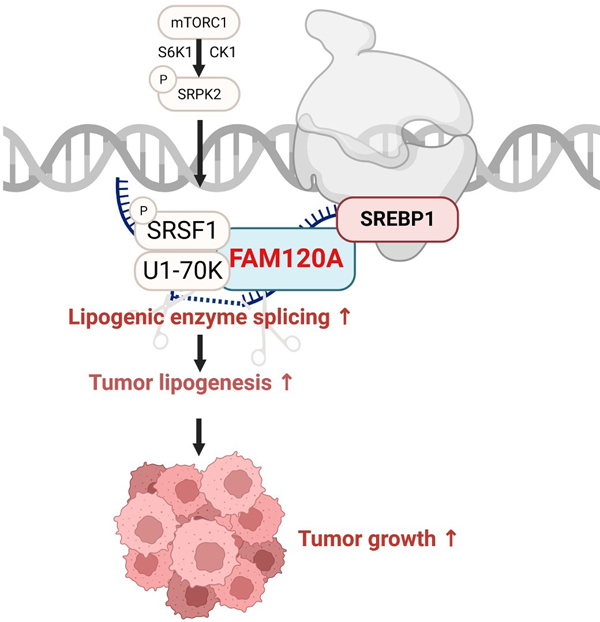
Summary
The mechanistic target of rapamycin complex 1 (mTORC1) is a master regulator of cell growth that stimulates macromolecule synthesis through transcription, RNA processing, and post-translational modification of metabolic enzymes. However, the mechanisms of how mTORC1 orchestrates multiple steps of gene expression programs remain unclear. Here, we identify FAM120A as a transcription co-activator that couples transcription and splicing of de novo lipid synthesis enzymes downstream of mTORC1-SRPK2 signaling. The mTORC1-activated SRPK2 phosphorylates splicing factor SRSF1, enhancing its binding to FAM120A. FAM120A directly interacts with a lipogenic transcription factor SREBP1 at active promoters, thereby bridging the newly transcribed lipogenic genes from RNA polymerase II to the SRSF1 and U1-70K-containing RNA splicing machinery. This mTORC1-regulated, multi-protein complex promotes efficient splicing and stability of lipogenic transcripts, resulting in fatty acid synthesis and cancer cell proliferation. These results elucidate FAM120A as a critical transcription co-factor that connects mTORC1-dependent gene regulation programs for anabolic cell growth.
Graphical Abstract
eTOC blurb.
Cho et al. report FAM120A as a transcription co-activator that connects transcription and splicing of de novo lipid synthesis enzymes downstream of mTORC1. Upon mTORC1 activation, SRPK2 phosphorylates splicing factor SRSF1, inducing its binding with FAM120A. Through its interaction with SRSF1, the SREBP1-associated FAM120A promotes efficient splicing of lipogenic genes and facilitates lipid metabolism of cancer cells. These findings reveal FAM120A as a critical downstream of mTORC1 in anabolic gene expression program and cell growth.
INTRODUCTION
Cancer cells rewire metabolic pathways to support increased demands for continuous proliferation1,2. mTORC1 signaling, which is activated in almost all human cancers, is one of the key mechanisms responsible for such metabolic rewiring. In normal cells, mTORC1 works as a molecular rheostat to promote macromolecule synthesis upon environmental cues such as growth factors and nutrients. It does so by activating metabolic enzymes directly or through the regulation of metabolic gene expression3,4. For instance, upon insulin stimulation, mTORC1 induces de novo lipid synthesis by upregulating the expression and activity of sterol regulatory element-binding proteins (SREBP), a master transcription factor of lipogenic enzymes5–8. In cancer cells, however, overactive mTORC1 signaling constitutively promotes SREBP-dependent lipogenic enzyme expression for tumor cell growth and proliferation 9,10.
In addition to regulating transcriptional programs, mTORC1 also activates metabolic enzymes through post-transcriptional RNA processing11–14. Previously, we found that mTORC1-regulated S6 protein kinase 1 (S6K1) phosphorylates serine/arginine-rich protein kinase 2 (SRPK2) to stimulate its nuclear translocation and phosphorylation of downstream serine/arginine-rich splicing factor 1 (SRSF1)12. Such phosphorylation facilitates SRSF1’s interaction with U1 small nuclear ribonucleoprotein 70 kDa (U1-70K), a core spliceosome protein that initiates assembly of the spliceosome complex on target pre-mRNAs 15–17. Serine/arginine-rich (SR) proteins are additional components of the spliceosome complex that potentiate co-transcriptional splicing18–21. For example, SRSF2 directly interacts with RNA polymerase II (RNA pol II) and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) for efficient splicing of mitochondrial biogenesis genes22. Interestingly, the mTORC1-SRPK2-dependent activation of SRSF1 induces lipogenic gene splicing, but the underlying mechanism for this specificity is unclear.
In this study, we identified family with sequence similarity 120A (FAM120A) as a transcription co-factor and RNA binding protein (RBP) that determines the specificity of genes spliced downstream of mTORC1-SRPK2 signaling. Upon mTORC1 activation, the SRPK2-phosphorylated SRSF1 binds with FAM120A and U1-70K, forming a multiprotein complex with SREBP1, a lipogenic gene transcription factor, and RNA Pol ll to mediate selective splicing of lipogenic genes. Our findings provide mechanistic insights into how oncogenic mTORC1 signaling coordinates the interactions among transcription factors, RBPs, and target gene transcripts to promote cancer cell growth by connecting transcription and pre-mRNA splicing to the translation of metabolic enzymes.
RESULTS
mTORC1-SRPK2 signaling promotes SRSF1 interaction with FAM120A
To investigate how mTORC1-SRPK2 signaling regulates lipogenic mRNA splicing via SRSF1, we previously performed a proteomics-based SRSF1 interactome analysis with or without torin1 (mTOR catalytic inhibitor). Most SRSF1-bound proteins were well-known RNA processing proteins including a spliceosome protein, U1-70K, which we characterized as a key mTORC1-dependent SRSF1 interactor in our previous study12. Additionally, we found FAM120A, whose peptide abundance in the SRSF1-bound fraction was decreased by torin1 (Figure 1A). FAM120A is an RNA binding protein that locates in the nucleus and associates with RNA pol II and spliceosome proteins23–26. Co-immunoprecipitation (co-IP) analysis confirmed FAM120A binding with SRSF1, which was suppressed by torin1 (Figure 1B). Rapamycin, an mTORC1-specific inhibitor, also blunted their association (Figure 1C).
Figure 1. mTORC1-SRPK promotes interaction of SRSF1 with FAM120A.
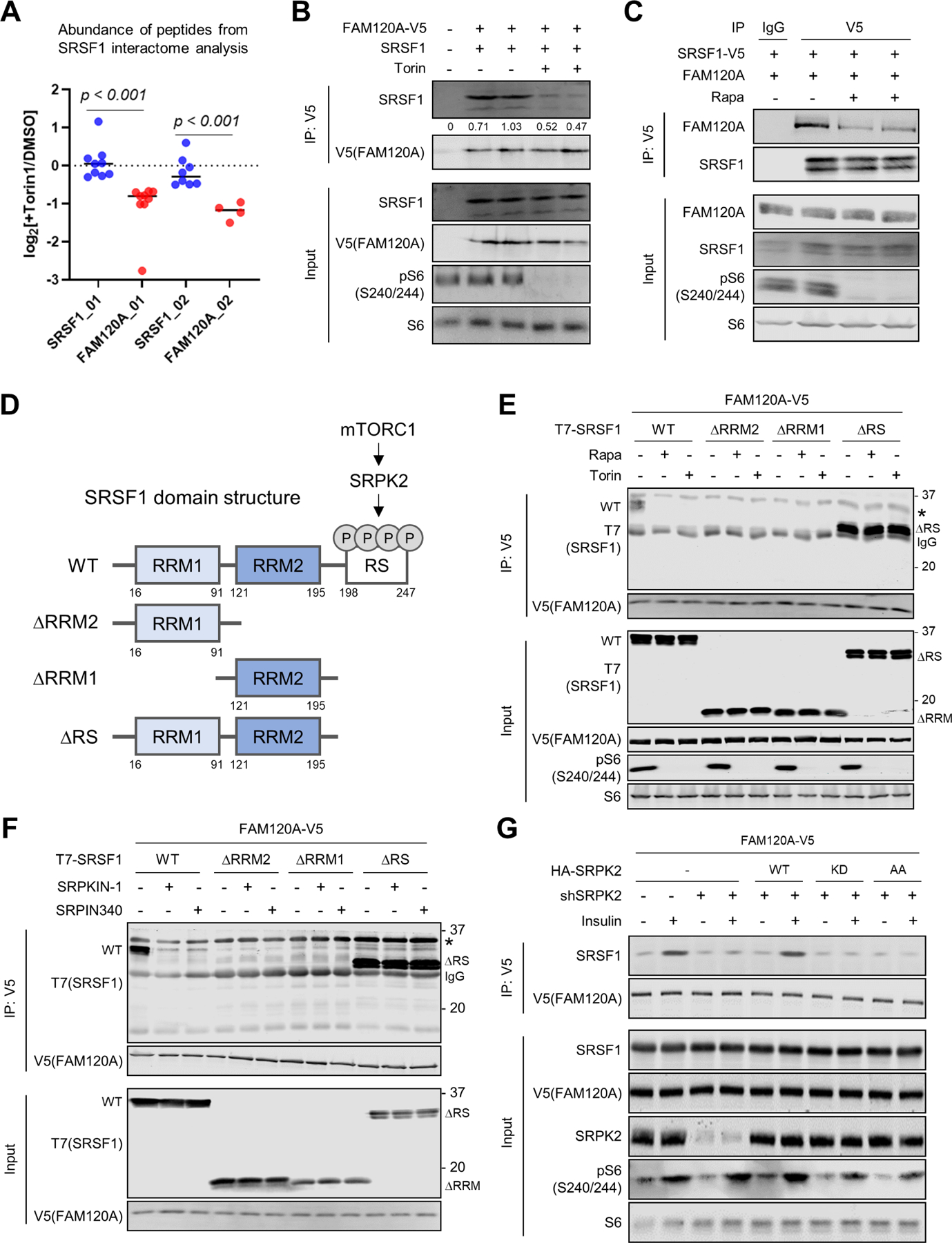
(A) Mass spectrometry analysis of peptides identified in the SRSF1 interactome. HEK293E cells expressing SRSF1-V5 were treated with DMSO or torin1 (250 nM) for 4 hr, and SRSF1-binding proteins were co-immunoprecipitated (Co-IP) by anti-V5 antibody. Each dot represents abundance of individual peptides (N = 4~9 for each protein) in log2[Torin1/DMSO]. Data are reanalyzed from a previous publication12.
(B, C) Co-IP analysis of HEK293E cells expressing FAM120A-V5 and SRSF1 (B) or FAM120A and SRSF1-V5 (C). Cells were treated with torin1 (250 nM) or rapamycin (100 nM) for 4 hr. 1% of total cell lysate was loaded as an input.
(D) Schematic diagram describing mTORC1-SRPK2-dependent phosphorylation of SRSF1 and the domains of wild-type (WT) and mutant SRSF1 constructs. RRM: RNA recognition motif. RS: Arginine/serine-rich domain.
(E, F) Co-IP analysis of HEK293E cells expressing FAM120A-V5 and T7-SRSF1 (WT or mutants). Cells were treated with torin1 (250 nM) (E) or SRPK inhibitors (SRPKIN-1, 5 µM; SRPIN340, 30 µM) (F) for 4 hr. 1% of total cell lysate was loaded as an input. IgG light chain (IgG, 25 kDa) and non-specific bands (asterisks) are indicated.
(G) Co-IP analysis of HEK293E cells expressing FAM120A-V5 and HA-SRPK2 (WT (wild type), KD (kinase dead, SRPK2-K110M), or AA (non-phosphorylatable, SRPK2-S394A/S397A)). Endogenous SRPK2 was knocked down by shRNA targeting 3’UTR of SRPK2. Cells were serum starved overnight followed by insulin (100 nM) stimulation for 4 hr. 1% of total cell lysate was loaded as an input.
See also Supplemental Figure S1.
SRSF1 contains two RNA recognition motifs (RRM1 and RRM2) and a C-terminal arginine/serine (RS)-rich domain (Figure 1D)27. Co-IP experiments revealed that SRSF1 mutants lacking either of the two RRM domains (SRSF1-ΔRRM1 or SRSF1-ΔRRM2) do not interact with FAM120A, suggesting that RNA binding is essential for the association of these two proteins (Figures 1E and S1A). In contrast, deletion of the RS domain of SRSF1 (SRSF1-ΔRS) dramatically enhanced the binding between SRSF1 and FAM120A that was insensitive to rapamycin or torin1, indicating an inhibitory function of RS domain in FAM120A-SRSF1 interaction (Figures 1E and S1A). The RS domain of SRSF1 has been shown to form a folded structure with the RRM1/2 domains15,28 and phosphorylation of the RS domain by SRPK opens this folded structure and promotes the binding of SRSF1 with other proteins12,15,29,30. Indeed, suppression of SRSF1 phosphorylation by inhibitors targeting mTORC1 (rapamycin and torin1) or SRPK (SRPKIN-1 and SRPIN340)31,32 decreased interaction of full-length SRSF1 (SRSF1-WT) with FAM120A while the interaction of RS domain-lacking mutant (SRSF1-ΔRS) with FAM120A was not affected by these inhibitors (Figures 1E, 1F, S1A and S1B). Furthermore, insulin-induced SRSF1 phosphorylation and SRSF1-FAM120A association was suppressed by knockdown of endogenous SRPK2, which was restored by re-expression of wild type SRPK2 but not by kinase dead (KD, SRPK2-K110M) or mTORC1-dependent phosphorylation site-lacking (AA, SRPK2-S494A/S497A) mutants12 (Figures 1G and S1C). These results demonstrate that when mTORC1 is activated, SRPK2 facilitates the association of FAM120A with SRSF1’s RRM domain by phosphorylating its RS domain (Figure 1D).
FAM120A promotes lipogenic enzyme expression
Given the interaction of FAM120A with RNA pol II24 and mTORC1-SRPK2-dependent interaction of FAM120A with SRSF1 (Figure 1), we reasoned that FAM120A would be involved in the regulation of gene expression downstream of mTORC1-SRPK2. We performed RNA-seq analysis in a kidney angiomyolipoma patient-derived cell line (LAM 621–101 or LAM cells where mTORC1 is activated due to TSC2 deficiency)33 after transfection with siRNAs targeting control or FAM120A (Figure 2A). To identify the genes downstream of mTORC1-SRPK2-dependent regulation of FAM120A, we compared FAM120A knockdown RNA-seq results with our previous whole transcriptome microarray data from LAM cells treated with rapamycin or SRPK2 knockdown (GSE104335)12. Among the 6 genes commonly downregulated in these three conditions, 5 were lipid metabolism regulators, including ATP citrate lyase (ACLY), acyl-CoA synthetase short chain family member 2 (ACSS2), fatty acid binding protein 3 (FABP3), hydroxysteroid 11-beta dehydrogenase 1 (HSD11B1), and mevalonate diphosphate decarboxylase (MVD) (Figures 2B and 2C). Pathway enrichment analysis of the genes downregulated by siFAM120A also ranked lipogenesis as one of the top FAM120A downstream biological processes (Figure 2D). Since de novo fatty acid and cholesterol synthesis enzymes were the major target of mTORC1-SRPK2-SRSF1 signaling12, we further looked into the level of individual genes in FAM120A RNA-seq and found that FAM120A depletion significantly decreased the expression of enzymes in de novo lipid synthesis pathway (Figures 2E and 2F).
Figure 2. FAM120A is required for the expression of lipogenic enzymes.
