracking the nuclear position over time using manual tracking. Nuclear shape measurements with the GFP.NLS were measured using the same semiautomatic analysis method described for dsRED.NLS above. Then, the average movement and shape change were binned based on the presence and position of flash for each movie.
Border cell size
To measure border cell size, cytoplasmic GFP in FlpOut clones expressing UAS-white RNAi or UAS-Lam RNAi were automatically segmented using a Matlab code and analyzed for area in ImageJ using the analyzed particles. Only clones that were distinguishable from other clones in a cluster were used for segmentation.
Border cell migration indexes
To measure border cell migration defects, all stage 10 egg chambers were identified by onset of centripetal cell migration and the border cell cluster was detected with E-cadherin or phalloidin staining. The position of the border cell cluster was manually scored based on positions (0%: still attached, <50% migrated relative to distance between anterior and oocyte boundary, >50% migrated relative to anterior:oocyte, and 100%: at oocyte boundary). Then, the position of the cluster was recorded for each stage egg chamber. RNAi and overexpression experiments were performed at least three times independently with three independent genetic crosses and with the control line (UAS-whiteRNAi or UAS-LacZ) crossed in parallel.
Migration speed and directionality index
To measure the average speed of each border cell cluster, the z-slices from time-lapse series were max-projected. Then, the egg chamber was aligned over time points to correct for drift using the StackReg plugin in ImageJ. The manual tracking plugin in ImageJ was used to track border cell cluster positions over time.
The directionality index was measured as previously reported (Cai et al., 2014). The net distance traveled over the time-lapse movie was divided by the sum of the distances moved at each time point.
Lamin staining and border cell number
In stage 10 egg chambers, the number of border cells was manually counted using Hoechst and border cell markers (E-cadherin or Phalloidin). These cell number counts were limited to 30-μm z-sections, which usually covered the entire cluster. Lamin depletion was categorized as “none” (complete rim around the Hoechst signal), “partial” (incomplete rim around Hoechst), or “full” (no detectable lamin at the nuclear rim). The percent of border cells with full, partial, or no knockdown was divided by the total number of border cells in the cluster. The averages from each LamRNAi line are shown in Fig. S2. Then, the averages were calculated for completed migration or incomplete migration in the Lam2RNAi (line #45635; Fig. S2 H).
STAT reporter quantification
To measure STAT activity in stage 8 egg chambers, we crossed c306Gal4/Y; 10XSTATGFP (STAT92e)/CyO to UAS-LamRNAi or UAS-wRNAi lines. We performed immunostaining and imaged the anterior of stage 8 egg chambers. All egg chambers were imaged using the same conditions. For each egg chamber, we drew a small region of interest in each of the two cells adjacent to the polar cells in ImageJ and acquired the average fluorescent intensity and then subtracted a box in the nurse cells as background. Then, averages were normalized to the average of the UAS-wRNAi control to show the relative change in the fluorescence of the STAT reporter from the control.
Notch responsive element quantification
To measure Notch activity, we generated a fly co-expressing UAS-LamRNAi (line #107419) or UAS-wRNAi with a Notch Responsive Element-Red Reporter (NRE-RR; Zacharioudaki and Bray, 2014) and crossed these flies to a hsFlpAyGal4;UAS-GFP line. We heat-shocked progeny as described above to generate clones. Only clusters with GFP+ clones were used for analysis. For measurements of NRE-RR fluorescent intensity, a box overlaying the nucleus was drawn in the center of each border cell and subtracted the average fluorescent intensity of a box drawn in the nurse cells (background). For each cluster, the average fluorescence intensity (F.I.) of the GFP+ clones was divided by the average fluorescent intensity of the GFP clones in the same cluster.
E-cadherin and F-actin quantification
For the total mean levels, each border cell cluster still juxtaposed to the anterior was traced and the mean F.I. was measured for F-actin and E-cadherin. Circularity of the cluster was measured from these same traces of the cluster.
For the peripheral E-cadherin analysis, FlpOUT clones with wildtype (WT) and LamRNAi clones were analyzed. The mean of 3-pixel-thick lines traced around the periphery of the GFP+ clone (UAS-LamRNAi) was divided by the mean of a 3-pixel-thick line around the periphery of a GFP− (WT) clone in the same cluster.
