Abstract
Pancreatic cancer (PC) remains one of the most challenging diseases, with a very poor 5-year overall survival of around 11.5%. Kirsten rat sarcoma virus (KRAS) mutation is seen in 90%-95% of PC patients and plays an important role in cancer cell proliferation, differentiation, metabolism, and survival, making it an essential mutation for targeted therapy. Despite extensive efforts in studying this oncogene, there has been little success in finding a drug to target this pathway, labelling it for decades as “undruggable”. In this article we summarize some of the efforts made to target the KRAS pathway in PC, discuss the challenges, and shed light on promising clinical trials.
Keywords: Kirsten rat sarcoma virus, Targeted therapy, Pancreatic cancer, Drug resistance, Next generation sequencing, Clustered regularly interspaced short palindromic repeats
Core Tip: Kirsten rat sarcoma virus (KRAS) mutation is the hallmark of pancreatic cancer (PC) and an important therapeutic target. Approaches to target this oncogene has been challenging. We herein discuss the role of KRAS in development of PC, efforts made to target this pathway, and ongoing clinical trials.
INTRODUCTION
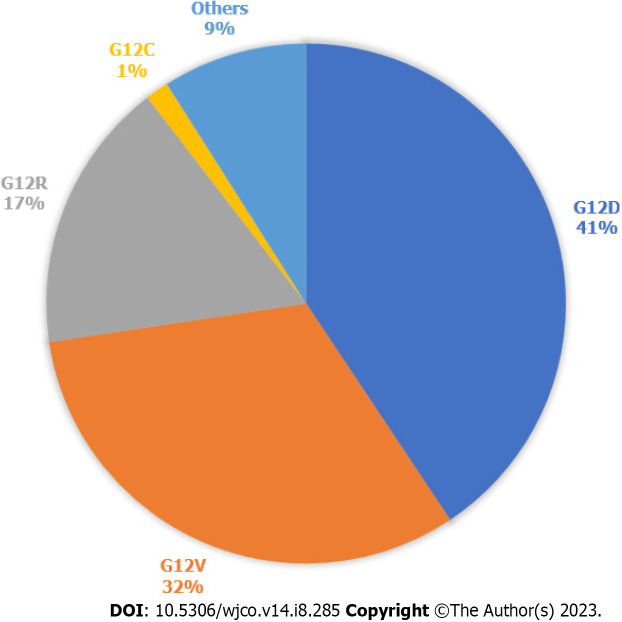
In 2022, there was an estimated 62210 new pancreatic cancer (PC) cases and 49830 estimated deaths. PC is the fourth leading cause of cancer death in the United States[1]. PC is driven primarily by mutations in the Kirsten rat sarcoma virus (KRAS) gene, cyclin-dependent kinase inhibitor 2A, tumor protein 53, and mothers against decapentaplegic protein homolog 4. KRAS is one of the most frequently mutated oncogenes in human cancers. It is seen in more than 90% of PCs and more than 40% of colorectal and lung cancers[2]. 93% of all KRAS mutations occur at codon 12 (G12) with other common mutation sites at G13 and Q61. Missense mutation in glycine residues of G12 result in amino acid substitution, glycine substituted with aspartic acid (G12D), with valine (G12V), or with cysteine (G12C)[3]. The predominant mutation in PC is G12D followed by G12V (Figure 1)[4], but in lung cancer G12C is the most common. KRAS plays a major role in the development of PC and, as a result, there have been significant efforts to target the mutated KRAS pathway.
Figure 1.
Kirsten rat sarcoma virus mutations in pancreatic cancer. Types of Kirsten rat sarcoma virus (KRAS) mutations seen in pancreatic cancer, according to data publicly available on cBioPortal. 812 samples with altered KRAS collected from 5 pancreatic cancer studies. Others are A11T, A146T, A18V, G12A, G12I, G12L, G12S, G13C, G13D, G13H, G13P, G13R, L23V, Q61H, Q61K, Q61R.
BACKGROUND
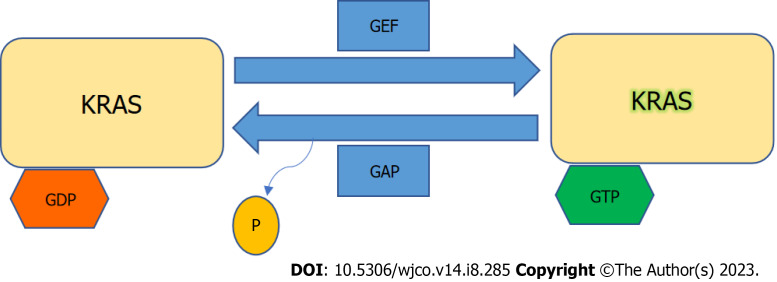
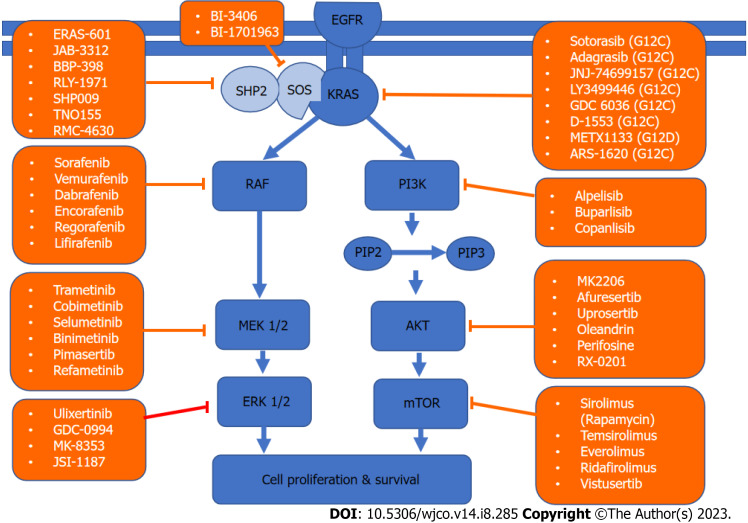
KRAS is a member of the rat sarcoma viral oncogene family (RAS), in addition to Neuroblastoma rat sarcoma virus and Harvey rat sarcoma virus. Identified in 1982, the KRAS is located on the short arm of chromosome 12[5,6]. It encodes two protein isoforms, KRAS-4B and KRAS-4A. Those are found in the inner side of the plasma membrane[7], and act as guanosine triphosphate (GTP)-binding proteins (G proteins), they bind guanine nucleotides that belong to the family of GTP-bound regulatory protein phosphatases (GTPase). An upstream signal e.g., epidermal growth factor receptor (EGFR) stimulates the dissociation of guanosine diphosphate (GDP) from the GDP-bound G protein form, and allows the binding of GTP[8]. RAS functions as a binary switch, determined by two regulatory proteins called guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAP)[9]. KRAS binds to GDP in resting state due to its intrinsic GTPase activity. But with relevant stimuli, GEFs turn on signaling by catalyzing the exchange from a KRAS G-protein-bound GDP to GTP[10] (Figure 2). KRAS proteins can be activated by tyrosine kinase receptors, growth factors, chemokines, or calcium. This in turn activates multiple signaling pathways including the rapidly accelerated fibrosarcoma (RAF)-mitogen-activated protein kinase (MAPK)-extracellular regulated protein kinases (ERK) (MAPK/ERK; MEK) signaling pathway, the phosphoinositide 3-kinase (PI3K)-protein kinase (AKT)-mammalian target of rapamycin (mTOR) signaling pathway, and others. These pathways result in cell proliferation and DNA synthesis (Figure 3).
Figure 2.