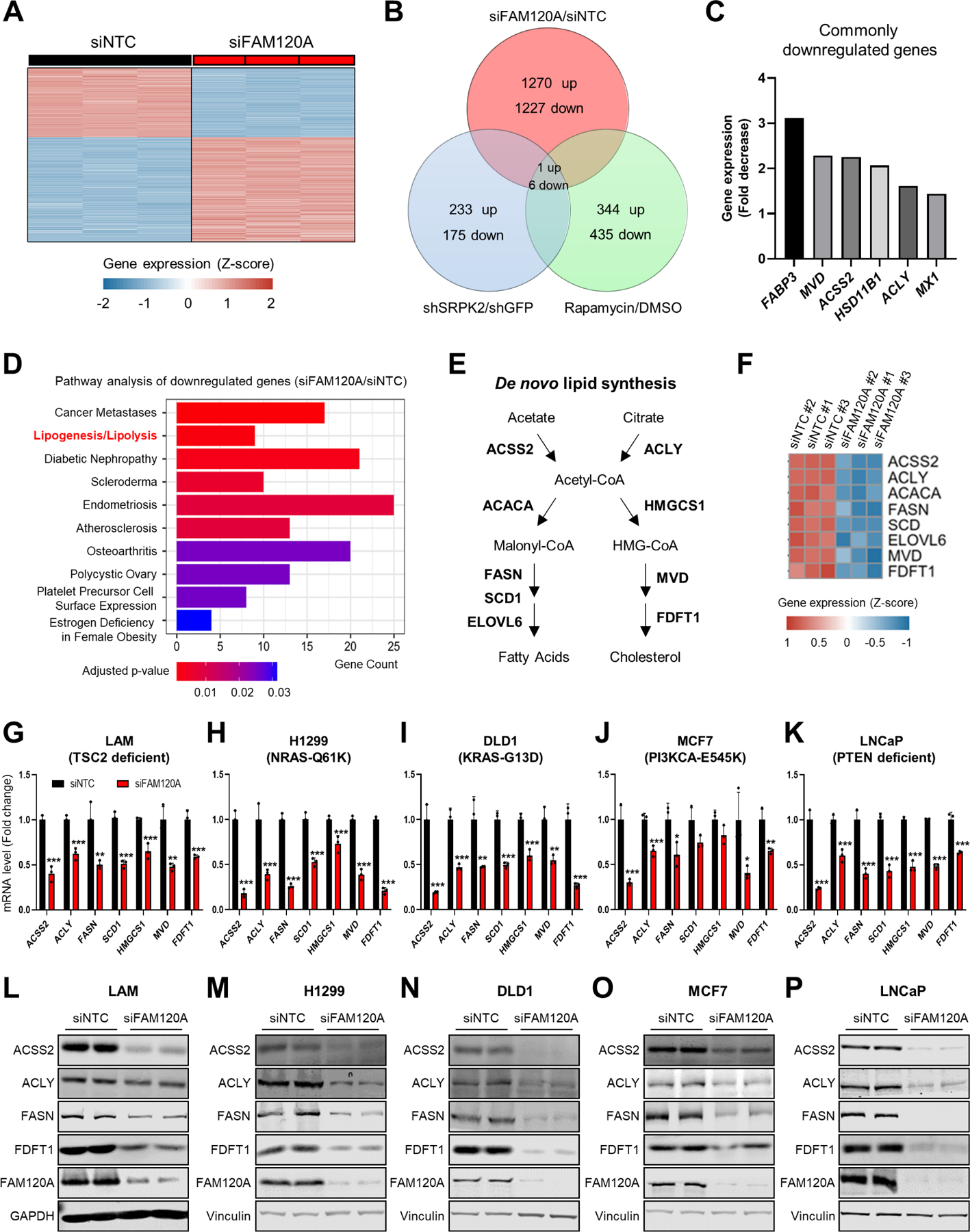
(A) Heatmap of z-scores for differentially expressed gene levels (cutoff: |Log2(Fold Change or FC)| ≥ 1, q-value < 0.005) in LAM 621–101 cells (hereinafter referred to as LAM cells) transfected with siRNAs targeting FAM120A or non-targeting control (NTC, hereinafter referred to as control) (GSE207172). N = 3. Same RNA-seq results are used in (A-F).
(B) Venn diagram analysis of the differentially regulated genes (cutoff: Linear fold change ≥ 1.5) identified from the whole-transcriptome microarray and RNA-seq analyses in LAM cells. The gene expression analyses were conducted on cells treated with rapamycin (20 nM) or vehicle for 24 hr, on cells stably expressing shRNAs targeting SRPK2 or GFP (GSE104335)12, or on cells transfected with siRNAs targeting FAM120A or control (GSE207172).
(C) Fold decrease of 6 commonly downregulated genes from the Venn diagram analysis in (B). Fold decrease values from siNTC vs. siFAM120A RNAseq analysis are shown.
(D) Pathway analysis of downregulated genes by FAM120A knockdown.
(E) Schematic of de novo lipogenesis.
(F) Heatmap of z-scores for the lipogenic gene levels decreased by FAM120A knockdown.
(G-K) QPCR analysis of LAM (G), H1299 (H), DLD1 (I), MCF7 (J), and LNCaP (K) cells transfected with siRNAs targeting FAM120A or control and serum starved overnight. N = 3. Data are represented as mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001.
(L-P) Immunoblot analysis of LAM (L), H1299 (M), DLD1 (N), MCF7 (O), and LNCaP (P) cells transfected with siRNAs targeting FAM120A or control and serum-starved overnight.
See also Supplemental Figure S1.
To examine whether FAM120A-dependent lipogenic gene regulation is relevant in other cancers, we knocked down FAM120A in a panel of mTORC1-overactive cancer cell lines derived from diverse tissues of origin and different genetic drivers such as H1299 (NRAS-mutated non-small cell lung cancer)34, DLD1 (KRAS-mutated colorectal adenocarcinoma)35, MCF7 (PI3K-mutated breast cancer)36, and LNCaP (PTEN-deficient prostate cancer)37. Across all these cells, FAM120A depletion decreased lipogenic enzymes at both mRNA and protein levels (Figures 2G–2P and S1D-S1H). In contrast, expression of another metabolic enzyme known to be regulated by mTORC1 for nucleotide synthesis, ribose 5-phosphate isomerase A (RPIA)10,38, was not regulated by FAM120A (Figure S1D-S1H), indicating a specific role of FAM120A in regulating lipogenic enzyme expression.
FAM120A promotes RNA splicing and stability of lipogenic enzymes
Next, we sought to elucidate the underlying mechanisms for FAM120A-dependent lipogenic gene regulation. Since FAM120A binds to phosphorylated SRSF1 (Figure 1) and exists in the spliceosome complex23–26, we speculated that FAM120A may regulate lipogenic gene expression via RNA splicing. To assess splicing changes, we performed deep RNA-seq of LAM cells knocked down with FAM120A or SRSF1. Differentially expressed gene analysis identified a total of 2,895 genes whose expression is significantly altered in siFAM120A or siSRSF1 cells compared to control (siNTC) (cutoff: |Log2(Fold Change or FC)| ≥ 0.5 and q-value < 0.01). Among these, 1,585 genes were downregulated by both FAM120A and SRSF1 knockdown (Figures 3A and S2A; Tables S1A and S1B). Pathway enrichment analysis revealed that lipid synthesis-related pathways including cholesterol biosynthesis (q-value = 6.45E-07), activation of gene expression by SREBP (q-value = 1.07E-06), and oleate biosynthesis (q-value = 0.00822) were among the top biological processes of the genes commonly regulated by FAM120A and SRSF1 (Figure S2B and Table S1C).
Figure 3. FAM120A is required for the splicing and stability of lipogenic genes.
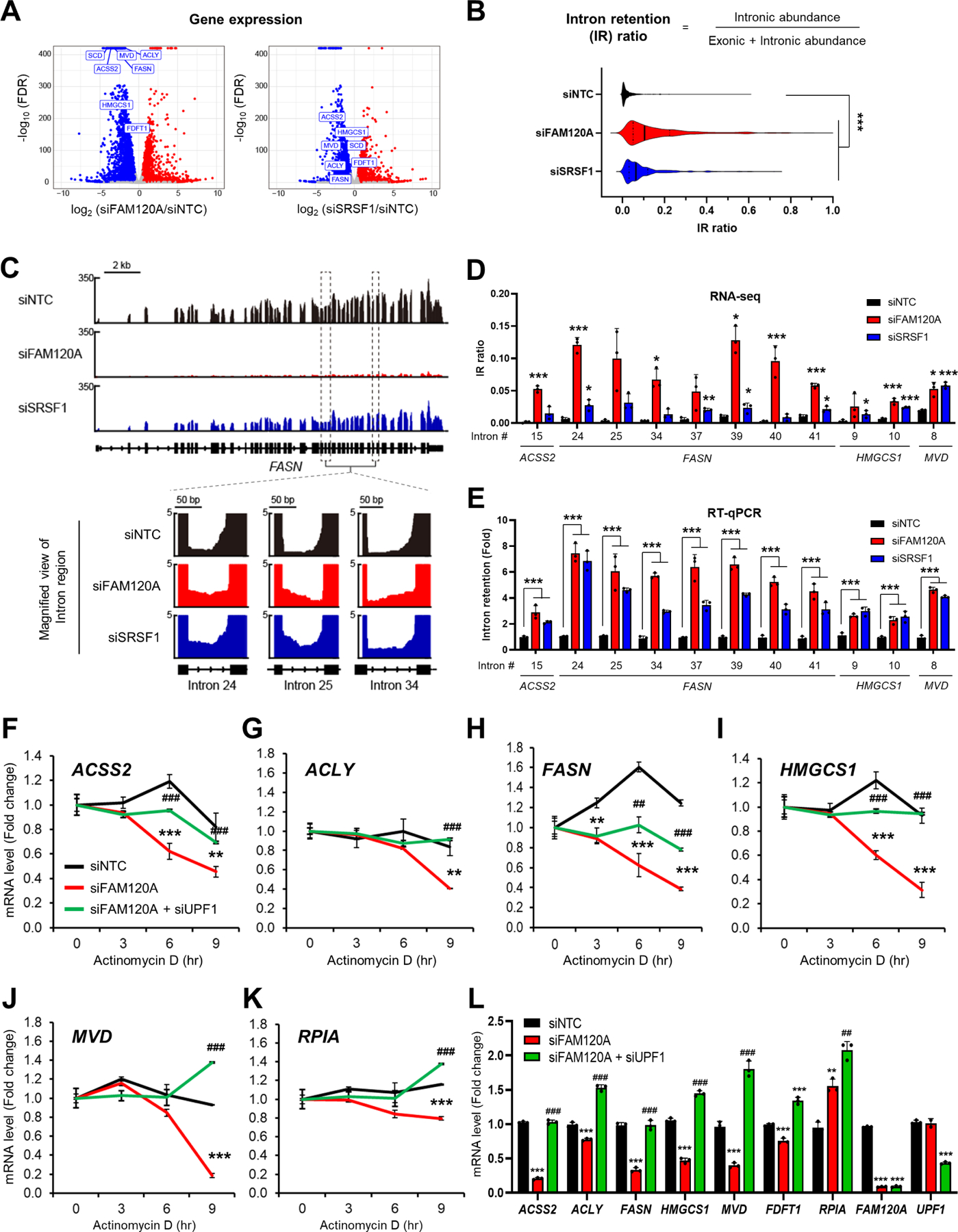
(A) Volcano plot of gene expression from RNA-seq results in LAM cells transfected with siRNAs targeting FAM120A, SRSF1 or control with overnight serum starvation (GSE229657). Lipogenic genes are highlighted in rectangles. N = 3. Same RNA-seq results are used in (A-D).
(B) Violin plot of 408 intron loci that show significant intron retention (IR) in siFAM120A and siSRSF1 compared to control.
(C) (Upper) Representative genome browser example of RNA-seq on FASN1 locus. (Lower) Zoom-in view of intron loci of FASN that are retained by FAM120A or SRSF1 knockdown.
(D) Individual IR ratio calculated from IRFinderS of lipogenic genes from RNA-seq data.
(E) Validation of intron retention by qPCR analysis of LAM cells transfected with siRNAs targeting FAM120A, SRSF1 or control. Intron retention = (Expression of intron-included region) / (Expression of intron-excluded region).
(F-L) QPCR analysis of LAM cells transfected with siRNAs targeting FAM120A, UPF1, or control, and serum starved overnight. Actinomycin D (Act D, 5 μg/mL) was treated for the indicated time points for mRNA stability analysis (F-K). N = 3. p-value (*) was calculated between siNTC and siFAM120A. p-value (#) was calculated between siFAM120A and siFAM120A+siUPF1. Data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ##p < 0.01, and ###p < 0.001.
See also Supplemental Figure S2 and Supplemental Table S1.
For comprehensive evaluation of splicing events regulated by FAM120A and SRSF1, we analyzed the RNA-seq data using Vertebrate Alternative Splicing and Transcription Tools (VAST-TOOLS)39. We identified 11,237 differential splicing events by FAM120A knockdown and 5,844 by SRSF1 knockdown, which includes intron retention, alternative 5’ splicing site, alternative 3’ splicing site, and micro-exon splicing. Among these, intron retention was the most prevalent splicing events by comprising 85% of the differential splicing events in siFAM120A and 73% in siSRSF1 (Figure S2C). To quantitate intron retention events more precisely, we calculated intron retention ratio (IR ratio = intronic abundance / (exonic + intronic abundance)) using IRFinder-S software40, which demonstrated a significant increase of IR by FAM120A or SRSF1 knockdown (Figure 3B). From the cutoff of q-value < 0.1 and Log2FC > 0, we selected 9,589 (siFAM120A/siNTC) and 2,849 (siSRSF1/siNTC) displaying significant intron retention events. Finally, by comparing the 1,893 intron retention events identified in both siFAM120A and siSRSF1 conditions and 1,585 commonly downregulated genes, we found 408 intron retention events in 186 genes (Figures S2D and S2E; Table S1D). Pathway enrichment analysis of these final candidate genes revealed that intron retention is enriched in lipid metabolism genes such as cholesterol metabolism (q-value = 0.01145), omega-9 fatty acid synthesis (q-value = 0.01541), and metabolism of lipids (q-value = 0.03637)), suggesting that FAM120A and SRSF1 control expression of lipid metabolism genes by promoting splicing of their introns (Figure S2F and Table S1E).
From in-depth analysis of RNA-seq results on lipogenic genes, we found significantly increased intron retention by FAM120A or SRSF1 knockdown in FASN, ACSS2, MVD, and HMGCS1 (Figures 3C, 3D, S2E and Table S1D). We validated these IR events by qPCR using the primers specifically targeting the introns identified from RNA-seq (Figure 3E). Failed splicing and intron retention lead to degradation of transcripts via nonsense-mediated decay (NMD), which we previously found as the mechanism for mTORC1-SRPK2-SRSF1-dependent regulation of lipogenic enzymes12,41,42. To examine whether the same mechanism underlies in FAM120A-dependent regulation of lipogenic genes, we assessed RNA stability by actinomycin D assay43. FAM120A knockdown accelerated degradation of lipogenic gene transcripts including ACSS2, ACLY, FASN, HMGCS1, and MVD (Figures 3F–3J). In contrast, the stability of RPIA, which is not regulated by mTORC1-SRPK2-FAM120A-SRSF1-dependent splicing, was not dramatically changed (Figure 3K). Suppression of NMD by knocking down a key NMD factor, up-frameshift 1 (UPF1), restored lipogenic gene RNA stability (Figures 3F–3J) as well as their expression levels in FAM120A knockdown cells (Figure 3L). Therefore, FAM120A induces expression of lipogenic enzymes by promoting efficient intron splicing and stability of their RNA.
FAM120A interacts with SRSF1 through lipogenic gene transcripts
Among the FAM120 family proteins, only FAM120A has been identified to be associated with spliceosomes23,25,26 and contains an RNA binding domain (RBD) (Figure 4A)44,45. Therefore, we reasoned that RBD would play a critical role in FAM120A-dependent lipogenic gene splicing. Indeed, removal of RBD (FAM120A-ΔRBD) completely abolished its binding with lipogenic gene transcripts (Figure 4B; Figure S3A). FAM120A-ΔRBD failed to bind with SRSF1 (Figure 4C), suggesting that RNA mediates FAM120A-SRSF1 interaction. In line with this notion, treatment of cell lysates with ribonucleases RNase A/T1 disrupted the interaction between FAM120A and SRSF1 (Figure 4D). Removal of SRSF1 RNA binding domain (SRSF1-ΔRRM) (Figures 1E and S1A) or ribonuclease treatment to SRSF1-ΔRS also abrogated the binding of SRSF1 with FAM120A (Figure 4D), confirming RNA as an essential component in FAM120A-SRSF1 interaction. In the absence of FAM120A, SRSF1 did not bind with lipogenic transcripts (Figures 4E and S3B), indicating that FAM120A is required for the association of SRSF1 with lipogenic genes.
