Abstract
An inherent problem in the diagnostic PCR assay is the presence of ill-defined inhibitors of amplification which may cause false-negative results. Addition of an amplifiable fragment of foreign DNA in the PCR to serve as a hybrid internal control (HIC) would allow for a simple way to identify specimens containing inhibitors. Two oligonucleotide hybrid primers were synthesized to contain nucleic acid sequences of the Chlamydia pneumoniae 16S rRNA primers in a position flanking two primers that target the sequences of a 650-bp lambda phage DNA segment. By using the hybrid primers, hybrid DNA comprising a large sequence of lambda phage DNA flanked by short pieces of chlamydia DNA was subsequently generated by PCR, cloned into a plasmid vector, and purified. Plasmids containing the hybrid DNA were diluted and used as a HIC by adding them to each C. pneumoniae PCR test. Consequently, C. pneumoniae primers were able to amplify both chlamydia DNA and the HIC DNA. The production of a 689-bp HIC DNA band on an acrylamide gel indicated that the specimen contained no inhibitors and that internal conditions were compatible with PCR. Subsequently, a biotinylated RNA probe for the HIC was transcribed from a nested sequence of the HIC and was used for its hybridization. Detection of the HIC DNA-RNA hybrid was achieved by enzyme immunoassay (EIA). This PCR-EIA system with a HIC was initially tested with 12 previously PCR-positive and 14 previously PCR-negative specimens. Of the 12 PCR-positive specimens, 11 were reconfirmed as positive; 1 had a negative HIC value, indicating inhibition. Of the 14 previously PCR-negative specimens, 13 were confirmed as true negative; 1 had a negative HIC value, indicating inhibition. The assay was then used with 237 nasopharyngeal specimens from patients with pneumonia. Twenty-one of 237 (8.9%) were positive for C. pneumoniae, and 42 (17.7%) were found to inhibit the PCR. Specimens showing inhibitory activity were diluted 1:10 and were retested. Ten specimens were still inhibitory to the PCR and required further DNA purification. No additional positive samples were detected and 3 nasopharyngeal specimens remained inhibitory to PCR. Coamplification of a HIC DNA can help confirm true-negative PCR results by ruling out the presence of inhibitors of DNA amplification.
Since the recognition of Chlamydia pneumoniae by Grayston and coworkers (15, 19) in the mid-1980s as a significant respiratory pathogen, it has subsequently been associated with community-acquired pneumonia, sinusitis, pharyngitis, and bronchitis (4, 11–14, 18). In addition, C. pneumoniae has been linked to asthma, acute chest syndrome of sickle cell anemia, human immunodeficiency virus infection, Guillain-Barré syndrome, endocarditis, and more recently, coronary artery disease (1, 16, 20). Chronic persistent respiratory infections have been reported (17), and there is also evidence that C. pneumoniae can occasionally be identified by culture or serology from asymptomatic healthy individuals (10, 18).
Despite the association of C. pneumoniae with a growing list of diseases, culturing of the organism continues to pose a challenge for many clinical laboratories. DNA amplification-based diagnostic assays are being used more frequently by research laboratories to identify C. pneumoniae. Recently, ill-defined inhibitors have been recognized as obstacles to the improvement in performance of nucleic acid amplification assays (6, 21).
A common and efficient method of attenuating inhibitory substances in clinical specimens is resuspension of the centrifuged cellular component in a working buffer, rendering the majority of clinical specimens relatively free of inhibitors prior to amplification. Amplification assays like PCR (Chlamydia AMPLICOR; Roche Diagnostic Systems, Branchburg, N.J.) and ligase chain reaction (Abbott Laboratories, Abbott Park, Ill.) use this simple processing method in which urine specimens are resuspended in their respective buffers prior to the cell lysis step. Unfortunately, a small number of specimens undergoing similar routine processing continue to inhibit the amplification reaction, and positive specimens may go undetected because of false-negative results (2, 11). We developed a PCR-enzyme immunoassay (PCR-EIA) using a hybrid internal control that can simultaneously identify C. pneumoniae DNA and assess the potential inhibitory nature of a clinical specimen to DNA amplification. We used a hybrid lambda phage DNA fragment as a hybrid internal control for coamplification with C. pneumoniae. Specific recognition of specimens harboring inhibitors can greatly increase the reliability of a diagnostic PCR assay by identifying only those specimens that require more stringent DNA extraction procedures prior to the PCR assay, thus minimizing false-negative results.
