Abstract
The present study was designed to identify and comparatively investigate 35 beta-hemolytic streptococci isolated from stranded harbor porpoises or from animals caught in fishing nets of the North and Baltic seas. According to biochemical and serological data and to lectin agglutination tests with the lectin of Arachis hypogaea, all 35 isolates could be classified in Lancefield’s serological group L and could be identified as Streptococcus dysgalactiae subsp. dysgalactiae. All 35 group L streptococci were uniformly sensitive to most of the antibiotics tested. To further analyze the epidemiological relationship, the isolates were subjected to macrorestriction analysis of their chromosomal DNA by pulsed-field gel electrophoresis. Digestion of the chromosomal DNA with the restriction enzymes SmaI and ApaI revealed that most of the group L streptococci seemed to be apparently identical or related. These results indicate that one clone or at least related group L streptococcal clones play an important role for infections of harbor porpoises of the North and Baltic seas. This might possibly be caused by a direct transfer of the bacteria from animal to animal.
The harbor porpoise (Phocoena phocoena) is the most-seen representative of the small cetaceans in the North and Baltic seas (3, 8). In connection with the seal epidemic in the North and Baltic seas in the years 1988 and 1989, an increased number of small cetacean carcasses on the German coast was registered. The collected data showed a dramatic decrease in the harbor porpoise population since the beginning of this century. This could be seen especially in the Baltic Sea (3, 16).
Bacteriological investigations of animals caught in fishing nets and stranded small cetaceans of the North and Baltic seas resulted in the isolation of various bacteria and frequently in the isolation of alpha- and beta-hemolytic streptococci. The beta-hemolytic streptococci could be isolated, partly in pure culture, from abscesses, from bronchopneumonia patients, and often from patients with septicemia causing suppurative leptomeningitis, pyelonephritis, myocarditis, and osteomyelitis (17). The isolated beta-hemolytic streptococci seemed to be one of the major causative agents of these diseases. However, in these studies the beta-hemolytic streptococci had not been identified to species level. In addition, nothing is known about the properties and epidemiological relationship of these bacteria. The present study was designed to identify and further characterize beta-hemolytic streptococci isolated from stranded harbor porpoises and from harbor porpoises caught in fishing nets in the North and Baltic seas.
MATERIALS AND METHODS
Bacterial isolates.
A total of 35 beta-hemolytic streptococci isolated from various organs of 16 harbor porpoises were investigated in this study. Among the 35 cultured streptococci 33 were isolated from 14 harbor porpoises of the North Sea and 2 were from two harbor porpoises from the Baltic Sea. The animal designations, the tissues from which the beta-hemolytic streptococci were isolated, and the places and dates of discovery of the harbor porpoises are summarized in Table 1. The bacteria were cultivated on Columbia sheep blood agar plates (Oxoid, Wesel, Germany) and in Todd-Hewitt broth (THB) (Gibco Europe, Karlsruhe, Germany) as fluid media. The growth intensity after cultivation on blood agar plates is shown in Table 1. None of the cultures required primary cultivation in THB to be recovered. Among the 35 isolates, 9 were isolated in pure culture and 16 predominated. For comparative purposes streptococci of serological group L isolated from pigs (n = 2), bovines (n = 2), and humans (n = 2) and the group L-streptococcal reference culture ATCC 9932 (MS 210) were included (18).
TABLE 1.