For the distribution of F-actin, a 10-pixel-thick line scan was drawn across the cluster from front to back and the levels were measured. Lines were normalized and binned to a position where the top 10% of the line was deemed the front, the 11–89% was deemed the middle, and the last 10% was deemed the back for each trace.
GCaMP imaging and quantification
GCaMP signal was monitored every 5 s over a total of 5 min as described previously to monitor GCaMP flashes (Sahu et al., 2017). A region of interest was drawn in one z-plane at the front of the border cell cluster and the average F.I. was measured over time. As a positive control, a region of interest was drawn in a follicle cell displaying GCaMP flashes. The average F.I. and the maximum fold change (F.I._time1−F.I_.time2)/F.I._time1) was measured from those values.
Photoactivation experiments
For photoactivation of GFP-ɑtubulin, a region of interest (ROI) was drawn in the protrusion of a leading cell or on one side of the nucleus in a posterior follicle cell in ZEN. Photoactivation was performed on a Zeiss 800 with 405 lasers at 20% power for three iterations every 2 s. Photoactivation started after 6 s and all movies were captured over 120 s total. Then, a back and front ROI were drawn in ImageJ and the mean was measured for each time point. Data were normalized to the front:back ratio of the first time point for Fig. 6 C. To measure the time constant, we fit each curve of the mean intensity over time to a Boltzmann sigmoidal function as done previously (Petrie et al., 2014) using Prism software. The slope of the curve is the time constant and inversely proportional to diffusion. All slopes were included in the figure despite the R2 value, but removing data that did not fit the curve R2 < 0.8 did not change the results.
Statistical analysis and data presentation
Data points, middle bars, and error bars are specified in the legends but generally refer to the mean ± SEM with each data point plotted as a dot. Experimental replicates are reported in Table S2 and sample sizes (e.g., number of egg chambers, number cells, number nuclei) are specified in figures and/or legends. Inclusion criteria: all undamaged samples, designated egg chamber stage, and in-focus images were included for analysis. Exclusion criteria: damaged, out of focus, or incorrect stage time points and images. Nuclear shape analyses from live imaging were blinded. All other measurements were not blinded but performed in a systematic manner to minimize bias. Power calculations were not generally performed, but the aim was to perform experiments in triplicate to have sufficient power for statistical testing. Genetic crosses were performed at least in duplicate. Flies that were used to establish genetic crosses and flies used for experiments were selected randomly from vials of the appropriate age and genotype, but there were no procedures that required randomization of subjects. Data were analyzed for normality using a Shapiro-Wilk normality test in Prism. A two-sided t test or one-way ANOVA (parametric test) was performed if data passed the normality test. If data did not pass the normality test, a nonparametric test was performed in Prism. Statistical tests used and sample size are reported in figure legends. Fluorescence images were acquired and processed exactly the same between control and experimental conditions. Figure legends include additional information on data representation for each specific experiment.
Online supplemental material
Fig. S1 shows nuclear shape dynamics relative to protrusion and cluster dynamics in border cells. Fig. S2 shows LamC and Lam are depleted with RNAi lines, Lam is required in outer border cells, and LBR overexpression is required for nuclear shape and for migration in outer border cells. Fig. S3 shows border cell organization and gene expression in Lam-depleted clusters versus controls. Fig. S4 shows border cell polarity and protrusion and nuclear dynamics in control and Lam-depleted clusters. Fig. S5 shows iPLA2 mutants and clusters depleted of LINC complex components complete migration and also shows examples of myosin II and calcium dynamics in border cell clusters. Video 1 shows border cell delamination. Video 2 shows leading nuclei deform rapidly. Video 3 shows leader cells undergo the most deformation while polar cells do not have major shape changes. Video 4 shows control clusters delaminate and form a main protrusion while Lam-depleted clusters fail to invade and stabilize a protrusion. Video 5 shows protrusion expansion in controls versus protrusion narrowing in Lam-depleted clusters relative to nuclear dynamics. Video 6 shows photoactivation of PA-GFP-tubulin in a leading border cell and follicle cell. Video 7 shows GCaMP6s in border cells and follicle cells. Video 8 shows myosin II dynamics in control and Lam-depleted clusters. Video 9 shows myosin II dynamics correlate with nuclear dynamics. Video 10 shows strong sqh RNAi results in failed delamination, long-lived protrusions, and lack of nuclear movement. Table S1 lists D. melanogaster stocks. Table S2 reports genotypes, experimental conditions, and replicates for figures. Table S3 reports key antibodies and chemical reagents.
