Abstract
B- and T-cell receptors (BCRs, Antibodies, and TCRs, or AIRs, adaptive immune receptors) are the means by which the adaptive immune system recognizes foreign and self-antigens, playing an integral part in host defense, as well as the emergence of autoimmunity. Importantly, the interaction between AIRs and their cognate antigens defies a simple key-in-lock paradigm and is instead a complex many-to-many mapping between an individual’s massively diverse AIR repertoire, and a similarly diverse antigenic space. Understanding how adaptive immunity balances specificity with epitopic coverage is a key challenge for the field and terms such as broad specificity, cross-reactivity, and polyreactivity remain ill-defined and are used inconsistently. In this Immunology Notes and Resources article, a group of experimental, structural, and computational immunologists define commonly used terms associated with AIR binding, describe methodologies to study these binding modes, as well as highlight the implications of these different binding modes for therapeutic design.
Introduction
Adaptive immune receptors (AIRs) play a critical role in the generation of adaptive immune responses via interactions with their cognate antigens. The specific binding region of an AIR is known as the paratope, whereas the region of the antigen the paratope interacts with is known as the epitope. B cell receptors (BCRs) and their secreted counterparts, antibodies, recognize native three-dimensional epitopes, whereas T cell receptors (TCRs) bind to linear peptide fragments from larger proteins presented in complex with Major Histocompatibility Complexes (MHCs), also called Human Leukocyte Antigens (HLAs) in humans, or lipid and small molecule antigens presented by CD1 and MHC class I-related protein (MR1). AIRs are classically known for highly specific interactions with their cognate antigens. However, AIRs demonstrate multiple different binding modes that facilitate binding to both closely and distantly related antigens, as well as non-specific binding to unrelated antigens. While experimental and computational approaches to study AIR binding have considerably increased our understanding of their properties, it remains a challenge to clearly delineate the complexity of AIR binding into distinct phenomena. Here, we review current progress and remaining challenges in studying AIR binding and propose coherent definitions for important terms relating to specificity and cross-reactivity for both BCRs and TCRs, incorporating perspectives ranging from fundamental immunology to therapeutics and diagnostics design.
Genetic, Structural, and Binding features of AIRs
3D structure of BCRs and TCRs
The diversity, and thus antigen recognition breadth, of AIR repertoires is influenced by the statistics of variable (V), diversity (D), and joining (J) genes segment recombination (1). Specific germline genes (and alleles), as well as their frequencies, have been linked to neutralizing antibody classes (2–6), and autoantigen-specific binding in autoimmunity (7, 8). For BCRs, somatic hypermutation (SHM) of antibody genes add another layer of genetic complexities that can lead to distinct binding modes. Importantly, V(D)J gene usage shape the 3D structure of AIRs as well as the diversity of loops responsible for antigen recognition (9).
BCRs are transmembrane proteins located on the surface of B cells and are responsible for recognizing diverse antigens (10, 11). Structurally, a BCR can be subdivided into a membrane-bound immunoglobulin (mIg) and signaling domains (10) that are responsible for antigen-binding and B cell activation, respectively, resulting in the production of plasma cells, which secrete large quantities of secreted immunoglobulins (i.e., antibodies) (10). The classical IgG structure is composed of two identical sets of paired heavy and light chains, and the stem of the antibody, also known as the crystallizable fragment region (Fc), is responsible for interacting with other components of the immune system through Fc receptor binding (12, 13). The antigen-binding fragment (Fab) arms of the Y-shaped immunoglobulin structure are composed of heavy and light chains that can be subdivided into a constant (Fc) and a variable (Fv) domain (13, 14).
Genetically the variable region is comprised of recombined VDJ genes for the heavy chain and a recombined VJ genes for the light chain. Structurally, the variable fragment is comprised of six hypervariable loops, known as the complementarity determining region (CDR) loops, where the sequence and structural diversity of the antibody is concentrated (15). Together with the relative orientation of the heavy and the light chain (VH-VL) (16, 17), the CDR loops shape the antigen binding site, also known as the paratope (18). The CDR loops are separated by structurally conserved framework regions (FR), that display less sequence variability and are responsible for maintaining the immunoglobulin β-sheet structure to support antigen binding by the CDR loops, although the FR regions can contribute to antigen binding (19). Additionally, certain framework residues can influence the CDR loop conformations, thereby contributing indirectly to antigen recognition (20–22).
TCRs are transmembrane heterodimers consisting of disulfide-linked α and β chains, analogous to the heavy and light chain in antibodies, and the signal transducing invariant CD3 dimers (23). TCR signaling in canonical αβ TCRs is initiated by the receptor recognizing peptides (up to 24 residues long) bound to MHC proteins (pMHC) (24). Both α and β chains are involved in pMHC binding and can each be subdivided into a constant and variable domain (25). Similar to BCRs, each variable domain of the TCR is comprised of three CDR loops. However, the divergent antigen recognition needs of TCRs and BCRs have selected for structural differences in their recognition modes (26, 27). TCRs, with the exception of noncanonical T cell subsets such as Mucosal-Associated Invariant T cells and γδ T cells (28), have evolved to recognize small but highly diverse peptides in the context of pMHC presentation. Consequently, most TCRs bind pMHCs in a canonical binding mode using both α and β chains with a conserved docking polarity. While CDR1 and CDR2 loops predominantly interact with the MHC molecule, peptide recognition is primarily achieved by the CDR3 loops (reviewed in (29, 30)). While TCRs that bind with a non-canonical reverse docking polarity have been observed, recent studies suggest that canonical docking is essential for signaling and thus T cell activation (31).
In contrast, the binding modes of BCRs and antigens are highly diverse (reviewed in (32)), although each CDR loop still adopts characteristic canonical structures upon antigen binding, with the exception of the CDRH3 (15, 33). Unlike TCRs, BCR affinity can be improved by SHMs at positions that optimize the paratope, including positions that are not directly contacting antigen. For example, non-paratope SHMs can modulate the flexibility of CDR loops, which in turn can influence binding affinity (34, 35). Furthermore, such mutations can affect antigen binding affinity by modulating the interactions between the heavy and light variable domains (36–39).
Binding affinities and avidities
Antigen-binding affinity is defined as the strength of the molecular interaction between a single paratope and a single epitope. BCRs have bivalent binding potential, as a single BCR possesses two Fabs arms, each Fab possessing identical antigen-binding affinities. Furthermore, secreted antibodies can form structures with up to 12 identical Fab arms, such as for IgM isotype antibodies. Avidity is the combined binding affinity of the BCR, antibody, or TCR to multiple copies of an epitope, whether on the surface of the cell or in solution. As such, avidity is frequently assessed through the apparent affinity of multivalent AIRs to represent their physiologically relevant binding strengths more accurately.
Both the affinity and avidity of receptor binding play critical roles in cellular activation and differentiation, B cell selection, T cell tolerance, and antibody function. Avidity of BCRs and TCRs is achieved by the clustering of immune receptors within lipid rafts, which increases intracellular signaling (40, 41). The strength of signaling is dependent on both receptor affinity and avidity, which ultimately dictates cellular activation and differentiation. For example, B cells with low affinity BCRs are more likely to differentiate into germinal center B cells and memory B cells, whereas B cells with high affinity BCRs are more likely to differentiate into antibody secreting cells (42). In addition, T cells possessing TCRs with excessively high apparent affinity to self-antigens presented in the thymus during thymic selection are more likely to be deleted or differentiate into regulatory T cells (Treg; reviewed in (43)). Antibody isotypes can compensate for lower affinity with higher avidity. For example, bivalent IgG isotype antibodies possess lower binding valency but often high affinity, owing in part to substantial SHM, whereas pentameric IgM isotype (the basal isotype of antibodies) antibodies often possess lower affinity and little-to-no SHM, but can still be potently neutralizing due to their potential for highly avid binding (44–48). Therefore, affinity and avidity play critical roles in B and T cell activation, as well as in antibody function.
Ultimately, whether a BCR or TCR binds its cognate antigen(s) and mediates effector functions is dependent upon its antigen binding affinity and avidity. The dissociation constant (KD) between TCRs and pMHCs is typically in the micromolar (μM) range (29), with T cell responses reported for TCR-pMHC interactions with KDs as high as ~1 mM (49), whereas the KD between BCRs/antibodies and antigens can often reach the nanomolar (nM) and picomolar (pM) range (50). The higher affinity of BCRs can at least be partly explained by the affinity maturation process via SHM, which is absent in mammalian TCRs. Moreover, high affinity (KD < 100 nM) T cells clones are purged from TCR repertoires during thymic selection, presumably to limit the potential for autoreactivity (51). Although the monomeric affinities between AIRs and their cognate antigens have been well-studied, the physiological apparent affinities necessary for T cell and B cell activation remain ill-defined. For TCRs, this understanding may be improved by assays that assess TCR-pMHC interactions in their native membrane-bound context, as these measurements have been found to correlate better with T cell activation than kinetics measurements in solution (52).
Modes of AIR binding
Specificity for AIRs can be defined in relation to their antigen binding properties. However, owing to structural differences in the antigens bound by BCRs/antibodies and TCRs, these definitions should be tailored to each immune receptor class (Table 1).
Table 1:
Binding mode definitions.
| Term | Definition | Examples |
|---|---|---|
| Monospecificity | Monospecific binding of AIRs refers to their capacity to bind to a single epitope with a consistent binding interface and binding mode within the apparent affinity ranges for BCRs (KD range ≈ pM-nM) and TCRs (KD range ≈ μM-mM) | Antibodies targeting variable epitopes of SARS-CoV-2 RBD |
| Broad specificity | Broadly reactive specificities react with epitopes that are generally well conserved but have either minor residue changes or changes to the local structure | Broadly neutralizing antibodies against HIV or influenza viruses; TCRs cross-reactive with nonapeptide and decapeptide of MART-1 |
| Cross-reactive | Recognition of a discrete motif that is shared in antigens that otherwise share little-to-no similarity | BCR and TCR binding to Streptococcal M protein and cardiac myosin; HIV and HCV binding antibodies |
| Polyreactive (BCR) | Binding to many distinct antigens with diverse motifs using multiple antibody paratopes that interact with diverse epitopes found on different antigen classes (e.g., protein, lipid, glycan) | Antibodies against influenza HA stalk domain; anti-HIV MPER antibodies |
| Promiscuity (TCR) | Binding to vast numbers of unrelated peptide antigens | TCR 1E6 binding to recognize over 1 million distinct decameric peptides in the context of HLA-A*02:01 |
Monospecificity
Monospecific binding of AIRs refers to the capacity of a receptor to only bind to a single epitope with a consistent binding interface and mode within the apparent affinity ranges described above. This definition implies that a monospecific receptor should not bind to mutated forms of the epitope but includes binding to variants of the same antigen provided that the epitope is unaltered. As such, monospecificity refers to the classic lock-and-key mode of AIR binding to antigen. However, true monospecificity according to this strict definition is exceedingly rare, as both antibodies and TCRs often recognize at least some point-mutated epitopes (53–60). In fact, it is believed that, due to the vast excess of potential pMHC antigens relative to the number of T cells in the naive repertoire, all T cells must recognize more than one pMHC to prevent gaps in immunosurveillance (61, 62).