Summary of 35 beta-hemolytic streptococci isolated from 16 harbor porpoises from the North and Baltic seasa
| Animal | Place of discoveryb | Date of discovery (day.mo.yr) | Nature of animal when foundc | Streptococcus
|
Restriction pattern after digestion with:
|
||
|---|---|---|---|---|---|---|---|
| Tissue of origin | Growthd | SmaI | ApaI | ||||
| A | List on Sylt | 24.02.1991 | S | Lung | + | S I | A II |
| B | List on Sylt | 09.08.1991 | S | Lung | + | S I | A II |
| F | List on Sylt | 24.02.1992 | S | Kidney | +++ | S II | A III |
| J | Kampen on Sylt | 16.07.1994 | N | Intestine | + | S Ie | A Ie |
| Liver | + | S I | A I | ||||
| Lung | + | S I | A I | ||||
| K | Kampen on Sylt | 15.07.1994 | S | Intestine | + | S I | A I |
| Kidney | ++ | S I | A I | ||||
| Lung | +++ | S I | A I | ||||
| Spleen | ++ | S I | A I | ||||
| I | Wenningstedt on Sylt | 19.06.1994 | S | Kidney | ++ | S I | A I |
| Liver | + | S I | A I | ||||
| Lung | +++ | S I | A I | ||||
| Lymph node | +++ | S I | A I | ||||
| M | Westerland on Sylt | 18.07.1995 | S | Lung | + | S III | A IV |
| N | Westerland on Sylt | 04.03.1995 | S | Lung | ++ | S IV | A V |
| D | Baakdeel on Sylt | 06.01.1992 | S | Kidney | +++ | S I | A VI |
| E | Rantum on Sylt | 23.02.1992 | N | Kidney | + | S II | A III |
| G | Hörnum on Sylt | 18.04.1993 | S | Kidney | +++ | S I | A I |
| Lung | +++ | S I | A I | ||||
| L | Nieblum on Föhr | 08.09.1994 | S | Kidney | ++ | S V | A VII |
| Lung | + | S V | A VII | ||||
| P | Amrum | 18.07.1995 | S | Intestine | +++ | S I | A VI |
| Intestine (lymph node) | +++ | S I | A VI | ||||
| Kidney | +++ | S I | A VI | ||||
| Liver | +++ | S I | A VI | ||||
| Lung | +++ | S I | A VI | ||||
| Spleen | +++ | S I | A VI | ||||
| O | Friedrichskoog | 01.06.1995 | S | Kidney | +++ | S I | A VI |
| Lung | +++ | S I | A VI | ||||
| Spleen | +++ | S I | A VI | ||||
| Muscle | +++ | S I | A VI | ||||
| C | Laboe | 05.12.1991 | N | Lung | ++ | S VI | A VIII |
| H | Lübeck-Travemünde | 01.06.1994 | S | Kidney | + | S VII | A IX |
| Lung | + | S VII | A IX | ||||
DNA restriction patterns of chromosomal DNA after digestion with endonucleases SmaI and ApaI are also shown.
List, Kampen, Wennigstedt, Westerland, Baakdeel, Rantum, and Hörnum (all on the island of Sylt) as well as Nieblum on Föhr, Amrum, and Friedrichskoog are all on the North Sea. Laboe and Lübeck-Travemünde are on the Baltic Sea.
S, stranded; N, caught in fishing nets.
Growth intensity after cultivation on blood agar (+, slight; ++, moderate; +++, excessive).
For differences, see Results section.
Biochemical properties.
The biochemical properties of the beta-hemolytic streptococci were determined with a commercial identification system (rapid ID 32 Strep; bioMérieux, Nürtingen, Germany). The test kit, containing 32 different reactions (arginine dihydrolase, β-glucosidase, β-galactosidase, β-glucuronidase, α-galactosidase, alkaline phosphatase, ribose, mannitol, sorbitol, lactose, trehalose, raffinose, sucrose, arabinose, arabitol, cyclodextrin, acetoin production, alanine-phenylalanine-proline arylamidase, β-galactosidase, pyroglutamic acid arylamidase, N-acetyl-β-glucosaminidase, glycyl-tryptophane arylamidase, hydrolysis of hippurate, glycogen, pullulan, maltose, melibiose, melezitose, methyl-β-d-glucopyranoside, tagatose, β-mannosidase, and urease) was used according to the instructions of the manufacturer.
Serogrouping.
With the commercial grouping kit Streptex (Murex, Burgwedel, Germany) the β-hemolytic cultures could be investigated for serogroup A, B, C, D, F, and G. In addition, the group-specific antigen was extracted by autoclaving the bacteria and the extracts were subsequently tested by agar gel diffusion with specific antiserum prepared against a group L streptococcal reference culture (18).
Lectin agglutination.
The lectin from Arachis hypogaea (Sigma, Deisenhofen, Germany) was suspended in phosphate-buffered saline (PBS) at a final concentration of 0.5 g/ml. The streptococci were cultivated in 5 ml of THB for 24 h at 37°C, centrifuged, washed in PBS, and suspended in 200 μl of PBS. To prevent self-agglutination the bacteria were pretreated with trypsin (5 μg/200 μl of bacterial suspension) for 1 h at 37°C, washed, and resuspended in PBS. For lectin agglutination tests 20 μl of the lectin preparation was mixed with 20 μl of streptococcal suspension on microscopic slides (13). The slides were rotated and examined for agglutination reaction.