Kirsten rat sarcoma virus activation. Kirsten rat sarcoma virus is activated when guanine nucleotide exchange factor displaces guanosine diphosphate from nucleotide binding site allowing guanosine triphosphate (GTP) binding and inactivated upon GTP hydrolysis by intrinsic GTP-bound regulatory protein phosphatases (GTPase) activity enhanced by GTPase activating protein. GTP: Guanosine triphosphate; GAP: GTPase activating protein; GDP: Guanosine diphosphate; GEF: Guanine nucleotide exchange factor; KRAS: Kirsten rat sarcoma virus.
Figure 3.
Kirsten rat sarcoma virus signaling network and targeted therapy. A schematic of the two major Kirsten rat sarcoma virus pathways driving cell survival and drugs that target them. KRAS: Kirsten rat sarcoma virus; AKT: Protein kinase; EGFR: Epidermal growth factor receptor; PIP: Prolactin-induced protein; ERK: Extracellular regulated protein kinases; MEK: Mitogen-activated protein kinase/extracellular regulated protein kinases; mTOR: Mammalian target of rapamycin; PI3K: Phosphoinositide 3-kinase; RAF: Rapidly accelerated fibrosarcoma; SHP2: Src homology-2 domain-containing protein tyrosine phosphatase-2; SOS: Son of sevenless.
Precursor lesions of pancreatic ductal adenocarcinoma (PDAC) include pancreatic intraepithelial neoplasia (PanIN), intraductal papillary mucinous neoplasm (IPMN), and mucinous cystic neoplasm[11,12]. KRAS mutation was detected in 36% of PanIN-1A lesions and 87% of PanIN-2-3 lesions[13]. It was also found in 61% of patients with IPMN[14]. To study the role of KRAS in PC progression, scientists developed transgenic mice with inducible KRASG12D. Induction of oncogenic KRASG12D altered normal epithelium and led to the development of precancerous lesions; on the other hand, inactivation of KRASG12D in precursor lesions and during cancer progression led to disease regression[15]. These studies confirm the early role of KRAS mutation in the initiation and progression of precursor lesions into invasive PDAC as well as the correlation between frequency of KRAS mutation and degree of dysplasia.
KRAS mutation drives PC progression by resistance to apoptosis, induction of autophagy[16], immune evasion by downregulating major histocompatibility complex class I on tumor cells[17], and stimulating angiogenesis, resulting in cell survival and tumor progression.
TARGETED THERAPY
Upstream regulators
Some of the key regulators of KRAS include Son of Sevenless (SOS) and Src homology phosphatase 2 (SHP2). SOS is a GEF that activates KRAS, and SHP2 is a protein tyrosine phosphatase encoded by PTPN11 that also promotes RAS activation, inhibiting either can delay tumor progression[18,19].
BI-3406 inhibits the interaction between KRAS and SOS1 which has been shown to cause tumor regression in KRAS-driven cancer cell models. Synergy was observed with SOS1/MEK inhibitors as this combination can counteract adaptive resistance to MEK inhibitors[20]. ERAS-601 is a small molecule allosteric inhibitor of SHP2 that stops KRAS from cycling into its GTP-active state, which inhibits cellular proliferation in multiple KRASG12C mutated tumor cell models[21]. Recently the Food and Drug Administration (FDA) granted fast track designation to BBP-398 (SHP2 inhibitor) in combination with Sotorasib for KRASG12C-mutated metastatic non-small-cell lung carcinoma (NSCLC). There is an ongoing trial to evaluate the safety and efficacy of this combination [national clinical trial (NCT) 05480865]. Combination of KRASG12C inhibitor (JAB-21822) and SHP2 inhibitor (JAB-3312) showed synergistic effect in KRASG12C-resistant tumor cells[22], currently in phase I/II trial for PDAC (NCT05288205).
MAPK/ERK pathway
The MAPK/ERK pathway was shown in Table 1.
Table 1.
Kirsten rat sarcoma virus-rapidly accelerated fibrosarcoma-mitogen-activated protein kinase/extracellular regulated protein kinases-extracellular regulated protein kinases pathway inhibitors
| Agent | FDA approved1 |
Clinical trials2
|
||
|
Conditions (phase)
|
Combination
|
NCT number
|
||
| SOS inhibitors | ||||
| BI-1701963 | N/A | Advanced solid tumors (I); advanced solid tumors (I); metastatic colorectal cancer (I) | Trametinib; BI 3011441; irinotecan | NCT04111458; NCT04835714; NCT04627142 |
| SHP2 inhibitors | ||||
| ERAS-601 | N/A | Advanced/ metastatic solid tumors (I) | Cetuximab, pembrolizumab | NCT04670679 |
| JAB-3312 | N/A | Advanced solid tumors (I); advanced solid tumors (I/II) | N/A; binimetinib, pembrolizumab, sotorasib, osimertinib | NCT04045496; NCT04720976 |
| BBP-398 (IACS-15509) | (+ sotorasib) fast track designation for metastatic NSCLC | Advanced solid tumor (I); advanced NSCLC (I); advanced solid tumors (I); advanced solid tumors (I) | N/A; nivolumab; N/A; sotorasib | NCT05621525; NCT05375084; NCT04528836; NCT05480865 |
| RLY-1971 | N/A | Advanced/metastatic solid tumors (I) | N/A | NCT04252339 |
| TNO155 | N/A | Advanced solid tumors (I); advanced solid tumors (I) | EGF816 (nazartinib); spartalizumab, ribociclib | NCT03114319; NCT04000529 |
| RMC-4630 | N/A | Relapsed/refractory solid tumors (I); NSCLC (II); metastatic KRAS mutant cancers (I); relapsed/refractory solid tumors, locally advanced/metastatic EGFR positive NSCLC (I/II) | N/A; sotorasib LY3214996; cobimetinib, osimertinib | NCT03634982; NCT05054725; NCT04916236; NCT03989115 |
| Direct KRAS inhibitors | ||||
| G12C | ||||
| Sotorasib (AMG 510, Lumakras) | Advanced NSCLC | Colorectal cancer (III); advanced solid tumors (Ib/II) | Panitumumab; N/A | NCT05198934; NCT04185883 |
| Adagrasib (MRTX849, Krazati) | Locally advanced or metastatic NSCLC | Metastatic PC (Ib); colorectal cancer (I); solid tumors (I/II); advanced solid tumors (I); advanced/metastatic cancers (I/II) | N/A; cetuximab and irinotecan; N/A; BI-1701963; TNO155 | NCT05634525; NCT05722327; NCT05162443; NCT04975256; NCT04330664 |
| JNJ-74699157 | N/A | Advanced solid tumors (I) | N/A | NCT04006301 |
| LY3499446 | N/A | Advanced solid tumors (I/II) | Abemaciclib, cetuximab, erlotinib, docetaxel | NCT04165031 |
| GDC 6036 | N/A | Advanced/metastatic solid tumors (I) | Atezolizumab, cetuximab, bevacizumab, erlotinib, GDC-1971, inavolisib | NCT04449874 |
| D-1553 | N/A | Advanced/metastatic solid tumors (I/II); NSCLC (I/II) | N/A; N/A | NCT04585035; NCT05383898 |