Figure 4. FAM120A interacts with SRSF1 through lipogenic gene RNAs.
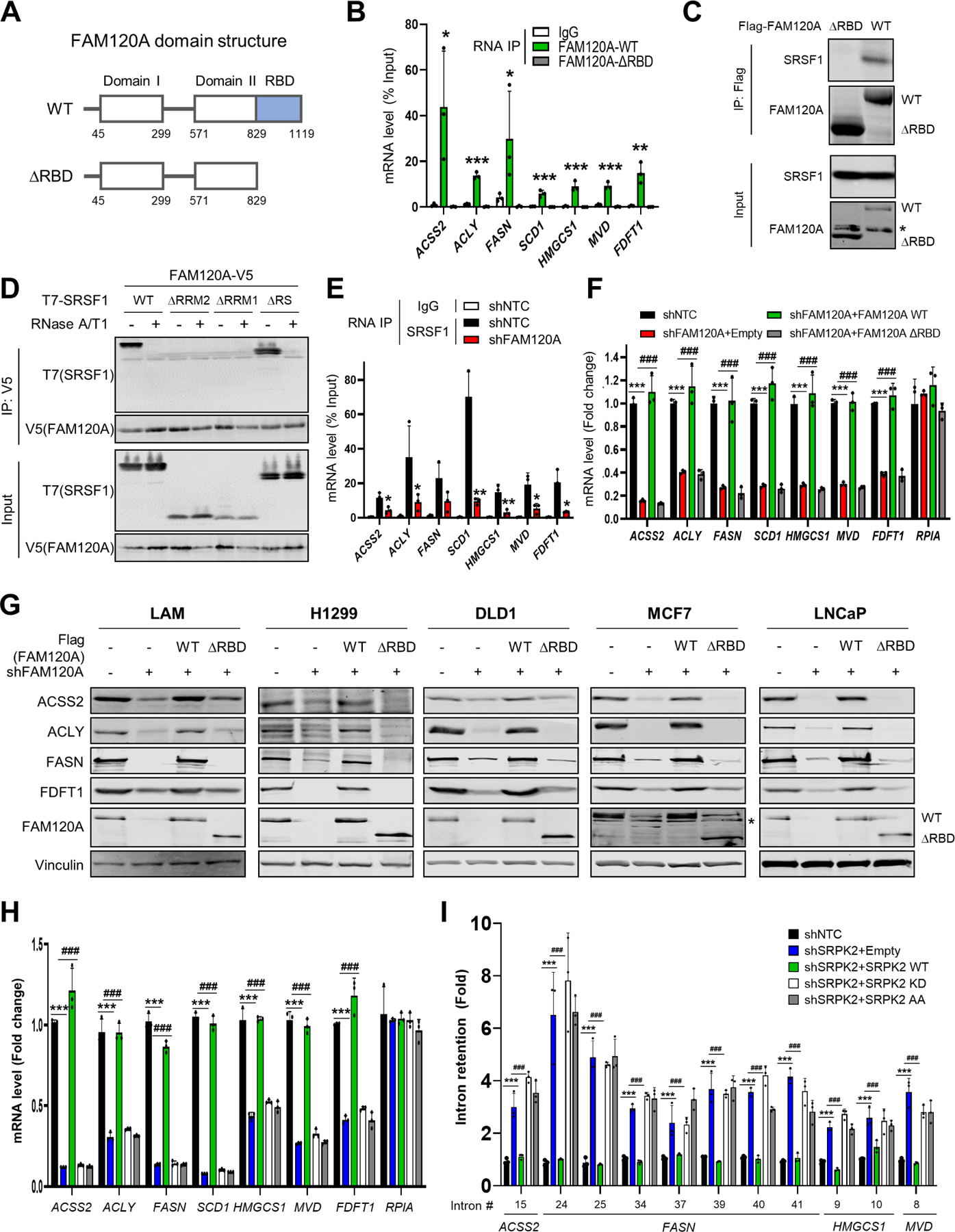
(A) Schematic diagram of the domains in wild-type (WT) and RNA binding domain (RBD)-deleted (∆RBD) FAM120A.
(B) RNA immunoprecipitation qPCR (RIP-qPCR) analysis of LAM cells expressing FAM120A WT or ∆RBD. IP was performed with IgG or anti-FAM120A antibodies. The graph shows qPCR analysis of RIP products. N = 3. p-value was calculated between IgG and FAM120A-WT.
(C) Co-IP analysis of HEK293E cells expressing SRSF1 with Flag-FAM120A WT or ∆RBD. 1% of total cell lysate was loaded as an input. Asterisk indicates a non-specific band.
(D) Co-IP analysis of HEK293E cells expressing FAM120A-V5 and T7-SRSF1 (WT or mutants). Cell lysates were treated with RNases before IP. 1% of total cell lysate was loaded as an input.
(E) RIP-qPCR analysis of LAM cells stably expressing shRNAs targeting FAM120A or control. Immunoprecipitation was performed with IgG or anti-SRSF1 antibodies. The graph shows qPCR analysis of RIP products. N = 3. p-value was calculated between SRSF1-IP samples from shNTC and shFAM120A.
(F) QPCR analysis of LAM cells stably expressing FAM120A WT or ∆RBD in FAM120A knockdown cells. Cells were serum-starved overnight. N = 3.
(G) Immunoblot analysis of LAM, H1299, and DLD1 cells stably expressing FAM120A WT or ∆RBD in FAM120A knockdown cells. Cells were serum-starved overnight.
(H-I) QPCR analysis of LAM cells stably expressing HA-SRPK2 (WT (wild type), KD (kinase dead, SRPK2-K110M), or AA (non-phosphorylatable, SRPK2-S394A/S397A)). Endogenous SRPK2 was knocked down by shRNA targeting 3’UTR of SRPK2. Cells were serum-starved overnight. Graphs show mRNA levels (H) and intron retention (I). N = 3.
Data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, and ###p < 0.001.
See also Supplemental Figure S3.
We then evaluated the functional significance of the binding between FAM120A, SRSF1, and lipogenic transcripts in lipogenic gene expression. The expression of lipogenic enzyme mRNA and protein levels was decreased by FAM120A knockdown in multiple cancer cell lines (Figures 2G–2P), which was rescued by wild type FAM120A but not by RNA binding-deficient FAM120A-ΔRBD mutant (Figures 4F, 4G and S3C-S3F). Abrogation of FAM120A-SRSF1 interaction by knocking down SRPK2 and re-expression of mutant SRPK2 proteins led to intron retention and decreased expression of lipogenic genes (Figures 4H, 4I and S3G). Collectively, these results suggest that FAM120A facilitates recruitment of lipogenic transcripts to phosphorylated SRSF1, which is essential for efficient splicing and expression of lipogenic enzymes.
FAM120A recruits SRSF1 to lipogenic gene promoter
To regulate transcription and pre-mRNA splicing, SR proteins are recruited to gene promoters and active transcription sites22,46–48. By analyzing previously published ChIP-seq data46, we found that Srsf1 is enriched near the promoter of lipogenic genes including Acss2, Acly, and Fasn (Figures 5A–5C). ChIP-qPCR confirmed that SRSF1 binds lipogenic gene promoters in diverse cancer cells. However, binding of SRSF1 to these promoters was almost completely abolished by FAM120A knockdown (Figures 5D–5F and S4A-S4C), indicating that FAM120A is essential for bringing SRSF1 to these promoters. To understand how FAM120A achieves the specific recruitment of SRSF1 to lipogenic gene promoters, we examined interaction of FAM120A with several lipogenic transcription factors, including SREBP1/2, carbohydrate-response element-binding protein (ChREBP), liver X receptor (LXR) α/β, peroxisome proliferator-activated receptor (PPAR) α/γ, p300, and X-box binding protein 1 (XBP1). Among these, only SREBP1 co-immunoprecipitated with FAM120A (Figure 5G). Furthermore, the binding of SREBP1 at lipogenic gene promoters and their promoter activities were substantially decreased by FAM120A depletion (Figures 5H–5J, S4D and S4E) while SREBP1/2 depletion abolished SRSF1 recruitment to lipogenic gene promoters (Figures 5K–5M and S4F-S4H). Therefore, FAM120A is the key mediator that brings SREBP1 and SRSF1 together to the lipogenic genes (Figure S6).
Figure 5. FAM120A bridges SRSF1 to SREBP1 to induce lipogenic enzyme expression.
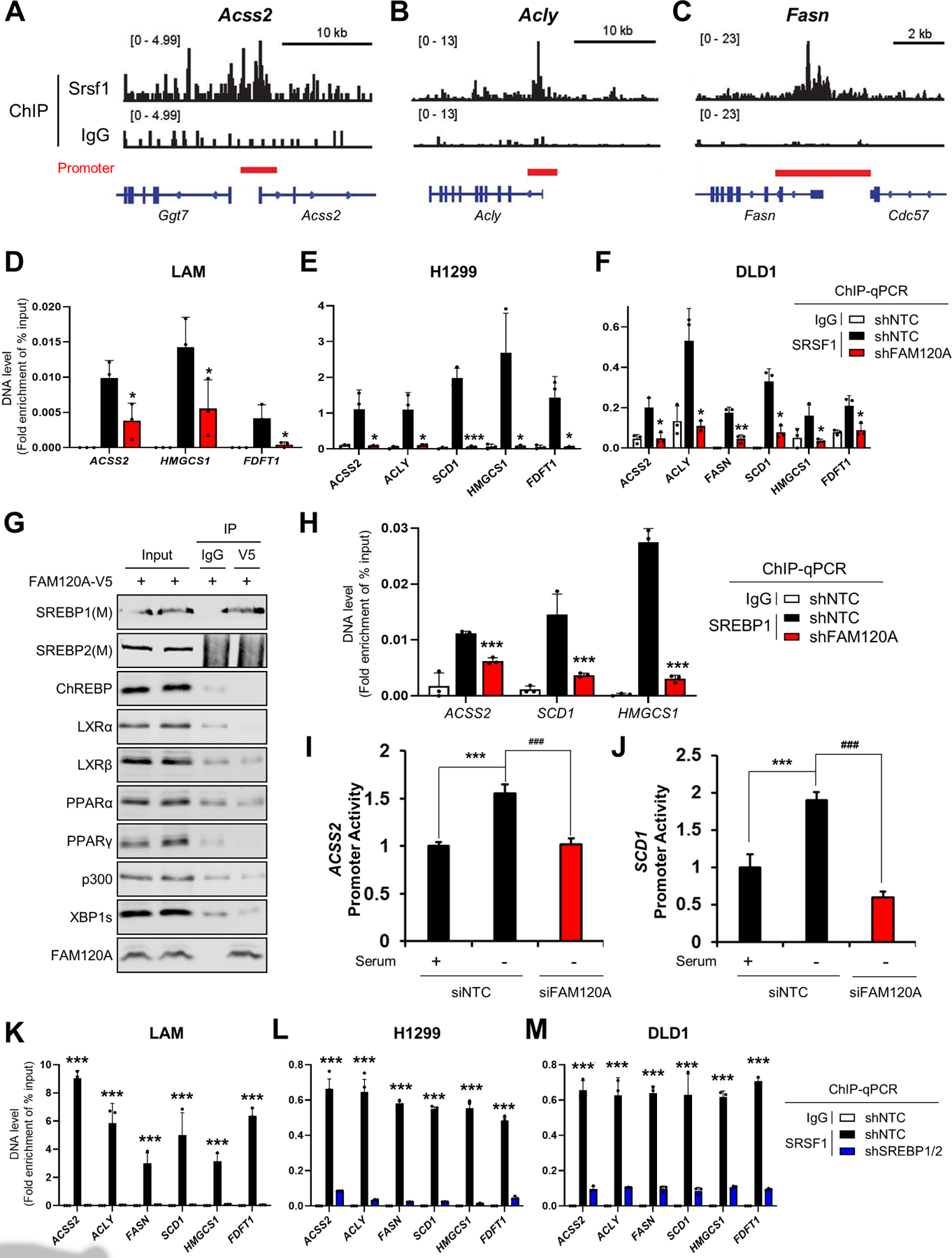
(A-C) Analysis of Srsf1 chromatin immunoprecipitation (ChIP)-seq results (GSE45517)46 in MEF cells near the transcription start sites of Acss2 (A), Acly (B), and Fasn (C).
(D-F) ChIP-qPCR analysis of SRSF1 in target gene promoters. Immunoprecipitation was performed using IgG or anti-SRSF1 antibodies in LAM (D), H1299 (E), and DLD1 (F) cells stably expressing shRNAs targeting FAM120A or control. Cells were serum-starved overnight. p-value was calculated between shNTC and shFAM120A. N = 3.
(G) Co-IP analysis of transcription factors in HEK293E cells expressing FAM120A-V5. 1% of total cell lysate was loaded as an input.
(H) ChIP-qPCR analysis of SREBP1 on target gene promoters. Immunoprecipitation was performed using IgG or anti-SREBP1 antibodies in LAM cells stably expressing shRNAs targeting FAM120A or control. Cells were serum-starved overnight. p-value was calculated between shNTC and shFAM120A. N = 3.
(I, J) Promoter activity analysis of ACSS2 (H) and SCD1 (I) in LAM cells transfected with promoter constructs and siRNAs targeting FAM120A or control. N = 3.
(K-M) ChIP-qPCR analysis of SRSF1 in target gene promoters. Immunoprecipitation was performed using IgG or anti-SRSF1 antibodies in LAM (K), H1299 (L), and DLD1 (M) cells stably expressing shRNAs targeting SREBP1/2 or control. Cells were serum-starved overnight. p-value was calculated between shNTC and shSREBP1/2. N = 3.
Data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, and ###p < 0.001.
See also Supplemental Figure S4.
FAM120A connects transcription and splicing machineries to promote efficient splicing of lipogenic genes
Next, we sought to further delve into the nature of SREBP1-FAM120A-SRSF1 complex. We and others have previously identified U1-70K as the key spliceosome protein recruited to the target genes upon the phosphorylation of SRSF1 by mTORC1-SRPK212,15. Knockdown of U1-70K led to the intron retention of lipogenic genes (Figures 6A and S5A), similar to knockdown of FAM120A and SRSF1 (Figure 3). On the other hand, in the cells depleted with the transcription factor SREBP1/2, FAM120A knockdown did not induce intron retention (Figures 6B and S5B), indicating that transcription is pre-requisite for the splicing of lipogenic genes. Immunoprecipitation of endogenous FAM120A confirmed that RNA pol II and U1-70K associate with the SREBP1-FAM120A-SRSF1 complex (Figure 6C). Ribonuclease treatment abolished the binding among these proteins (Figure 6C), showing that RNA is a key component of this multi-protein co-transcriptional splicing complex. Notably, while deletion of RNA binding domain of SRSF1 (SRSF1-ΔRRM) abolished its interaction with FAM120A, the binding between FAM120A and SREBP1 was intact (Figure 6D). In contrast, deletion of the RNA binding domain of FAM120A (FAM120A-ΔRBD) dissociated binding of all three proteins (Figure 6E), hence FAM120A bridges the binding between SREBP1 and SRSF1. Indeed, SRSF1 did not directly bind with SREBP1 (Figure S5C).
Figure 6. FAM120A promotes efficient splicing of lipogenic genes by associating with transcriptional and splicing machineries.