Antibiotic susceptibility.
The determination of antibiotic susceptibilities was performed according to the recommendations of the Bundesinstitut für gesundheitlichen Verbraucherschutz und Veterinärmedizin, Berlin, Germany. According to the instructions, four to five colonies of the bacteria were inoculated in 3 ml of THB for 2 h at 37°C. Approximately 0.1 ml of this suspension was transferred onto Müller-Hinton agar (Oxoid) containing 5% sheep blood. The paper discs used contained bacitracin (0.04 IU), bacitracin (10 IU), cefoxitin (30 μg), chloramphenicol (30 μg), clindamycin (2 μg), erythromycin (15 μg), gentamicin (10 μg), minocycline (30 μg), neomycin (30 μg), nitrofurantoin (100 μg), penicillin G (10 IU), sulfamethoxazole-trimethoprim (23.75 and 1.25 μg, respectively), and tetracycline (30 μg).
Pulsed-field gel electrophoresis.
The preparation and digestion of genomic DNA were performed according to the methods of Maslow et al. (11) and Thiele et al. (25). As described previously (21) the bacteria were cultivated in 40 ml of THB, centrifuged, washed in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]), resuspended in TE buffer (5% transmission at 620 nm), and mixed with the same volume of 1% low-melting-point and low-gelling-point InCert agarose (FMC Bio Products, Rockland, Maine). This mixture was solidified for 10 min on ice in special molds. The cells were lysed by subsequent incubation of the blocks in 200 μl of ES buffer (0.5 M EDTA [pH 8.0], 1% N-lauroylsarcosine [Sigma]) for 30 min at room temperature followed by an incubation for 1 h at 37°C in the same buffer containing 10 μg of mutanolysin (10 U/μl; Sigma). After overnight treatment with proteinase K (final concentration, 0.5 μg/ml; Boehringer, Mannheim, Germany) in 200 μl of TE buffer at 56°C, the agarose blocks were washed twice for 30 min each time in TE buffer, incubated with phenylmethylsulfonyl fluoride (final concentration, 1.0 mM in 200 μl of TE buffer; Sigma) twice for 1 h each time at 56°C, and washed in TE buffer twice for 30 min each time. The genomic DNA of the isolates was digested with the endonucleases SmaI and ApaI (Stratagene, Heidelberg, Germany). For digestion with SmaI each agarose block was incubated with 4 μl of SmaI (20 U) suspended in 20 μl of 10× universal buffer (Stratagene) and 176 μl of twice-distilled water for 5 h at 25°C. For digestion with ApaI each agarose block was incubated with 1.5 μl of ApaI (15 U) suspended in 20 μl of 10× universal buffer (Stratagene) and 178.5 μl of twice-distilled water overnight at 30°C. Samples were electrophoresed at 14°C through 1% chromosomal-grade agarose (Bio-Rad Laboratories, Munich, Germany) (in a 13-by-14-by-0.55-cm gel) containing 0.5 μg of ethidium bromide (Sigma) per ml in 0.5× TBE buffer (45 mM Tris, 45 mM borate, 1.0 mM EDTA [pH 8.3]) in a CHEF-DR II PFGE cell (Bio-Rad). Lambda DNA/HindIII fragments at 0.1 to 200 kb (Sigma) and lambda DNA concatemers at 50 to 1,000 kb (Sigma) served as standards. The running conditions were as follows. Switch time ramping for SmaI-digested agarose blocks was 0.1 to 11 s for 8 h at 5 V/cm and then 9 to 34 s for 17 h at 6 V/cm. For ApaI-digested agarose blocks, switch time ramping was 0.1 to 10 s for 13 h at 6 V/cm and then 5 to 8 s for 7 h at 6 V/cm.
Statistics.
The BMDP Statistical Software package (5) was used for the estimation of the genetic relationship of the beta-hemolytic streptococci, and the program PlotIt-Graphics and Statistics (6) was used for the presentation of the genetic relationship in a dendrogram.
RESULTS
All 35 isolates investigated in the present study were beta-hemolytic streptococci and could be classified biochemically and serologically into Lancefield’s serological group L.