| G12D | ||||
| MRTX1133 | N/A | Pancreatic, lung, and colorectal cancers (I/II) | N/A | Enters phase I in 2023 |
| Tricomplex inhibitors | ||||
| RMC-6236 | N/A | Advanced solid tumors (I) | N/A | NCT05379985 |
| RMC-6291 | N/A | Advanced solid tumors (I) | N/A | NCT05462717 |
| RAF inhibitors | ||||
| Sorafenib (BAY43-9006, NEXAVAR) | Unresectable HCC; advanced RCC; thyroid cancer | PC that cannot be removed by surgery (II); unresectable PC (I); metastatic PC (II) | Erlotinib; gemcitabine, sorafenib and radiotherapy; alone or with gemcitabine | NCT00837876; NCT00375310; NCT00114244 |
| Vemurafenib (PLX4032, RG7204, RO5185426, ZELBORAF) | BRAF V600E melanoma, ECD | PC (II) | Sorafenib | NCT05068752 |
| Dabrafenib (GSK2118436, TAFINLAR) | (+ Trametinib) BRAF V600E or V600K melanoma, NSCLC, anaplastic thyroid cancer, solid tumors | Colorectal cancer (II); advanced/metastatic BRAF V600 colorectal cancer (I) | Trametinib + PDR001; trametinib, LTT462, LXH254, TNO155, spartalizumab, tislelizumab | NCT03668431; NCT04294160 |
| Encorafenib (BRAFTOVI) | BRAF V600E metastatic colorectal cancer | Localized colon or upper rectum cancer with BRAF V600E mutation (II) | Cetuximab | NCT05706779 |
| Regorafenib (BAY 73-4506, STIVARGA) | Metastatic colorectal cancer; advanced GIST | Solid tumors (II) | Nivolumab | NCT04704154 |
| Lifirafeni (BGB-283) | N/A | Advanced or refractory solid tumors (I/II) | Mirdametinib | NCT03905148 |
| Paradox breakers | ||||
| PLX7904/ PLX8394 (PB04) | N/A | Advanced cancers (I/IIa) | N/A | NCT02012231 |
| Pan-RAF inhibitors | ||||
| LY3009120 | N/A | Advanced cancer (I) | N/A | NCT02014116 |
| MLN2480 (BIIB-024, TAK580, Tovorafenib) | N/A | Relapsed or refractory solid tumors followed by a dose expansion in participants with metastatic melanoma (I); advanced non-hematologic malignancies (I) | N/A; MLN0128 or alisertib, or paclitaxel, or cetuximab, or irinotecan | NCT01425008; NCT02327169 |
| HM95573 (Belvarafenib) | N/A | Locally advanced or metastatic solid tumors (I) | Cobimetinib or cetuximab | NCT03284502 |
| BMS-908662 (XL281) | N/A | Advanced or metastatic colorectal cancer (I/II); advanced solid tumors (I) | Alone or with cetuximab; N/A | NCT01086267; NCT00451880 |
| MEK inhibitors | ||||
| Trametinib (GSK1120212, JTP-74057) | (+Dabrafenib) BRAF V600E or V600K melanoma, NSCLC, anaplastic thyroid cancer, solid tumors | Cancers with BRAF V600E mutations (II); solid tumors (I); PC (II); metastatic PC (II); biliary tract cancer (II) | Dabrafenib; gemcitabine; SBRT + pembrolizumab; gemcitabine; N/A | NCT04439292; NCT01428427; NCT02704156; NCT01231581; NCT01943864 |
| Cobimetinib (XL-518, GDC-0973, RG7421, Cotellic) | Histiocytic neoplasms, melanoma | PC (I); locally advanced or metastatic PC (I) | N/A; calaspargase Pegol-mknl | NCT04005690; NCT05034627 |
| Selumetinib (AZD6244, ARRY-142886, Koselugo) | Pediatric neurofibromatosis type 1 | Advanced or metastatic PC who have failed first line gemcitabine (II); locally advanced or metastatic pancreatic cancer with KRAS G12R mutations (II); metastatic pancreatic cancer previously treated with chemotherapy (II); locally advanced or metastatic PC (II) | N/A; N/A; MK2206 (Akt inhibitor) or mFOLFOX; erlotinib hydrochloride | NCT00372944; NCT03040986; NCT01658943; NCT01222689 |
| Binimetinib (ARRY-438162, ARRY-162, MEK162, MektoviI) | Unresectable or metastatic melanoma with a BRAF V600E mutation | Advanced BRAF mutant cancers (I/II); PC with somatic BRAF V600E mutation (II); advanced solid tumors harboring RAS or BRAFV60330E mutations (I) | Encorafenib; Encorafenib; RAF 265 | NCT03843775; NCT04390243; NCT01352273 |
| Pimasertib (AS703026, SAR24550, EMD1036239, MSC1936369B) | N/A | PC (I/II) | Gemcitabine | NCT01016483 |
| Refametinib (RDEA119, BAY86-9766) | N/A | Advanced or metastatic cancer (I); RAS-mutant hepatocellular carcinoma (II); advanced cancer (Ib) | Regorafenib; N/A; copanlisib | NCT02168777; NCT01915589; NCT01392521 |
| E6201 (ER 806201) | N/A | BRAF V600 mutated metastatic melanoma (I); advanced solid tumors (I) | Dabrafenib; N/A | NCT05388877; NCT00794781 |
| PD-0325901 (Mirdametinib) | N/A | Advanced cancer (I) | PF-05212384 or Irinotecan | NCT01347866 |
| AZD8330 (ARRY-424704, ARRY-704) | N/A | Advanced malignancies (I) | N/A | NCT00454090 |
| GDC-0623 (RG7420, G-868) | N/A | Locally advanced or metastatic solid tumors (I) | N/A | NCT01106599 |
| RO4987655 (CH4987655, RG7167) | N/A | Advanced solid tumors (I) | N/A | NCT00817518 |
| RO5126766 (CH5126766, RG7304) | N/A | Advanced solid tumors (I) | N/A | NCT00773526 |
| TAK733 | N/A | Advanced nonhematologic malignancies (I) | N/A | NCT00948467 |
| ERK inhibitors | ||||
| Ulixertinib (BVD-523) | N/A | Advanced pancreatic and other solid tumors (I); metastatic PC (I); advanced MAPK pathway-altered malignancies | Palbociclib; Nab-paclitaxel and gemcitabine; N/A | NCT03454035; NCT02608229; NCT04566393 |
| GDC-0994 (RG7842) | N/A | Locally advanced or metastatic solid tumors (I) | N/A | NCT01875705 |
| MK-8353 (SCH900353) | N/A | Advanced/metastatic solid tumors (I); advanced malignancies (I) | Selumetinib; pembrolizumab | NCT03745989; NCT02972034 |
| JSI-1187 | N/A | Advanced solid tumors with MAPK pathway mutations (I) | Alone or with dabrafenib | NCT04418167 |
| ERAS-007 | N/A | Advanced or metastatic solid tumors (I/II); advanced gastrointestinal malignancies (I/II) | ERAS-601; encorafenib, cetuximab, palbociclib | NCT04866134; NCT05039177 |
| Menin inhibitor | ||||
| BMF-219 | N/A | NSCLC, pancreatic, colorectal cancers (I) | N/A | NCT05631574 |
FDA: Food and Drug Administration; SOS: Son of Sevenless; KRAS: Kirsten rat sarcoma virus; HCC: Hepatocellular carcinoma; RCC: Renal cell carcinoma; ECD: Erdheim-Chester disease; GIST: Gastrointestinal stroma tumors; PC: Pancreatic cancer; RAF: Rapidly accelerated fibrosarcoma; RAS: Rat sarcoma viral oncogene family; MAPK: Mitogen-activated protein kinases; NSCLC: Non-small-cell lung carcinoma; SHP2: Src homology-2 domain-containing protein tyrosine phosphatase-2; NCT: National clinical trial; MEK: Mitogen-activated protein kinase/extracellular regulated protein kinases; N/A: Not applicable.