Supplementary Material
shows D. melanogaster stocks.
shows fly genotypes and experimental replicates.
shows key antibodies and chemicals.
Acknowledgments
We would like to thank the Bloomington Drosophila Stock Center, the Vienna Drosophila Resource Center for the Drosophila stocks. We would like to thank Georg Krohne (University of Würzburg, Würzburg, Germany) for the Lamin B receptor antibody, Lori Wallrath (University of Iowa, Iowa City, IA) for the UAS-LamC fly stock, and Jörg Großhans (University of Göttingen, Göttingen, Germany) for the UAS-Lam fly stock. We would like to thank Carinna Tran, Emory Campbell, and Jim Mondo for their technical assistance, Joseph Campanale and Maddalena Nano for their careful reading of the manuscript, and all of the members of the Montell lab for their feedback on this work.
This work was supported by National Institutes of Health grant R01GM073164 to D.J. Montell, and training of L. Penfield was supported in part by the American Cancer Society Postdoctoral Fellowship (PF-22-091-01-MM).
Author contributions: L. Penfield and D.J. Montell designed the research; L. Penfield performed the research; L. Penfield analyzed data; and L. Penfield and D.J. Montell wrote the paper.
Data availability
Data are shown in the figures and supplemental files, and additional data files are available upon request.
References
- Aranjuez, G., Burtscher A., Sawant K., Majumder P., and McDonald J.A.. 2016. Dynamic myosin activation promotes collective morphology and migration by locally balancing oppositional forces from surrounding tissue. Mol. Biol. Cell. 27:1898–1910. 10.1091/mbc.e15-10-0744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaker, G., Ramel D., Wculek S.K., González-Gaitán M., and Emery G.. 2010. Spatial restriction of receptor tyrosine kinase activity through a polarized endocytic cycle controls border cell migration. Proc. Natl. Acad. Sci. USA. 107:22558–22563. 10.1073/pnas.1010795108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle, C., Assanah M.C., Monzo P., Vallee R., Rosenfeld S.S., and Canoll P.. 2008. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 19:3357–3368. 10.1091/mbc.e08-03-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyaeva, V., Wachner S., Gyoergy A., Emtenani S., Gridchyn I., Akhmanova M., Linder M., Roblek M., Sibilia M., and Siekhaus D.. 2022. Fos regulates macrophage infiltration against surrounding tissue resistance by a cortical actin-based mechanism in Drosophila. PLoS Biol. 20:e3001494. 10.1371/journal.pbio.3001494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David, G., Miller E., and Steinhauer J.. 2015. Drosophila spermatid individualization is sensitive to temperature and fatty acid metabolism. Spermatogenesis. 5:e1006089. 10.1080/21565562.2015.1006089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie, C.A., and Sanders M.M.. 1993. A cDNA from Drosophila melanogaster encodes a lamin C-like intermediate filament protein. J. Cell Sci. 104:1263–1272. 10.1242/jcs.104.4.1263 [DOI] [PubMed] [Google Scholar]
- Cai, D., Chen S.-C., Prasad M., He L., Wang X., Choesmel-Cadamuro V., Sawyer J.K., Danuser G., and Montell D.J.. 2014. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 157:1146–1159. 10.1016/j.cell.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Cuenca, F.J., Janota C.S., and Gomes E.R.. 2018. Dealing with the nucleus during cell migration. Curr. Opin. Cell Biol. 50:35–41. 10.1016/j.ceb.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Campanale, J.P., Mondo J.A., and Montell D.J.. 2022. A Scribble/Cdep/Rac pathway controls follower-cell crawling and cluster cohesion during collective border-cell migration. Dev. Cell. 57:2483–2496.e4. 10.1016/j.devcel.2022.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, N.Y., Yang Y., Weston T.A., Belling J.N., Heizer P., Tu Y., Kim P., Edillo L., Jonas S.J., Weiss P.S., et al. 2019. An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc. Natl. Acad. Sci. USA. 116:25870–25879. 