Abstract
Vancomycin-resistant enterococci (VRE) pose an emerging health risk, but little is known about the precise epidemiology of the genes coding for vancomycin resistance. To determine whether the bacterial flora of consumer poultry serves as a gene reservoir, the level of contamination of poultry products with VRE was determined. VRE were genotyped by pulsed-field gel electrophoresis (PFGE), and transposon structure mapping was done by PCR. The vanX-vanY intergenic regions of several strains were further analyzed by sequencing. A total of 242 of 305 (79%) poultry products were found to be contaminated with VRE. Of these VRE, 142 (59%) were high-level-vancomycin-resistant Enterococcus faecium strains (VREF). PFGE revealed extensive VREF heterogeneity. Two genotypes were found nationwide on multiple occasions: type A (22 of 142 VREF [15%]) and type B (14 of 142 VREF [10%]). No PFGE-deduced genetic overlap was found when VREF from humans were compared with VREF from poultry. Two vanA transposon types were identified among poultry strains. In 59 of 142 (42%) of the poultry VREF, the size of the intergenic region between vanX and vanY was ∼1,300 bp. This transposon type was not found in human VREF. In contrast, all human strains and 83 of 142 (58%) of the poultry VREF contained an intergenic region 543 bp in size. Sequencing of this 543-bp intergenic vanX-vanY region demonstrated full sequence conservation. Though preliminary, these data suggest that dissemination of the resistance genes carried on transposable elements may be of greater importance than clonal dissemination of resistant strains. This observation is important for developing strategies to control the spread of glycopeptide resistance.
Colonization and infection by vancomycin-resistant enterococci (VRE) have been reported for hospitalized patients and for communities of various European countries, including France (7, 31), the United Kingdom (11, 24), Belgium (22), and The Netherlands (21). VRE pose a health risk, especially in patients with severe underlying disease or immunosuppression. In the United States, the prevalence of VRE in hospitalized patients is rising and hospital outbreaks of clonally related VRE have been described (9, 13, 18, 32). In contrast, the prevalence of VRE in hospitals in Europe remains low and a high degree of heterogeneity is observed among the VRE strains. Bates et al. (5) suggested that European VRE might be more widely disseminated than originally supposed. Furthermore, there are cases on record of the isolation of VRE from animals and from environmental sources in many European countries (5, 16, 29, 37). Paradoxically, VRE have not yet been recovered from animal and environmental sources in the United States (14, 36). The spread of vancomycin resistance is of considerable concern. Noble et al. (33) reported in vitro conjugative transfer of high-level vancomycin resistance from Enterococcus faecalis to Staphylococcus aureus. In response, the Hospital Infection Control Practices Advisory Committee in collaboration with the Centers for Disease Control and Prevention has developed recommendations to prevent the spread of VRE (23). Others have proposed control measures in case vancomycin-resistant S. aureus eventually arises (20). Recently, scientists from Japan and the United States have reported that S. aureus intermediately resistant to vancomycin has been isolated from patients (10, 15), although this resistance has been shown not to be mediated by the vanA, vanB, or vanC gene (30).
The increasing use of antimicrobial agents in human medicine and as animal growth promoters has been related to the emergence of VRE (32). In Europe, antimicrobial agents are widely used as feed additives for growth promotion in animal husbandry (38). Avoparcin is a glycopeptide antibiotic used for this purpose in poultry, and it appears to be associated with the emergence of resistance to glycopeptides in general (4, 5, 25). Enterococci belong to the natural intestinal flora of poultry. It is, thus, not unlikely that transmission of VRE occurs through human contact with poultry meat contaminated with resistant bacteria. However, such a route of transmission of VRE from poultry to humans has not been unequivocally documented so far. We determined the level of contamination of poultry products with VRE. The VRE isolated from poultry products were compared with a collection of VRE isolated from humans (21) with regard to their overall genome structures and eventual polymorphism in Tn1546, the transposon encoding high-level glycopeptide resistance.
(Parts of this study were presented at the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy, which was held in Toronto, Ontario, Canada, from 28 September to 1 October 1997 [21a].)
MATERIALS AND METHODS
Poultry products.
A total of 305 poultry products (whole chickens, chicken legs, chicken breasts, or other chicken parts) from either butchers, supermarkets, shop poulterers, or market poulterers were collected by Dutch food inspection services in the following cities: Den Haag, Maastricht, Alkmaar, Amsterdam, Nijmegen, Rotterdam, Leeuwarden, Den Bosch, Goes, Zutphen, and Groningen, The Netherlands. The sampling period was from June until September 1996.
Isolation of VRE.