KRAS
Direct inhibition of the KRAS protein remains a challenge, due to its small size of 21 kDa and the lack of hydrophobic pockets on its surface. Those pockets, if found, can then be blocked by small molecules and ultimately disrupt its interaction with other proteins[23]. Several attempts have been made to directly target KRAS, but results were non-satisfactory[24-26]. Only recently AMG 510 (sotorasib) was developed to target G12C mutation in NSCLC without inhibiting wild-type KRAS[27]. Adagrasib (MRTX849) which is also a KRASG12C inhibitor is well tolerated, and preliminary results showed partial response in 50% of patients with PDAC harboring this mutation[28]. However, KRASG12C only occurs in 1%-2% PC and attempts to target more common KRAS isoforms have failed. One promising compound is MRTX1133, a small molecule that selectively inhibits KRASG12D by preventing SOS-catalyzed nucleotide exchange. Subsequently, it promotes tumor regression in immunocompetent PC models and alters the tumor microenvironment by increasing tumor associated macrophages (TAM) and tumor-infiltrating cytotoxic T-cells. MRTX1133 is expected to enter phase I trial in 2023[29,30]. Other agents inhibiting G12D in the pre-clinical phase include BI-KRASG12D, JAB-22000, and ERAS-4. A new category of drugs called tricomplex inhibitors has shown promising results in pre-clinical models of KRASG12V mutant cancers[31] and in a phase I trial RMC-6236 in KRASG12-mutant advanced solid tumors excluding G12C (NCT05379985). A recent study was able to selectively target KRASG12R using a small molecule electrophile[32]. Due to the challenging nature of direct KRAS inhibition focus was shifted on downstream signaling, knowing that some of the challenges include compensation by other pathways, and that inhibiting multiple pathways can result in toxicity[33].
Multiple mechanisms are implicated in the inevitable drug resistance seen with KRAS inhibitors, either by activation of wild-type KRAS which is mediated by receptor tyrosine kinase[34], synthesizing new KRASG12C proteins in response to MAPK suppression[35], or developing secondary mutations in KRAS inhibitor binding pocket[36].
RAF
With regards to drugs targeting the MAPK pathway, sorafenib was the first RAF inhibitor to be FDA-approved, initially for advanced renal cell carcinoma, followed by unresectable/metastatic hepatocellular carcinoma and metastatic differentiated thyroid cancer[37]. In a phase II trial combining sorafenib and erlotinib, 12 of the first 15 patients required dose delays or reductions due to toxicity, and the study failed to reach its primary endpoint of 8-week progression-free survival (PFS)[38]. A second-generation of RAF inhibitors (e.g., vemurafenib and dabrafenib) was proven to be effective in BRAF V600E mutant metastatic melanoma[39]. Dabrafenib in combination with trametinib received a tumor agnostic accelerated approval for treatment of unresectable/metastatic solid tumors with BRAF V600E mutation that progressed on prior treatment[40]. Unfortunately, vemurafenib and dabrafenib were not as effective in KRAS-mutant cancers, due to compensatory ERK activation that led to enhanced tumor growth[41,42]. A third generation of RAF inhibitors called “paradox breakers” (PLX7904 and PLX8394) also blocks MEK-ERK1/2, which can overcome this resistance mechanism[43], Unfortunately, a phase I/II trial to evaluate the safety of PLX8394 was terminated due to low accrual. Recently, another group called “pan-RAF inhibitors” (LY3009120, MLN2480, and HM95573) entered phase I trials. LY3009120 is a kinase inhibitor that showed efficacy in inhibiting mutated KRAS and BRAF in preclinical models of colorectal cancer with minimal paradoxical MAPK activation[44,45], however, a phase I trial in advanced cancers was terminated early due to lack of sufficient clinical efficacy (NCT02014116). MLN2480 (tovorafenib) showed an acceptable safety profile[46], and HM95573 (belvarafenib) was well tolerated and showed anti-tumor activity in advanced solid tumors with RAS or RAF mutations[47]. The Yes-associated protein (YAP) is a transcription coregulator downstream from KRAS that promotes cell proliferation[48]. Combining LY3009120 and YAP-inhibitor (verteporfin) showed anti-tumor effect in vivo and in vitro by blocking compensatory activation of AKT pathway[49].
MEK
As mentioned above, trametinib is a MEK1/2 inhibitor FDA approved in combination with dabrafenib (RAF-inhibitor) as a tumor agnostic drug[50]. Trametinib was studied in combination with gemcitabine in a placebo controlled clinical trial for untreated metastatic PDAC. Unfortunately, it did not show improvement in overall survival (OS), PFS, or overall response rate (ORR)[51]. This is potentially due to a compensatory mechanism called autophagy, initiated through activation of the AKT pathway[52]. A Phase II trial of selumetinib (MEK1/2 kinase inhibitor) in PC did not show any significant difference in OS when compared to capecitabine[53], another phase II study of selumetinib targeting only PC patients with KRASG12R mutation after at least two lines of prior systemic chemotherapy did not improve ORR, however, three patients had stable disease for ≥ 6 months[54]. A phase I/II trial studied the selective MEK1/2 inhibitor pimasertib in combination with gemcitabine vs gemcitabine alone in patients with metastatic PC. Despite the promising safety and efficacy of this combination, it did not improve PFS or OS[55]. Unfortunately, in whole there was no observed clinical benefit of MEK inhibitors in the multiple trials done in PC.
ERK
After resistance to BRAF and MEK inhibitors, the next downstream target is ERK. SCH772984[56] is a selective inhibitor of ERK1/2 that showed tumor regression in xenograft models refractory to BRAF and MEK inhibitors. Similar effects were seen with ulixertinib[57]. A phase Ib trial combining ERK1/2 inhibitor (GDC-0994) and MEK inhibitor (cobimetinib) in advanced solid tumors was terminated due to tolerability issues[58]. The ERK1/2 inhibitor JSI-1187-01 demonstrated pre-clinical efficacy in tumor models with MAPK pathway mutations, as well as synergy with BRAF inhibitors[59], and is being studied in a phase I trial (NCT04418167).
PI3K-AKT-mTOR-pathway
The PI3K-AKT-mTOR-pathway was shown in Table 2. One of the postulated reasons EGFR inhibitors and other targeted therapies develop resistance is the hyper-activation of PI3K-AKT-mTOR pathway, which can drive cancer progression and survival. PI3K is overexpressed in around 50% of patients with PC[60], and AKT2 is amplified in 10%-20% of PDAC[61]. TAM plays a role in the development of PC[62] by creating an immune-suppressive microenvironment, minimizing the antitumor effect of T-cells[63]. PI3K helps drive this immune suppression, so its inhibition can restore immune response against cancer cells as well as potentiate the effect of chemotherapy[64]. Additionally, AKT mediates an anti-apoptotic effect and plays a role in chemoresistance[65]. Phosphatase and tensin homolog is a tumor suppressor of the AKT/mTOR pathway, its loss has been implicated in PC development, recurrence, and prognosis[66], as well as acceleration of KRASG12D-induced PDAC in mice[67]. An in vivo study tested PI3Kα-specific inhibitor (BYL) in combination with an EGFR inhibitor (erlotinib) and showed reduced tumor volume and apoptosis in PDAC cell lines[68]. Currently a clinical trial combining gedatolisib (PI3K/mTOR inhibitor) with palbociclib (CDK4/6 inhibitor) in advanced squamous cell cancers of the lung, pancreas, and solid tumors is recruiting (NCT03065062). A phase I/II trial studied the safety and efficacy of combining everolimus (mTOR inhibitor), cetuximab (EGFR inhibitor), and capecitabine, however, the combination resulted in significant epidermal and mucosal toxicities with minimal efficacy[69].