10.1073/pnas.1917225116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, M.A., Mandigo T.R., Camuglia J.M., Vazquez G.A., Anderson A.J., Hudson C.H., Hanron J.L., and Folker E.S.. 2017. Emery-Dreifuss muscular dystrophy-linked genes and centronuclear myopathy-linked genes regulate myonuclear movement by distinct mechanisms. Mol. Biol. Cell. 28:2303–2317. 10.1091/mbc.e16-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp, M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., and Hodzic D.. 2006. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 172:41–53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W., Guo X., Cao Y., Mondo J.A., Campanale J.P., Montell B.J., Burrous H., Streichan S., Gov N., Rappel W.-J., and Montell D.J.. 2020. Tissue topography steers migrating Drosophila border cells. Science. 370:987–990. 10.1126/science.aaz4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, P.M., Denais C., Bakshi M.C., and Lammerding J.. 2014. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 7:293–306. 10.1007/s12195-014-0342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson, P.M., and Lammerding J.. 2014. Broken nuclei--lamins, nuclear mechanics, and disease. Trends Cell Biol. 24:247–256. 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais, C.M., Gilbert R.M., Isermann P., McGregor A.L., te Lindert M., Weigelin B., Davidson P.M., Friedl P., Wolf K., and Lammerding J.. 2016. Nuclear envelope rupture and repair during cancer cell migration. Science. 352:353–358. 10.1126/science.aad7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchek, P., Somogyi K., Jékely G., Beccari S., and Rørth P.. 2001. Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell. 107:17–26. 10.1016/s0092-8674(01)00502-5 [DOI] [PubMed] [Google Scholar]
- Ferrera, D., Canale C., Marotta R., Mazzaro N., Gritti M., Mazzanti M., Capellari S., Cortelli P., and Gasparini L.. 2014. Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. FASEB J. 28:3906–3918. 10.1096/fj.13-247635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, P., and Gilmour D.. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Friedl, P., Wolf K., and Lammerding J.. 2011. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 23:55–64. 10.1016/j.ceb.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga, T.A., and Rørth P.. 2002. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat. Cell Biol. 4:715–719. 10.1038/ncb848 [DOI] [PubMed] [Google Scholar]
- Graham, D.M., Andersen T., Sharek L., Uzer G., Rothenberg K., Hoffman B.D., Rubin J., Balland M., Bear J.E., and Burridge K.. 2018. Enucleated cells reveal differential roles of the nucleus in cell migration, polarity, and mechanotransduction. J. Cell Biol. 217:895–914. 10.1083/jcb.201706097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampoelz, B., Mackmull M.-T., Machado P., Ronchi P., Bui K.H., Schieber N., Santarella-Mellwig R., Necakov A., Andrés-Pons A., Philippe J.M., et al. 2016. Pre-assembled nuclear pores insert into the nuclear envelope during early development. Dev. Cell. 166:664–678. 10.1016/j.cell.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, T., Swift J., Irianto J., Shin J.-W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C.D.P., Ivanovska I.L., and Discher D.E.. 2014. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 204:669–682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer, M.W. 2010. The nuclear envelope. Cold Spring Harb. Perspect. Biol. 2:a000539. 10.1101/cshperspect.a000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, K., Dreger C.K., Olins A.L., Olins D.E., Shultz L.D., Lucke B., Karl H., Kaps R., Müller D., Vayá A., et al. 2002. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huët anomaly). Nat. Genet. 31:410–414. 10.1038/ng925 [DOI] [PubMed] [Google Scholar]
- Hutchison, C.J. 2014. B-type lamins in health and disease. Semin. Cell Dev. Biol. 29:158–163. 10.1016/j.semcdb.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irianto, J., Xia Y., Pfeifer C.R., Athirasala A., Ji J., Alvey C., Tewari M., Bennett R.R., Harding S.M., Liu A.J., et al. 2017. DNA damage follows repair factor depletion and portends genome variation in cancer cells after pore migration. Curr. Biol. 27:210–223. 10.1016/j.cub.2016.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed, Z., and Mofrad M.R.. 2019. The nucleus feels the force, LINCed in or not!. Curr. Opin. Cell Biol. 58:114–119. 10.1016/j.ceb.2019.02.012 [DOI] [PubMed] [Google Scholar]