Approximately 250 g of each poultry product was rinsed in 250 ml of buffered peptone water (Oxoid, Basingstoke, England). After overnight incubation of the buffered peptone water at 37°C, 1 ml was used to inoculate 9 ml of Enterococcosel (BBL Microbiology Systems, Cockeysville, Md.) supplemented with 6 mg of vancomycin per liter, which was then incubated at 37°C for 24 to 48 h (25). All esculin-positive broth cultures were subcultured on a kanamycin-esculin azide agar (Oxoid) (1). Presumptive identifications of the Enterococcus spp. were made on the basis of colony morphology, results of Gram staining, and the presence or absence of catalase and pyrase (Dryslide Pyrkit; Difco Laboratories, Detroit, Mich.). Definitive identifications were made with Accuprobe (GenProbe, San Diego, Calif.) and RAPID ID32 STREP (BioMérieux, ’s Hertogenbosch, The Netherlands) test kits. The identification strips were read after 5 and 24 h of incubation at 37°C. All strains containing the vanC1 gene were identified as Enterococcus gallinarum (27). Strains were stored at −80°C in medium containing 15% glycerol.
Additional enterococcal strains.
Nineteen vancomycin-resistant Enterococcus faecium strains (VREF), one vancomycin-resistant E. faecalis strain from hospitalized patients, and four VREF from nonhospitalized patients (21) were also included in the study. All strains were highly resistant to both vancomycin and teicoplanin and possessed the vanA gene (see below). E. faecium BM4147 (vanA), E. faecalis V583 (vanB), E. faecalis ATCC 19433, E. faecalis ATCC 29212, E. gallinarum BM4147 (vanC1), Enterococcus casseliflavus CCUG 18657 (vanC2), Streptococcus bovis ATCC 33317, and S. aureus ATCC 29213 were used as reference strains.
Antimicrobial susceptibility tests.
All enterococcal strains described above were tested for vancomycin and teicoplanin resistance on a Mueller-Hinton agar (Difco Laboratories) with E-test strips (AB BIODISK, Solna, Sweden) according to the instructions of the manufacturer. All plates were incubated at 37°C and read after 24 h.
DNA isolation.
DNAs were isolated according to the method of Boom et al. (8). In brief, all VRE strains were grown overnight at 37°C on brucella blood agar plates. Ten colonies were mixed and suspended in 75 μl of TEG buffer (25 mM Tris-HCl [pH 8.0], 10 mM EDTA, 50 mM glucose). A lysozyme solution (75 μl of 10 mg/ml) was added, and this mixture was incubated for 1 h at 37°C. Guanidine-hydrothiocyanate was added for cell lysis, and Celite (Janssen Pharmaceuticals, Beerse, Belgium) was used for DNA binding. DNAs were washed and finally eluted from Celite with 10 mM Tris-HCl (pH 8.0) by incubation at 56°C for 10 min. DNA concentrations were estimated by electrophoresis on 1% agarose gels (Hispanagar; Sphaero Q, Leiden, The Netherlands) containing ethidium bromide in the presence of known quantities of lambda DNA as references.
vanA, vanB, vanC1, and vanC2 PCR.
Diagnostic PCR assays targeting the various resistance genes were performed as described by Dutka-Malen et al. (19). Approximately 10 to 100 ng (10 μl) of DNA was added to a PCR mixture (90 μl) containing 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 2.5 mM MgCl2, 0.01% gelatin, 0.1% Triton X-100, 0.2 mM deoxyribonucleotide, and 1.2 U of Taq DNA polymerase (Sphaero Q). Four different primer couples (vanA, vanB, vanC1, and vanC2 [see Table 1 for their DNA sequences]) were used in the assay (50 pmol of each individual primer per reaction mixture). Amplification of DNA was performed in a Biomed (Theres, Germany) thermocycler (model 60), with predenaturation at 94°C for 2 min, followed by 30 cycles of 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C. Amplicons were analyzed by electrophoresis on 1% agarose gels (Hispanagar; Sphaero Q) containing ethidium bromide in the presence of a 100-bp DNA ladder (Gibco/BRL Life Technologies, Breda, The Netherlands).
TABLE 1.