Table 2.
Phosphoinositide 3-kinase-protein kinase-mammalian target of rapamycin-pathway inhibitors
|
Agent
|
Combination
|
Phase
|
NCT number1 |
| PI3K inhibitors (p110α) isoform | |||
| Alpelisib (BYL719) | Gemcitabine and abraxane | I | NCT02155088 |
| Buparlisib (BKM120) | mFOLFOX6; trametinib (MEKi) | I; I | NCT01571024; NCT01155453 |
| Pan-PI3K inhibitors | |||
| Copanlisib(BAY 80–6946) | N/A | I | NCT00962611 |
| PI3K and mTOR inhibitors | |||
| Voxtalisib (SAR245409, XL765) | N/A | I | NCT00485719 |
| Dactolisib(NVP-BEZ235) | MEK162 (MEKi) | I | NCT01337765 |
| Gedatolisib (PF-05212384, PKI-587) | Palbociclib (CDKi) | I | NCT03065062 |
| Pan-Akt inhibitors | |||
| MK2206 | Monotherapy; dinaciclib (CDKi); selumitinib (MEKi) vs mFOLFOX6 | I; I; II | NCT00848718; NCT01783171; NCT01658943 |
| Afuresertib (GSK2110183) | Trametinib (MEKi); N/A | I; II | NCT01476137; NCT01531894 |
| Uprosertib (GSK2141795) | Trametinib (MEKi) | I | NCT01138085 |
| Oleandrin (PBI-05204) | N/A | II | NCT02329717 |
| Perifosine | N/A | II; II | NCT00053924; NCT00059982 |
| RX-0201 | Gemcitabine | II | NCT01028495 |
| Rapalogs (mTORC1 inhibitors) | |||
| Sirolimus (rapamycin) | Sunitinib (RTKi); N/A; metformin; vismodegib (SMOi) | I; II; I/II; I | NCT00583063; NCT00499486; NCT02048384; NCT01537107 |
| Temsirolimus (CCI-779, Torisel) | Lenalidomide; gemcitabine; nivolumab (PD-1i) | I; I; I/II | NCT01183663; NCT00593008; NCT02423954 |
| Everolimus (RAD001) | Sorafenib (RTKi); trametinib (MEKi); gemcitabine; cetuximab (EGFRi) and capecitabine; N/A | I; I; I/II; I/II; II | NCT00981162; NCT00955773; NCT00560963; NCT01077986; NCT00409292 |
| Ridafirolimus | Bevacizumab (VEGFRi) | I | NCT00781846 |
| mTORC1/2 inhibitors | |||
| Vistusertib (AZD2014) | N/A; selumitinib (MEKi); olaparib (PARPi) | I; II; II | NCT01026402; NCT02583542; NCT02576444 |
PI3K: Phosphoinositide 3-kinase; NCT: National clinical trial; MEKi: Mitogen-activated protein kinase/ extracellular regulated protein kinases inhibitor; CDKi: Cyclin-dependent kinase inhibitor; RTKi: Receptor tyrosine kinase inhibitor; SMOi: Smoothened inhibitor; PD-1i: Programmed cell death receptor-1 inhibitor; EGFRi: Epidermal growth factor receptor inhibitor; VEGFRi: Vascular endothelial growth factor receptor inhibitor; mTOR: mammalian target of rapamycin; PARPi: Poly (ADP-ribose) polymerase inhibitor; N/A: Not applicable.
Small interfering RNA, MicroRNA, and clustered regularly interspaced short palindromic repeats
Pre-clinical studies show that small interfering RNAs (siRNAs) have potential in cancer treatment. To deliver siRNAs to target cancer cells, scientists devised two unique methods, one utilized nanoparticle[70] to target lung cancer cells and another study used a biodegradable polymeric matrix (LODER) to carry the anti KRASG12D siRNA. This resulted in the decrease of KRAS levels and inhibited cell proliferation[71]. MicroRNAs (miRNA) regulate cell proliferation and contribute to PC development. Depending on their role they can act as tumour suppressor or oncogenic miRNAs[72,73]. MRX34 (miRNA-34 mimic) was used in a phase I clinical trial that utilized lipid-based vesicles (NOV40) as a delivery vector, for treating patients with advanced solid tumors. miRNA-96 directly targets KRAS oncogene decreasing PC cell invasion and slowing tumor growth both in vivo and in vitro[74]. Clustered regularly interspaced short palindromic repeat (CRISPR) is currently being studied in KRAS-mutated cancers. This technology is being harnessed to target inactivated tumor suppressor genes or overactive oncogenes. In a 2018 study CRISPR-Cas13a was developed to target KRASG12D mRNA. Subsequently, it also suppressed downstream ERK and AKT proteins resulting in apoptosis and significant tumor suppression in vivo and in vitro[75]. Two phase I trials utilizing the CRISPR platform are currently ongoing in PC (NCT04426669 and NCT04842812).
CONCLUSION
KRAS mutation remains the hallmark genetic aberration leading to PC. Although several studies have demonstrated positive preclinical results, the resulting clinical trial results have been largely disappointing. As we continue to have a deeper understanding of the KRAS pathway, resistance mechanisms, and the role and function of the immune system; we get closer to developing effective therapies to outsmart the scourge that is PC. Ongoing clinical trials targeting more common KRAS mutations in PC will hopefully lead to more effective therapy and change the outcomes for the thousands of patients affected by this disease every year.
ACKNOWLEDGEMENTS
Dr. Babiker is a Paul Calabresi Scholar at the Mayo Clinic Cancer Center and acknowledges K-12 grant Program, K12CA090628.
Footnotes
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: April 12, 2023
First decision: June 21, 2023
Article in press: July 27, 2023
Specialty type: Oncology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gao W, China; Luchini C, Italy; Pan Y, China S-Editor: Lin C L-Editor: A P-Editor: Xu ZH
Contributor Information
Ahmed Elhariri, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States.
Ahmed Alhaj, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States. Ahmed.Elhariri@bcm.edu.
Daniel Ahn, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Arizona, Mayo Clinic Cancer Center, Phoenix, AZ 85054, United States.
Mohamad Bassam Sonbol, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Arizona, Mayo Clinic Cancer Center, Phoenix, AZ 85054, United States.
Tanios Bekaii-Saab, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Arizona, Mayo Clinic Cancer Center, Phoenix, AZ 85054, United States.
Christina Wu, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Arizona, Mayo Clinic Cancer Center, Phoenix, AZ 85054, United States.
Michael Scott Rutenberg, Department of Radiation-Oncology, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States.
John Stauffer, Department of Surgical Oncology, Hepatopancreatobiliary Surgery, Mayo Clinic Florida, Jacksonville, FL 32224, United States.
Jason Starr, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States.
Umair Majeed, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States.
Jeremy Jones, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States.
Mitesh Borad, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Arizona, Mayo Clinic Cancer Center, Phoenix, AZ 85054, United States.