More practically, monospecific AIRs may be defined as those with very limited breadth, where binding is lost upon comparatively minor mutation of the epitope relative to broadly specific AIRs (discussed below). Examples of monospecific antibodies according to this less stringent definition include those against the variable epitopes of coronavirus receptor-binding domain and influenza virus hemagglutinin (HA) head domain. Antibodies against these domains are functionally limited in their binding breadth against viral variants and on a molecular-level can be shown to lose binding affinity upon a relatively modest accumulation of mutations (53–58).
It should be noted that most monospecific AIRs are isolated following immunization, infection, or in diseased states, and screened only against antigens present in these states and their variants. As a result, receptors identified in this manner as monospecific may nevertheless bind to distinct unrelated antigens and may display features of cross-reactive and polyreactive receptors (described below) when assayed outside of those conditions.
Broad specificity
Broad specificity, also called broad reactivity, refers to AIRs that bind to multiple variants of a conserved epitope on polymorphic antigens (Fig. 1A). Broadly specific AIRs react with epitopes that are generally well conserved but have either minor residue or structural changes. As such, this definition includes binding to partially conserved epitopes on highly homologous but evolutionarily distinct proteins, including cross-species orthologs and proteins conserved between virus families. Unlike monospecific AIRs, broadly specific AIRs are able to retain physiologically relevant affinities for their epitopes in the presence of these changes.
Figure 1:
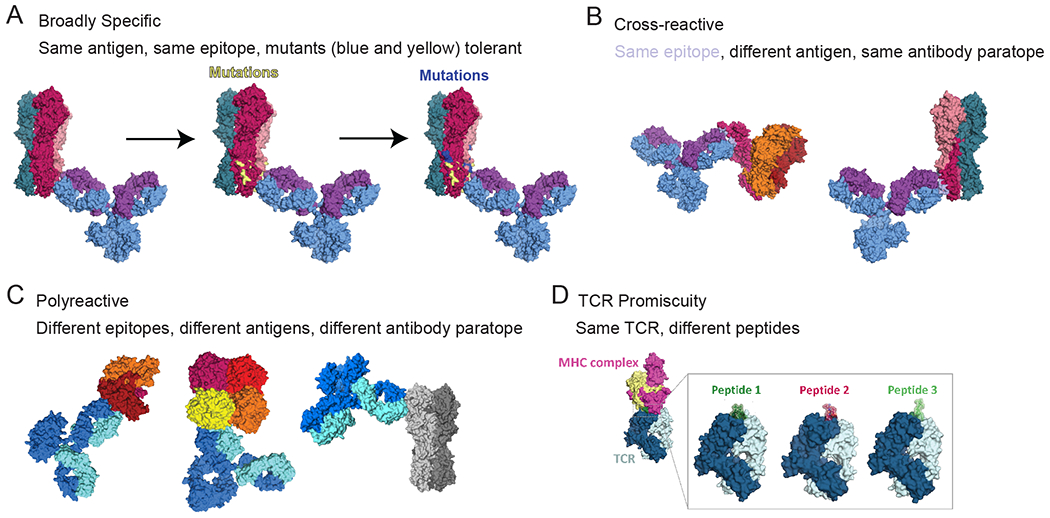
AIR binding modes. (A) BCR/antibody broadly specific binding (PDB accession codes: 1IGY and 7T3D). (B) BCR/antibody cross-reactive binding (PDB accession codes: 1IGY, 5UTY and 7T3D). (C) BCR/antibody polyreactivity binding (PDB accession codes: 1IGY, 6DZL, 6Q23, 7T3D). (D) TCR promiscuous binding (PDB accession codes: 3QDG, 7N5C, 4MAY).
Examples of broadly reactive antibodies include antibodies against the influenza virus HA stalk (stem) domain (63) and CD4 binding site of HIV (64). To accommodate binding to distinct variants of these partially conserved epitopes, broadly reactive antibodies often utilize multiple modes of recognition. For example, three mAbs that bind the HA stalk domain, PN-SIA28 (65), FI6v3 (66), and MEDI8852 (67), each exhibit multiple binding modes to recognize antigenic variants. In addition, broadly reactive antibodies often possess variable affinities for distinct variants of a partially conserved antigen. For example, broadly specific antibodies against HA antibodies often possess higher affinity for past influenza strains, relative to current circulating viruses, due to iterative rounds of affinity maturation against past strains (55, 57, 58).
For TCRs, this definition can be applied to TCRs that bind highly similar pMHC antigens, such as two length variants of a single peptide, or peptide variants that differ only in their MHC contacts, so long as the local structure of the peptide backbone and remaining side chains remains unaltered. Examples of broadly specific TCRs would therefore include MEL5, which binds the nonapeptide (AAGIGILTV) and decapeptide (EAAGIGILTV) of MART-1 presented on HLA-A*02:01 (68), as well as the MART-1 A27L heteroclitic decapeptide variant (ELAGIGILTV), which possesses enhanced binding to its presenting HLA allele (HLA-A*02:01) (69).
Cross-reactivity
We propose restricting the definition of cross-reactivity to recognition of a discrete motif that is shared in antigens that otherwise share little-to-no homology (Fig. 1B). This can include binding to structurally similar motifs in distinct antigens, including examples of molecular mimicry, wherein immune responses to infectious agents can potentiate autoimmunity due to cross-reactivity with unrelated autoantigens (70). One of the earliest and most clinically significant examples of this was the identification of antibodies and T cells that cross-reacted with group A streptococcal M protein and cardiac myosin (71, 72), which is implicated in the development of rheumatic fever. B and T cells recognized motifs shared between this bacterial antigen and self-antigen, including a repeated seven amino acid motif found in a variety of self-antigens including myosin, tropomyosin, and vimentin (73).
In addition, numerous anti-viral cross-reactive antibodies have been found to target glycan moieties on viral surface glycoproteins. One such antibody, 2G12, was initially discovered as an HIV antibody that binds to oligomannose cluster (74–76), but has also been shown to bind influenza HA with nM affinity (77). Similarly, a recent study has identified another glycan-directed antibody, mAb688, that can bind to at least four highly divergent viruses (hepatitis C virus [HCV], HIV, influenza, SARS-CoV-2) (78). The same study also discovered HIV/HCV cross-reactive antibodies that are independent of glycan for binding (78), implying a different structural motif shared across viruses. Moreover, antibodies against the HIV membrane proximal external region possess the capacity to cross-react with the viral membrane (79–81).
As noted above, pMHC cross-reactivity (also referred to as binding degeneracy) is an ingrained feature of TCRs owing to the vast diversity of potential peptide antigens that can be presented by MHC molecules (62). In addition, highly similar peptides may be frequently derived from highly dissimilar antigens due to shared statistical biases between proteomes (82), or molecular mimicry between foreign and self-antigens (61, 83). Cross-reactivity can also be applied to the binding of TCRs to highly distinct peptides presented by the same MHC allele, such as for the binding of 2C TCR to H-2Kb presenting DEV8 (EQYKFYSV) and SIY (SIYRYYGL) peptides, facilitated by a high degree of plasticity in the CDR loops upon binding (84, 85).
Furthermore, cross-reactivity is enhanced in high-affinity TCRs, often due to excess binding affinity to the MHC protein, independent of the bound peptide, resulting in a loss of peptide specificity (86). As such, TCR cross-reactivity has been a major impediment to the development of TCRs for adoptive therapy for cancer, wherein cross-reactivity can result in autoimmunity. The most notable example was the engineered affinity-enhanced TCR MAG-IC3 targeting an epitope from the cancer-testis antigen MAGE-A3 (EVDPIGHLY) presented by HLA-A*01:01, which led to the death of two patients in a clinical study (87). The likely source of this off-target toxicity was later traced back to cross-reactivity with a highly similar but unrelated peptide (ESDPIVAQY) derived from striated muscle-specific protein titin (88).
Notably, the true number of cross-reactive BCRs and TCRs, as well as the full extent of their cross-reactivity, is not known, as there is a near infinite number of antigen combinations to be analyzed. Therefore, the contributions of cross-reactivity to protection against infectious diseases and cancer as well as the initiation and exacerbation of inflammatory diseases is not well understood.
Polyreactivity (BCR)
BCRs and antibodies that are capable of binding to many distinct antigens with diverse motifs are defined as polyreactive (Fig. 1C). Unlike broadly reactive or cross-reactive antibodies, polyreactive antibodies are predicted to have multiple antibody paratopes that interact with diverse epitopes found on different antigen classes (e.g., protein, lipid, glycan). Polyreactivity is also a phenomenon distinct from polyspecificity, which represents an extreme of cross-reactivity, with binding to many highly divergent antigens with meaningful affinities (89). Polyreactive antibodies generally exhibit weak interactions with many distinct motifs based largely on hydrophobicity and charge (reviewed in (90)). For example, polyreactivity to DNA is associated with hydrogen bonding surface patches on the antibody (91), and polyreactivity to negatively charged biological components, such as heparin and cell membranes, is associated with positively charged surface patches on the antibody (92).
Conformational variability of the binding site is required to accommodate distinct antigens. Therefore, polyreactive antibodies reveal a multitude of weakly populated conformations, each of which able to recognize different binding partners (56, 93–95). This view of the antigen binding site provides a direct link of polyreactivity with flexibility (94–96). On the other hand, specificity can be understood as a reduction of conformational states, which is accompanied by an increase in probability towards the conformation responsible for binding a specific antigen. Polyreactive antibodies tend to follow the conformational selection paradigm, as all conformations pre-exist in solution and the state probabilities shift upon binding to the respective antigens (97). Therefore, while monospecificity, broad specificity, cross-reactivity, and polyspecificity are primarily driven by the nature of the epitope, polyreactivity is primarily driven by inherent properties of the paratope.
Many polyreactive antibodies possess both highly specific binding interactions with cognate antigen(s) as well as polyreactivity as a secondary feature to augment their function. Notably, numerous broadly protective antibodies against influenza viruses and HIV possess polyreactivity, which is linked to improved antibody binding avidity and neutralizing capacity (56, 80, 98, 99). In some cases, polyreactive antibodies can have strong cross-reactive antibody binding to multiple distinct antigens, as well as possess lower affinity polyreactive binding to a variety of diverse antigens, such as highly mutated polyreactive anti-HIV antibodies identified during acute infection frequently bind gut microbiota (100–104).
Promiscuity (TCR)
The extent of TCR-pMHC cross-reactivity can be so substantial that TCR binding can be described as promiscuous, with cross-reactivity to vast numbers of unrelated peptide antigens from highly divergent antigens (Fig. 1D; reviewed in (105)). This highly degenerate or promiscuous binding occurs when the CDR loops of the TCR make few contacts with the peptide sidechains, and therefore the TCR tolerates many amino acid substitutions at the remaining peptide positions (62). The most well studied example is the autoimmune TCR 1E6, which was isolated from a patient with type 1 diabetes and recognizes a preproinsulin peptide (PPI15-24) presented on HLA-A*02:01. This TCR was found to recognize over one million distinct decameric peptides in the context of this single HLA allele with up to 7 out of 10 peptide residues altered, due to TCR binding to only a small 3 amino acid motif in the center of the peptide (62, 106). In general, this highly degenerate binding is often associated with autoimmune TCRs, although all TCRs display a degree of cross-reactivity in their binding (107).