- Johnston, I.D., McCluskey D.K., Tan C.K.L., and Tracey M.C.. 2014. Mechanical characterization of bulk Sylgard 184 for microfluidics and microengineering. J. Micromech. Microeng. 24:035017. 10.1088/0960-1317/24/3/035017 [DOI] [Google Scholar]
- Jung, H.-J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S.J., Yang S.H., Barnes R.H. II, Hong J., Sun T., Pleasure S.J., et al. 2012. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl. Acad. Sci. USA. 109:E423–E431. 10.1073/pnas.1111780109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalukula, Y., Stephens A.D., Lammerding J., and Gabriele S.. 2022. Mechanics and functional consequences of nuclear deformations. Nat. Rev. Mol. Cell Biol. 23:583–602. 10.1038/s41580-022-00480-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, M.C., Kaluarachchi C.P., Lansakara T.I., Mellentine S.Q., Lan Y., Tivanski A.V., and Tootle T.L.. 2021. Fascin limits myosin activity within Drosophila border cells to control substrate stiffness and promote migration. Elife. 10:e69836. 10.7554/eLife.69836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding, J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., and Lee R.T.. 2004. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 113:370–378. 10.1172/JCI19670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.-P., Alisafaei F., Adebawale K., Chang J., Shenoy V.B., and Chaudhuri O.. 2021. The nuclear piston activates mechanosensitive ion channels to generate cell migration paths in confining microenvironments. Sci. Adv. 7:eabd4058. 10.1126/sciadv.abd4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S.H., Hale C.M., Panorchan P., Khatau S.B., George J.P., Tseng Y., Stewart C.L., Hodzic D., and Wirtz D.. 2007. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys. J. 93:2542–2552. 10.1529/biophysj.106.102426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Böhme, B., Wismar J., Fuchs S., Reifegerste R., Buchner E., Betz H., and Schmitt B.. 1997. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 137:1001–1016. 10.1083/jcb.137.5.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, G., Lee P.-T., Chen K., Mao D., Tan K.L., Zuo Z., Lin W.-W., Wang L., and Bellen H.J.. 2018. Phospholipase PLA2G6, a parkinsonism-associated gene, affects Vps26 and Vps35, retromer function, and ceramide levels, similar to α-synuclein gain. Cell Metab. 28:605–618.e6. 10.1016/j.cmet.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Lomakin, A.J., Cattin C.J., Cuvelier D., Alraies Z., Molina M., Nader G.P.F., Srivastava N., Sáez P.J., Garcia-Arcos J.M., Zhitnyak I.Y., et al. 2020. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science. 370:eaba2894. 10.1126/science.aba2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J., Zhou P., Guo X., Wang D., and Chen J.. 2019. The polarity protein Dlg5 regulates collective cell migration during Drosophila oogenesis. PLoS One. 14:e0226061. 10.1371/journal.pone.0226061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder, P., Aranjuez G., Amick J., and McDonald J.A.. 2012. Par-1 controls myosin-II activity through myosin phosphatase to regulate border cell migration. Curr. Biol. 22:363–372. 10.1016/j.cub.2012.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini, F.J., and Valdeolmillos M.. 2010. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J. Neurosci. 30:8660–8670. 10.1523/JNEUROSCI.1962-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J.A., Khodyakova A., Aranjuez G., Dudley C., and Montell D.J.. 2008. PAR-1 kinase regulates epithelial detachment and directional protrusion of migrating border cells. Curr. Biol. 18:1659–1667. 10.1016/j.cub.2008.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor, A.L., Hsia C.-R., and Lammerding J.. 2016. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr. Opin. Cell Biol. 40:32–40. 10.1016/j.ceb.