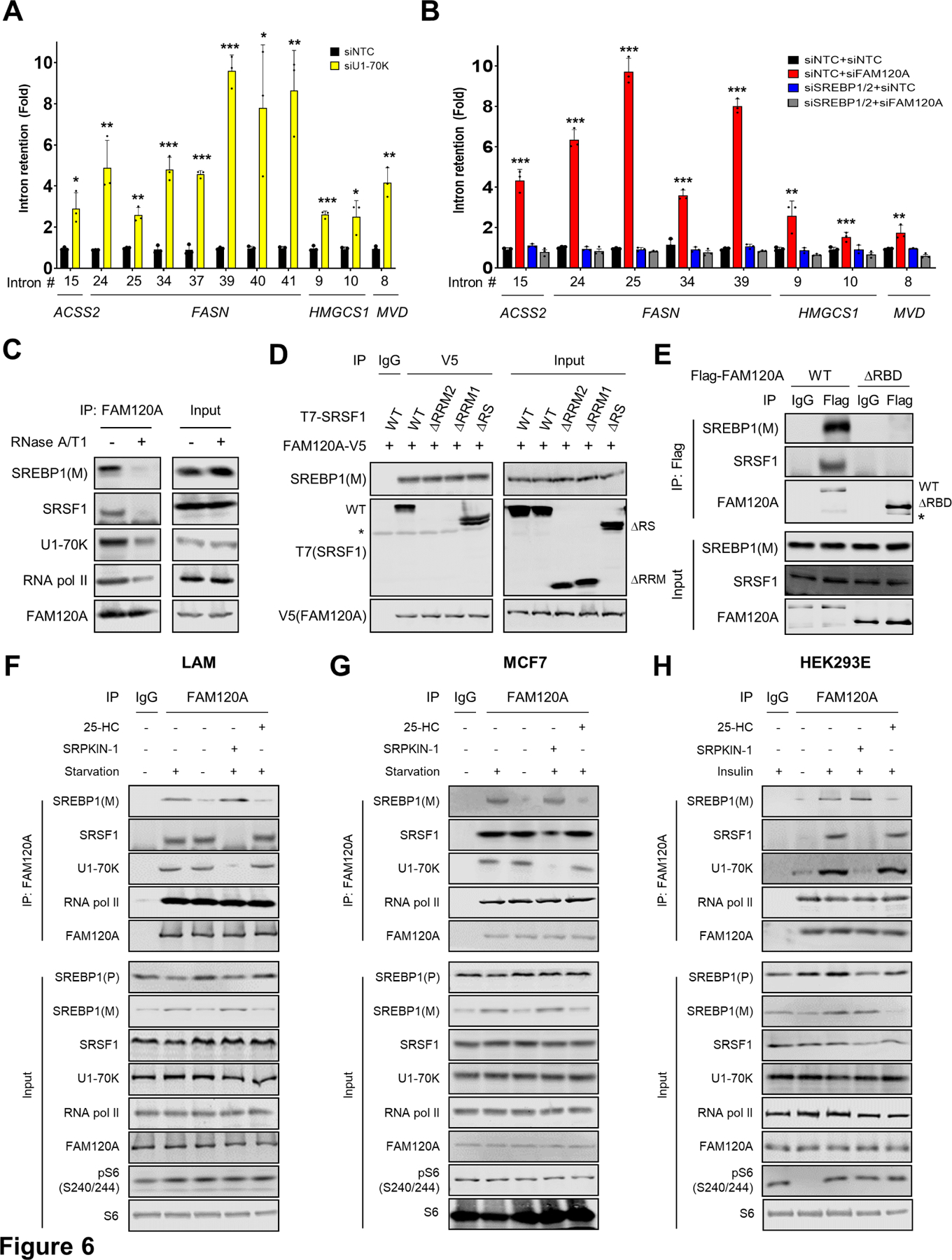
(A, B) QPCR analysis of intron retention in LAM cells transfected with siRNAs targeting U1-70K or control (A), and FAM120A, SREBP1/2, SREBP1/2+FAM120A or control (B). Cells were serum starved overnight. N = 3. Data are represented as mean ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001. p-value was calculated between siNTC and siFAM120A (B). N = 3.
(C) Co-IP analysis of HEK293E cells. Immunoprecipitation was performed using anti-FAM120A antibodies. Cell lysates were treated with RNases before IP. 1% of total cell lysate was loaded as an input.
(D) Co-IP analysis of HEK293E cells expressing FAM120A-V5 and T7-SRSF1 (WT or mutants). 1% of total cell lysate was loaded as an input. Asterisk indicates a non-specific band.
(E) Co-IP analysis of HEK293E cells expressing Flag-FAM120A WT or ∆RBD. 1% of total cell lysate was loaded as an input. Asterisk indicates a non-specific band.
(F-H) Co-IP analysis of LAM (F) and MCF7 (G) cells serum starved overnight (to induce SREBP cleavage) and treated with SREBP cleavage inhibitor (25-HC, 10 µM), or SRPK inhibitor (SRPKIN-1, 5 µM) for 4 hr (F, G); HEK239E cells serum starved overnight followed by insulin stimulation (100 nM) (to induce SREBP cleavage) with or without 25-HC (10 µM) or SRPKIN-1 (5 µM) for 4 hr (H). Note that serum starvation decreases mTORC1 activity in HEK293E cells (H) but does not affect constitutively active mTORC1 activity in LAM and MCF7 cancer cells (F, G). Immunoprecipitation was performed using IgG or anti-FAM120A antibodies. 1% of total cell lysate was loaded as an input. SREBP1(P), precursor SREBP1; SREBP1(M), cleaved and matured SREBP1.
See also Supplemental Figure S5.
Given that the transcriptionally active nuclear form of SREBP1, SREBP1(M), was identified to interact with FAM120A (Figure 5G), we examined whether SREBP1 processing affects its interaction with FAM120A or vice versa. Depletion of cholesterol by serum starvation induces cleavage of the precursor SREBP1(P) to the mature SREBP1(M) in cancer cells (Figures 6F and 6G), while insulin promotes SREBP1(M) production in non-cancerous HEK293E cells49 (Figure 6H). In both cases, inhibition of SREBP1 cleavage by 25-hydroxycholesterol (25-HC) treatment50 prevented SREBP1(M) formation and its interaction with FAM120A (Figures 6F–6H). However, neither FAM120A knockdown nor the RNA binding-defective FAM120A mutant (FAM120A-ΔRBD) decreased SREBP1(M) levels (Figures 6E, S5C and S5D), demonstrating that FAM120A does not affect SREBP1 cleavage.
Since SREBP1-FAM120A interaction was independent of the SRSF1-FAM120A interaction (Figure 6D), we reasoned that the formation of SREBP1-FAM120A-RNA pol II transcription complex would be independent of splicing complex formation. Unphosphorylated SRSF1 cannot bind with FAM120A (Figures 1E–1G and S1A-S1C) or spliceosome protein U1-70K12,15. However, suppression of SRSF1 phosphorylation by SRPK inhibitor (SRPKIN-1) did not disrupt the interaction of FAM120A with SREBP1 and RNA pol II (Figures 6F–6H). Blocking SREBP1 cleavage by 25-HC treatment did not affect the binding of FAM120A with SRSF1 and RNA pol II, either (Figures 6F–6H). Together, these findings suggest that FAM120A bridges the two independent transcription vs. splicing complexes to promote efficient splicing of newly transcribed lipogenic genes (Figure S6).
FAM120A is essential for lipid production and tumor growth
To examine the functional relevance of FAM120A-dependent lipogenic gene regulation, we measured levels of the four most abundant fatty acids (C16:0, C16:1, C18:0, and C18:1) produced by de novo lipid synthesis using liquid chromatography-mass spectrometry (LC-MS)51. The knockdown of FAM120A markedly depleted these fatty acids in LAM and H1299 cells (Figures 7A-7C, S7A, and S7B). Consistent with the notion that these fatty acids are critical for phospholipid membrane synthesis during cell proliferation52, FAM120A knockdown suppressed proliferation of various cancer cell lines (Figures 7D-7H and S7C-S7G). Removal of exogenous lipid supply via lipoprotein-deficient serum (LPDS)53 augmented growth defects in FAM120A knockdown cells, which was rescued by supplementation of exogenous lipoproteins or fatty acids (Figures 7I-7M and S7H-S7O). Finally, FAM120A-depleted cells showed strongly reduced xenograft tumor growth in vivo and re-expression of wild-type FAM120A but not FAM120A-ΔRBD mutant partially rescued tumor growth (Figures 7N, 7O, and S7P). Together, these data show that FAM120A-dependent lipogenic gene expression is critical for fatty acid synthesis and growth of tumor cells.
Figure 7. FAM120A RNA binding is essential for lipogenesis and tumor growth.
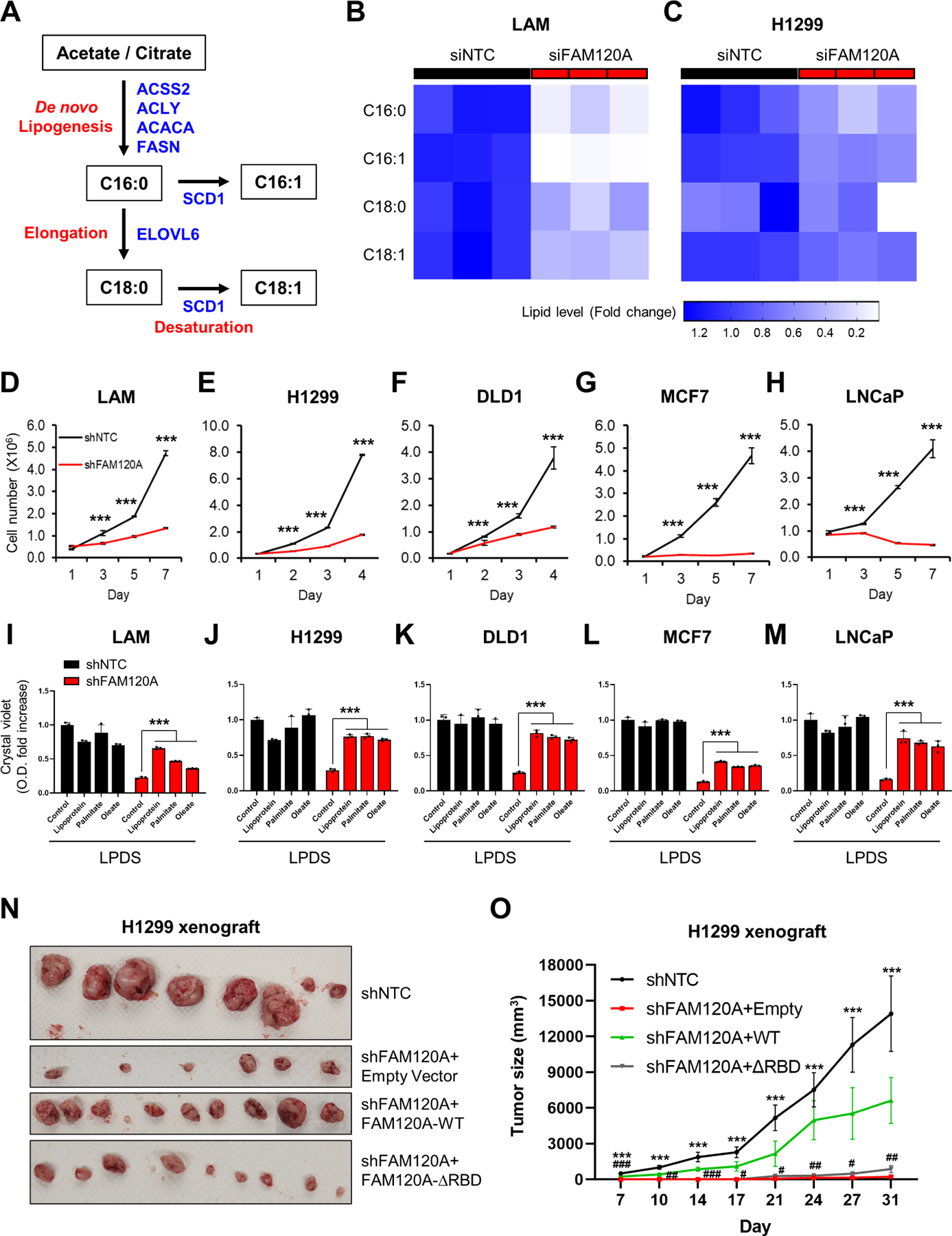
(A) Schematic of de novo fatty acid synthesis pathway.
(B, C) Heatmap of fatty acid levels. LC-MS analysis was performed on LAM cells transfected with siRNAs targeting FAM120A or control (B) and H1299 cells stably expressing shRNAs targeting FAM120A or control (C) with overnight serum starvation. N = 3.
(D-H) Cell proliferation analysis of LAM (D), H1299 (E), DLD1 (F), MCF7 (G), and LNCaP (H) cells stably expressing shRNAs targeting FAM120A or control. N = 3. Data are represented as mean ± SD.
(I-M) Crystal violet (CV) analysis of LAM (I), H1299 (J), DLD1 (K), MCF7 (L), and LNCaP (M) cells stably expressing shRNAs targeting FAM120A or control. Cells were grown in lipoprotein-deficient serum (LPDS)-containing media supplemented with lipoprotein (25 μg/ml), palmitate-albumin (10 μM, palmitate), oleate-albumin (50 μM, oleate), or fatty acid-free albumin (25 μM, control). The graph shows the quantified absorbance of solubilized CV stain. N = 3. Data are represented as mean ± SD.
(N-O) Xenograft tumor growth assay of H1299 cells stably expressing shNTC or shFAM120A with FAM120A WT or ∆RBD. N ≥ 4. Data are represented as mean ± SEM. p-value (*) was calculated between shNTC and shFAM120A+Empty. p-value (#) was calculated between shFAM120A+Empty and shFAM120A+WT. Data are represented as mean ± SD. ***p < 0.001, #p < 0.05, ##p < 0.01, and ###p < 0.001.
See also Supplemental Figure S7.
DISCUSSION
Here, we discovered FAM120A as a unique factor that bridges transcription and splicing of lipogenic genes for tumor cell growth. As a transcription co-factor and RNA binding protein (RBP), FAM120A binds to both transcription factor SREBP and splicing factor SRSF1 to connect efficient splicing of newly transcribed lipogenic gene transcripts. This function of FAM120A is stimulated by the mTORC1-activated SRPK2 signaling in various cancers (Figure S6).
Splicing is carried out by a small nuclear ribonucleoprotein (snRNP) complex composed of small nuclear RNAs and RNPs54. While the core spliceosome complex (U1, U2, U4, U5, and U6 snRNPs) recognizes exon-intron borders and splices introns, additional RBPs are involved in determining the target gene specificity and direction of the splicing events. SR proteins are the splicing inducers that bind to exonic splicing enhancer (ESE) sequences for the recruitment of spliceosome complex55. Association of SR proteins with the spliceosome proteins is a highly regulated process, and we previously showed that mTORC1-SRPK2-dependent phosphorylation of SRSF1 promotes its interaction with U1-70K12, a U1 snRNP protein involved in spliceosome formation initiation15,56. In this study, we demonstrate that mTORC1-SRPK2 drives the selectivity of SRSF1-dependent splicing toward de novo lipid synthesis genes through an SREBP1-interacting transcription co-factor, FAM120A. SREBP1 is a transcription factor that recruits transcription preinitiation complex proteins including Pol II, TATA box-binding protein (TBP), and other general transcription factors such as transcription initiation factor IIA (TFIIA) and TFIIB. Once Pol II is released from the transcription initiation stage, Pol II enters an active elongation stage and moves along the gene body with transcription elongation associated proteins including positive-transcription elongation factor b (P-TEFb) and RNA processing proteins57–59. Therefore, it is unlikely that SREBP1 piggybacks with Pol II and SRSF1 through the transcript during transcription elongation. Instead, our proposed mechanism highlights a key role for FAM120A in coordinating transcription initiation and splicing of target genes by promoting the formation of distinct complexes that augment the efficiency of these processes. In the case of lipogenic gene transcription, one complex is composed of FAM120A-Pol II-SREBP1, while the other responsible for the splicing of newly transcribed genes contains FAM120A-Pol II-U1-70K-SRSF1. Notably, the two complexes form independently of each other, but they cooperate for the FAM120A-mediated co-transcriptional splicing of lipogenic genes (Figures 6 and S6). This provides a bona fide mechanism for the efficient coupling of transcription, splicing, and stabilization of SREBP1-regulated lipogenic genes.
As a master regulator of cell growth, mTORC1 is acutely activated by nutrients and growth factors to coordinate macromolecule synthesis for cell growth. However, when mTORC1 is aberrantly and constitutively activated, as in many human cancers, its activation leads to overgrowth and proliferation60,61. Given the vastly broad effect of mTORC1 in biological processes, it may be important to target specific downstream signaling effectors for therapeutic purposes to avoid severe toxicities of mTORC1 targeting drugs62. Our data suggests that suppressing lipogenic gene splicing via FAM120A binding inhibitors or SRPK inhibitors would target a possible metabolic vulnerability. Splicing inhibitors have been in clinical trials based on the findings that splicing is overactivated in tumor cells63. Since splicing is such a fundamental process that occurs across all mammalian tissues and cell types, splicing inhibitors caused toxic side effects in patients64,65. On the other hand, selectively targeting the SRPK2-SRSF1-FAM120A axis could be an alternative therapeutic strategy to preferentially suppress lipogenic enzymes with low toxicity. In fact, suppressing fatty acid synthesis by a second generation fatty acid synthase (FASN) inhibitor was well tolerated since de novo lipid synthesis is almost inactive in many cells except for the hepatocytes and adipocytes66–68. Looking forward, it will be exciting to investigate if small molecule inhibitors, oligonucleotides, and RNA-PROTACs (Proteolysis Targeting Chimeric)69–71 that block FAM120A function and its interaction with lipogenic transcripts (e.g., via its RNA binding domain) become important therapies for the treatment of mTORC1-driven cancers.