Resources and technologies to study AIR-antigen interactions
Numerous assays have been developed to understand AIR binding interactions. Here, we review traditional and emerging approaches to measure AIR binding breadth, with emerging technologies highlighted in Table 2.
Table 2:
Emerging technologies to study AIR binding modes.
| Approach | Readouts | References |
|---|---|---|
| Cryo-Electron Microscopy Polyclonal Epitope Mapping | Paratope:Epitope binding of polyclonal antibodies | (113) |
| Machine learning | AIR:Antigen binding; paratope:epitope prediction; affinity | (25, 119–125) |
| Molecular Dynamics and Structure Prediction | Confirmational AIR landscape predictions and probabilities; AIR:Antigen binding; paratope:epitope prediction | (25, 135–137, 140) |
| LIBRA-seq | Paired Gene expression, VDJ, and antigen binding; B cells only | (143) |
| Ig-Seq | Monoclonal serum specificities and clonal diversity | (151, 152, 156–159) |
| RAPTR | High-throughput screening of TCR-pMHC or BCR:Antigen interactions | (167) |
Kinetics/Affinity measurements
To measure binding kinetics of BCRs and antibodies, surface plasmon resonance (SPR) and biolayer Interferometry (BLI) are typically used and are considered the gold-standard for calculation of accurate affinity measurements and kinetics. Similarly, the kinetics of TCR-pMHC interactions are frequently measured via SPR, with recent experimental advances allowing precise measurements of low-affinity interactions (49). However, these measurements require laborious reformatting of TCRs into soluble formats and do not incorporate the contributions of CD4 and CD8 coreceptor binding to avidity (108). In contrast, TCR avidity is often measured in situ and includes the contributions of coreceptor binding, either using fluorescently-labeled pMHC multimers (e.g., tetramers) or monomers, providing ‘structural avidity’ via coreceptor binding (108). In addition, kinetics measurements obtained by these ‘three-dimensional’ assay formats have been shown to correlate less accurately with T cell activation and functional responses than ‘two-dimensional’ assay formats in which both reagents are anchored into apposing membranes (reviewed in (109)).
Structural Biology
X-ray crystallography and cryo-electron microscopy (cryo-EM) are commonly used for structural characterizations of antibody-antigen complexes, although other techniques such as nuclear magnetic resonance (NMR) are also used. Negative stain EM can also analyze heterogeneous and polyclonal samples, which has led to development of EM-based polyclonal epitope mapping (EMPEM) (110, 111), a technique that has been applied to numerous infectious diseases, including influenza viruses, HIV, and coronaviruses (CoVs) (112–114). Cryo-EMPEM can be used to generate high-resolution antibody maps and was recently used to generate maps under 4Å of polyclonal sera against HIV envelope protein (113).
In contrast to antibody-antigen complex structures, TCR-pMHC complex structures have historically been characterized by X-ray crystallography instead of cryo-EM, owing in part to the relatively small size of TCR-pMHC complexes. While most structural characterizations of BCRs and TCRs focus on their soluble forms without the transmembrane domain, recent studies have reported the cryo-EM structures of full length BCR (10, 115, 116), full-length TCR-CD3 complex (23), and the membrane-bound TCR-CD3-pMHC complex (24). These structures have provided important mechanistic insights into BCR and TCR signaling.
Drawbacks of these methods include that protein samples for X-ray crystallography and cryo-EM often need to be at a high concentration that may not be physiologically realistic. In addition, X-ray crystallography and cryo-EM only have limited ability to capture structural dynamics.
Molecular dynamics
The ability of AIRs to recognize and bind a variety of antigens is ultimately governed by dynamics, as their three-dimensional structures fluctuate constantly (18, 117). Therefore, structural and dynamic characterization of AIRs is important for understanding their biophysical properties and consequently antigen recognition (118). Molecular dynamics (MD) simulations can be used to characterize the conformational diversity of proteins by modeling conformational ensembles in solution (117). MD simulations capture the movements of atoms over time, following Newton’s second law of motion. The resulting time-resolved motions can be used to reconstruct thermodynamics and kinetics of the captured conformational rearrangements, making it a powerful tool to elucidate biophysical properties of antigen recognition by antibodies (Fig. 2A) (97). Additionally, MD simulations allow the characterization of conformational consequences of point mutations that are otherwise hard to predict from single-static structures and models (20).
Figure 2:
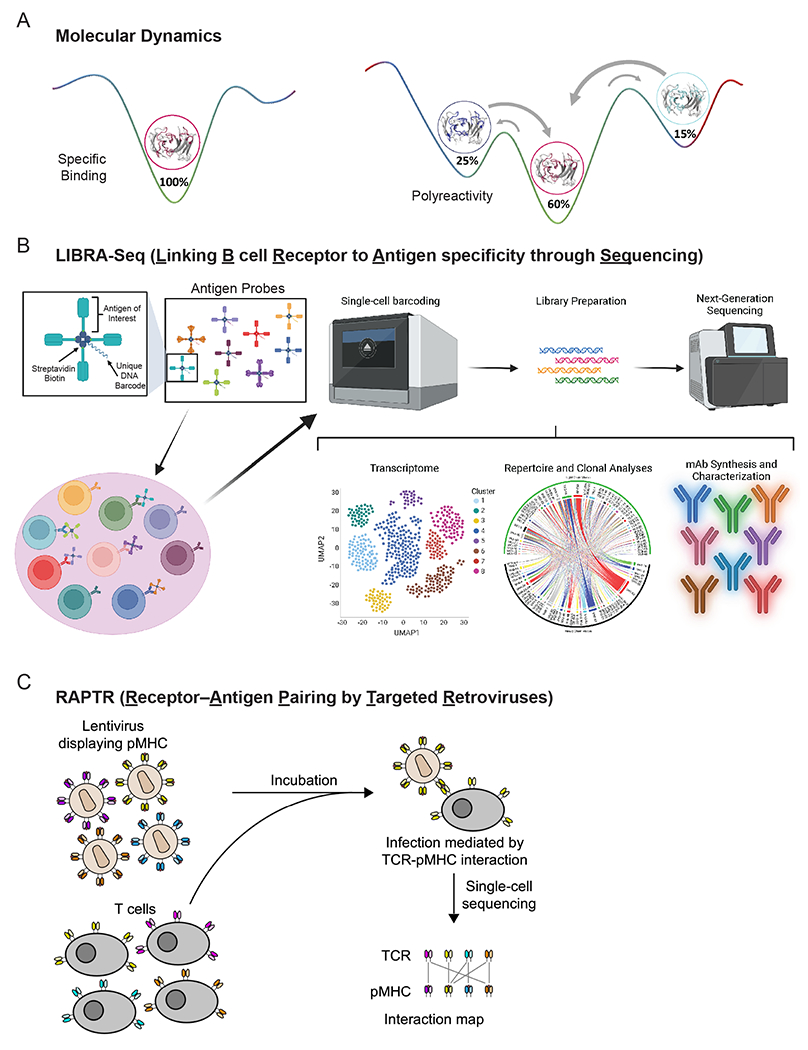
Emerging technologies to study AIR binding modes. (A) Different antibody conformations as indicator of antigen-specific binding (left) and polyreactivity (right) using molecular dynamics. (B) Depiction of LIBRA-seq workflow and utility. Panel was made in part using Biorender.com. (C) Depiction of RAPTR and application to screening TCR:pMHC libraries.
Machine Learning
Machine learning tasks focused on predicting AIR-antigen binding may take several forms such as classification of binders versus non-binders, prediction (or optimization) of affinity, and the prediction of paratope-epitope interactions. These problems may be formulated using sequence-only, structure-only, or hybrid approaches. Importantly, it was shown that combining sequence and structural information increases paratope-epitope interaction prediction accuracy (25, 119–122). Protein language models, which learn non-linear protein sequence similarity by training on million or even billions of sequences, have also shown a remarkable capacity to capture structure-based patterns from sequence-based data (123–125). However, we do not yet understand the underlying interaction rules of AIR-antigen binding, particularly the long-range sequence and structure-based patterns that are causally linked with binding. One promising approach to overcome data-limitations in machine-learning is to guide them through physics-based modeling of receptor-ligand binding energy landscapes. This technique can build upon the successes of statistical biophysics models of TCR-pMHC interactions (126–129), which have explained how thymic selection shapes the HIV-reactive T cell repertoire (127) and the breadth of TCRs binding a common pMHC (129). Ultimately, elucidating the rules of AIR-antigen binding may enable in silico screening of the binding behaviors discussed in this text (e.g., monospecificity, cross-reactivity, polyreactivity).
Structure prediction
Predicting protein structures directly from sequences has been one of the grand challenges in structural biology/computational chemistry (130). AlphaFold2 revolutionized the field by combining co-evolutionary information with machine learning to predict protein structures (131). However, predicting the structures of the variable domains of TCRs and antibodies due to their high diversity in length and structure, in particular the heavy chain CDR3 loop, still remains challenging (97, 132–134). Therefore, various antibody- and TCR- specific tools have been developed to advance structure prediction and consequently enhance the predictive power of the respective models (25, 135–137).
The performance of the available prediction tools has improved tremendously, resulting in structure models with high accuracy, reflected in low overall root-mean-square deviations relative to their X-ray crystal structures (25, 132, 135). However, special care has to be taken when basing experiments and conclusions on antibody models, as some of these structure models can suffer from physical inaccuracies, such as cis-amide bonds, D-amino acids or steric clashes (132). These issues can strongly deteriorate the quality of predictions that rely on accurate side-chain and backbone conformations such as antibody-antigen docking or surface property predictions (138, 139). Despite this, a recent study compared the predicted structure of a broadly neutralizing antibody binding to influenza virus HA to that generated via cryo-EM and found high accuracy in this prediction (140).
Broadly reactive and cross-reactive antibody binding
Testing the binding breadth of antibody responses is critical for understanding immunity against antigenically variable pathogens as well as antibodies against unknown antigens. Various assays have been developed to assess and define the cross-reactivity of antibodies, with various throughputs and resolutions. At low throughput, antibodies can be tested in tissue cross-reactivity assays, which rely on ex vivo immunohistochemistry to identify binding to sites not associated with an antibody’s target of interest, or binding to its target of interest outside of the desired tissue (141). Importantly, these assays capture binding to the target of interest and potential off-targets in their natively expressed form, and therefore are often used to support first-in-human trials. However, they are incompatible with widely expressed targets and cannot resolve putative off-targets. In addition, a yeast-based platform has been recently described that allows for the enrichment and identification of putative off-targets in a library-based format (142).
An emerging technology in identifying BCR binding breadth is LIBRA-seq (Linking B cell Receptor to Antigen specificity through Sequencing), which uses antigen probes linked to unique DNA barcodes that are detected using barcoding microfluidic technologies, such as 10x Genomics (143). LIBRA-seq allows for the simultaneous analysis of the transcriptome, antibody gene usage, and specificity of individual B cells (Fig. 2B) and has been used to investigate binding breadth of antibodies targeting HIV, influenza viruses, and SARS-CoV-2 (143–145). Noteworthy, 10x Genomics has also generated a related product line known as BEAM (Barcode Enabled Antigen Mapping) that is able to link B cell specificity and T cell specificity to gene expression and V(D)J gene usage.