MATERIALS AND METHODS
Generation of the hybrid internal control.
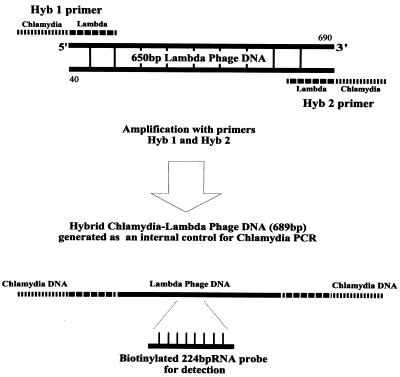
To make the melting temperature of the hybrid internal control DNA comparable to the melting temperature of the C. pneumoniae 16S rRNA gene, a 650-bp lambda phage DNA segment (sequence positions 40 to 690) with a G+C content 61% similar to that of the C. pneumoniae gene was selected for use as the hybrid internal control (Fig. 1). Hybrid primers Hyb1 and Hyb2 were synthesized with previously described C. pneumoniae 16S rRNA primers (7), CpnA (sense) and CpnB (antisense), external to the corresponding 20-bp sense and antisense primer sequences of the lambda phage template, respectively: CpnA (sense), 5′-TGACAACTGTAGAAATACAGC-3′ (chlamydia sequence); CpnB (antisense), 5′-CGCCTCTCTCCTATAAAT 3′ (chlamydia sequence); Hyb1 (sense), 5′-TGACAACTGTAGAAATACAGCTTCCGGTTTAAGGCGTTTCC 3′ (boldface indicates outer, chlamydia sequence and regular type indicates inner, lambda sequence); and Hyb2 (antisense), 5′-CGCCTCTCTCCTATAAATTCATCCAGCGCGGCTGCTTT-3′ (boldface indicates outer, chlamydia sequence and regular type indicates inner, lambda sequence).
FIG. 1.
Generation of hybrid chlamydia-lambda phage DNA for the internal control using primers Hyb1 and Hyb2. Primer positions are indicated by the discontinuous lines. The resultant PCR product of 689 bp, which was used as a hybrid internal control, originates from a large segment of lambda phage DNA (650 bp) and is flanked by two smaller sequences of C. pneumoniae DNA (18 and 21 bp). The flanking sequences are complementary to chlamydia primers CpnA and CpnB and are consequently targeted by them. The position of the RNA probe for the subsequent EIA detection of the internal control is also indicated.
Hybrid DNA was amplified from primers Hyb1 and Hyb2, with the lambda phage DNA as template (Biolabs, Beverly, Mass.). The resultant 689-bp hybrid fragment consisted of a large sequence of lambda phage (650 bp) flanked at both ends by two short C. pneumoniae sequences (18 and 21 bp). These flanking chlamydia sequences are complementary to the primers CpnA and CpnB, respectively, and are consequently targets for these primers. To improve its storage, the lambda phage-chlamydia hybrid DNA was then cloned into a plasmid vector of Escherichia coli (plasmid pCR 2.1 [3.9 kb]; Original TA Cloning Kit; Invitrogen, San Diego, Calif.). After exponential growth in E. coli culture, plasmids containing the hybrid insert were purified with the Wizard miniprep DNA kit (Promega, Madison, Wis.).
The optimal dilution of plasmid was determined by titration, in order to minimize competition between the chlamydia DNA target and the hybrid internal control DNA target during PCR amplification. A series of 10-fold dilutions of the plasmid solution through a dilution of 10−9 was amplified with chlamydia primers CpnA and CpnB. The titers were analyzed by ethidium-stained polyacrylamide gel electrophoresis. The plasmid dilution presenting the faintest positive band (10−8 dilution, ≈50 fg DNA or ≈2,000 copies of the insert) was selected as the optimal amount of the hybrid internal control. Five microliters of the 10−8 plasmid dilution containing cloned hybrid lambda DNA was then added as a hybrid internal control to each C. pneumoniae PCR, including the negative controls.
Lambda RNA probe generation and titration.