Hani Babiker, Division of Hematology-Oncology, Department of Medicine, Mayo Clinic Florida, Mayo Clinic Cancer Center, Jacksonville, FL 32224, United States.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther. 2021;6:386. doi: 10.1038/s41392-021-00780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veluswamy R, Mack PC, Houldsworth J, Elkhouly E, Hirsch FR. KRAS G12C-Mutant Non-Small Cell Lung Cancer: Biology, Developmental Therapeutics, and Molecular Testing. J Mol Diagn. 2021;23:507–520. doi: 10.1016/j.jmoldx.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride OW, Swan DC, Tronick SR, Gol R, Klimanis D, Moore DE, Aaronson SA. Regional chromosomal localization of N-ras, K-ras-1, K-ras-2 and myb oncogenes in human cells. Nucleic Acids Res. 1983;11:8221–8236. doi: 10.1093/nar/11.23.8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen X, Niu N, Xue J. Oncogenic KRAS triggers metabolic reprogramming in pancreatic ductal adenocarcinoma. J Transl Int Med. 2022:AOP. doi: 10.2478/jtim-2022-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos E, Nebreda AR. Structural and functional properties of ras proteins. FASEB J. 1989;3:2151–2163. doi: 10.1096/fasebj.3.10.2666231. [DOI] [PubMed] [Google Scholar]
- 8.Takai Y, Kaibuchi K, Kikuchi A, Kawata M. Small GTP-binding proteins. Int Rev Cytol. 1992;133:187–230. doi: 10.1016/s0074-7696(08)61861-6. [DOI] [PubMed] [Google Scholar]
- 9.Drugan JK, Rogers-Graham K, Gilmer T, Campbell S, Clark GJ. The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J Biol Chem. 2000;275:35021–35027. doi: 10.1074/jbc.M004386200. [DOI] [PubMed] [Google Scholar]
- 10.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Singh M, Maitra A. Precursor lesions of pancreatic cancer: molecular pathology and clinical implications. Pancreatology. 2007;7:9–19. doi: 10.1159/000101873. [DOI] [PubMed] [Google Scholar]
- 12.Bian Y, Jiang H, Zheng J, Shao C, Lu J. Basic Pancreatic Lesions: Radiologic-pathologic Correlation. J Transl Int Med. 2022;10:18–27. doi: 10.2478/jtim-2022-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löhr M, Klöppel G, Maisonneuve P, Lowenfels AB, Lüttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JH, Kim Y, Choi JW, Kim YS. KRAS, GNAS, and RNF43 mutations in intraductal papillary mucinous neoplasm of the pancreas: a meta-analysis. Springerplus. 2016;5:1172. doi: 10.1186/s40064-016-2847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins MA, Bednar F, Zhang Y, Brisset JC, Galbán S, Galbán CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira A, Pereira F, Reis C, Oliveira MJ, Sousa MJ, Preto A. Crucial Role of Oncogenic KRAS Mutations in Apoptosis and Autophagy Regulation: Therapeutic Implications. Cells. 2022;11 doi: 10.3390/cells11142183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dias Carvalho P, Guimarães CF, Cardoso AP, Mendonça S, Costa ÂM, Oliveira MJ, Velho S. KRAS Oncogenic Signaling Extends beyond Cancer Cells to Orchestrate the Microenvironment. Cancer Res. 2018;78:7–14. doi: 10.1158/0008-5472.CAN-17-2084. [DOI] [PubMed] [Google Scholar]
- 18.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, Görgülü K, Dantes Z, Wörmann SM, Diakopoulos KN, Karpathaki AF, Kowalska M, Kaya-Aksoy E, Song L, van der Laan EAZ, López-Alberca MP, Nazaré M, Reichert M, Saur D, Erkan MM, Hopt UT, Sainz B Jr, Birchmeier W, Schmid RM, Lesina M, Algül H. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 19.Kessler D, Gerlach D, Kraut N, McConnell DB. Targeting Son of Sevenless 1: The pacemaker of KRAS. Curr Opin Chem Biol. 2021;62:109–118. doi: 10.1016/j.cbpa.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann MH, Gmachl M, Ramharter J, Savarese F, Gerlach D, Marszalek JR, Sanderson MP, Kessler D, Trapani F, Arnhof H, Rumpel K, Botesteanu DA, Ettmayer P, Gerstberger T, Kofink C, Wunberg T, Zoephel A, Fu SC, Teh JL, Böttcher J, Pototschnig N, Schachinger F, Schipany K, Lieb S, Vellano CP, O'Connell JC, Mendes RL, Moll J, Petronczki M, Heffernan TP, Pearson M, McConnell DB, Kraut N. BI-3406, a Potent and Selective SOS1-KRAS Interaction Inhibitor, Is Effective in KRAS-Driven Cancers through Combined MEK Inhibition. Cancer Discov. 2021;11:142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin L, Patel R, Zhang J, Nevarez R, Congdon T, Brail L, Shoemaker B. Abstract 2670: ERAS-601, a potent allosteric inhibitor of SHP2, synergistically enhances the efficacy of sotorasib/adagrasib and cetuximab in NSCLC, CRC, and HNSCC tumor models. Cancer Res. 2022;82 Suppl 12:2670. [Google Scholar]
- 22.Wang P, Zheng Q, Kang D, Sun X, Zhu S, Wang Y, Long W, Lin Y. 30P Investigation of KRAS G12C inhibitor JAB-21822 as a single agent and in combination with SHP2 inhibitor JAB-3312 in preclinical cancer models. Ann Oncol. 2022;33 Suppl 9:S1441. [Google Scholar]
- 23.Wang W, Fang G, Rudolph J. Ras inhibition via direct Ras binding--is there a path forward? Bioorg Med Chem Lett. 2012;22:5766–5776. doi: 10.1016/j.bmcl.2012.07.082. [DOI] [PubMed] [Google Scholar]
- 24.Taveras AG, Remiszewski SW, Doll RJ, Cesarz D, Huang EC, Kirschmeier P, Pramanik BN, Snow ME, Wang YS, del Rosario JD, Vibulbhan B, Bauer BB, Brown JE, Carr D, Catino J, Evans CA, Girijavallabhan V, Heimark L, James L, Liberles S, Nash C, Perkins L, Senior MM, Tsarbopoulos A, Webber SE. Ras oncoprotein inhibitors: the discovery of potent, ras nucleotide exchange inhibitors and the structural determination of a drug-protein complex. Bioorg Med Chem. 1997;5:125–133. doi: 10.1016/s0968-0896(96)00202-7. [DOI] [PubMed] [Google Scholar]
- 25.Peri F, Airoldi C, Colombo S, Martegani E, van Neuren AS, Stein M, Marinzi C, Nicotra F. Design, synthesis and biological evaluation of sugar-derived Ras inhibitors. Chembiochem. 2005;6:1839–1848. doi: 10.1002/cbic.200400420. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann C, Block C, Geisen C, Haas K, Weber C, Winde G, Möröy T, Müller O. Sulindac sulfide inhibits Ras signaling. Oncogene. 1998;17:1769–1776. doi: 10.1038/sj.onc.1202085. [DOI] [PubMed] [Google Scholar]