Another emerging technology for analyzing binding breadth and cross-reactivity is the use of PhIP-seq (Phage Immunoprecipitation sequencing) (146, 147). PhIP-seq utilizes phage display of proteomic scale peptide libraries, with peptides up to 90 amino acids in length. Using polyclonal or monoclonal antibodies for immunoprecipitation, antibody binding breadth, cross-reactivity, and potentially polyreactivity can be assessed. PhIP-seq has been applied to various diseases widely, including identifying antibody targets of autoreactive antibodies (148), identification of broadly reactive anti-coronavirus antibodies (149), and determining the seroprevalence of antibodies against human viruses (150). However, phage-displayed peptides represent linear epitopes and lack post-translational modifications, and therefore do not accurately mimic native epitopes for which antibodies were raised. Moreover, PhIP-seq has largely been applied to polyclonal antibodies, which makes it unclear if individual antibodies possess cross-reactivity/broad reactivity or if individual antibodies against antigens are present.
Ig-Seq, also known as Ab-seq, combines proteomics and next-generation sequencing to connect polyclonal antibody specificities to monoclonal B cell clones that can be further tested for broad reactivity, cross reactivity, and polyreactivity (151–155). Polyclonal serum antibodies are subjected to mass spectrometry to determine the composition of clonotypes. In parallel, B cells are sequenced to generate a database of distinct clonotypes and for mAb production. B cells against a new trimer interface epitope of influenza HA (156) as well as antibodies against an egg-glycan (157) have been discovered using this technique. Ig-Seq elegantly allows for tracking of clonotypes overtime that can be linked to epitope specificity, binding breadth, function, and disease state (158, 159).
TCR cross-reactivity and promiscuity
Various platforms have been developed to assess TCR cross-reactivity and promiscuity. However, owing to the degenerate nature of TCR-pMHC interactions and the vast diversity of pMHCs, these assays must sample immense target spaces, utilize computational predictions, or both (60, 160, 161). Assessments of single amino acid variant (X-scan) libraries centered around a cognate peptide can provide insight into the key peptide contacts and the tolerance of the TCR to mutation at those positions (59). To assess TCR recognition more comprehensively, diverse (>1E8 variants) pMHC libraries can be expressed recombinantly in yeast- or phage-display systems, and motifs generated from these libraries can then similarly be used to predict TCR off-targets (61, 162). However, these systems cannot differentiate between activating and non-activating TCR-pMHC interactions (163). Therefore, mammalian display systems including SABR (164), MCR (165), T-Scan (166) have been developed to assess and define these interactions at high-throughput for both class I and II pMHCs.
A recently developed technique called RAPTR (Receptor–Antigen Pairing by Targeted Retroviruses) allows for high-throughput screening of TCR-pMHC interactions (167). RAPTR involves pseudotyping lentivirus with pMHC, which then can infect T cells with TCR that binds to the pMHC on the lentivirus, allowing a library-on-library screen (Fig. 2C). RAPTR has also been successfully applied to study BCR-antigen interactions (167).
Polyreactivity (BCR)
Analysis of polyreactive binding requires the use of diverse antigens. The most widely used and affordable assay is the polyreactive ELISA panel, which uses diverse antigens composed of distinct antigen types (56, 168, 169). Expanded panels frequently include HEp-2 binding to investigate self-reactive antibody binding to subcellular structures (58, 168). Moreover, soluble cell membrane and cytosolic protein extracts, baculovirus particles, and heat-shock proteins have also been used to determine polyreactive antibody binding (170–172). However, these approaches are limited to the antigens tested and therefore likely do not recapitulate the full antigen-binding landscape of polyreactive antibodies.
Limitations of AIR binding and therapeutic potential
The specificity of antibodies and TCRs is an important consideration for their development as therapeutics, as it shapes both their efficacy and safety in the clinic. Polyreactive antibodies have poor serum half-lives and biodistibutions, and may have increased propensities for immunogenicity, due to increased undesirable cell interactions, leading to higher internalization and clearance rates (89, 173). Separately, cross-reactivity can result in off-target toxicities, such as the emergence of capillary hemangiomas in patients treated with the anti-PD1 antibody camrelizumab due to cross-reactivity with pro-angiogenic receptors such as VEGFR2 (174). Similarly, off-target cross-reactivities can have disastrous consequences for TCRs. As noted above, clinical trials for a transgenic TCR T cell therapy utilizing an affinity matured TCR MAG-IC3 were halted due to severe cardiac toxicities (87). As such, it is of critical importance to both assess and understand the specificity of antibodies and TCRs to assess their capacity to be safely taken forward into the clinic.
Conclusions
We define commonly used terms to describe AIR binding modes, how these terms apply to discrete AIR classes (e.g., BCRs and antibodies versus TCRs), and methodologies to study these binding modes. While we intend to present consensus terminology for AIR binding modes, several outstanding questions remain relating to the physiologic roles of these binding modes on the generation of adaptive immune responses.
What are the physiologically relevant binding affinities for these various binding modes that are necessary for the activation and function of B and T cells?
What thresholds distinguish monospecificity from broad specificity, broad-specificity from cross-reactivity, cross-reactivity versus polyspecificity, polyspecificity from polyreactivity, and degeneracy from promiscuity?
What is the evolutionary significance of AIR binding modes? How does this relate to functional immune responses against foreign antigen and self-antigens?
While we do not put forward the answers to these questions here, we review emerging technologies that may shed light on these questions and highlight the importance for future studies to take into consideration how AIR repertoires, affinities, and binding modes shape protective and pathogenic immune responses.
Funding:
M.L.F.-Q. is supported by the APART-MINT PostDoc fellowship of the Austrian Academy of Sciences (No. 11985). J.J.G. is supported by National Institute of Allergy and Infectious Diseases (NIAID) grant numbers R00AI159136 and 75N93019C00051 and the Michelson Prize: Next Generation Grant. N.C.W. is supported by National Institutes of Health (NIH) DP2 AT011966, R01 AI167910, and the Searle Scholars Program. V.G. acknowledges funding from The Leona M. and Harry B. Helmsley Charitable Trust (#2019PG-T1D011), UiO World-Leading Research Community, UiO:LifeScience Convergence Environment Immunolingo, EU Horizon 2020 iReceptorplus (#825821), a Norwegian Cancer Society Grant (#215817), Research Council of Norway projects (#300740, #331890, #311341).
Abbreviations used in this article:
- BCR
B cell receptor
- TCR
T cell receptor
- AIR
adaptive immune receptor
- MHC
Major Histocompatibility Complex
- HLA
Human Leukocyte Antigens
- MR1
MHC class I-related protein
- V
variable gene
- D
diversity gene
- J
joining gene
- SHM
somatic hypermutation
- mIg
membrane-bound immunoglobulin
- Fc
crystallizable fragment region
- Fab
antigen-binding fragment
- Fv
variable domain
- CDR
complementarity determining region
- VH
heavy chain
- VL
light chain
- FR
framework region
- pMHC
peptide bound MHC
- Tregs
regulatory T cells
- KD
dissociation constant
- HA
hemagglutinin
- SPR
surface plasmon resonance
- BLI
biolayer interferometry
- Cryo-EM
cryogenic electron microscopy
- NMR
nuclear magnetic resonance
- EMPEM
electron microscopy based polyclonal epitope mapping
- CoV
coronaviruses
- MD
molecular dynamics
Footnotes
Conflict of interest statement: C.G.R. is an employee of Adimab, LLC, and may hold shares of Adimab, LLC. N.C.W consults for HeliXon. V.G. declares advisory board positions in aiNET GmbH, Enpicom B.V, Absci, Omniscope, and Diagonal Therapeutics. VG is a consultant for Adaptyv Biosystems, Specifica Inc, Roche/Genentech, Immunai, and LabGenius.