To accommodate the large numbers of clinical specimens, a hybridization method and an EIA for the detection of amplified hybrid internal control DNA were developed. An RNA probe for the PCR product of the hybrid internal control was generated. Similar to the methods outlined by Gaydos et al. (7), a paired set of nested primers, primers T7-lambda 1 (containing the 22-mer T7-transcription promoter region) and lambda 2, were synthesized to be used in the amplification of a 202-bp nested lambda phage DNA fragment, which was needed for the transcription of a 224-mer biotinylated-UTP (Promega) lambda phage RNA probe: T7-lambda 1, 5′-TTAATACGACTCACTATAGGGTAGCTGGCTGACATTTTCGGT-3′ (boldface indicates the T7 promoter region and regular type indicates the sense sequence), and lambda 2, 5′-CAACCTCCCGGCGCAGCTTT-3′ (antisense sequence).
The resultant RNA transcript-probe was then titrated against chlamydia and lambda DNA products generated by PCR to achieve maximum sensitivity for the EIA detection of the hybrid internal control while minimizing the background signal. RNA probe dilutions of 2, 1, 0.5, and 0.25 μl per ml of hybridization buffer were assessed, and 0.5 μl/ml was chosen as the optimal RNA probe concentration for use against the amplified hybrid internal control DNA. Final detection of the DNA-RNA hybrid was performed by EIA, analogous to the methods used by Coutle et al. (5).
Specimen preparation for PCR.
Three hundred microliters of chlamydia samples or patient specimens was pelleted at 13,000 × g, resuspended in 300 μl of PCR buffer (10 mM Tris [pH 8.3], 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin), and then treated with 100 μg of proteinase K (Sigma Chemical Co., St. Louis, Mo.) per ml and 0.5% each Nonidet P-40 and Tween 20 (Sigma). The specimens were incubated at 60°C either for 1 h or overnight, depending on the amount of cellular debris, and were then heated at 100°C for 10 min. Positive specimens were regularly incorporated to control for gross target loss. In addition, negative specimens were included to control for interspecimen contamination.
PCR.
Diagnostic PCR for C. pneumoniae was performed as described previously by Gaydos et al. (9) with their original primers CpnA and CpnB. Thermocycling parameters were programmed for denaturation at 94°C for 15 s, annealing at 55°C for 15 s, and extension at 75°C for 35 s. Fifty milliliters of processed sample was used in a total PCR volume of 100 μl. In addition, the hybrid internal control DNA (in the form of purified plasmid containing the hybrid lambda phage insert) was included in the cocktail preparation for the PCRs (5 μl/reaction mixture). Since the hybrid internal control contains flanking sequences specific to the 16S rRNA gene of C. pneumoniae, chlamydia primers CpnA and CpnB targeted the hybrid internal control, in addition to their primary function of targeting C. pneumoniae DNA in respiratory specimens.
Hybridization and EIA.
Two hybridizations and EIAs were performed: one for amplified C. pneumoniae DNA and another one for the hybrid internal control amplified DNA (representative EIA data are presented in Table 1). The titration of the chlamydia-specific RNA probe for hybridization with chlamydia PCR products was performed as described previously by Gaydos et al. (8). Hybridizations were performed in microamp tubes (Perkin-Elmer, Norwalk, Conn.), with the RNA probe for C. pneumoniae amplicons in one plate (8) and the 202-bp RNA probe designed for the hybrid internal control amplicons in another plate. Microfluor plates (Dynatech, Chantilly, Va.) were coated overnight with goat antibiotin. Hybridization products were added, and the mixture was incubated for 1 h at 37°C (to capture the biotinylated RNA probe) and then washed with phosphate-buffered saline buffer. The bound DNA-RNA hybrid was detected with an anti-DNA-RNA monoclonal antibody conjugated to alkaline phosphatase (7, 8). Methylumbelliferyl phosphate in diethanolamine buffer was used as the substrate, and the reaction was measured in a fluorometer (Dynatech). The cutoff value for C. pneumoniae was calculated to be 5 standard deviations above the mean value (fluorescent units) for six chlamydia-negative controls. The cutoff value for the hybrid internal control was calculated to be 3 standard deviations above the mean value for three wells that did not contain an internal control (7).
TABLE 1.