Nucleotide sequences of PCR primers
| Test | Primer paira | Nucleotide sequence | Size of PCR product (bp) | Reference |
|---|---|---|---|---|
| Diagnostic PCR | A1 | 5′-GGGAAAACGACAATTGC-3′ | 732 | 19 |
| A2 | 5′-GTACAATGCGGCCGTTA-3′ | |||
| B1 | 5′-ATGGGAAGCCGATAGTC-3′ | 635 | 19 | |
| B2 | 5′-GATTTCGTTCCTCGACC-3′ | |||
| C1 | 5′-GGTATCAAGGAAACCTC-3′ | 822 | 19 | |
| C2 | 5′-CTTCCGCCATCATAGCT-3′ | |||
| D1 | 5′-CTCCTACGATTCTCTTG-3′ | 439 | 19 | |
| D2 | 5′-CGAGCAAGACCTTTAAG-3′ | |||
| Transposon mapping of structural genes | VanR | 5′-AGCGATAAAATACTTATTGTGGA-3′ | 645 | 28 |
| VanR1 | 5′-CGGATTATCAATGGTGTCGTT-3′ | |||
| VanS | 5′-AACGACTATTCCAAACTAGAAC-3′ | 1,094 | 28 | |
| VanS1 | 5′-GCTGGAAGCTCTACCCTAAA-3′ | |||
| VanH | 5′-ATCGGCATTACTGTTTATGGAT-3′ | 943 | 28 | |
| VanH1 | 5′-TCCTTTCAAAATCCAAACAGTTT-3′ | |||
| VanA | 5′-ATGAATAGAATAAAAGTTGCAATAC-3′ | 1,029 | 28 | |
| VanA1 | 5′-CCCCTTTAACGCTAATACGAT-3′ | |||
| VanY | 5′-ACTTAGGTTATGACTACGTTAAT-3′ | 866 | 28 | |
| VanY1 | 5′-CCTCCTTGAATTAGTATGTGTT-3′ | |||
| Orf1A | 5′-AGGGCGACATATGGTGTAACA-3′ | 844 | 28 | |
| Orf1A1 | 5′-GGGCGACGGTACAACATCTT-3′ | |||
| Orf1B | 5′-TGGTGGCTCCTTTTCCCAGTTC-3′ | 1,007 | 28 | |
| Orf1B1 | 5′-CGTCCTGCCGACTATGATTATTT-3′ | |||
| Orf1C | 5′-ACCGTTTTTGCAGTAAGTCTAAAT-3′ | 1,066 | 28 | |
| Orf1C1 | 5′-AAACGGGATTTAGAAATAGTTAAT-3′ | |||
| Orf2D | 5′-CCATTTCTGTATTTTCAATTTATTA-3′ | 925 | 28 | |
| Orf2D1 | 5′-CATAGTTATCACCCTTTCACATA-3′ | |||
| Orf2E | 5′-TTGCGGAAAATCGGTTATATTC-3′ | 540 | 28 | |
| Orf2E1 | 5′-AGCCCTAGATACATTAGTAATT-3′ | |||
| Transposon mapping of intergenic regions | VanXY1 | 5′-AATAGCTATTTTGATTTCCCCGTTA-3′ | 543 | 6 |
| VanXY2 | 5′-TCCTGAGAAAACAGTGCTTCATTAA-3′ | |||
| VanSH1 | 5′-TAGGGTAGAGCTTCCAGCGATTGC-3′ | 311 | 6 | |
| VanSH2 | 5′-CTCATCCTGCTCACATCCATAAACA-3′ | |||
| VanYZ1 | 5′-GTTTCCCGGATCAACACATACTA-3′ | 336 | 6 | |
| VanYZ2 | 5′-CCCAGTAGCAGTAAATGGAGTCA-3′ |
Primers D1 and D2 are specific for the vanC2 gene. Orf1 is transposase, and Orf2 is resolvase.
Transposon mapping by PCR.
. To study heterogeneity of the VanA-encoding transposon Tn1546, regions of potentially various lengths within the 10,801-bp genetic element were studied by PCR (see Table 1 for primer sequences) (6, 28). Trial experiments were performed for E. faecium and E. faecalis only, and selection of a limited number of strains derived from either humans or poultry was at random. PCR was performed as described above. Whenever differences were detected in amplicon size, all additional E. faecalis and E. faecium strains harboring the vanA gene were investigated.
PFGE.