References
- 1.Chi X, Li Y, and Qiu X. 2020. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology 160: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikocziova I, Greiff V, and Sollid LM. 2021. Immunoglobulin germline gene variation and its impact on human disease. Genes Immun 22: 205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sangesland M, Torrents de la Pena A, Boyoglu-Barnum S, Ronsard L, Mohamed FAN, Moreno TB, Barnes RM, Rohrer D, Lonberg N, Ghebremichael M, Kanekiyo M, Ward A, and Lingwood D. 2022. Allelic polymorphism controls autoreactivity and vaccine elicitation of human broadly neutralizing antibodies against influenza virus. Immunity 55: 1693–1709 e1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avnir Y, Watson CT, Glanville J, Peterson EC, Tallarico AS, Bennett AS, Qin K, Fu Y, Huang CY, Beigel JH, Breden F, Zhu Q, and Marasco WA. 2016. IGHV1-69 polymorphism modulates anti-influenza antibody repertoires, correlates with IGHV utilization shifts and varies by ethnicity. Sci Rep 6: 20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez OL, Safonova Y, Silver CA, Shields K, Gibson WS, Kos JT, Tieri D, Ke H, Jackson KJL, Boyd SD, Smith ML, Marasco WA, and Watson CT. 2022. Genetic variation in the immunoglobulin heavy chain locus shapes the human antibody repertoire. bioRxiv: 2022.2007.2004.498729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee JH, Toy L, Kos JT, Safonova Y, Schief WR, Havenar-Daughton C, Watson CT, and Crotty S. 2021. Vaccine genetics of IGHV1-2 VRC01-class broadly neutralizing antibody precursor naive human B cells. NPJ Vaccines 6: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parks T, Mirabel MM, Kado J, Auckland K, Nowak J, Rautanen A, Mentzer AJ, Marijon E, Jouven X, Perman ML, Cua T, Kauwe JK, Allen JB, Taylor H, Robson KJ, Deane CM, Steer AC, Hill AVS, and Pacific N Islands Rheumatic Heart Disease Genetics. 2017. Association between a common immunoglobulin heavy chain allele and rheumatic heart disease risk in Oceania. Nat Commun 8: 14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo B, Dobritzsch D, Ge C, Ekman D, Xu B, Lindh I, Forster M, Uysal H, Nandakumar KS, Schneider G, and Holmdahl R. 2014. Epitope-specific antibody response is controlled by immunoglobulin V(H) polymorphisms. J Exp Med 211: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Lazikani B, Lesk AM, and Chothia C. 1997. Standard conformations for the canonical structures of immunoglobulins. J Mol Biol 273: 927–948. [DOI] [PubMed] [Google Scholar]
- 10.Dong Y, Pi X, Bartels-Burgahn F, Saltukoglu D, Liang Z, Yang J, Alt FW, Reth M, and Wu H. 2022. Structural principles of B cell antigen receptor assembly. Nature 612: 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkitaraman AR, Williams GT, Dariavach P, and Neuberger MS. 1991. The B-cell antigen receptor of the five immunoglobulin classes. Nature 352: 777–781. [DOI] [PubMed] [Google Scholar]
- 12.Chiu ML, Goulet DR, Teplyakov A, and Gilliland GL. 2019. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies (Basel) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feige MJ, Hendershot LM, and Buchner J. 2010. How antibodies fold. Trends Biochem Sci 35: 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder HW Jr., and Cavacini L. 2010. Structure and function of immunoglobulins. J Allergy Clin Immunol 125: S41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weitzner BD, Dunbrack RL Jr., and Gray JJ. 2015. The origin of CDR H3 structural diversity. Structure 23: 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunbar J, Fuchs A, Shi J, and Deane CM. 2013. ABangle: characterising the VH-VL orientation in antibodies. Protein Eng Des Sel 26: 611–620. [DOI] [PubMed] [Google Scholar]
- 17.Bujotzek A, Lipsmeier F, Harris SF, Benz J, Kuglstatter A, and Georges G. 2016. VH-VL orientation prediction for antibody humanization candidate selection: A case study. MAbs 8: 288–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez-Quintero ML, Georges G, Varga JM, and Liedl KR. 2021. Ensembles in solution as a new paradigm for antibody structure prediction and design. MAbs 13: 1923122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teplyakov A, Obmolova G, Malia TJ, Luo J, Muzammil S, Sweet R, Almagro JC, and Gilliland GL. 2016. Structural diversity in a human antibody germline library. MAbs 8: 1045–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Quintero ML, Kroell KB, Hofer F, Riccabona JR, and Liedl KR. 2021. Mutation of Framework Residue H71 Results in Different Antibody Paratope States in Solution. Front Immunol 12: 630034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tramontano A, Chothia C, and Lesk AM. 1990. Framework residue 71 is a major determinant of the position and conformation of the second hypervariable region in the VH domains of immunoglobulins. J Mol Biol 215: 175–182. [DOI] [PubMed] [Google Scholar]
- 22.Krauss J, Arndt MA, Zhu Z, Newton DL, Vu BK, Choudhry V, Darbha R, Ji X, Courtenay-Luck NS, Deonarain MP, Richards J, and Rybak SM. 2004. Impact of antibody framework residue VH-71 on the stability of a humanised anti-MUC1 scFv and derived immunoenzyme. Br J Cancer 90: 1863–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong D, Zheng L, Lin J, Zhang B, Zhu Y, Li N, Xie S, Wang Y, Gao N, and Huang Z. 2019. Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 573: 546–552. [DOI] [PubMed] [Google Scholar]
- 24.Susac L, Vuong MT, Thomas C, von Bulow S, O’Brien-Ball C, Santos AM, Fernandes RA, Hummer G, Tampe R, and Davis SJ. 2022. Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC. Cell 185: 3201–3213 e3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley P 2023. Structure-based prediction of T cell receptor:peptide-MHC interactions. Elife 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leem J, de Oliveira SHP, Krawczyk K, and Deane CM. 2018. STCRDab: the structural T-cell receptor database. Nucleic Acids Res 46: D406–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Quintero ML, Pomarici ND, Loeffler JR, Seidler CA, and Liedl KR. 2020. T-Cell Receptor CDR3 Loop Conformations in Solution Shift the Relative Valpha-Vbeta Domain Distributions. Front Immunol 11: 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schattgen SA, and Thomas PG. 2018. Bohemian T cell receptors: sketching the repertoires of unconventional lymphocytes. Immunol Rev 284: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szeto C, Lobos CA, Nguyen AT, and Gras S. 2020. TCR Recognition of Peptide-MHC-I: Rule Makers and Breakers. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundberg EJ, Deng L, and Mariuzza RA. 2007. TCR recognition of peptide/MHC class II complexes and superantigens. Semin Immunol 19: 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zareie P, Szeto C, Farenc C, Gunasinghe SD, Kolawole EM, Nguyen A, Blyth C, Sng XYX, Li J, Jones CM, Fulcher AJ, Jacobs JR, Wei Q, Wojciech L, Petersen J, Gascoigne NRJ, Evavold BD, Gaus K, Gras S, Rossjohn J, and La Gruta NL. 2021. Canonical T cell receptor docking on peptide-MHC is essential for T cell signaling. Science 372. [DOI] [PubMed] [Google Scholar]
- 32.Sela-Culang I, Kunik V, and Ofran Y. 2013. The structural basis of antibody-antigen recognition. Front Immunol 4: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.North B, Lehmann A, and Dunbrack RL Jr. 2011. A new clustering of antibody CDR loop conformations. J Mol Biol 406: 228–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu NC, Andrews SF, Raab JE, O’Connell S, Schramm CA, Ding X, Chambers MJ, Leung K, Wang L, Zhang Y, Mascola JR, Douek DC, Ledgerwood JE, McDermott AB, and Wilson IA. 2020. Convergent Evolution in Breadth of Two V(H)6-1-Encoded Influenza Antibody Clonotypes from a Single Donor. Cell Host Microbe 28: 434–444 e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, Gnanapragasam PN, Oliveira TY, Seaman MS, Kwong PD, Bjorkman PJ, and Nussenzweig MC. 2013. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 153: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuda K, Sakamoto K, Kojima M, Aburatani T, Ueda T, and Ueda H. 2006. The role of interface framework residues in determining antibody V(H)/V(L) interaction strength and antigen-binding affinity. FEBS J 273: 2184–2194. [DOI] [PubMed] [Google Scholar]
- 37.Khalifa MB, Weidenhaupt M, Choulier L, Chatellier J, Rauffer-Bruyere N, Altschuh D, and Vernet T. 2000. Effects on interaction kinetics of mutations at the VH-VL interface of Fabs depend on the structural context. J Mol Recognit 13: 127–139. [DOI] [PubMed] [Google Scholar]
- 38.Koenig P, Lee CV, Walters BT, Janakiraman V, Stinson J, Patapoff TW, and Fuh G. 2017. Mutational landscape of antibody variable domains reveals a switch modulating the interdomain conformational dynamics and antigen binding. Proc Natl Acad Sci U S A 114: E486–E495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warszawski S, Borenstein Katz A, Lipsh R, Khmelnitsky L, Ben Nissan G, Javitt G, Dym O, Unger T, Knop O, Albeck S, Diskin R, Fass D, Sharon M, and Fleishman SJ. 2019. Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLoS Comput Biol 15: e1007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez PA, Carreno LJ, Coombs D, Mora JE, Palmieri E, Goldstein B, Nathenson SG, and Kalergis AM. 2005. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc Natl Acad Sci U S A 102: 4824–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ketchum C, Miller H, Song W, and Upadhyaya A. 2014. Ligand mobility regulates B cell receptor clustering and signaling activation. Biophys J 106: 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor JJ, Pape KA, Steach HR, and Jenkins MK. 2015. Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naive B cell. Science 347: 784–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer E, and Naeher D. 2009. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat Rev Immunol 9: 207–213. [DOI] [PubMed] [Google Scholar]