- 27.Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li S, Long YO, Thach C, Liu Y, Zarieh A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P. Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor. Cell. 2018;172:578–589.e17. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Ou SI, Jänne PA, Leal TA, Rybkin II, Sabari JK, Barve MA, Bazhenova L, Johnson ML, Velastegui KL, Cilliers C, Christensen JG, Yan X, Chao RC, Papadopoulos KP. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients With Advanced KRAS(G12C) Solid Tumors (KRYSTAL-1) J Clin Oncol. 2022;40:2530–2538. doi: 10.1200/JCO.21.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Allen S, Blake JF, Bowcut V, Briere DM, Calinisan A, Dahlke JR, Fell JB, Fischer JP, Gunn RJ, Hallin J, Laguer J, Lawson JD, Medwid J, Newhouse B, Nguyen P, O'Leary JM, Olson P, Pajk S, Rahbaek L, Rodriguez M, Smith CR, Tang TP, Thomas NC, Vanderpool D, Vigers GP, Christensen JG, Marx MA. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J Med Chem. 2022;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 30.Kemp SB, Cheng N, Markosyan N, Sor R, Kim IK, Hallin J, Shoush J, Quinones L, Brown NV, Bassett JB, Joshi N, Yuan S, Smith M, Vostrejs WP, Perez-Vale KZ, Kahn B, Mo F, Donahue TR, Radu CG, Clendenin C, Christensen JG, Vonderheide RH, Stanger BZ. Efficacy of a Small-Molecule Inhibitor of KrasG12D in Immunocompetent Models of Pancreatic Cancer. Cancer Discov. 2023;13:298–311. doi: 10.1158/2159-8290.CD-22-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koltun E, Cregg J, Rice MA, Whalen DM, Freilich R, Jiang J, Hansen R, Bermingham A, Knox, Dinglasan J, Seamon K, Blaj C, Chang SS, Liu Y, Huang J, Chou KJ, McDowell L, Lee BJ, Wildes D, Wang Z, Singh M, Gill AL, Smith JA. Abstract 1260: First-in-class, orally bioavailable KRASG12V(ON) tri-complex inhibitors, as single agents and in combinations, drive profound anti-tumor activity in preclinical models of KRASG12V mutant cancers. Cancer Res. 2021;81 Suppl 13:1260. [Google Scholar]
- 32.Zhang Z, Morstein J, Ecker AK, Guiley KZ, Shokat KM. Chemoselective Covalent Modification of K-Ras(G12R) with a Small Molecule Electrophile. J Am Chem Soc. 2022;144:15916–15921. doi: 10.1021/jacs.2c05377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jørgensen F, Hesse B, Grønbaek P, Fogh J, Haunsø S. Abnormal oesophageal function in patients with non-toxic goiter or enlarged left atrium, demonstrated by radionuclide transit measurements. Scand J Gastroenterol. 1989;24:1186–1192. doi: 10.3109/00365528909090785. [DOI] [PubMed] [Google Scholar]
- 34.Ryan MB, Fece de la Cruz F, Phat S, Myers DT, Wong E, Shahzade HA, Hong CB, Corcoran RB. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res. 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue JY, Zhao Y, Aronowitz J, Mai TT, Vides A, Qeriqi B, Kim D, Li C, de Stanchina E, Mazutis L, Risso D, Lito P. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, Johnson ML, Heist RS, Patil T, Riely GJ, Jacobson JO, Yang X, Persky NS, Root DE, Lowder KE, Feng H, Zhang SS, Haigis KM, Hung YP, Sholl LM, Wolpin BM, Wiese J, Christiansen J, Lee J, Schrock AB, Lim LP, Garg K, Li M, Engstrom LD, Waters L, Lawson JD, Olson P, Lito P, Ou SI, Christensen JG, Jänne PA, Aguirre AJ. Acquired Resistance to KRAS(G12C) Inhibition in Cancer. N Engl J Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, Schwartz B, Simantov R, Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 38.Cardin DB, Goff L, Li CI, Shyr Y, Winkler C, DeVore R, Schlabach L, Holloway M, McClanahan P, Meyer K, Grigorieva J, Berlin J, Chan E. Phase II trial of sorafenib and erlotinib in advanced pancreatic cancer. Cancer Med. 2014;3:572–579. doi: 10.1002/cam4.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salama AKS, Li S, Macrae ER, Park JI, Mitchell EP, Zwiebel JA, Chen HX, Gray RJ, McShane LM, Rubinstein LV, Patton D, Williams PM, Hamilton SR, Armstrong DK, Conley BA, Arteaga CL, Harris LN, O'Dwyer PJ, Chen AP, Flaherty KT. Dabrafenib and Trametinib in Patients With Tumors With BRAF(V600E) Mutations: Results of the NCI-MATCH Trial Subprotocol H. J Clin Oncol. 2020;38:3895–3904. doi: 10.1200/JCO.20.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callahan MK, Rampal R, Harding JJ, Klimek VM, Chung YR, Merghoub T, Wolchok JD, Solit DB, Rosen N, Abdel-Wahab O, Levine RL, Chapman PB. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367:2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, Roden C, Chalk CJ, Ardlie K, Palescandolo E, Piris A, MacConaill LE, Robert C, Hofbauer GF, McArthur GA, Schadendorf D, Garraway LA. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J Clin Oncol. 2012;30:316–321. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basile KJ, Le K, Hartsough EJ, Aplin AE. Inhibition of mutant BRAF splice variant signaling by next-generation, selective RAF inhibitors. Pigment Cell Melanoma Res. 2014;27:479–484. doi: 10.1111/pcmr.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peng SB, Henry JR, Kaufman MD, Lu WP, Smith BD, Vogeti S, Rutkoski TJ, Wise S, Chun L, Zhang Y, Van Horn RD, Yin T, Zhang X, Yadav V, Chen SH, Gong X, Ma X, Webster Y, Buchanan S, Mochalkin I, Huber L, Kays L, Donoho GP, Walgren J, McCann D, Patel P, Conti I, Plowman GD, Starling JJ, Flynn DL. Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers. Cancer Cell. 2015;28:384–398. doi: 10.1016/j.ccell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Vakana E, Pratt S, Blosser W, Dowless M, Simpson N, Yuan XJ, Jaken S, Manro J, Stephens J, Zhang Y, Huber L, Peng SB, Stancato LF. LY3009120, a panRAF inhibitor, has significant anti-tumor activity in BRAF and KRAS mutant preclinical models of colorectal cancer. Oncotarget. 2017;8:9251–9266. doi: 10.18632/oncotarget.14002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasco DW, Olszanski AJ, Patnaik A, Espino G, Neuwirth R, Faucette S, Bargfrede M, Gangolli EA, Walker RM, Kneissl M, Bozon V. MLN2480, an investigational oral pan-RAF kinase inhibitor, in patients (pts) with relapsed or refractory solid tumors: Phase I study. J Clin Oncol. 2013;31 Suppl 15:2547. [Google Scholar]
- 47.Kim TW, Lee J, Shin SJ, Kim JS, Kim YJ, Han HS, Lee SJ, Lim HS, Hong YH, Noh YS, Kyoung Y, Han O, Yoon J, Lim JA, Kim SR. Belvarafenib, a novel pan-RAF inhibitor, in solid tumor patients harboring BRAF, KRAS, or NRAS mutations: Phase I study. J Clin Oncol. 2019;37 Suppl 15:3000. [Google Scholar]
- 48.Zhang W, Nandakumar N, Shi Y, Manzano M, Smith A, Graham G, Gupta S, Vietsch EE, Laughlin SZ, Wadhwa M, Chetram M, Joshi M, Wang F, Kallakury B, Toretsky J, Wellstein A, Yi C. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7:ra42. doi: 10.1126/scisignal.2005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao X, Wang X, Fang L, Lan C, Zheng X, Wang Y, Zhang Y, Han X, Liu S, Cheng K, Zhao Y, Shi J, Guo J, Hao J, Ren H, Nie G. A combinatorial strategy using YAP and pan-RAF inhibitors for treating KRAS-mutant pancreatic cancer. Cancer Lett. 2017;402:61–70. doi: 10.1016/j.canlet.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Vranic S, Basu GD, Hall DW, Gatalica Z. Tumor-Type Agnostic, Targeted Therapies: BRAF Inhibitors Join the Group. Acta Med Acad. 2022;51:217–231. doi: 10.5644/ama2006-124.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Infante JR, Somer BG, Park JO, Li CP, Scheulen ME, Kasubhai SM, Oh DY, Liu Y, Redhu S, Steplewski K, Le N. A randomised, double-blind, placebo-controlled trial of trametinib, an oral MEK inhibitor, in combination with gemcitabine for patients with untreated metastatic adenocarcinoma of the pancreas. Eur J Cancer. 2014;50:2072–2081. doi: 10.1016/j.ejca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 52.Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, Burrell LD, Lum DH, Whisenant JR, Gilcrease GW 3rd, Cavalieri CC, Rehbein KM, Cutler SL, Affolter KE, Welm AL, Welm BE, Scaife CL, Snyder EL, McMahon M. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bodoky G, Timcheva C, Spigel DR, La Stella PJ, Ciuleanu TE, Pover G, Tebbutt NC. A phase II open-label randomized study to assess the efficacy and safety of selumetinib (AZD6244 [ARRY-142886]) versus capecitabine in patients with advanced or metastatic pancreatic cancer who have failed first-line gemcitabine therapy. Invest New Drugs. 2012;30:1216–1223. doi: 10.1007/s10637-011-9687-4. [DOI] [PubMed] [Google Scholar]