Ten colonies of an overnight culture, grown on a blood agar plate, were mixed and suspended in 100 μl of EET buffer (100 mM Na2EDTA, 10 mM EGTA, 10 mM Tris-HCl [pH 8.0]). This suspension was mixed with 100 μl of 1% agarose (Incert Agarose; FMC, Rockland, Maine) and pipetted into small plug molds. The cells suspended in the agarose plugs were lysed by incubation for 4 h at 37°C in 1 ml of EET buffer containing 10 mg of lysozyme (Sigma, Instruchemie, Hilversum, The Netherlands). Next, the lysis solution was replaced by a 1-ml EET buffer solution containing 1 mg of proteinase K (dissolved in 10 mM NaCl–10 mM Tris-HCl [pH 8.0]–1% sodium dodecyl sulfate), which was then incubated at 37°C for 16 h. The plugs were then washed six times (30 min each time at room temperature) with T10E1 solution (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Plugs were then stabilized twice for 30 min in 120 μl of 1× restriction buffer solution, and approximately 40 U of the restriction enzyme SmaI (Boehringer Mannheim, Mannheim, Germany) was added (incubation, 16 h at 25°C). Electrophoresis (1% SeaKem agarose in 0.5× Tris-borate-EDTA) was performed with a Bio-Rad contour-clamped homogeneous electric field mapper, programmed in the auto-algorithm mode (block 1 run time, 8 h; switch time, 0.5 to 15 s; block 2 run time, 10 h; switch time, 15 to 30 s). The gels were stained with ethidium bromide for 15 min and then destained in distilled water for 1 h before being photographed under UV irradiation. The gels were inspected visually by two different investigators. The pulsed-field gel electrophoresis (PFGE) patterns were interpreted according to the guidelines of Tenover et al. (35). Isolates that differed by one to three bands, consistent with a single differentiating genetic event, were assigned a numbered subtype. Four or more band differences between two strains defined a different genotype. Genotypes determined for all VREF isolated from chickens were compared with the PFGE characteristics determined for VREF isolated from humans (21). Since the interpretative guidelines brought forward by Tenover et al. (35) are mainly for outbreak investigations, additional comparisons were performed. Data obtained from a randomly selected group of 48 human- and poultry-derived VRE strains were studied in more detail with Gelcompar software (Applied Maths, Gent, Belgium). The PFGE patterns were scanned, and Dice analysis of peak positions was executed. Unweighted pair group method using arithmetic averages was applied, and the band-width tolerance was set critically at 1.2%.
Cloning and sequencing.
For several strains, the amplicon derived from the vanX-vanY intergenic region was cloned into plasmid pCR1 (Invitrogen, Leek, The Netherlands) according to the manufacturer’s instructions. Clones containing a correctly sized insert were sequenced by using cycle sequencing technology and a sequencing machine (model 373; Applied Biosystems Inc., Warrington, United Kingdom). Raw sequence data were edited with 373 software (Applied Biosystems Inc.).
RESULTS
VRE screening and antimicrobial susceptibility testing.
Table 2 summarizes all data gathered from the chicken specimens. Apparently, 242 of 305 (79%) of the poultry samples studied contained VRE. Out of these, 142 of 242 (59%) were identified as VREF, which were found nationwide in all of the participating centers. Thirty-six of 242 VRE strains (15%) were identified as Enterococcus durans, 34 of 242 VRE strains (14%) were identified as Enterococcus hirae, and 27 of 242 VRE strains (11%) were identified as E. gallinarum. E. faecalis was found only three times (1%). For all VREF and vancomycin-resistant E. faecalis strains, vancomycin MICs were ≥256 μg/ml and teicoplanin MICs were 16 to ≥256 μg/ml, which is indicative of the VanA phenotype. For vancomycin-resistant E. gallinarum, vancomycin MICs were 8 to 16 μg/ml and teicoplanin MICs were 1 to 3 μg/ml, the VanC phenotype. For the 70 strains classified as E. hirae or E. durans MICs ranged from 16 to ≥256 μg/ml for vancomycin and from 2 to ≥256 μg/ml for teicoplanin. All those VRE, except vancomycin-resistant E. gallinarum, harbored the vanA gene. Strains containing the vanB or vanC2 gene were not found.
TABLE 2.
Numbers and percentages of VRE isolated from 305 poultry products by 11 health inspectorates in The Netherlands in the period from June to September 1996
| Region of the food inspection department in The Netherlands | No. of poultry products | No. of poultry products with VRE | No. (%) of poultry products with VRE of type:
|
||||
|---|---|---|---|---|---|---|---|
| E. faecium | E. durans | E. hirae | E. gallinarum | E. faecalis | |||
| Den Haag | 34 | 18 | 15 (44) | 3 (9) | |||
| Maastricht | 40 | 33 | 16 (40) | 4 (10) | 6 (15) | 7 (18) | |
| Alkmaar | 22 | 17 | 11 (50) | 2 (9) | 4 (18) | ||
| Amsterdam | 47 | 35 | 19 (40) | 4 (9) | 5 (11) | 5 (11) | 2 (4) |
| Nijmegen | 25 | 21 | 6 (24) | 5 (20) | 3 (12) | 6 (24) | 1 (4) |
| Rotterdam | 17 | 12 | 5 (29) | 4 (24) | 3 (18) | ||
| Leeuwarden | 32 | 20 | 13 (41) | 2 (6) | 4 (13) | 1 (3) | |
| Den Bosch | 16 | 16 | 10 (63) | 3 (19) | 3 (19) | ||
| Goes | 25 | 25 | 15 (60) | 5 (20) | 1 (4) | 4 (16) | |
| Zutphen | 23 | 21 | 15 (65) | 1 (4) | 1 (4) | 4 (17) | |
| Groningen | 24 | 24 | 17 (71) | 3 (13) | 4 (17) | ||
| Total (%)a | 305 | 242 (79) | 142 (47) | 36 (12) | 34 (11) | 27 (9) | 3 (1) |
Percentage of all 305 products.