- 44.Callegari I, Schneider M, Berloffa G, Muhlethaler T, Holdermann S, Galli E, Roloff T, Boss R, Infanti L, Khanna N, Egli A, Buser A, Zimmer G, Derfuss T, and Sanderson NSR. 2022. Potent neutralization by monoclonal human IgM against SARS-CoV-2 is impaired by class switch. EMBO Rep 23: e53956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hale M, Netland J, Chen Y, Thouvenel CD, Smith KN, Rich LM, Vanderwall ER, Miranda MC, Eggenberger J, Hao L, Watson MJ, Mundorff CC, Rodda LB, King NP, Guttman M, Gale M, Abraham J, Debley JS, Pepper M, and Rawlings DJ. 2022. IgM antibodies derived from memory B cells are potent cross-variant neutralizers of SARS-CoV-2. J Exp Med 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thouvenel CD, Fontana MF, Netland J, Krishnamurty AT, Takehara KK, Chen Y, Singh S, Miura K, Keitany GJ, Lynch EM, Portugal S, Miranda MC, King NP, Kollman JM, Crompton PD, Long CA, Pancera M, Rawlings DJ, and Pepper M. 2021. Multimeric antibodies from antigen-specific human IgM+ memory B cells restrict Plasmodium parasites. J Exp Med 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hopp CS, Sekar P, Diouf A, Miura K, Boswell K, Skinner J, Tipton CM, Peterson ME, Chambers MJ, Andrews S, Lu J, Tan J, Li S, Doumbo S, Kayentao K, Ongoiba A, Traore B, Portugal S, Sun PD, Long C, Koup RA, Long EO, McDermott AB, and Crompton PD. 2021. Plasmodium falciparum-specific IgM B cells dominate in children, expand with malaria, and produce functional IgM. J Exp Med 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh T, Hwang KK, Miller AS, Jones RL, Lopez CA, Dulson SJ, Giuberti C, Gladden MA, Miller I, Webster HS, Eudailey JA, Luo K, Von Holle T, Edwards RJ, Valencia S, Burgomaster KE, Zhang S, Mangold JF, Tu JJ, Dennis M, Alam SM, Premkumar L, Dietze R, Pierson TC, Ooi EE, Lazear HM, Kuhn RJ, Permar SR, and Bonsignori M. 2022. A Zika virus-specific IgM elicited in pregnancy exhibits ultrapotent neutralization. Cell 185: 4826–4840 e4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettmann J, Huhn A, Shah E. Abu, Kutuzov MA, Wilson DB, Dustin ML, Davis SJ, van der Merwe PA, and Dushek O. 2021. The discriminatory power of the T cell receptor. Elife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foote J, and Eisen HN. 1995. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A 92: 1254–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foote J, and Eisen HN. 2000. Breaking the affinity ceiling for antibodies and T cell receptors. Proc Natl Acad Sci U S A 97: 10679–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, and Zhu C. 2010. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 464: 932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greaney AJ, Starr TN, Barnes CO, Weisblum Y, Schmidt F, Caskey M, Gaebler C, Cho A, Agudelo M, Finkin S, Wang Z, Poston D, Muecksch F, Hatziioannou T, Bieniasz PD, Robbiani DF, Nussenzweig MC, Bjorkman PJ, and Bloom JD. 2021. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat Commun 12: 4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE Jr., and Bloom JD. 2021. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29: 44–57 e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guthmiller JJ, Han J, Li L, Freyn AW, Liu STH, Stovicek O, Stamper CT, Dugan HL, Tepora ME, Utset HA, Bitar DJ, Hamel NJ, Changrob S, Zheng NY, Huang M, Krammer F, Nachbagauer R, Palese P, Ward AB, and Wilson PC. 2021. First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes. Sci Transl Med 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guthmiller JJ, Lan LY, Fernandez-Quintero ML, Han J, Utset HA, Bitar DJ, Hamel NJ, Stovicek O, Li L, Tepora M, Henry C, Neu KE, Dugan HL, Borowska MT, Chen YQ, Liu STH, Stamper CT, Zheng NY, Huang M, Palm AE, Garcia-Sastre A, Nachbagauer R, Palese P, Coughlan L, Krammer F, Ward AB, Liedl KR, and Wilson PC. 2020. Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses. Immunity 53: 1230–1244 e1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dugan HL, Guthmiller JJ, Arevalo P, Huang M, Chen YQ, Neu KE, Henry C, Zheng NY, Lan LY, Tepora ME, Stovicek O, Bitar D, Palm AE, Stamper CT, Changrob S, Utset HA, Coughlan L, Krammer F, Cobey S, and Wilson PC. 2020. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee JH, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, and Wilson PC. 2015. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 7: 316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luksza M, Sethna ZM, Rojas LA, Lihm J, Bravi B, Elhanati Y, Soares K, Amisaki M, Dobrin A, Hoyos D, Guasp P, Zebboudj A, Yu R, Chandra AK, Waters T, Odgerel Z, Leung J, Kappagantula R, Makohon-Moore A, Johns A, Gill A, Gigoux M, Wolchok J, Merghoub T, Sadelain M, Patterson E, Monasson R, Mora T, Walczak AM, Cocco S, Iacobuzio-Donahue C, Greenbaum BD, and Balachandran VP. 2022. Neoantigen quality predicts immunoediting in survivors of pancreatic cancer. Nature 606: 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vazquez-Lombardi R, Jung JS, Schlatter FS, Mei A, Mantuano NR, Bieberich F, Hong KL, Kucharczyk J, Kapetanovic E, Aznauryan E, Weber CR, Zippelius A, Laubli H, and Reddy ST. 2022. High-throughput T cell receptor engineering by functional screening identifies candidates with enhanced potency and specificity. Immunity 55: 1953–1966 e1910. [DOI] [PubMed] [Google Scholar]
- 61.Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, and Garcia KC. 2014. Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157: 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sewell AK 2012. Why must T cells be cross-reactive? Nat Rev Immunol 12: 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, Kong WP, Leung K, Narpala SN, Prabhakaran MS, Yang ES, Zhang B, Zhang Y, Asokan M, Boyington JC, Bylund T, Darko S, Lees CR, Ransier A, Shen CH, Wang L, Whittle JR, Wu X, Yassine HM, Santos C, Matsuoka Y, Tsybovsky Y, Baxa U, Program NCS, Mullikin JC, Subbarao K, Douek DC, Graham BS, Koup RA, Ledgerwood JE, Roederer M, Shapiro L, Kwong PD, Mascola JR, and McDermott AB. 2016. Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses. Cell 166: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, and Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329: 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, Wang F, Yin L, Jiang H, Lu X, Bi Y, Zhang W, Shi Y, Burioni R, Tong Z, Song H, Qi J, and Gao GF. 2022. Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes. Nat Commun 13: 7603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, and Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333: 850–856. [DOI] [PubMed] [Google Scholar]
- 67.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, Vorlaender MK, Bianchi S, Guarino B, De Marco A, Vanzetta F, Agatic G, Foglierini M, Pinna D, Fernandez-Rodriguez B, Fruehwirth A, Silacci C, Ogrodowicz RW, Martin SR, Sallusto F, Suzich JA, Lanzavecchia A, Zhu Q, Gamblin SJ, and Skehel JJ. 2016. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166: 596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madura F, Rizkallah PJ, Legut M, Holland CJ, Fuller A, Bulek A, Schauenburg AJ, Trimby A, Hopkins JR, Wells SA, Godkin A, Miles JJ, Sami M, Li Y, Liddy N, Jakobsen BK, Loveridge EJ, Cole DK, and Sewell AK. 2019. TCR-induced alteration of primary MHC peptide anchor residue. Eur J Immunol 49: 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madura F, Rizkallah PJ, Holland CJ, Fuller A, Bulek A, Godkin AJ, Schauenburg AJ, Cole DK, and Sewell AK. 2015. Structural basis for ineffective T-cell responses to MHC anchor residue-improved “heteroclitic” peptides. Eur J Immunol 45: 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cusick MF, Libbey JE, and Fujinami RS. 2012. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol 42: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krisher K, and Cunningham MW. 1985. Myosin: a link between streptococci and heart. Science 227: 413–415. [DOI] [PubMed] [Google Scholar]
- 72.Huber SA, and Cunningham MW. 1996. Streptococcal M protein peptide with similarity to myosin induces CD4+ T cell-dependent myocarditis in MRL/++ mice and induces partial tolerance against coxsakieviral myocarditis. J Immunol 156: 3528–3534. [PubMed] [Google Scholar]
- 73.Phillips GN Jr., Flicker PF, Cohen C, Manjula BN, and Fischetti VA. 1981. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci U S A 78: 4689–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, and Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70: 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murin CD, Julien JP, Sok D, Stanfield RL, Khayat R, Cupo A, Moore JP, Burton DR, Wilson IA, and Ward AB. 2014. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol 88: 10177–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seabright GE, Cottrell CA, van Gils MJ, D’Addabbo A, Harvey DJ, Behrens AJ, Allen JD, Watanabe Y, Scaringi N, Polveroni TM, Maker A, Vasiljevic S, de Val N, Sanders RW, Ward AB, and Crispin M. 2020. Networks of HIV-1 Envelope Glycans Maintain Antibody Epitopes in the Face of Glycan Additions and Deletions. Structure 28: 897–909 e896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee CD, Watanabe Y, Wu NC, Han J, Kumar S, Pholcharee T, Seabright GE, Allen JD, Lin CW, Yang JR, Liu MT, Wu CY, Ward AB, Crispin M, and Wilson IA. 2021. A cross-neutralizing antibody between HIV-1 and influenza virus. PLoS Pathog 17: e1009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pilewski KA, Wall S, Richardson SI, Manamela NP, Clark K, Hermanus T, Binshtein E, Venkat R, Sautto GA, Kramer KJ, Shiakolas AR, Setliff I, Salas J, Mapengo RE, Suryadevara N, Brannon JR, Beebout CJ, Parks R, Raju N, Frumento N, Walker LM, Fechter EF, Qin JS, Murji AA, Janowska K, Thakur B, Lindenberger J, May AJ, Huang X, Sammour S, Acharya P, Carnahan RH, Ross TM, Haynes BF, Hadjifrangiskou M, Crowe JE Jr., Bailey JR, Kalams S, Morris L, and Georgiev IS. 2023. Functional HIV-1/HCV cross-reactive antibodies isolated from a chronically co-infected donor. Cell Rep 42: 112044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, Burton DR, and Wilson IA. 2005. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22: 163–173. [DOI] [PubMed] [Google Scholar]
- 80.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, and Alam SM. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308: 1906–1908. [DOI] [PubMed] [Google Scholar]
- 81.Irimia A, Sarkar A, Stanfield RL, and Wilson IA. 2016. Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10. Immunity 44: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mayer A, Russo CJ, Marcou Q, Bialek W, and Greenbaum BD. 2022. How different are self and nonself? arXiv preprint arXiv:2212.12049. [Google Scholar]
- 83.Rojas M, Restrepo-Jimenez P, Monsalve DM, Pacheco Y, Acosta-Ampudia Y, Ramirez-Santana C, Leung PSC, Ansari AA, Gershwin ME, and Anaya JM. 2018. Molecular mimicry and autoimmunity. J Autoimmun 95: 100–123. [DOI] [PubMed] [Google Scholar]
- 84.Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, and Wilson IA. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science 279: 1166–1172. [DOI] [PubMed] [Google Scholar]
- 85.Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, and Wilson IA. 2000. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity 12: 251–261. [DOI] [PubMed] [Google Scholar]
- 86.Stone JD, and Kranz DM. 2013. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front Immunol 4: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Linette GP, Stadtmauer EA, Maus MV, Rapoport AP, Levine BL, Emery L, Litzky L, Bagg A, Carreno BM, Cimino PJ, Binder-Scholl GK, Smethurst DP, Gerry AB, Pumphrey NJ, Bennett AD, Brewer JE, Dukes J, Harper J, Tayton-Martin HK, Jakobsen BK, Hassan NJ, Kalos M, and June CH. 2013. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 122: 863–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, Bossi G, Vuidepot A, Powlesland AS, Legg A, Adams KJ, Bennett AD, Pumphrey NJ, Williams DD, Binder-Scholl G, Kulikovskaya I, Levine BL, Riley JL, Varela-Rohena A, Stadtmauer EA, Rapoport AP, Linette GP, June CH, Hassan NJ, Kalos M, and Jakobsen BK. 2013. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med 5: 197ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cunningham O, Scott M, Zhou ZS, and Finlay WJJ. 2021. Polyreactivity and polyspecificity in therapeutic antibody development: risk factors for failure in preclinical and clinical development campaigns. MAbs 13: 1999195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ausserwöger H, Schneider MM, Herling TW, Arosio P, Invernizzi G, Knowles TPJ, and Lorenzen N. 2022. Non-specificity as the sticky problem in therapeutic antibody development. Nature Reviews Chemistry 6: 844–861. [DOI] [PubMed] [Google Scholar]
- 91.Ausserwöger H, Krainer G, Welsh TJ, Sneideris T, Schneider MM, Invernizzi G, Herling TW, Lorenzen N, and Knowles TPJ. 2022. Surface interaction patches link non-specific binding and phase separation of antibodies. bioRxiv: 2022.2003.2007.483238. [Google Scholar]