Representative EIA results for coamplified C. pneumoniae and hybrid internal control PCR products
| Specimen |
C. pneumoniae EIA result (fluorescent units)a
|
Internal control EIA result (fluorescent units) | |
|---|---|---|---|
| No internal control | Yes internal control | ||
| A | 29 | 19 | 277 |
| B | 45 | 47 | 453 |
| C | 77 | 90 | 1,376 |
| D | 31 | 30 | 52b |
| E | 122 | 169 | 171 |
| F | 322 | 246 | 2,550 |
| G | 1,560 | 1,520 | 1,920 |
| H | 3,900 | 4,000 | 1,490 |
The cutoff value for C. pneumoniae was 100 fluorescent units. Yes and No int. control, an internal control was either included or not included in the PCR mixture, respectively.
Inhibitory activity was shown by this sample.
RESULTS
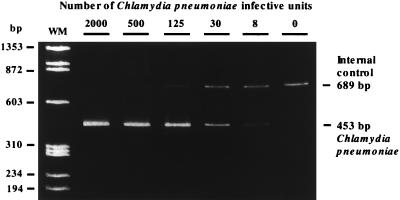
Coamplification of C. pneumoniae and the hybrid lambda phage internal control with chlamydia primers CpnA and CpnB was achieved because the hybrid internal control DNA exhibited sequences common to C. pneumoniae at its 3′ and 5′ ends. Therefore, the PCR amplification of C. pneumoniae cultures or inhibitory substance-free clinical specimens positive for C. pneumoniae exhibited two bands on ethidium-stained polyacrylamide gel electrophoresis: one band (463 bp) diagnostic for C. pneumoniae and a control band (689 bp) for the hybrid internal control. Inhibitory substance-free clinical specimens negative for C. pneumoniae contained only the control band of 689 bp, representing the amplified hybrid internal control. A positive hybrid internal control band indicated that no detectable inhibitors were present within the specimen and that biochemical conditions were optimal for PCR amplification. A gel showing the coamplification of both chlamydia DNA from culture and the hybrid internal control DNA is presented in Fig. 2.
FIG. 2.
Coamplification of C. pneumoniae DNA from culture and the hybrid internal control (≈2,000 copies per PCR mixture). The internal control bands (689 bp) become stronger with decreasing concentrations of chlamydia targets (453 bp). The sizes of the PCR products (in base pairs) and the weight markers (WM) are indicated.
Twenty-six specimens previously tested by PCR were used in a pilot study, including 12 positive specimens and 14 negative specimens. Eleven of these 12 specimens positive by PCR again tested positive for C. pneumoniae, and 1 specimen showed inhibition. Thirteen of the 14 specimens negative by PCR again tested negative for C. pneumoniae, but 1 specimen demonstrated inhibition. The inhibition was resolved after dilution of the specimens 1:10, and the PCR result was negative for both specimens (Table 2).
TABLE 2.
Hybrid internal control (HIC) results for 26 samples previously tested for C. pneumoniae by PCR and 237 previously untested respiratory samples
| Test result for respiratory specimens | No. of specimens | Routine sample processing
|
No. (%) HIC negative following:
|
||
|---|---|---|---|---|---|
| No. (%) HIC positive | No. (%) HIC negative | Further 1/10 dilution | Further DNA purification | ||
| C. pneumoniae PCR | |||||
| Positive | 12 | 11 (92) | 1 (8) | 0 (0) | —a |
| Negative | 14 | 13 (93) | 1 (7) | 0 (0) | — |
| Previously untested | 237 | 195 (82) | 42 (18) | 10 (4)b | 3 (1)b |
—, not further tested.
For 4 of the 42 inhibitory samples, insufficient volumes were available for further dilution and/or DNA purification; therefore, percentages are based on 233 total samples.
Previously untested nasopharyngeal (n = 237) swab samples from pneumonia patients were assayed (Table 2). Of these specimens, 21 (8.9%) were found to be positive for C. pneumoniae by our PCR-EIA, while 42 (17.7%) exhibited inhibition. Thirty-eight of these 42 specimens that inhibited the PCR and for which enough volume for retesting was available were diluted 1:10 in PCR buffer and were retested. Of these, 28 (73.7%) were no longer inhibitory and retested negative, indicating that they were true negative for C. pneumoniae by PCR. The remaining 10 (26.3%) specimens, which continued to show evidence of inhibition, were subjected to phenol-chloroform extraction and ethanol precipitation to further purify the DNA and were then retested. None tested positive for C. pneumoniae, but 3 of these 10 specimens continued to show inhibition.