PFGE.
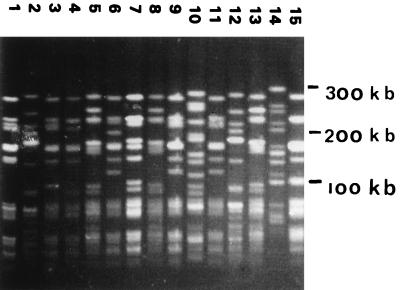
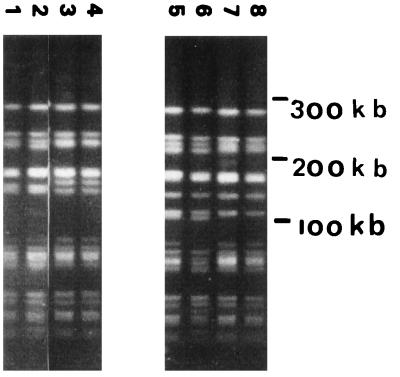
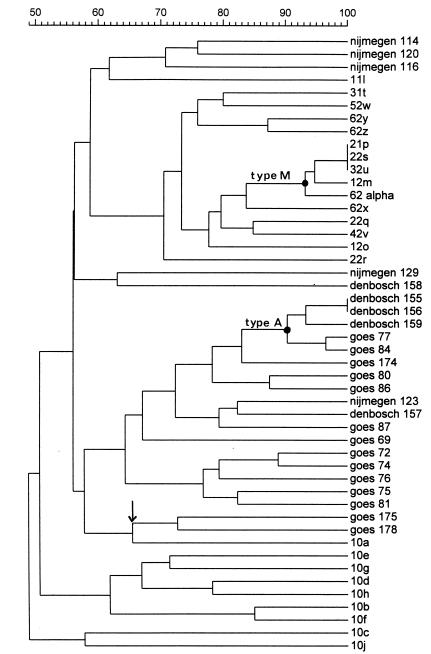
One hundred forty-two E. faecium and three E. faecalis isolates were analyzed by PFGE. Most of the PFGE banding patterns comprised 15 to 20 differently sized DNA fragments. The data revealed that two of three E. faecalis strains were genetically identical. Both strains originated from the same geographical region. One hundred different genotypes were identified in the group of VREF from poultry (for some examples of PFGE banding patterns, see Fig. 1). However, two genotypes of E. faecium, type A (22 of 142 VREF) and type B (14 of 142 VREF) (Fig. 2), were frequently found by 10 of 11 of the food inspection services. These two genotypes may represent Dutch epidemic VREF. When the poultry VREF were compared with patient VREF, however, no overlap in visually defined genotypes was identified by PFGE on the basis of the criteria of Tenover et al. (35). This result was essentially corroborated by Gelcompar analysis of the PFGE data of 48 strains (Fig. 3). The figure shows that the highest homology value between VRE from chicken and human is 60% (Goes 175 and Goes 178 versus 10a). Strains from the different origins present in a clustered fashion. The epidemic PFGE type A clusters at a high homology value (90% for Den Bosch 155 to Goes 84). The type that was encountered among humans relatively frequently (PFGE type M, described in reference 21) clustered as well. Finally, the figure shows that chicken strains mingle with respect to geographic origin. A total of 27 E. gallinarum strains could be identified on the basis of the characteristic PFGE patterns displaying DNA fragments smaller than 200 kb only (33).
FIG. 1.
PFGE patterns for 15 VREF isolated from poultry products collected by the Dutch Food Inspection Services in Zutphen, The Netherlands. Molecular lengths of the markers are indicated on the right.
FIG. 2.
PFGE patterns of two epidemic genotypes of VREF. Lanes 1 to 4, genotype A; lanes 5 to 8, genotype B. These genotypes were frequently found by most of the food inspection services. Molecular lengths of the markers are indicated on the right.
FIG. 3.
Phylogenetic tree constructed on the basis of several PFGE types of VREF derived from poultry products (originating from Den Bosch, Goes, and Nijmegen, The Netherlands) and humans (20). The arrow indicates the highest level of homology between VRE from poultry and humans (Goes 175 and Goes 178 versus 10a). Type A is the epidemic PFGE type among poultry clusters, and strains have a high level of homology (Den Bosch 155 to Goes 84). Type M is the type that was encountered among humans relatively frequently [20]).
Transposon mapping by PCR and sequencing.