LIMITATIONS OF THE STUDY
In this study, we found FAM120A as a bona fide effector in the mTORC1-SRPK2-SRSF1 signaling cascade that determines the downstream target specificity toward lipid metabolism genes (Figure S6). However, from the pathway enrichment analysis of transcriptome and splicing changes in FAM120A and SRSF1 knockdown cells, we also identified interferon signaling and immune response genes such as interferon (IFN) stimulated genes (ISG) and major histocompatibility complex (MHC)-mediated antigen processing genes as top biological process regulated by FAM120A and SRSF1 (Figure S2 and Table S1). The potential regulation of IFN pathway by FAM120A and SRSF1 is particularly interesting because IFN signaling was among the top biological pathways regulated by SRPK2 in our previous study12. The ISG gene, myxovirus resistance 1 (MX1), was indeed one of the commonly downregulated genes by rapamycin, SRPK2 knockdown, and FAM120A knockdown (Figure 2C). These observations suggest that interferon-mediated immune response could be another important biological function of mTORC1-SRPK2-SRSF1-FAM120A pathway. Like de novo lipid synthesis enzymes, ISG genes are known to be transcribed by master transcription factors such as signal transducer and activator of transcription (STAT) and interferon regulatory factor (IRF)72–74. Future studies will be required to identify the immune transcription factor that interacts with FAM120A for the coordination of efficient transcription and splicing of ISG genes. While traditional interferon signaling and immune research have been focused on anti-viral responses, recent studies found that programmed cell death ligand 1 (PDL1)-associated tumor resistance is linked to the sustained expression of ISGs including MXI175–77. Ultimately, understanding the role of SRPK2 and FAM120A in immune checkpoint control can potentially provide mechanistic insights for improving immunotherapeutic approaches and patient outcomes in various immune-related diseases and cancers.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John Blenis (job2064@med.cornell.edu).
Materials Availability
All reagents generated in this study are available from the Lead Contact without restriction.
Data and Code Availability
Sequencing data have been deposited at GEO with identifier GSE207172 and GSE229657. The original images have been deposited at Mendeley Data (DOI: 10.17632/rgfgx9xb8w.1). All data are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-SRPK2 | BD Biosciences | Cat#BD611118; RRID: AB_398429 |
| Rabbit polyclonal anti-ACLY | Cell Signaling Technology | Cat#4332; RRID: AB_2223744 |
| Rabbit monoclonal anti-FASN | Cell Signaling Technology | Cat#3180; RRID: AB_2100796 |
| Mouse monoclonal anti-S6 | Cell Signaling Technology | Cat#2317; RRID: AB_2238583 |
| Rabbit monoclonal anti-pS6(S240/S244) | Cell Signaling Technology | Cat#5364; RRID: AB_10694233 |
| Rabbit polyclonal anti-SCD1 | Cell Signaling Technology | Cat#2438; RRID: AB_823634 |
| Rabbit polyclonal anti-CHREBP | Novus | Cat# NB400-135;RRID:AB_10002435 |
| Rabbit polyclonal anti-FAM120A | GeneTex | Cat#GTX120824; RRID:AB_10723014 |
| Rabbit polyclonal anti-FAM120A | Thermo Fisher Scientific | Cat# A303-889A; RRID:AB_2620239) |
| Rabbit polyclonal anti-LXR alpha | Thermo Fisher Scientific | Cat#PA1-330;RRID:AB_325600 |
| Mouse monoclonal anti-SREBP2 | BD Biosciences | Cat#557037; RRID:AB_396560 |
| Rabbit polyclonal anti-LXR beta | Thermo Fisher Scientific | Cat#PA1-333;RRID:AB_2154770 |
| Rabbit polyclonal anti-PPAR alpha | Thermo Fisher Scientific | Cat#PA1-822A; RRID:AB_2165595 |
| Rabbit polyclonal anti-PPAR gamma | Abcam | Cat#ab209350; RRID:AB_2890099 |
| Rabbit monoclonal anti-p300 | Cell Signaling Technology | Cat#54062; RRID:AB_2799450 |
| Rabbit monoclonal anti-XBP-1s | Cell Signaling Technology | Cat#12782;RRID:AB_2687943 |
| Mouse monoclonal anti-GAPDH | Sigma-Aldrich | Cat#G8795; RRID: AB_1078991 |
| Rabbit polyclonal anti-ACSS2 | Sigma-Aldrich | Cat#HPA004141; RRID: AB_1078094 |
| Mouse monoclonal anti-V5 | Sigma-Aldrich | Cat#V8012; RRID:AB_261888 |
| Mouse monoclonal anti-Vinculin | Sigma-Aldrich | Cat#V9264; RRID: AB_10603627 |
| Rabbit polyclonal anti-RNA polymerase II | Abcam | Cat#ab26721; RRID:AB_777726 |
| Rabbit monoclonal anti-FDFT1 | Abcam | Cat#ab109723; RRID: AB_10859772 |
| Mouse monoclonal anti-U1–70k | Santa Cruz Biotechnology | Cat#sc-390899; RRID:AB_2801569 |
| Rabbit polyclonal anti-SREBP1 | Santa Cruz Biotechnology | Cat#sc-8984; RRID: AB_2194223 |
| Mouse monoclonal anti-SRSF1 | Santa Cruz Biotechnology | Cat#sc-33652; RRID: AB_628248 |
| Mouse monoclonal anti-T7 | EMD Millipore | Cat#69522; RRID:AB_11211744 |
| Mouse monoclonal anti-Phosphoepitope SR proteins | EMD Millipore | Cat#MABE50;RRID:AB_10807429 |
| Rabbit Normal IgG | Cell Signaling Technology | Cat#2729, RRID:AB_1031062 |
| Mouse Normal IgG | Thermo Scientific |
Cat#10-400-C; RRID:AB_2532980 |
| IRDye 800CW Donkey anti-Rabbit IgG | LI-COR Biosciences | Cat# 926-32213; RRID:AB_621848 |
| IRDye 680RD Donkey anti-Mouse IgG | LI-COR Biosciences | Cat# 926-68072; RRID:AB_10953628 |
| IRDye 680RD Detection Reagent | LI-COR Biosciences | Cat#:926-69100 |
| Bacterial and virus strains | ||
| E. coli BL21 | New England Biolabs | Cat#C2527 |
| Chemicals, peptides, and recombinant proteins | ||
| Rapamycin | Calbiochem | Cat#553210 |
| Torin1 | Tocris Bioscience | Cat#4247 |
| Actinomycin D | Sigma-Aldrich | Cat#A1410 |
| PhosSTOP | Roche | Cat#04906837001 |
| RNase inhibitor | Invitrogen | Cat#10777019 |
| RNase T1 | Thermo Fisher Scientific | Cat#AM2283 |
| RNase A | Thermo Fisher Scientific | Cat#12091-021 |
| Anti-V5 agarose affinity gel | Sigma-Aldrich | Cat#A7345 |
| Anti-Flag agarose affinity gel | Sigma-Aldrich | Cat# A2220 |
| Lipofectamine™ RNAiMAX Transfection Reagent | Invitrogen | Cat#13778075 |
| DNase I | Sigma-Aldrich | Cat#AMPD1 |
| TRIzol | Invitrogen | Cat#15596018 |
| Protein A/G magnetic beads | Thermo Fisher Scientific | Cat#88802 |
| Protein A/G PLUS-Agarose | Santa Cruz Biotechnology | Cat#sc-2003 |
| Proteinase K | New England Biolabs | Cat#P8107 |
| Lipofectamine 2000 reagent | Invitrogen | Cat#11668500 |
| Phenol:Chloroform:IAA, 25:24:1, pH 6.6 | Thermo Fisher Scientific | Cat#AM9730 |
| Insulin solution human | Sigma-Aldrich | Cat#I9278 |
| Crystal Violet | Sigma-Aldrich | Cat#C3886 |
| SRPIN340 | Selleck Chemicals | Cat#S7270 |
| SRPKIN-1 | MedChem Express | Cat#HY-116856 |
| Lipoprotein, low density from human plasma | Sigma-Aldrich | Cat#L7914 |
| Oleic acid-albumin | Sigma-Aldrich | Cat#O3008 |
| Fatty acid-free albumin | Sigma-Aldrich | Cat#A8806 |
| Lipoprotein-deficient serum | Alfa Aesar | Cat#BT-907 |
| Sodium palmitate | Sigma-Aldrich | Cat#P9767 |
| Methanol-free 10% formaldehyde | Polysciences | Cat#04018-1 |
| 25-Hydroxycholesterol | MedChem Express | Cat#HY-113134 |
| Critical commercial assays | ||
| PureLink RNA Mini kit | Ambion | Cat#12183018A |
| iScript cDNA synthesis kit | Bio-rad | Cat#170–8891BUN |
| Illumina® Stranded mRNA Prep Kit | Illumina | Cat#20040534 |
| Gateway LR clonase II enzyme mix | Invitrogen | Cat#11791 |
| QuickChange site-directed mutagenesis kit | Strategene | Cat#200521 |
| RNA Clean & Concentrator | Zymo Research | Cat#R1016 |
| Illumina® Stranded Total RNA Prep, Ligation with Ribo-Zero Plus | Illumina | Cat#20040529 |
| LightSwitch dual assay kit (promoter assay) | Active Motif | Cat#32035 |
| Chromatin IP DNA purification kit | Active Motif | Cat#58002 |
| SYBR Green PCR Master Mix | Life Technologies | Cat#4312704 |
| QuikChange II XL Site-Directed Mutagenesis Kit | Strategene | Cat#200521 |
| ChIP-IT Express Kit | Active Motif | Cat#53009 |
| Deposited data | ||
| Raw RNAseq data (siNTC and siFAM120A) | This paper | GSE207172 |
| Raw RNAseq data (siNTC, siFAM120, and siSRSF1) | This paper | GSE229657 |
| Raw data of Images and WB | This paper, Mendeley Data | DOI: 10.17632/rgfgx9xb8w.1 |
| Experimental models: Cell lines | ||
| Human: Renal angiomyolipoma-derived LAM 621–101 (TSC2−/−) | Drs. Jane Yu and Elizabeth Henske | Yu et al., 2004; RRID: CVCL_S879 |
| Human: HEK293E | ATCC | Cat#293 c18; RRID: CVCL_6974 |
| Human: HEK293T | GenHunter | Cat#Q401; RRID: CVCL_0063 |
| Human: MCF7 | ATCC | Cat#HTB-22; RRID: CVCL_0031 |
| Human: DLD1 | ATCC | Cat#CCL-221; RRID: CVCL_0248 |
| Human: NCI-H1299 | ATCC | Cat#CRL-5803; RRID: CVCL_0060 |
| Human: LNCaP | ATCC | Cat#CRL-1740; RRID:CVCL_1379 |
| Experimental models: Organisms/strains | ||
| nu/nu athymic mice | Envigo | Cat#069 |
| Oligonucleotides | ||
| siRNA negative control-#1(siNTC) | Sigma-Aldrich | SIC001 |
| siRNA negative control-#2(siNTC) | Sigma-Aldrich | SIC002 |
| siRNA (Human SRSF1) | Sigma-Aldrich | SASI_Hs01_00115059 |
| siRNA (Human SRSF1) | Sigma-Aldrich | SASI_Hs02_00342609 |
| siRNA (Human UPF1) | Sigma-Aldrich | SASI_Hs01_00101017 |
| siRNA (Human UPF1) | Sigma-Aldrich | SASI_Hs01_00101018 |
| siRNA (Human U1–70k/SNRNP70) | Sigma-Aldrich | SASI_Hs02_00335028 |
| siRNA (Human U1–70k/SNRNP70) | Sigma-Aldrich | SASI_Hs01_00165101 |
| siRNA (Human FAM120A) | Sigma-Aldrich | SASI_Hs01_00149752 |
| siRNA (Human FAM120A) | Sigma-Aldrich | SASI_Hs02_00345898 |
| siRNA (Human FAM120A) | Sigma-Aldrich | SASI_Hs02_00345899 |
| siRNA (Human SREBP1) | Sigma-Aldrich | SASI_Hs01_00051828 |
| siRNA (Human SREBP1) | Sigma-Aldrich | SASI_Hs01_00051829 |
| siRNA (Human SREBP1) | Sigma-Aldrich | SASI_Hs01_00051830 |
| siRNA (Human SREBP2) | Sigma-Aldrich | SASI_Hs01_00075424 |
| siRNA (Human SREBP2) | Sigma-Aldrich | SASI_Hs01_00075425 |
| siRNA (Human SREBP2) | Sigma-Aldrich | SASI_Hs01_00075426 |
| Primers for qPCR analysis, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| ACSS2 promoter construct | SwitchGear Genomics | Cat#S715263 |
| SCD1 promoter construct | SwitchGear Genomics | Cat#S714484 |
| pLX304-SRSF1-V5 | Harvard plasmid | Cat#HsCD00420441 |
| Lentiviral packaging and envelope plasmids | Dr. David Baltimore | N/A |
| pcDNA3-EGFP | Addgene | Cat#13031 |
| pKH3-HA-SRPK2-WT, K110M | Dr. John Blenis | Lee et al., 2007 |
| pLNCX-HA-SRPK2-S494A/S497A | Dr. John Blenis | Lee et al., 2007 |
| pLX304-FAM120A-V5 | Harvard plasmid | HsCD00440255 |
| pENTR1A-Flag-FAM120A(WT) | This paper | N/A |
| pENTR1A-Flag-FAM120A(∆RBD) | This paper | N/A |
| pLenti-CMV-Puro-DEST | Addgene | Cat#17452 |
| pENTR223-SRSF1 | Harvard plasmid | Cat#HsCD00367645 |
| pLenti-CMV-Puro-SRSF1 | This paper | N/A |
| pLenti-CMV-Puro-FAM120A | This paper | N/A |
| pLenti-CMV-Puro-Flag-FAM120A(WT) | This paper | N/A |
| pLenti-CMV-Puro-Flag-FAM120A(∆RBD) | This paper | N/A |
| pENTR223-FAM120A | Harvard plasmid | Cat#HsCD00438604 |
| T7-SRSF1(WT) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| T7-SRSF1(∆RRM1) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| T7-SRSF1(∆RRM2) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| T7-SRSF1(∆RS) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| pLKO.1-puro-Non-Mammalian shRNA Control | Sigma-Aldrich | SHC002 |
| pLKO.1-puro-shSRPK2 | Sigma-Aldrich | TRCN0000006274 |
| pLKO.1-puro-shFAM120A | Sigma-Aldrich | TRCN0000074918 |
| pLKO.1-puro-shSREBP1 | Sigma-Aldrich | TRCN0000055325 |
| pLKO.1-puro-shSREBP2 | Sigma-Aldrich | TRCN0000020667 |
| Software and algorithms | ||
| IRFinder-S | Lorenzi et. al., 2021 | https://europepmc.org/article/med/34749764 |
| HISAT2 v2.2.1 | Kim et. al., 2019 | https://www.nature.com/articles/s41587-019-0201-4 |
| StringTie2 v2.1.4 | Kovaka et. al., 2019 | https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1910-1 |
| DESeq2 (v1.30.1) | Love et. al., 2014 | https://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0550-8 |
| Enhanced Volcano v1.8.0 R package | Blighe, 2022 | https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html |
| Adobe Photoshop | Adobe | Adobe |
| Odyssey imaging system | LI-COR Biosciences | LI-COR Biosciences |
| enrichR v3.0 package | Webb et al., 2014 | https://git.ecdf.ed.ac.uk/sgrannem/pycrac |
| EI-MAVEN, Version 0.12.0 | Agrawal et al, 2019 | Elucidata |
| VAST-tools | Irimia et. al., 2014 | https://www.sciencedirect.com/science/article/pii/S0092867414015128?via%3Dihub |
| BioRender | BioRender | https://www.biorender.com/ |
Experimental Model and Study Participant Details
Details of cell lines used have been provided under section “Cell lines and cell culture” and details of mice used have been provided under “Mouse studies”.