DISCUSSION
The key feature of our new PCR-EIA system is the incorporation of a novel hybrid DNA fragment, the internal control, which can be coamplified along with the 16S rRNA gene of chlamydia by using the same primer set described earlier by Gaydos et al. (9) to specifically target C. pneumoniae. No other primers were needed for the actual chlamydia assay. The cotargeting ability of chlamydia primers CpnA and CpnB was entirely a reflection of the hybrid nature of the internal control, designed to possess DNA sequences from both lambda phage and chlamydia genomes. The hybrid internal control was first created in a single PCR with hybrid primers Hyb1 and Hyb2. The hybrid primers were designed to have four primer sequences that serve two distinct functions: the inner lambda sequences of the hybrid primers (i) initially targeted a 650-bp lambda phage segment for the separate generation of a 689-bp hybrid lambda phage-chlamydia fragment and (ii) concurrently ferried the outer chlamydia-specific sequences (18 and 21 bp) of the hybrid primers into the final hybrid product. Therefore, chlamydia primers CpnA and CpnB could specifically target these external sites of the hybrid internal control when it was added to our standard diagnostic PCR assay for C. pneumoniae. This new scheme does not alter the original concentrations of the previously optimized PCR components for the testing of C. pneumoniae, nor does it require a change of the targeting primers.
Because of the annealing competition between the chlamydia sites and the hybrid internal control sites for same primers CpnA and CpnB, no detectable hybrid internal control DNA was amplified by these primers when very high concentrations of chlamydia target DNA (500 to 2,000 infective units) were present in the sample. However, at the lower concentrations of chlamydia target DNA (8 to 125 infective units), the hybrid internal control was increasingly amplified, with the strongest amplification obtained in the absence of chlamydia DNA. The lack of amplification of the hybrid internal control is not an issue when specimens strongly positive for chlamydia are detected. When specimens are negative for chlamydia DNA, the strongest amplification of the hybrid internal control product is achieved, thus confirming the absence of PCR inhibitors in the processed samples.
It is unknown why 1 of the 12 previously PCR-positive specimens in the pilot study failed to test positive both for C. pneumoniae and for the hybrid internal control. Freeze-thawing or prolonged storage may have exhausted an already low concentration of the chlamydia target within the specimen. After a 10-fold dilution of the same processed specimen, the hybrid internal control was amplified by repeat testing, indicating that an inhibitor was present in this specimen.
From the routine processing of previously untested clinical specimens, our internal control-based PCR system for C. pneumoniae detected 42 of 237 (17.7%) specimens that contained inhibitors. Further dilution and/or DNA purification of only these specimens was required to remove the inhibitors and resulted in the successful amplification of the hybrid internal control for all but three clinical specimens, which remained inhibitory (6). This selection method was more routinely efficient and easier compared to retrospectively spiking all negative samples with chlamydia DNA and then retesting them all. As suggested by Verkooyen et al. (21), too much DNA used for spiking experiments can circumvent PCR inhibition but may fail to reveal baseline inhibition for amplifying the target of interest.
Inhibitors of PCR amplification have been noted in the past by comparing PCR methods to either culture or other “gold standard” evaluation methods (3, 21). However, there is concern when an amplification system has no comparison schemes for enabling the recognition of specimens containing inhibitors. Although several processing techniques have been recommended to reduce the effects of inhibitors, such as incubation of the sample at 4°C with a delay in testing, preparation of a 10-fold dilution, heating of the sample at 95°C for 10 min, freeze-thawing of the sample, or DNA extraction, the complete reliance on a diagnostic PCR assay necessitates use of a routine and efficient method to determine which specimens contain inhibitors that may prevent the identification of a true-positive sample (21).