- 92.Datta-Mannan A, Thangaraju A, Leung D, Tang Y, Witcher DR, Lu J, and Wroblewski VJ. 2015. Balancing charge in the complementarity-determining regions of humanized mAbs without affecting pI reduces non-specific binding and improves the pharmacokinetics. MAbs 7: 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Babor M, and Kortemme T. 2009. Multi-constraint computational design suggests that native sequences of germline antibody H3 loops are nearly optimal for conformational flexibility. Proteins 75: 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fernandez-Quintero ML, Loeffler JR, Bacher LM, Waibl F, Seidler CA, and Liedl KR. 2020. Local and Global Rigidification Upon Antibody Affinity Maturation. Front Mol Biosci 7: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fernandez-Quintero ML, Loeffler JR, Kraml J, Kahler U, Kamenik AS, and Liedl KR. 2018. Characterizing the Diversity of the CDR-H3 Loop Conformational Ensembles in Relationship to Antibody Binding Properties. Front Immunol 9: 3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thorpe IF, and Brooks CL 3rd. 2007. Molecular evolution of affinity and flexibility in the immune system. Proc Natl Acad Sci U S A 104: 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fernandez-Quintero ML, Kraml J, Georges G, and Liedl KR. 2019. CDR-H3 loop ensemble in solution - conformational selection upon antibody binding. MAbs 11: 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, Walker BD, Ho DD, Wilson PC, Seaman MS, Eisen HN, Chakraborty AK, Hope TJ, Ravetch JV, Wardemann H, and Nussenzweig MC. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prigent J, Jarossay A, Planchais C, Eden C, Dufloo J, Kok A, Lorin V, Vratskikh O, Couderc T, Bruel T, Schwartz O, Seaman MS, Ohlenschlager O, Dimitrov JD, and Mouquet H. 2018. Conformational Plasticity in Broadly Neutralizing HIV-1 Antibodies Triggers Polyreactivity. Cell Rep 23: 2568–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cram JA, Fiore-Gartland AJ, Srinivasan S, Karuna S, Pantaleo G, Tomaras GD, Fredricks DN, and Kublin JG. 2019. Human gut microbiota is associated with HIV-reactive immunoglobulin at baseline and following HIV vaccination. PLoS One 14: e0225622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, Gao F, Markowitz M, Heath SL, Bar KJ, Goepfert PA, Montefiori DC, Shaw GC, Alam SM, Margolis DM, Denny TN, Boyd SD, Marshal E, Egholm M, Simen BB, Hanczaruk B, Fire AZ, Voss G, Kelsoe G, Tomaras GD, Moody MA, Kepler TB, and Haynes BF. 2011. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med 208: 2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Planchais C, Kok A, Kanyavuz A, Lorin V, Bruel T, Guivel-Benhassine F, Rollenske T, Prigent J, Hieu T, Prazuck T, Lefrou L, Wardemann H, Schwartz O, Dimitrov JD, Hocqueloux L, and Mouquet H. 2019. HIV-1 Envelope Recognition by Polyreactive and Cross-Reactive Intestinal B Cells. Cell Rep 27: 572–585 e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trama AM, Moody MA, Alam SM, Jaeger FH, Lockwood B, Parks R, Lloyd KE, Stolarchuk C, Scearce R, Foulger A, Marshall DJ, Whitesides JF, Jeffries TL Jr., Wiehe K, Morris L, Lambson B, Soderberg K, Hwang KK, Tomaras GD, Vandergrift N, Jackson KJL, Roskin KM, Boyd SD, Kepler TB, Liao HX, and Haynes BF. 2014. HIV-1 envelope gp41 antibodies can originate from terminal ileum B cells that share cross-reactivity with commensal bacteria. Cell Host Microbe 16: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams WB, Liao HX, Moody MA, Kepler TB, Alam SM, Gao F, Wiehe K, Trama AM, Jones K, Zhang R, Song H, Marshall DJ, Whitesides JF, Sawatzki K, Hua A, Liu P, Tay MZ, Seaton KE, Shen X, Foulger A, Lloyd KE, Parks R, Pollara J, Ferrari G, Yu JS, Vandergrift N, Montefiori DC, Sobieszczyk ME, Hammer S, Karuna S, Gilbert P, Grove D, Grunenberg N, McElrath MJ, Mascola JR, Koup RA, Corey L, Nabel GJ, Morgan C, Churchyard G, Maenza J, Keefer M, Graham BS, Baden LR, Tomaras GD, and Haynes BF. 2015. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science 349: aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilson DB, Wilson DH, Schroder K, Pinilla C, Blondelle S, Houghten RA, and Garcia KC. 2004. Specificity and degeneracy of T cells. Mol Immunol 40: 1047–1055. [DOI] [PubMed] [Google Scholar]
- 106.Cole DK, Bulek AM, Dolton G, Schauenberg AJ, Szomolay B, Rittase W, Trimby A, Jothikumar P, Fuller A, Skowera A, Rossjohn J, Zhu C, Miles JJ, Peakman M, Wooldridge L, Rizkallah PJ, and Sewell AK. 2016. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J Clin Invest 126: 2191–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bentzen AK, and Hadrup SR. 2019. T-cell-receptor cross-recognition and strategies to select safe T-cell receptors for clinical translation. Immunooncol Technol 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hebeisen M, Allard M, Gannon PO, Schmidt J, Speiser DE, and Rufer N. 2015. Identifying Individual T Cell Receptors of Optimal Avidity for Tumor Antigens. Front Immunol 6: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Edwards LJ, Zarnitsyna VI, Hood JD, Evavold BD, and Zhu C. 2012. Insights into T cell recognition of antigen: significance of two-dimensional kinetic parameters. Front Immunol 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bianchi M, Turner HL, Nogal B, Cottrell CA, Oyen D, Pauthner M, Bastidas R, Nedellec R, McCoy LE, Wilson IA, Burton DR, Ward AB, and Hangartner L. 2018. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 49: 288–300 e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nogal B, Bianchi M, Cottrell CA, Kirchdoerfer RN, Sewall LM, Turner HL, Zhao F, Sok D, Burton DR, Hangartner L, and Ward AB. 2020. Mapping Polyclonal Antibody Responses in Non-human Primates Vaccinated with HIV Env Trimer Subunit Vaccines. Cell Rep 30: 3755–3765 e3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han J, Schmitz AJ, Richey ST, Dai YN, Turner HL, Mohammed BM, Fremont DH, Ellebedy AH, and Ward AB. 2021. Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Cell Rep 34: 108682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Antanasijevic A, Sewall LM, Cottrell CA, Carnathan DG, Jimenez LE, Ngo JT, Silverman JB, Groschel B, Georgeson E, Bhiman J, Bastidas R, LaBranche C, Allen JD, Copps J, Perrett HR, Rantalainen K, Cannac F, Yang YR, de la Pena AT, Rocha RF, Berndsen ZT, Baker D, King NP, Sanders RW, Moore JP, Crotty S, Crispin M, Montefiori DC, Burton DR, Schief WR, Silvestri G, and Ward AB. 2021. Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM. Nat Commun 12: 4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bangaru S, Antanasijevic A, Kose N, Sewall LM, Jackson AM, Suryadevara N, Zhan X, Torres JL, Copps J, de la Pena AT, Crowe JE Jr., and Ward AB. 2022. Structural mapping of antibody landscapes to human betacoronavirus spike proteins. Sci Adv 8: eabn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Su Q, Chen M, Shi Y, Zhang X, Huang G, Huang B, Liu D, Liu Z, and Shi Y. 2022. Cryo-EM structure of the human IgM B cell receptor. Science 377: 875–880. [DOI] [PubMed] [Google Scholar]
- 116.Ma X, Zhu Y, Dong D, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, and Huang Z. 2022. Cryo-EM structures of two human B cell receptor isotypes. Science 377: 880–885. [DOI] [PubMed] [Google Scholar]
- 117.Henzler-Wildman K, and Kern D. 2007. Dynamic personalities of proteins. Nature 450: 964–972. [DOI] [PubMed] [Google Scholar]
- 118.Kern D 2021. From structure to mechanism: skiing the energy landscape. Nat Methods 18: 435–436. [DOI] [PubMed] [Google Scholar]
- 119.Akbar R, Robert PA, Pavlovic M, Jeliazkov JR, Snapkov I, Slabodkin A, Weber CR, Scheffer L, Miho E, Haff IH, Haug DTT, Lund-Johansen F, Safonova Y, Sandve GK, and Greiff V. 2021. A compact vocabulary of paratope-epitope interactions enables predictability of antibody-antigen binding. Cell Rep 34: 108856. [DOI] [PubMed] [Google Scholar]
- 120.Eguchi RR, Choe CA, and Huang PS. 2022. Ig-VAE: Generative modeling of protein structure by direct 3D coordinate generation. PLoS Comput Biol 18: e1010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aguilar Rangel M, Bedwell A, Costanzi E, Taylor RJ, Russo R, Bernardes GJL, Ricagno S, Frydman J, Vendruscolo M, and Sormanni P. 2022. Fragment-based computational design of antibodies targeting structured epitopes. Sci Adv 8: eabp9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Robert PA, Akbar R, Frank R, Pavlović M, Widrich M, Snapkov I, Slabodkin A, Chernigovskaya M, Scheffer L, Smorodina E, Rawat P, Mehta BB, Vu MH, Mathisen IF, Prósz A, Abram K, Olar A, Miho E, Haug DTT, Lund-Johansen F, Hochreiter S, Haff IH, Klambauer G, Sandve GK, and Greiff V. 2022. Unconstrained generation of synthetic antibody–antigen structures to guide machine learning methodology for antibody specificity prediction. Nature Computational Science 2: 845–865. [DOI] [PubMed] [Google Scholar]
- 123.Shuai RW, Ruffolo JA, and Gray JJ. 2021. Generative Language Modeling for Antibody Design. bioRxiv: 2021.2012.2013.472419. [Google Scholar]
- 124.Chowdhury R, Bouatta N, Biswas S, Floristean C, Kharkar A, Roy K, Rochereau C, Ahdritz G, Zhang J, Church GM, Sorger PK, and AlQuraishi M. 2022. Single-sequence protein structure prediction using a language model and deep learning. Nat Biotechnol 40: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Madani A, Krause B, Greene ER, Subramanian S, Mohr BP, Holton JM, Olmos JL Jr., Xiong C, Sun ZZ, Socher R, Fraser JS, and Naik N. 2023. Large language models generate functional protein sequences across diverse families. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kosmrlj A, Jha AK, Huseby ES, Kardar M, and Chakraborty AK. 2008. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc Natl Acad Sci U S A 105: 16671–16676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, and Chakraborty AK. 2010. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature 465: 350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lin X, George JT, Schafer NP, Chau KN, Birnbaum ME, Clementi C, Onuchic JN, and Levine H. 2021. Rapid Assessment of T-Cell Receptor Specificity of the Immune Repertoire. Nat Comput Sci 1: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mayer A, and Callan CG Jr. 2023. Measures of epitope binding degeneracy from T cell receptor repertoires. Proc Natl Acad Sci U S A 120: e2213264120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Al-Lazikani B, Jung J, Xiang Z, and Honig B. 2001. Protein structure prediction. Curr Opin Chem Biol 5: 51–56. [DOI] [PubMed] [Google Scholar]
- 131.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, and Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fernández-Quintero ML, Kokot J, Waibl F, Fischer A-LM, Quoika PK, Deane CM, and Liedl KR. 2022. Challenges in antibody structure prediction. bioRxiv: 2022.2011.2009.515600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Finton KA, Friend D, Jaffe J, Gewe M, Holmes MA, Larman HB, Stuart A, Larimore K, Greenberg PD, Elledge SJ, Stamatatos L, and Strong RK. 2014. Ontogeny of recognition specificity and functionality for the broadly neutralizing anti-HIV antibody 4E10. PLoS Pathog 10: e1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Regep C, Georges G, Shi J, Popovic B, and Deane CM. 2017. The H3 loop of antibodies shows unique structural characteristics. Proteins 85: 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abanades B, Wong WK, Boyles F, Georges G, Bujotzek A, and Deane CM. 2022. ImmuneBuilder: Deep-Learning models for predicting the structures of immune proteins. bioRxiv: 2022.2011.2004.514231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wong WK, Marks C, Leem J, Lewis AP, Shi J, and Deane CM. 2020. TCRBuilder: multi-state T-cell receptor structure prediction. Bioinformatics 36: 3580–3581. [DOI] [PubMed] [Google Scholar]