Cell lines and cell culture
HEK293E and HEK293T cells were obtained from ATCC and GenHunter, respectively. Human renal angiomyolipoma-derived LAM 621–101 cell line (referred to as LAM cells in the manuscript) was provided by Dr. Henske (Harvard Medical School)33. H1299, DLD1, MCF7, and LNCaP cells were obtained from ATCC. Cells were grown in DMEM with 10% FBS at 37°C with 5% CO2 unless otherwise indicated.
Mouse studies
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at Weill Cornell Medicine. 6-week-old female athymic nu/nu mice were purchased from Envigo. 1×106 cells resuspended in 100 µl of PBS (phosphate-buffered saline): Growth Factor Reduced Matrigel (Corning) mixture (1:1) were subcutaneously inoculated into the posterior back regions of the mouse. Once the tumor is formed two to three weeks post-inoculation, tumor length and width were measured 3 times a week using a digital caliper. Tumor volume was calculated using the formula: Tumor volume = (Length × Width2)/2.
Method Details
Expression constructs
Expression constructs were transfected overnight using Lipofectamine 2000 (Invitrogen). pENTR223-FAM120A and pENTR223-SRSF1 were obtained from Harvard PlasmID. pENTR223-FAM120A and pENTR223-SRSF1 recombined with plx304-V5-DEST or pLenti-puromycin-DEST vector by LR reaction (Gateway LR clonase II enzyme mix, Invitrogen) to generate expression constructs. To generate FAM120A RNA deletion mutant construct, the coding sequence of human FAM120A with or without RNA binding domain (NM_014612.5) was cloned into pENTR1A vector (NotI and SalI). The SRSF1 WT and mutant constructs were a gift from Dr. Krainer (Cold Spring Harbor Laboratory)78.
siRNA oligonucleotides
30 nM siRNA (Sigma-Aldrich; see list in Key Resources Table) was transfected using Lipofectamine RNAiMAX reagent (Invitrogen).
Generation of stable cell lines
Lipofectamine 2000 reagent (Invitrogen) was used to co-transfect with the viral plasmid of interest with lentiviral packaging and envelope plasmids in HEK293T cells to generate lentivirus. After 60 hr of transfection, virus-containing supernatant was collected and treated to the cells with 8 µg/mL polybrene overnight. Cells infected with pLKO.1-puromycin shRNA constructs (Sigma-Aldrich; see list in Key Resources Table) or pLenti-puromycin infected cells were treated with 2 µg/mL (H1299, DLD1, MCF7, and LNCaP) or 10 µg/mL (LAM) puromycin for 2–3 days to select the infected cells. pLenti-plx304-V5 infected cells were selected with 10 µg/mL (H1299, DLD1, MCF7, and LNCaP) or 30 µg/mL (LAM) blasticidin. See construct list in Key Resources Table.
Crystal violet staining
4% methanol-free formaldehyde (Polysis) was used to fix the cells for 10 min and incubated with 0.1% crystal violet solution (Sigma-Aldrich) for 20 min at room temperature. The plates were washed with PBS 5 times, and the plates were scanned for image analysis after overnight drying. To quantify the staining intensity, crystal violet was eluted by incubating the cells with methanol for 20 min. Absorbance was measured at 570 nm (Envision plate reader, PerkinElmer).
Immunoblot analysis
After 2 times of cold PBS wash, cells were incubated on ice for 30 min with a RIPA (40 mM HEPES [pH7.4], 1 mM EDTA, 120 mM NaCl, 0.5 mM DTT, 0.1% Brij-35, 0.1% deoxycholate, and 0.5% NP-40) lysis buffer supplemented with protease inhibitors (250 μM PMSF, 5 μg/mL pepstatin A, 10 μg/mL leupeptin, and 5 μg/mL aprotinin) and phosphatase inhibitors (10 mM β-glycerophosphate, 1 mM NaF, and 1 mM Na3VO4) to lysis cells. Soluble cell lysates were obtained after centrifugation at 21,130 x g at 4°C for 30 min to clear the lysates. Protein concentration was measured with detergent compatible (DC) protein assay (Bio-rad) and the same amounts of proteins were boiled for 10 min with Laemmli sample buffer to denature the protein structure. Equal amounts of proteins (15–50 μg) were separated by SDS‐PAGE gels and the proteins were transferred to nitrocellulose membranes (Amersham Biosciences). The transferred nitrocellulose membrane was blocked with Odyssey blocking solution (LI-COR Biosciences) for an hour at room temperature, and further incubated with primary and IRDye secondary antibodies (see antibody list on Key Resources Table). Odyssey imaging system (LI-COR Biosciences) was used to detect and quantify immunoblot signals. Immunoblot images show the representative images of at least two independent experiments.
Quantitative RT-PCR (qPCR)
Total RNA was harvested using PureLink RNA isolation kit (Life Technologies) according to the manufacturer’s protocol. DNase I (Sigma-Aldrich) was treated to eliminate genomic DNA. cDNA was synthesized from total RNA by reverse-transcription reaction using iScript (Bio-rad). QuantStudio6 Real-Time PCR system (Life Technologies) was used to analyze the resulting cDNA using SYBR green master mix (Life Technologies). Delta-delta CT method was used to calculate relative mRNA levels by normalization of housekeeping genes ACTIN, GAPDH, PPIB, and TBP as controls. The primer list is in Supplemental Table S2.
mRNA stability assay
Actinomycin D (5 µg/mL) was treated to cells for the indicated time points. Total RNA was extracted using Purelink RNA miniprep kit and mRNA levels were analyzed by qPCR.
Luciferase promoter activity assay
LightSwitch dual assay system was used according to the manufacturer’s protocol (SwitchGear Genomics). Cells plated on a 6-well plate were transfected with siRNAs targeting FAM120A or non-targeting control through Lipofectamine RNAiMax transfection reagent. After 2 days, the cells were co-transfected with 1 μg of renillia construct containing the promoter of the interested gene and 0.2 μg of control cypridina construct using FuGENE HD transfection reagent (Promega). 24 hr after transfection, the activity of renilla and cypridina luciferases was measured (Envision plate reader, PerkinElmer). For calculation of the promoter activities, the renilla luciferase activity was normalized by the cypridina luciferase activity.
RNA immunoprecipitation qPCR (RIP-qPCR)
Cells on a 15-cm plate were washed twice with ice-cold PBS and harvested into 15 mL tube. Cells were collected by centrifugation at 1,000 x g at 4 °C for 5 min. Cell pellets were resuspended with 1 ml lysis buffer (1 mM EDTA [pH8.0], 50 mM HEPES-KOH [pH7.5], 140 mM NaCl, 0.1% Sodium deoxycholate, 1% Triton X-100) supplemented with protease inhibitors (250 μM PMSF, 5 μg/mL pepstatin A, 10 μg/mL leupeptin, 5 μg/mL aprotinin) and phosphatase inhibitors (10 mM β-glycerophosphate, 1 mM NaF, 1 mM Na3VO4), and RNase inhibitor (160 unit/mL, Invitrogen), and incubated on ice for 30 min. After centrifugation at 21,130 x g at 4°C for 30 min, the supernatant was pre-cleared by incubating with 50 µL protein A/G beads. The pre-cleared lysate was incubated with 5 µg of IgG (Mouse: Thermo Scientific, Rabbit: Cell Signaling Technology), anti-SRSF1, or anti-FAM120A antibody (Bethyl Laboratories) overnight at 4°C. Then, 50 µL protein A/G magnetic beads were added to the lysate and incubated at 4°C for 1.5 hr to pull down antibody-associated complexes. The beads were washed 3 times with lysis buffer and RNA was eluted by incubating with 150 µL lysis buffer containing 1% SDS and 1.2 mg/mL proteinase K (New England Biolabs) at 55°C for 30 min. The RNA elute was purified by phenonol/chroloform extraction and precipitated with ammonium acetate and ethanol. Input and immunoprecipitated RNAs were treated with DNase I (Sigma-Aldrich) and reverse-transcribed (iScript cDNA synthesis kit, Bio-rad), and the resulting cDNA was analyzed by qPCR. The amount of transcripts (%) bound to the antibody was calculated: 100 × 2^[Ct(input) - Ct(IP)].
Co-immunoprecipitation of proteins (Co-IP)
Cells on a 10-cm plate were harvested through centrifugation at 1,000 x g at 4°C for 5 min after three times washing with ice-cold PBS. One mL of lysis buffer (20 mM Tris HCl [pH8.0], 137 mM NaCl, 1% Nonidet P-40, 2 mM EDTA) supplemented with protease inhibitors (250 μM PMSF, 5 μg/mL pepstatin A, 10 μg/mL leupeptin, 5 μg/mL aprotinin) and phosphatase inhibitors (10 mM β-glycerophosphate, 1 mM NaF, 1 mM Na3VO4) was added, and incubated on ice for 30 min to lyse the cells with mechanical disruption by syringe homogenization. After centrifugation at 21,130 x g at 4°C for 30 min, protein concentration was measured, and 1 mg of lysates were incubated with anti-V5 (Sigma-Aldrich) or anti-Flag agarose affinity gels (Sigma-Aldrich) at 4°C for 1.5 h with gentle agitation to pull down immune complexes. For endogenous IP, 1mg of lysates were incubated with 4 µg of anti-FAM120A (Thermo Fisher Scientific) or rabbit IgG (Cell Signaling Technology) for overnight at 4°C h with gentle agitation to pull down immune complexes. 50 µL protein A/G beads were added to the lysate and incubated at 4°C with gentle agitation for 1.5 hr to pull down antibody-associated complexes. The immunoprecipitated beads were then washed 3 times with lysis buffer to remove non-specific binding and the immune complexes were eluted by boiling with 2X Laemmli sample buffer for 5 min. The boiled beads were centrifuged at 845 x g for 5 min and supernatants were subjected to immunoblot analysis.
Ribonuclease (RNase) treatment
Cells on a 10-cm plate were washed twice with ice-cold PBS and harvested by centrifugation at 1,000 x g at 4°C for 5 min. Cells were lysed and physically disrupted by syringe homogenization in one mL of lysis buffer (20 mM Tris HCl [pH8.0], 137 mM NaCl, 1% Nonidet P-40, 2 mM EDTA) supplemented with protease inhibitors (250 μM PMSF, 5 μg/mL pepstatin A, 10 μg/mL leupeptin, 5 μg/mL aprotinin) and phosphatase inhibitors (10 mM β-glycerophosphate, 1 mM NaF, 1 mM Na3VO4), and incubated on ice for 30 minutes. After centrifugation at 21,130 x g at 4°C for 30 min, the clarified lysate was treated with a mixture of RNase A (100 μg/ml) (Invitrongen) and RNase T1 (1 unit/µL) (Invitrongen) for 1 hr at room temperature prior to co-IP analysis.
Chromatin immunoprecipitation qPCR (ChIP-qPCR)
Cells were incubated with 1% formaldehyde to fix the cells at room temperature for 10 min followed by quenching with 0.125 M glycine for 5 min at room temperature. ChIP assays were performed using a commercially available kit (ChIP-IT Express, Active Motif). First, lysis buffer was used to lyse the cells using a Dounce homogenizer to disrupt cells for harvesting chromatin. Isolated chromatin was sheared by an enzyme for 15 min at 37°C (ChIP-IT Express Enzymatic Shearing Kit, Active Motif). Immunoprecipitation was performed using anti-SRSF1 (32–4500, Invitrogen) or anti-SREBP1 (sc-13551, Santa Cruz) antibodies. Elution buffer containing SDS buffer was used to elute complex from the beads and incubated with RNase and proteinase K after several washes. Reverse crosslinking was performed by incubation overnight at 65°C, and a Chromatin IP DNA purification kit (Active Motif) was used to purify ChIP DNA. QuantStudio6 Real-Time PCR system (Life Technologies) was used to analyze the resulting DNA samples by qPCR. The amount of DNA (%) bound to the antibody was calculated: 100 × 2^[Ct(input) - Ct(IP)].
Lipid supplementation
To generate albumin conjugated palmitate, 10 mL of sodium palmitate (10 mM) was mixed with 10 mL of fatty acid-free BSA (1.66 mM BSA solution) and incubated for 1 hr at 70°C. Cells stably expressing shRNAs targeting FAM120A or control were cultured in DMEM with 10% lipoprotein-deficient serum (LPDS) supplemented with lipoprotein (25 μg/mL), oleate-albumin (50 μM; oleate), palmitate-albumin (10 μM; palmitate), or fatty acid-free albumin (25 μM; control) to the media.
Mass spectrometry analysis of SRSF1 interactome
We previously reported the SRSF1 interactome using reductive dimethylation isotopic labeling to provide quantitative comparisons between treated (torin1) and untreated cells12. Those data were reanalyzed at the individual peptide level for the bait protein, SRSF1-V5, and FAM120A. Data were manually extracted from each of two replicate analyses for all quantified peptides. A stringent signal-to-noise (SN) filter was applied, retaining only peptide quantifications where the sum SN of treated and untreated samples was >100. Figure 1A shows the log2[+TORIN/DMSO] for all passing peptides for each protein. Statistical analysis was performed in Prism using unpaired t-tests.
Mass spectrometry analysis of saponified fatty acids
90% confluent cells on 60 mm cell culture dish were incubated with 0.5 ml of 0.3 M KOH in 90% methanol at 80°C for 1 hr in a 2 mL glass vial. Then, formic acid (100 μL) was added for neutralization. The saponified fatty acids were extracted by adding 1 mL of hexane. After vortexing, the top hexane layer was transferred to a new glass vial. Samples were then dried under a nitrogen gas stream and redissolved in 100 μL of 1:1 isopropanol:methanol for LC-MS analysis. Fatty acids were detected with a quadrupole orbitrap mass spectrometer (Thermo Fisher Q-Exactive) operating in a negative ion mode with electrospray ionization and used to scan from m/z 200 to 600 at 1 Hz, with a 140,000 resolution. LC separation was achieved on a Poroshell 120 EC-C18 column (2.1 × 150 mm2, 2.7 μm particle size; Agilent) using a gradient of solvent A (90:10 = water: methanol with 1 mM of ammonium acetate and 0.2% acetic acid) and solvent B (90:10 = methanol:isopropanol with 1 mM of ammonium acetate and 0.2% acetic acid). The LC gradient was 0 min, 25% B; 2 min, 25% B; 4 min, 65% B; 16 min, 100% B; 20 min, 100% B; 21 min, 25% B; 22 min, 25% B; and 25 min, 25% B. Flow rate was 150 μl/min. The autosampler temperature was 5°C and the injection volume was 3 μL. Mass spectrometry data was analyzed using the EI-MAVEN open source software 79 (Version 0.12.0, Elucidata). Briefly, LC-MS data was converted from .raw files to .mzXML and then loaded into the EI-MAVEN software. Using our annotated MS library, containing retention times and m/z values, we selected metabolite peaks of interest on EI-MAVEN. Peak shape and quality were evaluated and Area Top of the metabolites of interest were recorded.
RNA-seq sample preparation
For GSE207172, we used 100 ng of high-quality total RNA and the Illumina® TruSeq Stranded Total RNA with Ribo-Zero Globin kit (Cat# 20040529) from Illumina. For GSE229657, the Illumina® Stranded mRNA Prep Kit (Cat# 20040534) and 100 ng of high-quality total RNA were used. RNA-seq library was prepared by Weill Cornell Medicine Core facility for the Illumina NovaSeq 6000 system sequencer at paired-end 2×100 cycles.
RNA-seq and pathway enrichment analyses
RNA-seq in Figure 2 was performed by using the Illumina NovaSeq 6000 system with 40M 2×101 bp paired-end (PE) read. Using the HISAT2 v2.2.1 software80, the sequencing reads were mapped onto the human GRCh38/hg38 reference genome, from which the counts matrix was generated with the default parameters using StringTie2 v2.1.481. Differentially expressed genes (DEGs) were identified using the DESeq2 (v1.30.1)82 based on a negative binomial generalized linear model. The data visualization was performed by using the EnhancedVolcano v1.8.0 R package83. DEGs that satisfy |(Log2FC)| > 1 and adjusted p-value (q-value) < 0.005 were considered as statistically significant. Pathway enrichment was measured by Fisher’s exact test against Elsevier pathway collection using the enrichR v3.0 package84. RNA-seq data files are accessible through Gene Expression Omnibus (GEO) accession number GSE207172.