As more diagnostic systems become increasingly dependent on DNA amplification strategies, the problem of PCR inhibition will inevitably become more problematic (21). The concurrent amplification of a hybrid internal control in each PCR test provides a measure of confidence for negative results when conditions are compatible for amplification to occur in that reaction. We found the degree of inhibition in the clinical respiratory specimens (17.7%) to be similar to those reported for endocervical specimens (19%) but higher than those reported for urine specimens (1.8 to 2.6%) (2, 11, 21). Unfortunately, the actual causes and mechanisms of inhibition are still vague. The possible sources of inhibition have been speculated to include protein, organic chemicals, and/or ionic compounds (6, 21). The use of BLOTTO (bovine lacto transfer technique optimizer; essentially 10% skim milk) in the PCR was suggested by De Boer et al. (6) to prevent the effects of inhibitory compounds, such as polyphenolic molecules, found in DNA preparations extracted from plants. Skim milk has been primarily used in Western and Southern blots to prevent nonspecific attachments of proteins and nucleic acids to the nitrocellulose. Although it did not hamper our PCR, skim milk was empirically detrimental to the EIA detection end of our diagnostic system. This was probably due to an abundance of milk proteins that might have interacted with the EIA antibodies to impede their proper activity.
The use of an extraneous DNA fragment as a hybrid internal control for the nucleic acid amplification of C. pneumoniae confirmed that PCR-negative specimens did not contain inhibitors and therefore were truly negative. The dilution or DNA purification and retesting by PCR of only those specimens which showed inhibitory activity provides an efficient system compared to processing schemes which might involve the routine dilution and/or arduous DNA purification of all specimens prior to amplification. Also, while the use of sample dilutions was observed to reduce the level of inhibition by some specimens, a standard practice of diluting all clinical specimens could inadvertently reduce or eliminate target DNA below the level of detection by PCR for samples already containing low levels of DNA targets (21). Thus, the incorporation of a minimal concentration of a hybrid internal control which can be amplified along with the target of interest by using the same primers specific for the same target sites on both products, was an effective measure for maximizing the sensitivity and reliability of our PCR-based diagnostic assay for C. pneumoniae.
ACKNOWLEDGMENT
We thank Margaret Hammerschlag for providing the nasopharyngeal specimens from the patients with pneumonia.
REFERENCES
- 1.Augenbraun M H, Roblin P M, Chirgwin K, Landman D, Hammerschlag M R. Isolation of Chlamydia pneumoniae from lungs of patients infected with the human immunodeficiency virus. J Clin Microbiol. 1991;29:401–402. doi: 10.1128/jcm.29.2.401-402.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassiri M, Mardh P-A, Domeika M The European Chlamydia Group. Multiplex AMPLICOR PCR screening for Chlamydia trachomatis and Neisseria gonorrhoeae in women attending non-sexually transmitted disease clinics. J Clin Microbiol. 1997;35:2556–2560. doi: 10.1128/jcm.35.10.2556-2560.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauwens J E, Clark A M, Stamm W E. Diagnosis of Chlamydia trachomatis endocervical infections by a commercial polymerase chain reaction assay. J Clin Microbiol. 1993;31:3023–3027. doi: 10.1128/jcm.31.11.3023-3027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chirgwin K, Roblin P M, Gelling M, Hammerschlag M R, Schachter J. Infection with Chlamydia pneumoniae in Brooklyn. J Infect Dis. 1991;163:757–761. doi: 10.1093/infdis/163.4.757. [DOI] [PubMed] [Google Scholar]
- 5.Coutle F C, Yang B, Bobo L. Enzyme immunoassay for the detection of hybrids between PCR amplified HIV-1 DNA and a RNA probe: PCR-EIA. AIDS. 1990;6:775–784. doi: 10.1089/aid.1990.6.775. [DOI] [PubMed] [Google Scholar]