- 137.Ruffolo JA, Chu L-S, Mahajan SP, and Gray JJ. 2022. Fast, accurate antibody structure prediction from deep learning on massive set of natural antibodies. bioRxiv: 2022.2004.2020.488972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guest JD, Vreven T, Zhou J, Moal I, Jeliazkov JR, Gray JJ, Weng Z, and Pierce BG. 2021. An expanded benchmark for antibody-antigen docking and affinity prediction reveals insights into antibody recognition determinants. Structure 29: 606–621 e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fernandez-Quintero ML, Vangone A, Loeffler JR, Seidler CA, Georges G, and Liedl KR. 2022. Paratope states in solution improve structure prediction and docking. Structure 30: 430–440 e433. [DOI] [PubMed] [Google Scholar]
- 140.Guthmiller JJ, Han J, Utset HA, Li L, Lan LY, Henry C, Stamper CT, McMahon M, O’Dell G, Fernandez-Quintero ML, Freyn AW, Amanat F, Stovicek O, Gentles L, Richey ST, de la Pena AT, Rosado V, Dugan HL, Zheng NY, Tepora ME, Bitar DJ, Changrob S, Strohmeier S, Huang M, Garcia-Sastre A, Liedl KR, Bloom JD, Nachbagauer R, Palese P, Krammer F, Coughlan L, Ward AB, and Wilson PC. 2022. Broadly neutralizing antibodies target a haemagglutinin anchor epitope. Nature 602: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leach MW, Halpern WG, Johnson CW, Rojko JL, MacLachlan TK, Chan CM, Galbreath EJ, Ndifor AM, Blanset DL, Polack E, and Cavagnaro JA. 2010. Use of tissue cross-reactivity studies in the development of antibody-based biopharmaceuticals: history, experience, methodology, and future directions. Toxicol Pathol 38: 1138–1166. [DOI] [PubMed] [Google Scholar]
- 142.Wang EY, Dai Y, Rosen CE, Schmitt MM, Dong MX, Ferre EMN, Liu F, Yang Y, Gonzalez-Hernandez JA, Meffre E, Hinchcliff M, Koumpouras F, Lionakis MS, and Ring AM. 2022. High-throughput identification of autoantibodies that target the human exoproteome. Cell Rep Methods 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Setliff I, Shiakolas AR, Pilewski KA, Murji AA, Mapengo RE, Janowska K, Richardson S, Oosthuysen C, Raju N, Ronsard L, Kanekiyo M, Qin JS, Kramer KJ, Greenplate AR, McDonnell WJ, Graham BS, Connors M, Lingwood D, Acharya P, Morris L, and Georgiev IS. 2019. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell 179: 1636–1646 e1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dugan HL, Stamper CT, Li L, Changrob S, Asby NW, Halfmann PJ, Zheng NY, Huang M, Shaw DG, Cobb MS, Erickson SA, Guthmiller JJ, Stovicek O, Wang J, Winkler ES, Madariaga ML, Shanmugarajah K, Jansen MO, Amanat F, Stewart I, Utset HA, Huang J, Nelson CA, Dai YN, Hall PD, Jedrzejczak RP, Joachimiak A, Krammer F, Diamond MS, Fremont DH, Kawaoka Y, and Wilson PC. 2021. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity 54: 1290–1303 e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.He B, Liu S, Wang Y, Xu M, Cai W, Liu J, Bai W, Ye S, Ma Y, Hu H, Meng H, Sun T, Li Y, Luo H, Shi M, Du X, Zhao W, Chen S, Yang J, Zhu H, Jie Y, Yang Y, Guo D, Wang Q, Liu Y, Yan H, Wang M, and Chen YQ. 2021. Rapid isolation and immune profiling of SARS-CoV-2 specific memory B cell in convalescent COVID-19 patients via LIBRA-seq. Signal Transduct Target Ther 6: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MA, Church GM, Kesari S, Leproust EM, Solimini NL, and Elledge SJ. 2011. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol 29: 535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mohan D, Wansley DL, Sie BM, Noon MS, Baer AN, Laserson U, and Larman HB. 2018. PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes. Nat Protoc 13: 1958–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Larman HB, Laserson U, Querol L, Verhaeghen K, Solimini NL, Xu GJ, Klarenbeek PL, Church GM, Hafler DA, Plenge RM, Nigrovic PA, De Jager PL, Weets I, Martens GA, O’Connor KC, and Elledge SJ. 2013. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun 43: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shrock E, Fujimura E, Kula T, Timms RT, Lee IH, Leng Y, Robinson ML, Sie BM, Li MZ, Chen Y, Logue J, Zuiani A, McCulloch D, Lelis FJN, Henson S, Monaco DR, Travers M, Habibi S, Clarke WA, Caturegli P, Laeyendecker O, Piechocka-Trocha A, Li JZ, Khatri A, Chu HY, Collection MC−, Processing T, Villani AC, Kays K, Goldberg MB, Hacohen N, Filbin MR, Yu XG, Walker BD, Wesemann DR, Larman HB, Lederer JA, and Elledge SJ. 2020. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O’Connor KC, Walker B, Larman HB, and Elledge SJ. 2015. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348: aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wine Y, Boutz DR, Lavinder JJ, Miklos AE, Hughes RA, Hoi KH, Jung ST, Horton AP, Murrin EM, Ellington AD, Marcotte EM, and Georgiou G. 2013. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc Natl Acad Sci U S A 110: 2993–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lavinder JJ, Wine Y, Giesecke C, Ippolito GC, Horton AP, Lungu OI, Hoi KH, DeKosky BJ, Murrin EM, Wirth MM, Ellington AD, Dorner T, Marcotte EM, Boutz DR, and Georgiou G. 2014. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci U S A 111: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Snapkov I, Chernigovskaya M, Sinitcyn P, Le Quy K, Nyman TA, and Greiff V. 2022. Progress and challenges in mass spectrometry-based analysis of antibody repertoires. Trends Biotechnol 40: 463–481. [DOI] [PubMed] [Google Scholar]
- 154.Ionov S, and Lee J. 2022. An Immunoproteomic Survey of the Antibody Landscape: Insights and Opportunities Revealed by Serological Repertoire Profiling. Front Immunol 13: 832533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.de Graaf SC, Hoek M, Tamara S, and Heck AJR. 2022. A perspective toward mass spectrometry-based de novo sequencing of endogenous antibodies. MAbs 14: 2079449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee CH, Lavinder JJ, Murrin EM, Chrysostomou C, Hoi KH, Tsybovsky Y, Thomas PV, Druz A, Zhang B, Zhang Y, Wang L, Kong WP, Park D, Popova LI, Dekker CL, Davis MM, Carter CE, Ross TM, Ellington AD, Wilson PC, Marcotte EM, Mascola JR, Ippolito GC, Krammer F, Quake SR, Kwong PD, and Georgiou G. 2016. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 22: 1456–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jung J, Mundle ST, Ustyugova IV, Horton AP, Boutz DR, Pougatcheva S, Prabakaran P, McDaniel JR, King GR, Park D, Person MD, Ye C, Tan B, Tanno Y, Kim JE, Curtis NC, DiNapoli J, Delagrave S, Ross TM, Ippolito GC, Kleanthous H, Lee J, and Georgiou G. 2021. Influenza vaccination in the elderly boosts antibodies against conserved viral proteins and egg-produced glycans. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lee J, Paparoditis P, Horton AP, Fruhwirth A, McDaniel JR, Jung J, Boutz DR, Hussein DA, Tanno Y, Pappas L, Ippolito GC, Corti D, Lanzavecchia A, and Georgiou G. 2019. Persistent Antibody Clonotypes Dominate the Serum Response to Influenza over Multiple Years and Repeated Vaccinations. Cell Host Microbe 25: 367–376 e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bondt A, Hoek M, Tamara S, de Graaf B, Peng W, Schulte D, van Rijswijck DMH, den Boer MA, Greisch JF, Varkila MRJ, Snijder J, Cremer OL, Bonten MJM, and Heck AJR. 2021. Human plasma IgG1 repertoires are simple, unique, and dynamic. Cell Syst 12: 1131–1143 e1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gerber HP, Sibener LV, Lee LJ, and Gee MH. 2020. Identification of Antigenic Targets. Trends Cancer 6: 299–318. [DOI] [PubMed] [Google Scholar]
- 161.Joglekar AV, and Li G. 2021. T cell antigen discovery. Nat Methods 18: 873–880. [DOI] [PubMed] [Google Scholar]
- 162.Coles CH, Mulvaney RM, Malla S, Walker A, Smith KJ, Lloyd A, Lowe KL, McCully ML, Martinez Hague R, Aleksic M, Harper J, Paston SJ, Donnellan Z, Chester F, Wiederhold K, Robinson RA, Knox A, Stacey AR, Dukes J, Baston E, Griffin S, Jakobsen BK, Vuidepot A, and Harper S. 2020. TCRs with Distinct Specificity Profiles Use Different Binding Modes to Engage an Identical Peptide-HLA Complex. J Immunol 204: 1943–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, Birnbaum ME, Yang X, Su LF, Yu W, Dong S, Gee MH, Jude KM, Davis MM, Groves JT, Goddard WA 3rd, Heath JR, Evavold BD, Vale RD, and Garcia KC. 2018. Isolation of a Structural Mechanism for Uncoupling T Cell Receptor Signaling from Peptide-MHC Binding. Cell 174: 672–687 e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Joglekar AV, Leonard MT, Jeppson JD, Swift M, Li G, Wong S, Peng S, Zaretsky JM, Heath JR, Ribas A, Bethune MT, and Baltimore D. 2019. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat Methods 16: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kisielow J, Obermair FJ, and Kopf M. 2019. Deciphering CD4(+) T cell specificity using novel MHC-TCR chimeric receptors. Nat Immunol 20: 652–662. [DOI] [PubMed] [Google Scholar]
- 166.Kula T, Dezfulian MH, Wang CI, Abdelfattah NS, Hartman ZC, Wucherpfennig KW, Lyerly HK, and Elledge SJ. 2019. T-Scan: A Genome-wide Method for the Systematic Discovery of T Cell Epitopes. Cell 178: 1016–1028 e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Dobson CS, Reich AN, Gaglione S, Smith BE, Kim EJ, Dong J, Ronsard L, Okonkwo V, Lingwood D, Dougan M, Dougan SK, and Birnbaum ME. 2022. Antigen identification and high-throughput interaction mapping by reprogramming viral entry. Nat Methods 19: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, and Nussenzweig MC. 2003. Predominant autoantibody production by early human B cell precursors. Science 301: 1374–1377. [DOI] [PubMed] [Google Scholar]
- 169.Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, and Bendelac A. 2017. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu Y, Roach W, Sun T, Jain T, Prinz B, Yu TY, Torrey J, Thomas J, Bobrowicz P, Vasquez M, Wittrup KD, and Krauland E. 2013. Addressing polyspecificity of antibodies selected from an in vitro yeast presentation system: a FACS-based, high-throughput selection and analytical tool. Protein Eng Des Sel 26: 663–670. [DOI] [PubMed] [Google Scholar]
- 171.Hotzel I, Theil FP, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, Fielder P, Carter PJ, and Kelley RF. 2012. A strategy for risk mitigation of antibodies with fast clearance. MAbs 4: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kelly RL, Geoghegan JC, Feldman J, Jain T, Kauke M, Le D, Zhao J, and Wittrup KD. 2017. Chaperone proteins as single component reagents to assess antibody nonspecificity. MAbs 9: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shehata L, Maurer DP, Wec AZ, Lilov A, Champney E, Sun T, Archambault K, Burnina I, Lynaugh H, Zhi X, Xu Y, and Walker LM. 2019. Affinity Maturation Enhances Antibody Specificity but Compromises Conformational Stability. Cell Rep 28: 3300–3308 e3304. [DOI] [PubMed] [Google Scholar]
- 174.Finlay WJJ, Coleman JE, Edwards JS, and Johnson KS. 2019. Anti-PD1 ‘SHR-1210’ aberrantly targets pro-angiogenic receptors and this polyspecificity can be ablated by paratope refinement. MAbs 11: 26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