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A.K., Campanale J.P., Mondo J.A., and Montell D.J.. 2019a. Cell interactions in collective cell migration. Development. 146:dev172056. 10.1242/dev.172056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, A.K., Mondo J.A., Campanale J.P., and Montell D.J.. 2019b. Coordination of protrusion dynamics within and between collectively migrating border cells by myosin II. Mol. Biol. Cell. 30:2490–2502. 10.1091/mbc.E19-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell, D.J., Yoon W.H., and Starz-Gaiano M.. 2012. Group choreography: Mechanisms orchestrating the collective movement of border cells. Nat. Rev. Mol. Cell Biol. 13:631–645. 10.1038/nrm3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.M., and Montell D.J.. 1996. Cell type-specific roles for Cdc42, Rac, and RhoL in Drosophila oogenesis. J. Cell Biol. 133:617–630. 10.1083/jcb.133.3.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo, M., and Folker E.S.. 2018. Specialized positioning of myonuclei near cell-cell junctions. Front. Physiol. 9:1531. 10.3389/fphys.2018.01531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie, R.J., Koo H., and Yamada K.M.. 2014. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 345:1062–1065. 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky, R., and Großhans J.. 2018. Expression of lamina proteins Lamin and Kugelkern suppresses stem cell proliferation. Nucleus. 9:104–118. 10.1080/19491034.2017.1412028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro, E.M., and Montell D.J.. 2004. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development. 131:5243–5251. 10.1242/dev.01412 [DOI] [PubMed] [Google Scholar]
- Prüfert, K., Vogel A., and Krohne G.. 2004. The lamin CxxM motif promotes nuclear membrane growth. J. Cell Sci. 117:6105–6116. 10.1242/jcs.01532 [DOI] [PubMed] [Google Scholar]
- Raab, M., Gentili M., de Belly H., Thiam H.R., Vargas P., Jimenez A.J., Lautenschlaeger F., Voituriez R., Lennon-Duménil A.M., Manel N., and Piel M.. 2016. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 352:359–362. 10.1126/science.aad7611 [DOI] [PubMed] [Google Scholar]
- Renkawitz, J., Kopf A., Stopp J., de Vries I., Driscoll M.K., Merrin J., Hauschild R., Welf E.S., Danuser G., Fiolka R., and Sixt M.. 2019. Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature. 568:546–550. 10.1038/s41586-019-1087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, N., d’Água B.B., and Ridley A.J.. 2013. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer. 13:858–870. 10.1038/nrc3628 [DOI] [PubMed] [Google Scholar]
- Rørth, P. 2009. Collective cell migration. Annu. Rev. Cell Dev. Biol. 25:407–429. 10.1146/annurev.cellbio.042308.113231 [DOI] [PubMed] [Google Scholar]
- Rowat, A.C., Jaalouk D.E., Zwerger M., Ung W.L., Eydelnant I.A., Olins D.E., Olins A.L., Herrmann H., Weitz D.A., and Lammerding J.. 2013. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 288:8610–8618. 10.1074/jbc.M112.441535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu, A., Ghosh R., Deshpande G., and Prasad M.. 2017. A gap junction protein, Inx2, modulates calcium flux to specify border cell fate during Drosophila oogenesis. PLoS Genet. 13:e1006542. 10.1371/journal.pgen.1006542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J.-W., Spinler K.R., Swift J., Chasis J.A., Mohandas N., and Discher D.E.. 2013. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc. Natl. Acad. Sci. USA. 110:18892–18897. 10.1073/pnas.1304996110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver, D.L., Geisbrecht E.R., and Montell D.J.. 2005. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development. 132:3483–3492. 10.1242/dev.01910 [DOI] [PubMed] [Google Scholar]
- Silver, D.L., and Montell D.J.. 2001. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 107:831–841. 10.1016/s0092-8674(01)00607-9 [DOI] [PubMed] [Google Scholar]