The biochemical properties of all 35 isolates determined with the rapid ID 32 Strep system appeared to be identical in most of the reactions. All cultures were positive for arginine dihydrolase, β-glucuronidase, alkaline phosphatase, ribose, trehalose, sucrose, glycogen, pullulan, and maltose; were negative for β-glucosidase, β-galactosidase, α-galactosidase, pyroglutamic acid arylamidase, N-acetyl-β-glucosaminidase, glycyl-tryptophane arylamidase, β-mannosidase, urease, acetoin production, and hydrolysis of hippurate; and did not produce acid from mannitol, sorbitol, raffinose, l-arabinose, d-arabitol, melibiose, melezitose, methyl-β-d-glucopyranoside, and tagatose. Acid production from lactose could be observed for 33 cultures, and a positive cyclodextrin and a positive alanine-phenylalanine-proline arylamidase reaction could be observed for 14 and 33 cultures, respectively. Autoclaved extracts of all 35 isolates reacted with group L-specific antiserum. No reaction could be observed with the commercial grouping kit, including serogroups A, B, C, D, F, and G. In lectin agglutination tests all 35 isolates agglutinated with the lectin from A. hypogaea.
The determination of antibiotic susceptibility revealed that all 35 beta-hemolytic streptococci were sensitive to bacitracin (10 IU), cefoxitin, chloramphenicol, clindamycin, erythromycin, minocycline, nitrofurantoin, penicillin G, sulfamethoxazole-trimethoprim, and tetracycline and that most of the strains were sensitive to gentamicin (83%) and bacitracin (0.04 IU) (94%); all strains were resistant to neomycin.
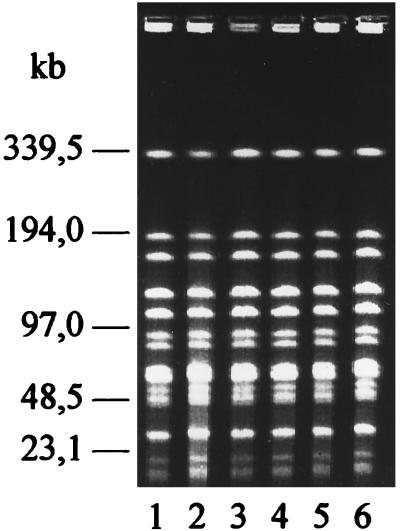
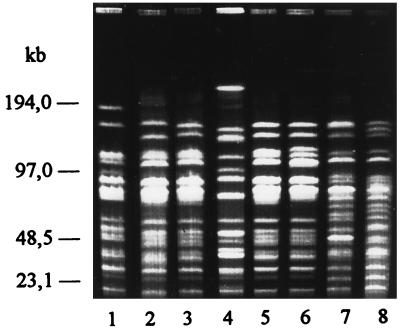
To further analyze the epidemiological relationship, the isolates were subjected to DNA fingerprinting. The digestion of the chromosomal DNA of the isolates with the endonucleases SmaI and ApaI and the separation of the fragments by pulsed-field gel electrophoresis (PFGE) revealed that the beta-hemolytic streptococci isolated from a single harbor porpoise had identical restriction patterns (Fig. 1) with one exception. The isolate from the intestine of harbor porpoise J differed in one and two fragments after digestion with SmaI and ApaI, respectively. To compare the DNA pattern of the group L streptococci obtained from the 16 harbor porpoises, one isolate from each animal was subjected to comparative PFGE. The digestion of the chromosomal DNA with the endonucleases SmaI and ApaI revealed seven (SI to SVII) (not shown data) and nine (AI to AIX) different restriction patterns, respectively (Fig. 2). The origin of the animals and their DNA pattern are summarized in Table 1.
FIG. 1.
PFGE restriction patterns of chromosomal DNA of group L streptococci isolated from six different organs of harbor porpoise P. Patterns were obtained after digestion of the DNA with endonuclease SmaI.
FIG. 2.
Restriction patterns of group L streptococci isolated from harbor porpoises as follows: A VI, harbor porpoises P, O, and D (lane 3, 5, and 6); A IV, harbor porpoise M (lane 1); A V, harbor porpoise N (lane 2); A VII, harbor porpoise L (lane 4); A VIII, harbor porpoise C (lane 7); and A IX, harbor porpoise H (lane 8).