All PCR tests for transposon mapping (Table 1) were done after a random selection of five human and five poultry strains. PCR products derived from structural Tn1546 genes for all human and poultry strains displayed identity in size after electrophoresis. The same conclusion was reached when the intergenic vanS-vanH and vanY-vanZ regions were amplified. However, the vanX-vanY intergenic region of two poultry strains was ∼1,300 bp in size whereas the size of the PCR product in the other three poultry strains and in the five human strains was approximately 540 bp. Subsequently, all VREF (142 poultry strains and 19 human strains) and all vancomycin-resistant E. faecalis strains (three poultry strains and one human strain) were analyzed with the vanX-vanY primer set. Both transposon types were found in all participating centers, indicating equal distributions of both of these transposon types. All human strains and 83 of the 142 (58%) isolated poultry VRE strains contained an intergenic region between vanX and vanY of approximately 540 bp. The 1,300-bp fragment was not found in human strains but was found in 59 of the 142 (42%) poultry strains. Sequencing of the 543-bp vanX-vanY intergenic regions of several VREF from poultry as well as from humans demonstrated full sequence conservation. With the larger vanX-vanY fragment, sequencing revealed the presence of IS1216V (40). This element was identified previously in the same location (GenBank accession no. L40841 [3]).
DISCUSSION
To the best of our knowledge this is the first systematic study from continental Europe reporting a high prevalence of VRE in consumer poultry at the retail level. Glycopeptide resistance in enterococci isolated from living poultry has been associated with the use of oral glycopeptide antibiotics in animal feed (4). High-level resistance to glycopeptides has been shown to be mediated by transferable plasmids that may harbor resistance determinants to other drugs as well (26). Therefore, other antimicrobial agents used as feed additives in veterinary medicine may also select for vancomycin resistance (33). Definition of causal relationships requires detailed studies of the development and spread of antibiotic resistance in poultry farms. Comparison of resistant microorganisms derived from poultry with those derived from humans may shed light on the role of poultry as a possible reservoir of VRE.
We found that 70% of the poultry products at the retail level were contaminated with VRE containing the vanA gene. The majority of these VRE were E. faecium. A study from the United Kingdom documented that 22 of 52 farm animals studied were colonized with VREF (5). In five uncooked chicken specimens, VREF were also identified. All strains possessed the vanA gene, which confers high-level resistance to vancomycin. A study from Manchester, United Kingdom, revealed that 90% of all uncooked chicken specimens contained VRE that were genetically distinguishable (12). The strains differed from clinical isolates but were capable of transferring the resistance trait by conjugation experiments. Others showed that vancomycin- and avoparcin-resistant E. faecium could be detected in five of eight conventional Danish poultry farms (29). On the other hand, among isolates from six ecology farms, no glycopeptide resistance was observed. In Belgium, about 7% of the animals investigated for VRE carriage (horses, dogs, pigs, and chickens) were colonized with VREF (16). Interestingly, VRE have so far not been recovered from animal sources in the United States, which may be related to the fact that glycopeptides are not licensed for use as feed additives in animal husbandry there (14, 35).
Twenty-seven of the poultry specimens contained E. gallinarum, a species which is rarely found in humans either as part of gut flora or as clinical isolates. However, we have observed an increase in the number of E. gallinarum strains isolated from clinical material in our hospital since the introduction of a screen agar containing 6 mg of vancomycin per liter (data not shown). These observations suggest that additional research into the relevance of E. gallinarum as a potential pathogen in humans is needed. As enterococci are not routinely identified to species level in many microbiology laboratories, E. gallinarum may well be underreported.
Two main routes of dissemination of vancomycin resistance genes can be envisaged. First, resistant strains may spread in a clonal fashion from one host to another. Second, the resistance determinant may be passed on to other bacterial strains through conjugation (2, 34).