- Stoletov, K., Kato H., Zardouzian E., Kelber J., Yang J., Shattil S., and Klemke R.. 2010. Visualizing extravasation dynamics of metastatic tumor cells. J. Cell Sci. 123:2332–2341. 10.1242/jcs.069443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift, J., Ivanovska I.L., Buxboim A., Harada T., Dingal P.C.D.P., Pinter J., Pajerowski J.D., Spinler K.R., Shin J.-W., Tewari M., et al. 2013. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 341:1240104. 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam, H.-R., Vargas P., Carpi N., Crespo C.L., Raab M., Terriac E., King M.C., Jacobelli J., Alberts A.S., Stradal T., et al. 2016. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun. 7:10997. 10.1038/ncomms10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, J.D., Hatch E.M., Anderson D.J., and Hetzer M.W.. 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 3:88–100. 10.4161/nucl.18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini, V., Pezzano F., Català Castro F., Häkkinen H.-M., Jiménez-Delgado S., Colomer-Rosell M., Marro M., Tolosa-Ramon Q., Paz-López S., Valverde M.A., et al. 2020. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science. 370:eaba2644. 10.1126/science.aba2644 [DOI] [PubMed] [Google Scholar]
- Wang, H., Qiu Z., Xu Z., Chen S.J., Luo J., Wang X., and Chen J.. 2018. aPKC is a key polarity determinant in coordinating the function of three distinct cell polarities during collective migration. Development. 145:dev158444. 10.1242/dev.158444 [DOI] [PubMed] [Google Scholar]
- Wang, X., Adam J.C., and Montell D.. 2007. Spatially localized Kuzbanian required for specific activation of Notch during border cell migration. Dev. Biol. 301:532–540. 10.1016/j.ydbio.2006.08.031 [DOI] [PubMed] [Google Scholar]
- Wang, X., He L., Wu Y.I., Hahn K.M., and Montell D.J.. 2010. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol. 12:591–597. 10.1038/ncb2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintner, O., Hirsch-Attas N., Schlossberg M., Brofman F., Friedman R., Kupervaser M., Kitsberg D., and Buxboim A.. 2020. A unified linear viscoelastic model of the cell nucleus defines the mechanical contributions of lamins and chromatin. Adv. Sci. 7:1901222. 10.1002/advs.201901222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., and Friedl P.. 2013. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201:1069–1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, K., Wu Y.I., Liu Y., Geiger J., Tam E., Overall C., Stack M.S., and Friedl P.. 2007. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat. Cell Biol. 9:893–904. 10.1038/ncb1616 [DOI] [PubMed] [Google Scholar]
- Wu, J.-W., Wang C.-W., Chen R.-Y., Hung L.-Y., Tsai Y.-C., Chan Y.-T., Chang Y.-C., and Jang A.C.-C.. 2022. Spatiotemporal gating of Stat nuclear influx by Drosophila Npas4 in collective cell migration. Sci. Adv. 8:eabm2411. 10.1126/sciadv.abm2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff, J.B., Jones J.G., Condeelis J.S., and Segall J.E.. 2000. A critical step in metastasis: In vivo analysis of intravasation at the primary tumor. Cancer Res. 60:2504–2511 [PubMed] [Google Scholar]
- Xie, W., Chojnowski A., Boudier T., Lim J.S.Y., Ahmed S., Ser Z., Stewart C., and Burke B.. 2016. A-Type lamins form distinct filamentous networks with differential nuclear pore complex associations. Curr. Biol. 26:2651–2658. 10.1016/j.cub.2016.07.049 [DOI] [PubMed] [Google Scholar]
- Yamada, K.M., and Sixt M.. 2019. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 20:738–752. 10.1038/s41580-019-0172-9 [DOI] [PubMed] [Google Scholar]
- Yamauchi, K., Yang M., Jiang P., Yamamoto N., Xu M., Amoh Y., Tsuji K., Bouvet M., Tsuchiya H., Tomita K., Moossa A.R., and Hoffman R.M.. 2005. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 65:4246–4252. 10.1158/0008-5472.CAN-05-0069 [DOI] [PubMed] [Google Scholar]
- Zacharioudaki, E., and Bray S.J.. 2014. Tools and methods for studying Notch signaling in Drosophila melanogaster. Methods. 68:173–182. 10.1016/j.ymeth.2014.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows D. melanogaster stocks.
shows fly genotypes and experimental replicates.
shows key antibodies and chemicals.
Data Availability Statement
Data are shown in the figures and supplemental files, and additional data files are available upon request.