- 6.De Boer S H, Ward L J, Li X, Chittaranjan S. Attenuation of PCR inhibition in the presence of plant compounds by addition of BLOTTO. Nucleic Acids Res. 1995;23:2567–2568. doi: 10.1093/nar/23.13.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaydos C A, Eiden J J, Oldach D, Mundy L M, Auwaerter P, Warner M L, Vance E, Burton A A, Quinn T C. Diagnosis of Chlamydia pneumoniae infection in patients with community acquired pneumonia by polymerase chain reaction enzyme immunoassay. Clin Infect Dis. 1994;19:157–160. doi: 10.1093/clinids/19.1.157. [DOI] [PubMed] [Google Scholar]
- 8.Gaydos C A, Fowler C L, Gill V J, Eiden J J, Quinn T C. Detection of Chlamydia pneumoniae by polymerase chain reaction-enzyme immunoassay in an immunocompromised population. J Clin Infect Dis. 1993;17:1718–1723. doi: 10.1093/clinids/17.4.718. [DOI] [PubMed] [Google Scholar]
- 9.Gaydos C A, Quinn T C, Eiden J J. Identification of Chlamydia pneumoniae by DNA amplification of the 16S rRNA gene. J Clin Microbiol. 1992;30:796–800. doi: 10.1128/jcm.30.4.796-800.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gnarpe J, Gnarpe H, Sundelof B. Endemic prevalence of Chlamydia pneumoniae in subjectively healthy persons. Scand J Infect Dis. 1991;23:387–388. doi: 10.3109/00365549109024328. [DOI] [PubMed] [Google Scholar]
- 11.Goessens W H F, Mouton J W, Van Der Meijden W I, Deelen S, Van Rijsoort-Vos T H, Toom N L, Verbrugh H A, Verkooyen R P. Comparison of three commercially available amplication assays, AMP CT, LCx, and COBAS AMPLICOR, for detection of Chlamydia trachomatis in first-void urine. J Clin Microbiol. 1997;35:2628–2633. doi: 10.1128/jcm.35.10.2628-2633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grayston J T, Aldous M B, Easton A, Wang S, Kuo C, Campbell L, Altman J. Evidence that Chlamydia pneumoniae causes pneumonia and bronchitis. J Infect Dis. 1994;168:1231–1235. doi: 10.1093/infdis/168.5.1231. [DOI] [PubMed] [Google Scholar]
- 13.Grayston J T, Campbell L A, Kuo C C, Mordhurst C H, Saikku P, Thom D H, Wang S P. A new respiratory tract pathogen: Chlamydia pneumoniae, strain TWAR. J Infect Dis. 1990;161:618–625. doi: 10.1093/infdis/161.4.618. [DOI] [PubMed] [Google Scholar]
- 14.Grayston J T, Diwan V K, Cooney M, Wang S P. Community and hospital acquired pneumonia associated with Chlamydia TWAR infection demonstrated serologically. Arch Intern Med. 1989;149:169–173. [PubMed] [Google Scholar]
- 15.Grayston J T, Kuo C C, Campbell L A, Wang S P. Chlamydia pneumoniae sp. nov. for Chlamydia sp. strain TWAR. Int J Syst Bacteriol. 1989;39:88–90. [Google Scholar]
- 16.Haidl S, Ivarsson S, Bjerre I, Persson K. Guillain-Barre syndrome after Chlamydia pneumoniae infection. N Engl J Med. 1992;326:576–577. [PubMed] [Google Scholar]
- 17.Hammerschlag M R, Chirgwin K, Roblin P M, Gelling M, Dumornay W, Mandel L, Smith P, Schachter J. Persistent infection with Chlamydia pneumoniae following acute respiratory illness. Clin Infect Dis. 1992;14:178–182. doi: 10.1093/clinids/14.1.178. [DOI] [PubMed] [Google Scholar]
- 18.Hyman C L, Roblin P M, Gaydos C A, Quinn T C, Schachter J, Hammerschlag M R. Prevalence of asymptomatic nasopharyngeal carriage of Chlamydia pneumoniae in subjectively healthy adults: assessment by polymerase chain reaction-enzyme immunoassay and culture. Clin Infect Dis. 1995;20:1174–1178. doi: 10.1093/clinids/20.5.1174. [DOI] [PubMed] [Google Scholar]
- 19.Marrie T J, Grayston J T, Wang S P, Kuo C C. Pneumonia associated with the TWAR strain of Chlamydia. Ann Intern Med. 1987;106:507–511. doi: 10.7326/0003-4819-106-4-507. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez J A Chlamydia pneumoniae/Atherosclerosis Study Group. Isolation of Chlamydia pneumoniae from the coronary artery of a patient with coronary atherosclerosis. Ann Intern Med. 1996;125:979–982. doi: 10.7326/0003-4819-125-12-199612150-00008. [DOI] [PubMed] [Google Scholar]
- 21.Verkooyen R P, Luijendijk A, Huisman W M, Goessens W H F, Kluytmans J A J W, Van Rijsoort-Vos J H, Verbrugh H A. Detection of PCR inhibitors in cervical specimens by using the amplicor Chlamydia trachomatis assay. J Clin Microbiol. 1996;34:3072–3074. doi: 10.1128/jcm.34.12.3072-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]