- 54.Kenney C, Kunst T, Webb S, Christina D Jr, Arrowood C, Steinberg SM, Mettu NB, Kim EJ, Rudloff U. Phase II study of selumetinib, an orally active inhibitor of MEK1 and MEK2 kinases, in KRAS(G12R)-mutant pancreatic ductal adenocarcinoma. Invest New Drugs. 2021;39:821–828. doi: 10.1007/s10637-020-01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Cutsem E, Hidalgo M, Canon JL, Macarulla T, Bazin I, Poddubskaya E, Manojlovic N, Radenkovic D, Verslype C, Raymond E, Cubillo A, Schueler A, Zhao C, Hammel P. Phase I/II trial of pimasertib plus gemcitabine in patients with metastatic pancreatic cancer. Int J Cancer. 2018;143:2053–2064. doi: 10.1002/ijc.31603. [DOI] [PubMed] [Google Scholar]
- 56.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, Long B, Liu J, Dinunzio E, Windsor W, Zhang R, Zhao S, Angagaw MH, Pinheiro EM, Desai J, Xiao L, Shipps G, Hruza A, Wang J, Kelly J, Paliwal S, Gao X, Babu BS, Zhu L, Daublain P, Zhang L, Lutterbach BA, Pelletier MR, Philippar U, Siliphaivanh P, Witter D, Kirschmeier P, Bishop WR, Hicklin D, Gilliland DG, Jayaraman L, Zawel L, Fawell S, Samatar AA. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 57.Germann UA, Furey BF, Markland W, Hoover RR, Aronov AM, Roix JJ, Hale M, Boucher DM, Sorrell DA, Martinez-Botella G, Fitzgibbon M, Shapiro P, Wick MJ, Samadani R, Meshaw K, Groover A, DeCrescenzo G, Namchuk M, Emery CM, Saha S, Welsch DJ. Targeting the MAPK Signaling Pathway in Cancer: Promising Preclinical Activity with the Novel Selective ERK1/2 Inhibitor BVD-523 (Ulixertinib) Mol Cancer Ther. 2017;16:2351–2363. doi: 10.1158/1535-7163.MCT-17-0456. [DOI] [PubMed] [Google Scholar]
- 58.Weekes C, Lockhart A, LoRusso P, Murray E, Park E, Tagen M, Singh J, Sarkar I, Mueller L, Dokainish H, Shapiro G, Burris H. A Phase Ib Study to Evaluate the MEK Inhibitor Cobimetinib in Combination with the ERK1/2 Inhibitor GDC-0994 in Patients with Advanced Solid Tumors. Oncologist. 2020;25:833–e1438. doi: 10.1634/theoncologist.2020-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li A, Jian S, Yuan X, Song F, Yang S, Du C, Tao Y, Wang L, Pan M, Dong P, Zhou J, Ge Z, Zhu Q, Hao W, Xu W, Zhang J, Li Q, Wang S. Abstract 4188: The ERK1/2 inhibitor, JSI-1187, demonstrates preclinical efficacy in tumor models with MAPK pathway mutations. Cancer Research. 2020;80 Suppl 16:4188. [Google Scholar]
- 60.Baer R, Cintas C, Therville N, Guillermet-Guibert J. Implication of PI3K/Akt pathway in pancreatic cancer: When PI3K isoforms matter? Adv Biol Regul. 2015;59:19–35. doi: 10.1016/j.jbior.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Cheng JQ, Ruggeri B, Klein WM, Sonoda G, Altomare DA, Watson DK, Testa JR. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci U S A. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liou GY, Storz P. Inflammatory macrophages in pancreatic acinar cell metaplasia and initiation of pancreatic cancer. Oncoscience. 2015;2:247–251. doi: 10.18632/oncoscience.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaneda MM, Cappello P, Nguyen AV, Ralainirina N, Hardamon CR, Foubert P, Schmid MC, Sun P, Mose E, Bouvet M, Lowy AM, Valasek MA, Sasik R, Novelli F, Hirsch E, Varner JA. Macrophage PI3Kγ Drives Pancreatic Ductal Adenocarcinoma Progression. Cancer Discov. 2016;6:870–885. doi: 10.1158/2159-8290.CD-15-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schlieman MG, Fahy BN, Ramsamooj R, Beckett L, Bold RJ. Incidence, mechanism and prognostic value of activated AKT in pancreas cancer. Br J Cancer. 2003;89:2110–2115. doi: 10.1038/sj.bjc.6601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foo WC, Rashid A, Wang H, Katz MH, Lee JE, Pisters PW, Wolff RA, Abbruzzese JL, Fleming JB. Loss of phosphatase and tensin homolog expression is associated with recurrence and poor prognosis in patients with pancreatic ductal adenocarcinoma. Hum Pathol. 2013;44:1024–1030. doi: 10.1016/j.humpath.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hill R, Calvopina JH, Kim C, Wang Y, Dawson DW, Donahue TR, Dry S, Wu H. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Res. 2010;70:7114–7124. doi: 10.1158/0008-5472.CAN-10-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong MH, Xue A, Julovi SM, Pavlakis N, Samra JS, Hugh TJ, Gill AJ, Peters L, Baxter RC, Smith RC. Cotargeting of epidermal growth factor receptor and PI3K overcomes PI3K-Akt oncogenic dependence in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2014;20:4047–4058. doi: 10.1158/1078-0432.CCR-13-3377. [DOI] [PubMed] [Google Scholar]
- 69.Kordes S, Richel DJ, Klümpen HJ, Weterman MJ, Stevens AJ, Wilmink JW. A phase I/II, non-randomized, feasibility/safety and efficacy study of the combination of everolimus, cetuximab and capecitabine in patients with advanced pancreatic cancer. Invest New Drugs. 2013;31:85–91. doi: 10.1007/s10637-012-9802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, Cai W, Chirino LM, Yang GR, Bronson R, Crowley DG, Sahay G, Schroeder A, Langer R, Anderson DG, Jacks T. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–E3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zorde Khvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E, Giladi H, Rivkin L, Simerzin A, Eliakim R, Khalaileh A, Hubert A, Lahav M, Kopelman Y, Goldin E, Dancour A, Hants Y, Arbel-Alon S, Abramovitch R, Galun E. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. 2018;10:59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W, Liu J, Yu J, Chen J. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 75.Zhao X, Liu L, Lang J, Cheng K, Wang Y, Li X, Shi J, Nie G. A CRISPR-Cas13a system for efficient and specific therapeutic targeting of mutant KRAS for pancreatic cancer treatment. Cancer Lett. 2018;431:171–181. doi: 10.1016/j.canlet.2018.05.042. [DOI] [PubMed] [Google Scholar]