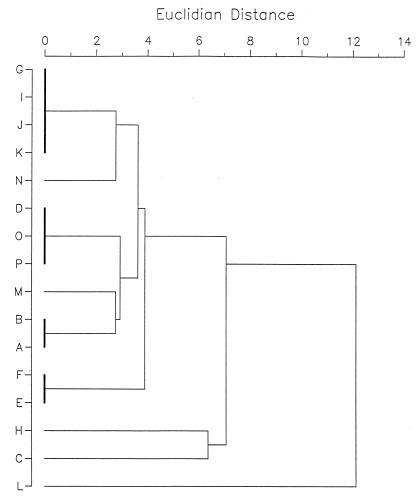
A dendrogram of similarity of the nine DNA restriction groups revealed after digestion of the chromosomal DNA with the endonuclease ApaI is presented in Fig. 3. According to this dendrogram analysis, the DNA profiles of all but one isolate (harbor porpoise L) from the harbor porpoises of the North Sea were related to each other and differed from DNA profiles of both strains isolated from harbor porpoises of the Baltic Sea (harbor porpoises C and H). The DNA restriction patterns of the group L streptococcal control strains appeared with unrelated DNA profiles (data not shown).
FIG. 3.
Dendrogram demonstrating genetic relationship of different DNA patterns of group L streptococci isolated from 16 harbor porpoises (listed on the left). Patterns were obtained after digestion of the chromosomal DNA with the restriction enzyme ApaI.
DISCUSSION
All beta-hemolytic streptococci investigated in the present study were isolated from stranded harbor porpoises or harbor porpoises caught in fishing nets in the North and Baltic seas. On the basis of their biochemical and serological properties, all 35 isolates belonged to Lancefield’s serological group L and could be identified as S. dysgalactiae subsp. dysgalactiae serovar L. The classification of group L as well as streptococci of serological group C of animal origin as S. dysgalactiae subsp. dysgalactiae was proposed by Vandamme et al. (26).
The group L streptococci could additionally be identified by lectin agglutination tests with the lectin from A. hypogaea. This has already been shown to be a useful additional test for presumptive identification of this species (13).
All group L streptococci seemed to be uniformly sensitive to most of the antibiotics tested in this study. According to previous studies group L streptococci isolated from various origins showed resistance to antibiotics such as tetracycline, minocycline, erythromycin, chloramphenicol, and gentamicin (18). Tetracycline- and minocycline-resistant group L streptococci seemed to posses the resistance genes tet(M) and tet(O) (22).
Streptococci of serological group L are well known as causative agents of bovine mastitis and various infections of pigs, dogs, poultry, and sheep, but they rarely cause infections in humans (2, 7, 9, 10, 19, 24). However, little is known about the isolation of streptococci of serological group L from harbor porpoises. Baker and Martin (1) investigated 41 harbor porpoises from British waters and could find group L streptococci as the cause of bacterial pneumonia. To our knowledge there are no further reports about the isolation of group L streptococci from sea animals.
The harbor porpoises of the present study were examined post mortem, and it was obvious that most of the animals, especially their respiratory systems, were infected by parasites. The bacteriological investigations revealed that besides beta-hemolytic streptococci other potentially pathogenic bacteria, such as Clostridium perfringens, Erysipelothrix rhusiopathiae, and Staphylococcus aureus, could be isolated from the organs of the animals. However, according to previous studies beta-hemolytic streptococci seemed to be the major pathogens causing septicemia, abscesses, or bronchopneumonia. These beta-hemolytic streptococci were isolated partly in pure culture (4, 15, 17).
To study their genetic relationship, the streptococci of the present study were further investigated by macrorestriction analysis of their chromosomal DNA after digestion with the restriction enzymes SmaI and ApaI by PFGE. This technique proved to be useful for molecular typing of animal-pathogenic Streptococcus equi subsp. zooepidemicus and Rhodococcus equi (20, 21).