In order to detect intron retention events more precisely, we did deep RNA sequencing in Figure 3 using the Illumina NovaSeq 6000 system with 100M 2×101 bp PE read. Analysis of deep RNA-seq was performed as previously described85. In brief, the quality of RNA-seq reads were checked using FastQC and the reads were aligned to the human GRCh38/hg38 reference genome using STAR (2.7.10b) with following parameters: --outFilterMismatchNmax 2-- outFilterMultimapNmax 10 --outSJfilterOverhangMin 30 888 --outSAMunmapped Within KeepPairs. Genome coverage files (bigwigs) were generated using bam2wig.py from RSeQC (v5.0.1) with following parameter: “1+-,1-+,2++,2--” -t 1000000000. Differential gene expression analysis was done by extracting counts using featureCounts (v2.0.3) and running DESeq2 (v1.34.0) to find differentially expressed genes with the cutoff of q-value < 0.01 and |(Log2FC)| ≥ 0.5. Pathway analyses were performed using ConsensusPathDB86 (KEGG, Reactome, Wikipathways, Biocarata, Ehmn, Humancyc and Inoh) and only pathways with q-value < 0.05 were considered. RNA-seq data files dataset generated in this study is accessible through Gene Expression Omnibus (GEO) accession number GSE229657.
Intron retention analysis
To examine the global splicing changes, raw fastq files from deep RNA-sequencing data were aligned to VAST-tools library (human vastdb.hs2.23.06.20) using VAST-tools (2.5.1) and splicing events were detected using VAST-tools tidy with following parameters: --min N 2--p_IR 0.1 -- min_SD 139. To precisely determine the intron retention events, we performed IRFinder-S (2.0.1) using BAM files from the STAR alignment step and calculated intron retention events with following parameters: -wl 1 -ir 0.01. Significantly increased intron retention events were selected with the cutoff of q-value < 0.1 and Log2FC > 040.
Analysis of published ChIP-seq and whole transcriptome microarray results
Previously published ChIP-seq dataset GSE4551746 and whole transcriptome microarray dataset GSE10433512 were obtained from GEO dataset.
Quantification and Statistical Analysis
LC-MS, qPCR, interactome, immunoblot, and crystal violet staining assays data are statistically analyzed using Student’s t-test and the graphs show mean ± SD. Tumor burdens from the xenograft experiment are statistically analyzed using Student’s t-test and the error bars show mean ± SEM. Detailed methods and p-value are described in the Figure Legends and Methods sections.
Supplementary Material
Excel spreadsheet: Supplemental Table S1. RNA-seq analysis of siFAM120A and siSRSF1 (Related to Figure 3).
(A) The alignment reads of RNA-seq data from LAM cells transfected with siRNAs targeting FAM120A, SRSF1, or control (NTC). Cells were serum-starved overnight before analysis. N = 3.
(B) List of 1,585 genes downregulated by both siFAM120A and siSRSF1 compared to siNTC.
(C) Pathway analysis for the commonly downregulated genes by siFAM120A and siSRSF1.
(D) List of 408 intron retention events identified in both siFAM120A and siSRSF1.
(E) Pathway analysis for the genes that show increased intron retentions and decreased gene expression by both siFAM120A and siSRSF1.
Excel spreadsheet: Supplemental Table S2. Primer list (Related to all Figures).
(A) Primers used for mRNA expression level test.
(B) Primers used for intron retention analysis.
(C) Primers used for RNA-IP qPCR experiments.
(D) Primers used for ChIP-qPCR experiments.
Highlights.
FAM120A couples transcription and splicing of lipogenic enzymes downstream of mTORC1
mTORC1-activated SRPK2 phosphorylates SRSF1 to induce FAM120-SRSF1 interaction
FAM120A directly interact with SREBP to bring lipogenic genes to RNA splicing machinery
mTORC1-driven RNA binding protein complex promotes lipogenic gene splicing and stability
ACKNOWLEDGEMENTS
We thank members of the Blenis and Lee laboratories for technical assistance and discussions. We are grateful to Drs. Elizabeth Henske, Adrian Krainer, and Yongsheng Shi for sharing reagents and scientific discussion. This research was supported by the following grants: NIH R01GM051405 (J.B.), R01CA046595 (J.B.), R01AA029124 (C.J.), K22CA234399 (G.L.), DOD TS200022 (G.L.), Mary Kay Ash Foundation (G.L.), and STARR Foundation (J.B). This work was also made possible in part through access to CFCCC GRTH core (P30CA062203). C.B.R. was supported by predoctoral fellowships from UCI IMSD (R25GM055246) and IDCR (T32CA009054) programs. Schematics are created using BioRender.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
J.B. is an advisory board member for Molecular Cell.
REFERENCES
- 1.Saxton RA, and Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell 169, 361–371. 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 2.Shimobayashi M, and Hall MN (2014). Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol 15, 155–162. 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, and Manning BD (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733. 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dibble CC, and Manning BD (2013). Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol 15, 555–564. 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J, Li E, Chen L, Zhang Y, Wei F, Liu J, Deng H, and Wang Y (2015). The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 524, 243–246. 10.1038/nature14557. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Brown MS, and Goldstein JL (2010). Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A 107, 3441–3446. 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen JL, Zhang Y, Bae S-H, Farooqi MS, Liang G, Hammer RE, Goldstein JL, and Brown MS (2012). Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. U.S.A 109, 16184–16189. 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. (2011). mTOR Complex 1 Regulates Lipin 1 Localization to Control the SREBP Pathway. Cell 146, 408–420. 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Nicholatos J, Dreier JR, Ricoult SJH, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, and Manning BD (2014). Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513, 440–443. 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. (2010). Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183. 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heintz C, Doktor TK, Lanjuin A, Escoubas C, Zhang Y, Weir HJ, Dutta S, Silva-García CG, Bruun GH, Morantte I, et al. (2017). Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature 541, 102–106. 10.1038/nature20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, et al. (2017). Post-transcriptional Regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 Signaling. Cell 171, 1545–1558.e18. 10.1016/j.cell.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, and Dikstein R (2015). Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21, 479–492. 10.1016/j.cmet.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Tang H-W, Hu Y, Chen C-L, Xia B, Zirin J, Yuan M, Asara JM, Rabinow L, and Perrimon N (2018). The TORC1-Regulated CPA Complex Rewires an RNA Processing Network to Drive Autophagy and Metabolic Reprogramming. Cell Metab 27, 1040–1054.e8. 10.1016/j.cmet.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho S, Hoang A, Sinha R, Zhong X-Y, Fu X-D, Krainer AR, and Ghosh G (2011). Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A 108, 8233–8238. 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, and Manley JL (1994). Protein–protein interactions and 5’-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124. 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 17.Wang H-Y, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, and Fu XD (1998). SRPK2: A Differentially Expressed SR Protein-specific Kinase Involved in Mediating the Interaction and Localization of Pre-mRNA Splicing Factors in Mammalian Cells. J Cell Biol 140, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das R, Dufu K, Romney B, Feldt M, Elenko M, and Reed R (2006). Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20, 1100–1109. 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, and Reed R (2007). SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26, 867–881. 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 20.de la Mata M, and Kornblihtt AR (2006). RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol 13, 973–980. 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 21.Sapra AK, Ankö M-L, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland E-M, and Neugebauer KM (2009). SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell 34, 179–190. 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 22.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, and Spiegelman BM (2000). Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6, 307–316. 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y-IG, Moore RE, Ge HY, Young MK, Lee TD, and Stevens SW (2007). Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res 35, 3928–3944. 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Möller A, Xie SQ, Hosp F, Lang B, Phatnani HP, James S, Ramirez F, Collin GB, Naggert JK, Babu MM, et al. (2012). Proteomic analysis of mitotic RNA polymerase II reveals novel interactors and association with proteins dysfunctional in disease. Mol Cell Proteomics 11, M111.011767. 10.1074/mcp.M111.011767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rappsilber J, Ryder U, Lamond AI, and Mann M (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res 12, 1231–1245. 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, and Black DL (2008). Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat. Struct. Mol. Biol 15, 183–191. 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S, and Krainer AR (2014). Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 12, 1195–1204. 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, and Fu X-D (2013). Regulation of Splicing by SR proteins and SR Protein-Specific Kinases. Chromosoma 122, 191–207. 10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao SH, and Manley JL (1997). Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev 11, 334–344. 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 30.Yeakley JM, Tronchère H, Olesen J, Dyck JA, Wang H-Y, and Fu X-D (1999). Phosphorylation Regulates In Vivo Interaction and Molecular Targeting of Serine/Arginine-rich Pre-mRNA Splicing Factors. Journal of Cell Biology 145, 447–455. 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, et al. (2006). Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proceedings of the National Academy of Sciences 103, 11329–11333. 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatcher JM, Wu G, Zeng C, Zhu J, Meng F, Patel S, Wang W, Ficarro SB, Leggett AL, Powell CE, et al. (2018). SRPKIN-1: A Covalent SRPK1/2 Inhibitor that Potently Converts VEGF from Pro-angiogenic to Anti-angiogenic Isoform. Cell Chem Biol 25, 460–470.e6. 10.1016/j.chembiol.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Astrinidis A, Howard S, and Henske EP (2004). Estradiol and tamoxifen stimulate LAM-associated angiomyolipoma cell growth and activate both genomic and nongenomic signaling pathways. Am. J. Physiol. Lung Cell Mol. Physiol 286, L694–700. 10.1152/ajplung.00204.2003. [DOI] [PubMed] [Google Scholar]
- 34.Krall EB, Wang B, Munoz DM, Ilic N, Raghavan S, Niederst MJ, Yu K, Ruddy DA, Aguirre AJ, Kim JW, et al. (2017). KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife 6, e18970. 10.7554/eLife.18970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J, Lee I, Han B, Park JO, Jang J, Park C, and Kang WK (2011). Effect of simvastatin on cetuximab resistance in human colorectal cancer with KRAS mutations. J Natl Cancer Inst 103, 674–688. 10.1093/jnci/djr070. [DOI] [PubMed] [Google Scholar]
- 36.Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, et al. (2005). Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res 7, R609–616. 10.1186/bcr1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. (2011). Reciprocal feedback regulation of I3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586. 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buj R, Chen C-W, Dahl ES, Leon KE, Kuskovsky R, Maglakelidze N, Navaratnarajah M, Zhang G, Doan MT, Jiang H, et al. (2019). Suppression of p16 Induces mTORC1-Mediated Nucleotide Metabolic Reprogramming. Cell Reports 28, 1971–1980.e8. 10.1016/j.celrep.2019.07.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irimia M, Weatheritt RJ, Ellis J, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O’Hanlon D, et al. (2014). A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523. 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenzi C, Barriere S, Arnold K, Luco RF, Oldfield AJ, and Ritchie W (2021). IRFinder-S: a comprehensive suite to discover and explore intron retention. Genome Biol 22, 307. 10.1186/s13059-021-02515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang Y-F, Imam JS, and Wilkinson MF (2007). The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76, 51–74. 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 42.Isken O, and Maquat LE (2008). The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet 9, 699–712. 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tani H, and Akimitsu N (2012). Genome-wide technology for determining RNA stability in mammalian cells: historical perspective and recent advantages based on modified nucleotide labeling. RNA Biol 9, 1233–1238. 10.4161/rna.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holden S, and Raymond FL (2003). The human gene CXorf17 encodes a member of a novel family of putative transmembrane proteins: cDNA cloning and characterization of CXorf17 and its mouse ortholog orf34. Gene 318, 149–161. 10.1016/s0378-1119(03)00770-4. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka M, Sasaki K, Kamata R, Hoshino Y, Yanagihara K, and Sakai R (2009). A novel RNA-binding protein, Ossa/C9orf10, regulates activity of Src kinases to protect cells from oxidative stress-induced apoptosis. Mol. Cell. Biol 29, 402–413. 10.1128/MCB.01035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, and Fu XD (2013). SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 153, 855–868. 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai MC, Teh BH, and Tarn WY (1999). A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J Biol Chem 274, 11832–11841. 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- 48.Pandit S, Wang D, and Fu X-D (2008). Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol 20, 260–265. 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horton JD, Goldstein JL, and Brown MS (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest 109, 1125–1131. 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, and Goldstein JL (2004). Cholesterol and 25-Hydroxycholesterol Inhibit Activation of SREBPs by Different Mechanisms, Both Involving SCAP and Insigs*. Journal of Biological Chemistry 279, 52772–52780. 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 51.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, and Rabinowitz JD (2013). Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A 110, 8882–8887. 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Menendez JA, and Lupu R (2007). Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7, 763–777. 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 53.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, Konisti S, Peck B, Miess H, East P, et al. (2013). Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab 1, 3. 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee Y, and Rio DC (2015). Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem 84, 291–323. 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu X-D, and Ares M (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15, 689–701. 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao SH, and Manley JL (1998). Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 17, 6359–6367. 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwak H, and Lis JT (2013). Control of transcriptional elongation. Annu Rev Genet 47, 483–508. 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.RNA Polymerase II Elongation Control | Annual Review of Biochemistry 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders A, Core LJ, and Lis JT (2006). Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7, 557–567. 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 60.Lam HC, Siroky BJ, and Henske EP (2018). Renal disease in tuberous sclerosis complex: pathogenesis and therapy. Nat Rev Nephrol 14, 704–716. 10.1038/s41581-018-0059-6. [DOI] [PubMed] [Google Scholar]
- 61.Mossmann D, Park S, and Hall MN (2018). mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757. 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 62.Soefje SA, Karnad A, and Brenner AJ (2011). Common toxicities of mammalian target of rapamycin inhibitors. Targ Oncol 6, 125–129. 10.1007/s11523-011-0174-9. [DOI] [PubMed] [Google Scholar]
- 63.Lee SC-W, and Abdel-Wahab O (2016). Therapeutic targeting of splicing in cancer. Nat Med 22, 976–986. 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujita K-I, Ishizuka T, Mitsukawa M, Kurata M, and Masuda S (2020). Regulating Divergent Transcriptomes through mRNA Splicing and Its Modulation Using Various Small Compounds. Int J Mol Sci 21, E2026. 10.3390/ijms21062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong DS, Kurzrock R, Naing A, Wheler JJ, Falchook GS, Schiffman JS, Faulkner N, Pilat MJ, O’Brien J, and LoRusso P (2014). A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs 32, 436–444. 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 66.Falchook G, Infante J, Arkenau H-T, Patel MR, Dean E, Borazanci E, Brenner A, Cook N, Lopez J, Pant S, et al. (2021). First-in-human study of the safety, pharmacokinetics, and pharmacodynamics of first-in-class fatty acid synthase inhibitor TVB-2640 alone and with a taxane in advanced tumors. EClinicalMedicine 34, 100797. 10.1016/j.eclinm.2021.100797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koundouros N, and Poulogiannis G (2020). Reprogramming of fatty acid metabolism in cancer. Br J Cancer 122, 4–22. 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fhu CW, and Ali A (2020). Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 25, 3935. 10.3390/molecules25173935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts TC, Langer R, and Wood MJA (2020). Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 19, 673–694. 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang L, Rowe RG, Jaimes A, Yu C, Nam Y, Pearson DS, Zhang J, Xie X, Marion W, Heffron GJ, et al. (2018). Small-Molecule Inhibitors Disrupt let-7 Oligouridylation and Release the Selective Blockade of let-7 Processing by LIN28. Cell Rep 23, 3091–3101. 10.1016/j.celrep.2018.04.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghidini A, Cléry A, Halloy F, Allain FHT, and Hall J (2021). RNA-PROTACs: Degraders of RNA-Binding Proteins. Angew Chem Int Ed Engl 60, 3163–3169. 10.1002/anie.202012330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Interferon-Stimulated Genes: What Do They All Do? | Annual Review of Virology 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 73. Transcriptional Regulation of Antiviral Interferon-Stimulated Genes - PMC. doi: 10.1016/j.tim.2017.01.001. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7127685/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. NLRC5: a key regulator of MHC class I-dependent immune responses | Nature Reviews Immunology. doi: 10.1038/nri3339. https://www.nature.com/articles/nri3339. [DOI] [PubMed] [Google Scholar]
- 75.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, Cucolo L, Lee DSM, Pauken KE, Huang AC, et al. (2016). Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 167, 1540–1554.e12. 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minn AJ, and Wherry EJ (2016). Combination Cancer Therapies with Immune Checkpoint Blockade: Convergence on Interferon Signaling. Cell 165, 272–275. 10.1016/j.cell.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 77.López-Soto A, Gonzalez S, and Folgueras AR (2017). IFN Signaling and ICB Resistance: Time is on Tumor’s Side. Trends Cancer 3, 161–163. 10.1016/j.trecan.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 78.Fregoso OI, Das S, Akerman M, and Krainer AR (2013). Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Mol Cell 50, 56–66. 10.1016/j.molcel.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agrawal S, Kumar S, Sehgal R, George S, Gupta R, Poddar S, Jha A, and Pathak S (2019). El-MAVEN: A Fast, Robust, and User-Friendly Mass Spectrometry Data Processing Engine for Metabolomics. In High-Throughput Metabolomics: Methods and Protocols Methods in Molecular Biology, D’Alessandro A, ed. (Springer; ), pp. 301–321. 10.1007/978-1-4939-9236-2_19. [DOI] [PubMed] [Google Scholar]
- 80.Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, and Pertea M (2019). Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biology 20, 278. 10.1186/s13059-019-1910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blighe K (2022). EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling [Google Scholar]
- 84.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research 44, W90–W97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mauger O, Lemoine F, and Scheiffele P (2016). Targeted Intron Retention and Excision for Rapid Gene Regulation in Response to Neuronal Activity. Neuron 92, 1266–1278. 10.1016/j.neuron.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 86.Kamburov A, Wierling C, Lehrach H, and Herwig R (2009). ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res 37, D623–D628. 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel spreadsheet: Supplemental Table S1. RNA-seq analysis of siFAM120A and siSRSF1 (Related to Figure 3).