Two major PFGE types of VRE have been identified among poultry-derived strains. Since these types were identified by all food inspection services, we are dealing with epidemic strains and not a local outbreak. Neither of these two types nor any of the other unique genotypes of VREF were found in fecal floras of patients screened for VREF carriage in The Netherlands (21). On the basis of these results, one could reject the hypothesis that direct horizontal transmission of VRE from poultry to humans via the food chain is a major transmission route, which is corroborated by more extensive phylogenetic analysis of the data (Fig. 3). Therefore, the answer to the question on the origin of human VRE remains obscure. An alternative route for introduction into humans may be by transmission of resistance-encoding genes rather than by transmission of resistant microorganisms. Several studies suggest that high-level resistance to glycopeptides in enterococci isolated in Europe and North America is mediated by transposons similar to Tn1546 (39). Mapping of the transposon as present in the poultry VRE by PCR revealed the presence of two distinct vanA types. Length variability was found in the vanX-vanY region. Among VREF from poultry, many strains, including epidemic VREF, carried an intergenic region between vanX and vanY of approximately 1,300 bp that was not encountered in the human strains. The other poultry strains and all human strains had identical vanX-vanY intergenic regions. This observation suggests that, for as yet unknown reasons, some sort of species barrier to the larger transposon type or limited conjugative transfer may exist. More Dutch VRE from humans should be investigated to confirm the data presented here. In contrast, another transposon type that is prevalent in many poultry strains and in all human strains may have spread from poultry to humans via the food chain. As we studied only a limited number of structural genes and intergenic regions, further detailed analysis of the vanA gene cluster is in progress to confirm that these transposons are related. The relationship between the vanA clusters of VRE isolated from humans and poultry was also determined by means of restriction fragment length polymorphism (RFLP) analysis of the Tn1546-like element. For this analysis, several human and poultry isolates were studied in detail. All human isolates showed the same RFLP type, as did some poultry isolates. The other isolates from poultry contained an RFLP type which was clearly distinct from the human RFLP type. (Work in collaboration with investigators at the National Institute of Public Health and the Environment, Bilthoven, The Netherlands, is still in progress [40].)
In conclusion, we report an extremely high prevalence of VRE in consumer poultry in The Netherlands. A high prevalence of a deviant transposon type is found in poultry VRE especially. Transmission of the resistance genes, rather than clonal dissemination of resistant microorganisms, may be the determining factor driving the spread of vancomycin resistance from poultry to humans. If this suggestion can be substantiated by additional research, this may have major implications for the development of strategies to control the spread of glycopeptide resistance among bacterial species pathogenic to humans. More information is needed to further clarify and quantify antibiotic resistance gene transfer from animals to humans.
ACKNOWLEDGMENTS
We gratefully acknowledge the Inspectorates for Health Protection of Alkmaar, Amsterdam, Den Haag, Rotterdam, Den Bosch, Goes, Groningen, Leeuwarden, Maastricht, Nijmegen, and Zutphen, The Netherlands, for their technical assistance. We thank Marian Humphrey for critically reading the English text.
REFERENCES
- 1.Anonymous. Kanamycin aesculin azide (KAA) agar. Int J Food Microbiol. 1987;5:731–734. [Google Scholar]
- 2.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bager F, Madsen M, Christensen J, Aarestrup F M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med. 1997;31:95–112. doi: 10.1016/s0167-5877(96)01119-1. [DOI] [PubMed] [Google Scholar]
- 5.Bates J, Jordens Z, Griffiths D T. Farm animals as a putative reservoir for vancomycin resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–516. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 6.Beighton, D., and M. Casewell. Personal communication.
- 7.Bingen E H, Denamur E, Lambert-Zechovsky N Y, Elion J. Evidence for the genetic unrelatedness of nosocomial vancomycin-resistant Enterococcus faecium strains in a pediatric hospital. J Clin Microbiol. 1991;29:1888–1892. doi: 10.1128/jcm.29.9.1888-1892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, Van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin in the United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. Staphylococcus aureus with reduced susceptibility to vancomycin—United States. Morbid Mortal Weekly Rep. 1997;46:765–766. [PubMed] [Google Scholar]
- 11.Chadwick P R, Oppenheim B A, Fox A, Woodford N, Morgenstern G R, Scarffe J H. Epidemiology of an outbreak due to glycopeptide resistant Enterococcus faecium on a leukaemia unit. J Hosp Infect. 1996;34:171–182. doi: 10.1016/s0195-6701(96)90063-8. [DOI] [PubMed] [Google Scholar]
- 12.Chadwick P R, Woodford N, Kacmarski E B, Gray S, Barrell R A, Oppenheim B A. Glycopeptide resistant enterococci isolated from uncooked meat. J Antimicrob Chemother. 1996;38:908–909. doi: 10.1093/jac/38.5.908. [DOI] [PubMed] [Google Scholar]