The macrorestriction analysis of the present investigation was performed with two restriction enzymes (SmaI and ApaI) to avoid epidemiologically misleading results. The present study’s investigation of group L streptococci revealed that DNA fragments obtained from isolates from different organs of one animal and from some of the isolates obtained from different animals produced apparently identical profiles, indicating that a single bacterial clone seems to be the causative agent. It was of interest that bacterial clones with identical profiles were mostly found among isolates from the same region (harbor porpoises A and B had identical profiles, as did harbor porpoises J and K) and/or among bacteria isolated at the same time (harbor porpoises E and F had identical profiles, as did harbor porpoises J and K). These findings can lead to the conclusion that direct transmission of the group L streptococci between the various animals occurred. However, group L streptococci with identical DNA profiles could also be found among isolates from animals which were found and investigated 3 1/2 years apart (harbor porpoises D, O, and P). The stability of the DNA profiles over a long period has already been reported by other authors (12, 14). According to the PFGE analysis criteria of Tenover et al. (23), most of the nonidentical group L streptococci isolates from harbor porpoises of the North Sea appeared to be closely related or at least possibly related; the two isolates from harbor porpoises from the Baltic Sea were closely related to each other but not to those from the North Sea. A comparison of the DNA profiles of group L streptococci isolated from harbor porpoises with DNA profiles of group L streptococci isolated from bovines, pigs, or humans revealed no genetic relationship. These results indicate that one group L streptococcal clone or at least closely related clones contribute to the infections of the harbor porpoises in the North and Baltic seas and that direct transfer from animal to animal seems to be a reasonable means of transmission.
ACKNOWLEDGMENTS
The collection of carcasses and samples as well as the pathological investigations were supported by grants from the Bundesministerium für Umwelt, Naturschutz und Reaktorsicherheit and the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie.
REFERENCES
- 1.Baker J R, Martin A R. Causes of mortality and parasites and incidental lesions in harbour porpoises (Phocoena phocoena) from British waters. Vet Rec. 1992;130:554–558. doi: 10.1136/vr.130.25.554. [DOI] [PubMed] [Google Scholar]
- 2.Barnham M, Neilson D J. Group L beta-haemolytic streptococcal infection in meat handlers: Another streptococcal zoonosis? Epidemiol Infect. 1987;99:257–264. doi: 10.1017/s0950268800067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohlken H, Benke H, Wulf J, editors. Untersuchungen über Bestand, Gesundheitszustand und Wanderungen der Kleinwalpopulationen (Cetacea) in deutschen Gewässern. Endbericht zum FE-Vorhaben des BMU, Nr. 10805017/11. Kiel, Germany: Institut für Haustierkunde, Universität Kiel; 1993. [Google Scholar]
- 4.Benke H, Siebert U. The current status of harbour porpoises (Phocoena phocoena) in German waters. Dublin: Internationale Walfangkommission; 1995. pp. 0–23. , SC/47/SM49. [Google Scholar]
- 5.Dixon W J. BMDP statistical software manual. 1 and 2. Berkeley: University of California Press; 1993. [Google Scholar]
- 6.Eisensmith S P. PlotIt-Graphics and Statistics. Haslett, Mich: Scientific Programming Enterprises; 1993. [Google Scholar]
- 7.Hahn G, Tolle A. Ergebnisse aus der Streptokokkenzentrale in Kiel von 1965–1977—ein Überblick. Zentbl Bakteriol Hyg Abt 1. 1979;244:427–428. [PubMed] [Google Scholar]
- 8.Hammond P S, Benke H, Berggren P, Borchers D L, Buchland S T, Collet A, Heide-Jørgensen M-P, Heimlich-Boran S, Hiby A R, Leopold M P, Oien N. Distribution and abundance of the harbour porpoise and other small cetaceans in the North Sea and adjacent waters. Life 92-2/UK/027. Final report to the European Commission, October 1995. Brussels, Belgium: European Commission; 1995. [Google Scholar]
- 9.Hinterdorfer F, Köfer J, Hahn G. Streptokokkenisolate aus Untersuchungsmaterial von Schweinen in der Steiermark. Wien Tieraeztl Monschr. 1990;77:161–163. [Google Scholar]