(A) The alignment reads of RNA-seq data from LAM cells transfected with siRNAs targeting FAM120A, SRSF1, or control (NTC). Cells were serum-starved overnight before analysis. N = 3.
(B) List of 1,585 genes downregulated by both siFAM120A and siSRSF1 compared to siNTC.
(C) Pathway analysis for the commonly downregulated genes by siFAM120A and siSRSF1.
(D) List of 408 intron retention events identified in both siFAM120A and siSRSF1.
(E) Pathway analysis for the genes that show increased intron retentions and decreased gene expression by both siFAM120A and siSRSF1.
Excel spreadsheet: Supplemental Table S2. Primer list (Related to all Figures).
(A) Primers used for mRNA expression level test.
(B) Primers used for intron retention analysis.
(C) Primers used for RNA-IP qPCR experiments.
(D) Primers used for ChIP-qPCR experiments.
Data Availability Statement
Sequencing data have been deposited at GEO with identifier GSE207172 and GSE229657. The original images have been deposited at Mendeley Data (DOI: 10.17632/rgfgx9xb8w.1). All data are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-SRPK2 | BD Biosciences | Cat#BD611118; RRID: AB_398429 |
| Rabbit polyclonal anti-ACLY | Cell Signaling Technology | Cat#4332; RRID: AB_2223744 |
| Rabbit monoclonal anti-FASN | Cell Signaling Technology | Cat#3180; RRID: AB_2100796 |
| Mouse monoclonal anti-S6 | Cell Signaling Technology | Cat#2317; RRID: AB_2238583 |
| Rabbit monoclonal anti-pS6(S240/S244) | Cell Signaling Technology | Cat#5364; RRID: AB_10694233 |
| Rabbit polyclonal anti-SCD1 | Cell Signaling Technology | Cat#2438; RRID: AB_823634 |
| Rabbit polyclonal anti-CHREBP | Novus | Cat# NB400-135;RRID:AB_10002435 |
| Rabbit polyclonal anti-FAM120A | GeneTex | Cat#GTX120824; RRID:AB_10723014 |
| Rabbit polyclonal anti-FAM120A | Thermo Fisher Scientific | Cat# A303-889A; RRID:AB_2620239) |
| Rabbit polyclonal anti-LXR alpha | Thermo Fisher Scientific | Cat#PA1-330;RRID:AB_325600 |
| Mouse monoclonal anti-SREBP2 | BD Biosciences | Cat#557037; RRID:AB_396560 |
| Rabbit polyclonal anti-LXR beta | Thermo Fisher Scientific | Cat#PA1-333;RRID:AB_2154770 |
| Rabbit polyclonal anti-PPAR alpha | Thermo Fisher Scientific | Cat#PA1-822A; RRID:AB_2165595 |
| Rabbit polyclonal anti-PPAR gamma | Abcam | Cat#ab209350; RRID:AB_2890099 |
| Rabbit monoclonal anti-p300 | Cell Signaling Technology | Cat#54062; RRID:AB_2799450 |
| Rabbit monoclonal anti-XBP-1s | Cell Signaling Technology | Cat#12782;RRID:AB_2687943 |
| Mouse monoclonal anti-GAPDH | Sigma-Aldrich | Cat#G8795; RRID: AB_1078991 |
| Rabbit polyclonal anti-ACSS2 | Sigma-Aldrich | Cat#HPA004141; RRID: AB_1078094 |
| Mouse monoclonal anti-V5 | Sigma-Aldrich | Cat#V8012; RRID:AB_261888 |
| Mouse monoclonal anti-Vinculin | Sigma-Aldrich | Cat#V9264; RRID: AB_10603627 |
| Rabbit polyclonal anti-RNA polymerase II | Abcam | Cat#ab26721; RRID:AB_777726 |
| Rabbit monoclonal anti-FDFT1 | Abcam | Cat#ab109723; RRID: AB_10859772 |
| Mouse monoclonal anti-U1–70k | Santa Cruz Biotechnology | Cat#sc-390899; RRID:AB_2801569 |
| Rabbit polyclonal anti-SREBP1 | Santa Cruz Biotechnology | Cat#sc-8984; RRID: AB_2194223 |
| Mouse monoclonal anti-SRSF1 | Santa Cruz Biotechnology | Cat#sc-33652; RRID: AB_628248 |
| Mouse monoclonal anti-T7 | EMD Millipore | Cat#69522; RRID:AB_11211744 |
| Mouse monoclonal anti-Phosphoepitope SR proteins | EMD Millipore | Cat#MABE50;RRID:AB_10807429 |
| Rabbit Normal IgG | Cell Signaling Technology | Cat#2729, RRID:AB_1031062 |
| Mouse Normal IgG | Thermo Scientific |
Cat#10-400-C; RRID:AB_2532980 |
| IRDye 800CW Donkey anti-Rabbit IgG | LI-COR Biosciences | Cat# 926-32213; RRID:AB_621848 |
| IRDye 680RD Donkey anti-Mouse IgG | LI-COR Biosciences | Cat# 926-68072; RRID:AB_10953628 |
| IRDye 680RD Detection Reagent | LI-COR Biosciences | Cat#:926-69100 |
| Bacterial and virus strains | ||
| E. coli BL21 | New England Biolabs | Cat#C2527 |
| Chemicals, peptides, and recombinant proteins | ||
| Rapamycin | Calbiochem | Cat#553210 |
| Torin1 | Tocris Bioscience | Cat#4247 |
| Actinomycin D | Sigma-Aldrich | Cat#A1410 |
| PhosSTOP | Roche | Cat#04906837001 |
| RNase inhibitor | Invitrogen | Cat#10777019 |
| RNase T1 | Thermo Fisher Scientific | Cat#AM2283 |
| RNase A | Thermo Fisher Scientific | Cat#12091-021 |
| Anti-V5 agarose affinity gel | Sigma-Aldrich | Cat#A7345 |
| Anti-Flag agarose affinity gel | Sigma-Aldrich | Cat# A2220 |
| Lipofectamine™ RNAiMAX Transfection Reagent | Invitrogen | Cat#13778075 |
| DNase I | Sigma-Aldrich | Cat#AMPD1 |
| TRIzol | Invitrogen | Cat#15596018 |
| Protein A/G magnetic beads | Thermo Fisher Scientific | Cat#88802 |
| Protein A/G PLUS-Agarose | Santa Cruz Biotechnology | Cat#sc-2003 |
| Proteinase K | New England Biolabs | Cat#P8107 |
| Lipofectamine 2000 reagent | Invitrogen | Cat#11668500 |
| Phenol:Chloroform:IAA, 25:24:1, pH 6.6 | Thermo Fisher Scientific | Cat#AM9730 |
| Insulin solution human | Sigma-Aldrich | Cat#I9278 |
| Crystal Violet | Sigma-Aldrich | Cat#C3886 |
| SRPIN340 | Selleck Chemicals | Cat#S7270 |
| SRPKIN-1 | MedChem Express | Cat#HY-116856 |
| Lipoprotein, low density from human plasma | Sigma-Aldrich | Cat#L7914 |
| Oleic acid-albumin | Sigma-Aldrich | Cat#O3008 |
| Fatty acid-free albumin | Sigma-Aldrich | Cat#A8806 |
| Lipoprotein-deficient serum | Alfa Aesar | Cat#BT-907 |
| Sodium palmitate | Sigma-Aldrich | Cat#P9767 |
| Methanol-free 10% formaldehyde | Polysciences | Cat#04018-1 |
| 25-Hydroxycholesterol | MedChem Express | Cat#HY-113134 |
| Critical commercial assays | ||
| PureLink RNA Mini kit | Ambion | Cat#12183018A |
| iScript cDNA synthesis kit | Bio-rad | Cat#170–8891BUN |
| Illumina® Stranded mRNA Prep Kit | Illumina | Cat#20040534 |
| Gateway LR clonase II enzyme mix | Invitrogen | Cat#11791 |
| QuickChange site-directed mutagenesis kit | Strategene | Cat#200521 |
| RNA Clean & Concentrator | Zymo Research | Cat#R1016 |
| Illumina® Stranded Total RNA Prep, Ligation with Ribo-Zero Plus | Illumina | Cat#20040529 |
| LightSwitch dual assay kit (promoter assay) | Active Motif | Cat#32035 |
| Chromatin IP DNA purification kit | Active Motif | Cat#58002 |
| SYBR Green PCR Master Mix | Life Technologies | Cat#4312704 |
| QuikChange II XL Site-Directed Mutagenesis Kit | Strategene | Cat#200521 |
| ChIP-IT Express Kit | Active Motif | Cat#53009 |
| Deposited data | ||
| Raw RNAseq data (siNTC and siFAM120A) | This paper | GSE207172 |
| Raw RNAseq data (siNTC, siFAM120, and siSRSF1) | This paper | GSE229657 |
| Raw data of Images and WB | This paper, Mendeley Data | DOI: 10.17632/rgfgx9xb8w.1 |
| Experimental models: Cell lines | ||
| Human: Renal angiomyolipoma-derived LAM 621–101 (TSC2−/−) | Drs. Jane Yu and Elizabeth Henske | Yu et al., 2004; RRID: CVCL_S879 |
| Human: HEK293E | ATCC | Cat#293 c18; RRID: CVCL_6974 |
| Human: HEK293T | GenHunter | Cat#Q401; RRID: CVCL_0063 |
| Human: MCF7 | ATCC | Cat#HTB-22; RRID: CVCL_0031 |
| Human: DLD1 | ATCC | Cat#CCL-221; RRID: CVCL_0248 |
| Human: NCI-H1299 | ATCC | Cat#CRL-5803; RRID: CVCL_0060 |
| Human: LNCaP | ATCC | Cat#CRL-1740; RRID:CVCL_1379 |
| Experimental models: Organisms/strains | ||
| nu/nu athymic mice | Envigo | Cat#069 |
| Oligonucleotides | ||
| siRNA negative control-#1(siNTC) | Sigma-Aldrich | SIC001 |
| siRNA negative control-#2(siNTC) | Sigma-Aldrich | SIC002 |
| siRNA (Human SRSF1) | Sigma-Aldrich | SASI_Hs01_00115059 |
| siRNA (Human SRSF1) | Sigma-Aldrich | SASI_Hs02_00342609 |
| siRNA (Human UPF1) | Sigma-Aldrich | SASI_Hs01_00101017 |
| siRNA (Human UPF1) | Sigma-Aldrich | SASI_Hs01_00101018 |
| siRNA (Human U1–70k/SNRNP70) | Sigma-Aldrich | SASI_Hs02_00335028 |
| siRNA (Human U1–70k/SNRNP70) | Sigma-Aldrich | SASI_Hs01_00165101 |
| siRNA (Human FAM120A) | Sigma-Aldrich | SASI_Hs01_00149752 |
| siRNA (Human FAM120A) | Sigma-Aldrich | SASI_Hs02_00345898 |
| siRNA (Human FAM120A) | Sigma-Aldrich | SASI_Hs02_00345899 |
| siRNA (Human SREBP1) | Sigma-Aldrich | SASI_Hs01_00051828 |
| siRNA (Human SREBP1) | Sigma-Aldrich | SASI_Hs01_00051829 |
| siRNA (Human SREBP1) | Sigma-Aldrich | SASI_Hs01_00051830 |
| siRNA (Human SREBP2) | Sigma-Aldrich | SASI_Hs01_00075424 |
| siRNA (Human SREBP2) | Sigma-Aldrich | SASI_Hs01_00075425 |
| siRNA (Human SREBP2) | Sigma-Aldrich | SASI_Hs01_00075426 |
| Primers for qPCR analysis, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| ACSS2 promoter construct | SwitchGear Genomics | Cat#S715263 |
| SCD1 promoter construct | SwitchGear Genomics | Cat#S714484 |
| pLX304-SRSF1-V5 | Harvard plasmid | Cat#HsCD00420441 |
| Lentiviral packaging and envelope plasmids | Dr. David Baltimore | N/A |
| pcDNA3-EGFP | Addgene | Cat#13031 |
| pKH3-HA-SRPK2-WT, K110M | Dr. John Blenis | Lee et al., 2007 |
| pLNCX-HA-SRPK2-S494A/S497A | Dr. John Blenis | Lee et al., 2007 |
| pLX304-FAM120A-V5 | Harvard plasmid | HsCD00440255 |
| pENTR1A-Flag-FAM120A(WT) | This paper | N/A |
| pENTR1A-Flag-FAM120A(∆RBD) | This paper | N/A |
| pLenti-CMV-Puro-DEST | Addgene | Cat#17452 |
| pENTR223-SRSF1 | Harvard plasmid | Cat#HsCD00367645 |
| pLenti-CMV-Puro-SRSF1 | This paper | N/A |
| pLenti-CMV-Puro-FAM120A | This paper | N/A |
| pLenti-CMV-Puro-Flag-FAM120A(WT) | This paper | N/A |
| pLenti-CMV-Puro-Flag-FAM120A(∆RBD) | This paper | N/A |
| pENTR223-FAM120A | Harvard plasmid | Cat#HsCD00438604 |
| T7-SRSF1(WT) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| T7-SRSF1(∆RRM1) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| T7-SRSF1(∆RRM2) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| T7-SRSF1(∆RS) | Dr. Adrian Krainer | Fregoso et al., 2013 |
| pLKO.1-puro-Non-Mammalian shRNA Control | Sigma-Aldrich | SHC002 |
| pLKO.1-puro-shSRPK2 | Sigma-Aldrich | TRCN0000006274 |
| pLKO.1-puro-shFAM120A | Sigma-Aldrich | TRCN0000074918 |
| pLKO.1-puro-shSREBP1 | Sigma-Aldrich | TRCN0000055325 |
| pLKO.1-puro-shSREBP2 | Sigma-Aldrich | TRCN0000020667 |
| Software and algorithms | ||
| IRFinder-S | Lorenzi et. al., 2021 | https://europepmc.org/article/med/34749764 |
| HISAT2 v2.2.1 | Kim et. al., 2019 | https://www.nature.com/articles/s41587-019-0201-4 |
| StringTie2 v2.1.4 | Kovaka et. al., 2019 | https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1910-1 |
| DESeq2 (v1.30.1) | Love et. al., 2014 | https://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0550-8 |
| Enhanced Volcano v1.8.0 R package | Blighe, 2022 | https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html |
| Adobe Photoshop | Adobe | Adobe |
| Odyssey imaging system | LI-COR Biosciences | LI-COR Biosciences |
| enrichR v3.0 package | Webb et al., 2014 | https://git.ecdf.ed.ac.uk/sgrannem/pycrac |
| EI-MAVEN, Version 0.12.0 | Agrawal et al, 2019 | Elucidata |
| VAST-tools | Irimia et. al., 2014 | https://www.sciencedirect.com/science/article/pii/S0092867414015128?via%3Dihub |
| BioRender | BioRender | https://www.biorender.com/ |