- 13.Chow J W, Kuritza A, Schlaes D A, Green M, Sahm D F, Zervos M J. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–1611. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coque T M, Tomayko J F, Ricke S V, Okhyusen P C, Murray B E. Vancomycin resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob Agents Chemother. 1996;40:2605–2609. doi: 10.1128/aac.40.11.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day M. Superbug spectre haunts Japan. New Sci. 1997;1997(5):521. [Google Scholar]
- 16.Devriese L A, Ieven M, Goossens H, Vandamme P, Pot B, Hommez J, Haesebrouck F. Presence of vancomycin-resistant enterococci in farm and pet animals. Antimicrob Agents Chemother. 1996;40:2285–2287. doi: 10.1128/aac.40.10.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donadedian S, Chow J W, Shlaes D M, Green M, Zervos M J. DNA hybridization and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J Clin Microbiol. 1995;33:141–145. doi: 10.1128/jcm.33.1.141-145.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunne W M, Wang W. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J Clin Microbiol. 1997;35:388–392. doi: 10.1128/jcm.35.2.388-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification of the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmond M B, Wenzel R P, Pasculle W. Vancomycin resistant Staphylococcus aureus: perspectives on measures needed for control. Ann Intern Med. 1996;123:329–334. doi: 10.7326/0003-4819-124-3-199602010-00008. [DOI] [PubMed] [Google Scholar]
- 21.Endtz H P, van den Braak N, van Belkum A, Kluytmans J A J W, Koeleman J G M, Spanjaard L, Voss A, Weersink A J L, Vandenbroucke-Grauls C M J E, Buiting A G M, van Duin A, Verbrugh H A. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Endtz H P, Van den Braak N, Van Belkum A, Van Keulen M, Vliegenthart J, Verbrugh H A. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. High prevalence and clonal spread of Tn1546, a transposon conferring resistance to vancomycin in enterococci from humans and consumer poultry, abstr. C-138; p. 70. [Google Scholar]
- 22.Gordts B, Van Landuyt H, Ieven M, Vandamme P, Goossens H. Vancomycin-resistant enterococci colonizing intestinal tracts of hospitalized patients. J Clin Microbiol. 1996;33:2842–2846. doi: 10.1128/jcm.33.11.2842-2846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hospital Infection Control Practices Advisory Committee. Recommendation for preventing the spread of vancomycin resistance: recommendation of the Hospital Infection Control Practices Advisory Committee (HICPAC) Am J Infect Control. 1995;23:87–94. doi: 10.1016/0196-6553(95)90104-3. [DOI] [PubMed] [Google Scholar]
- 24.Jordens J Z, Bates J, Griffiths D T. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother. 1994;35:515–528. doi: 10.1093/jac/34.4.515. [DOI] [PubMed] [Google Scholar]
- 25.Klare I, Heier H, Claus H, Reisbrodt R, Witte W. VanA mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol Lett. 1995;347:165–171. doi: 10.1111/j.1574-6968.1995.tb07353.x. [DOI] [PubMed] [Google Scholar]
- 26.Leclercq R, Derlot E, Weber M, Duval J, Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1989;33:10–15. doi: 10.1128/aac.33.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclercq R, Dutka-Malen S, Duval J, Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother. 1992;36:2005–2008. doi: 10.1128/aac.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miele A, Bandera M, Goldstein B P. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Møller Aarestrup F. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb Drug Resist. 1995;3:255–257. doi: 10.1089/mdr.1995.1.255. [DOI] [PubMed] [Google Scholar]
- 30.Moreira B, Boyle-Vavra S, deJonge B L M, Daum R S. Increased production of penicillin binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moulin F, Dumontier S, Saulnier P, Chachaty E, Loubeyre C, Brugieres L, Andremont A. Surveillance of intestinal colonisation and infection by vancomycin resistant enterococci in hospitalized cancer patients. Clin Microbiol Infect. 1996;2:192–201. doi: 10.1016/s1198-743x(14)65142-9. [DOI] [PubMed] [Google Scholar]
- 32.Murray B. What can we do about vancomycin-resistant enterococci? Clin Infect Dis. 1995;20:1134–1136. doi: 10.1093/clinids/20.5.1134. [DOI] [PubMed] [Google Scholar]
- 33.Noble W C, Virani Z, Cree R G A. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;93:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 34.Salyers A A, Amabile-Cuevas C F. Why are antibiotic resistance genes so resistant to elimination? Antimicrob Agents Chemother. 1997;41:2321–2325. doi: 10.1128/aac.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thal L A, Chow J W, Mahayni R, Bonilla H, Perri M B, Donabedian S A, Silverman J, Taber S, Zervos M J. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob Agents Chemother. 1995;39:2112–2115. doi: 10.1128/aac.39.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Belkum A, van den Braak N, Thomassen R, Endtz H. Vancomycin-resistant enterococci in cats and dogs. Lancet. 1996;348:1038–1039. doi: 10.1016/s0140-6736(05)64973-2. [DOI] [PubMed] [Google Scholar]
- 38.Van den Boogaard A E, Stobberingh E E. Time to ban all antibiotics as animal growth-promoting agents? Lancet. 1996;31:619. doi: 10.1016/s0140-6736(05)64838-6. [DOI] [PubMed] [Google Scholar]
- 39.Van Horn K G, Gedris C A, Rodney K M. Selective isolation of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:924–927. doi: 10.1128/jcm.34.4.924-927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willems, R. J. L. Personal communication.