- 10.Mantovani A, Restani R, Sciarra D, Simonella P. Streptococcus L infection in the dog. J Small Anim Pract. 1961;2:185–194. [Google Scholar]
- 11.Maslow J N, Slutsky A M, Arbeit R D. Application of pulsed-field gel electrophoresis to molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: American Society for Microbiology; 1993. pp. 563–572. [Google Scholar]
- 12.Sanches S, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, de Lencastre H. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus clone between Portugal and Spain. J Clin Microbiol. 1995;17:1243–1246. doi: 10.1128/jcm.33.5.1243-1246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaufuß P, Lämmler C, Niewerth B, Blobel H. Properties of L-streptococci in comparison with those of A-streptococci. Med Microbiol Immunol. 1987;176:169–173. doi: 10.1007/BF00193898. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz S, Liebisch B. Pulsed field gel electrophoretic identification of Salmonella enterica serovar typhimurium live vaccine strain Zoosoral H. Lett Appl Microbiol. 1994;19:469–472. doi: 10.1111/j.1472-765x.1994.tb00984.x. [DOI] [PubMed] [Google Scholar]
- 15.Siebert U, Frese K. Endbericht über das Teilprojekt ”Pathologisch-anatomische und histopathologische Untersuchungen an Kleinwalen“. In: Bohlken H, Benke H, Wulf J, editors. Untersuchungen über Bestand, Gesundheitszustand und Wanderungen der Kleinwalpopulationen (Cetacea) in deutschen Gewässern. Endbericht zum FE-Vorhaben des BMU, Nr. 10805017/11. Kiel, Germany: Institut für Haustierkunde der Universität Kiel; 1993. pp. 1–23. [Google Scholar]
- 16.Siebert U, Benke H, Schulze E, Sonntag R P. Über den Zustand der Kleinwale. In: Lozen J L, Lampe R, Mattäus W, Rachor E, Rumohr H, von Westernhagen H, editors. Warnsignale aus der Ostsee. Berlin, Germany: Parey Buchverlag; 1996. pp. 242–248. [Google Scholar]
- 17.Siebert U, Lick R, Weiss R, Frank H, Benke H, Frese K. Proceedings of the First Scientific Meeting of the European Association of Zoo- and Wildlife Veterinarians (EAZWV), Rostock, Germany. 1996. Post-mortem findings in small cetaceans from German waters of the North and Baltic seas; pp. 1–7. [Google Scholar]
- 18.Sippel K, Dülffer-Schneitzer B, Lämmler C. Characteristic properties of streptococci of serological group L. J Vet Med B. 1995;42:42–50. doi: 10.1111/j.1439-0450.1995.tb00680.x. [DOI] [PubMed] [Google Scholar]
- 19.Skovgaard N. Undersogelse vedrorende forekomst og oprindelse af Gr. L-streptokokker og stafylokokker i levnedsmidler. Nord Veterinaermed. 1967;6:240–248. [Google Scholar]
- 20.Soedarmanto I, Oliveira R, Lämmler C, Dürrling H. Identification and epidemiological relationship of Rhodococcus equi isolated from cases of lymphadenitis in cattle. Zentbl Bakteriol. 1997;286:457–467. doi: 10.1016/s0934-8840(97)80047-3. [DOI] [PubMed] [Google Scholar]
- 21.Soedarmanto I, Pasaribu F H, Wibawan I W T, Lämmler C. Identification and molecular characterization of serological group C streptococci isolated from diseased pigs and monkeys in Indonesia. J Clin Microbiol. 1996;34:2201–2204. doi: 10.1128/jcm.34.9.2201-2204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soedarmanto I, Schwarz S, Liebisch B, Lämmler C. Tetracycline resistance determinants among streptococci of serological group G and L. Vet Microbiol. 1995;45:331–337. doi: 10.1016/0378-1135(94)00140-r. [DOI] [PubMed] [Google Scholar]
- 23.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thal E, Moberg K. Serologische Gruppenbestimmung der bei Tieren vorkommenden ß-hämolysierenden Streptokokken. Nord Veterinaermed. 1953;5:835–846. [Google Scholar]
- 25.Thiele D, Willems H, Köpf G, Krauss H. Polymorphism in DNA restriction patterns of Coxiella burnettii isolates investigated by pulsed-field gel electrophoresis and image analysis. Eur J Epidemiol. 1993;29:419–425. doi: 10.1007/BF00157400. [DOI] [PubMed] [Google Scholar]
- 26.Vandamme P, Pot B, Falsen E, Kersters K, Devriese L A. Taxonomic study of Lancefield streptococcal groups C, G and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996;46:774–781. doi: 10.1099/00207713-46-3-774. [DOI] [PubMed] [Google Scholar]