Abstract
Prostate magnetic resonance imaging has become the imaging standard for prostate cancer in various clinical settings, with interpretation standardized according to the Prostate Imaging Reporting and Data System (PI-RADS). Each year, hundreds of scientific studies that report on the diagnostic performance of PI-RADS are published. To keep up with this ever-increasing evidence base, systematic reviews and meta-analyses are essential. As systematic reviews are highly resource-intensive, we investigated whether a machine learning framework can reduce the manual workload and speed up the screening process (title and abstract). We used search results from a living systematic review of the diagnostic performance of PI-RADS (1585 studies, of which 482 were potentially eligible after screening). A naïve Bayesian classifier was implemented in an active learning environment for classification of the titles and abstracts. Our outcome variable was the percentage of studies that can be excluded after 95% of relevant studies have been identified by the classifier (work saved over sampling: WSS@95%). In simulation runs of the entire screening process (controlling for classifier initiation and the frequency of classifier updating), we obtained a WSS@95% value of 28% (standard error of the mean ±0.1%). Applied prospectively, our classification framework would translate into a significant reduction in manual screening effort.
Patient summary
Systematic reviews of scientific evidence are labor-intensive and take a lot of time. For example, many studies on prostate cancer diagnosis via MRI (magnetic resonance imaging) are published every year. We describe the use of machine learning to reduce the manual workload in screening search results. For a review of MRI for prostate cancer diagnosis, this approach reduced the screening workload by about 28%.
Keywords: Automation, Machine learning, Meta-analysis, Prostate magnetic resonance imaging, Systematic review, Prostate Imaging Reporting and Data System
Prostate magnetic resonance imaging (MRI) has emerged as the core imaging modality in the diagnostic pathway for prostate cancer [1]. Interpretation of prostate MRI is standardized according to the Prostate Imaging Reporting and Data System (PI-RADS). PI-RADS involves a five-category risk stratification for the presence of clinically significant prostate cancer, with categories derived by combining various MRI descriptors [2]. Inclusion of prostate MRI in the diagnostic workup in various target populations is now strongly recommended by European and American urological guidelines. Every year, hundreds of original scientific papers are published that address the diagnostic performance of PI-RADS. To keep up with this ever-growing study pool, caregivers and patients must rely on results from systematic reviews and meta-analyses.
Systematic reviews and meta-analyses are highly resource-intensive, as all the search results from different databases must be checked for eligibility according to the article titles and abstracts. This is usually performed by two independent reviewers [3]. PI-RADS is intended as a living document (ie, one that evolves over time) and v2.1 is now the current version [2]. Maintaining an up-to-date evidence synthesis of diagnostic performance therefore requires a continuous effort. Our group has committed to conducting an ongoing living systematic review of PI-RADS (PROSPERO: CRD42022343931) for this reason [4].
Here we describe the framework for and results from a Bayesian machine learning approach for screening article titles and abstracts using the data from this living review. We hypothesize that machine learning could save valuable workload and facilitate faster updating of the review.
We used search results from MEDLINE, Embase and Cochrane Central from the ongoing review [4]. After exclusion of duplicates, two reviewers independently evaluated 1585 papers for further consideration using information provided in the title and abstract. After completion, discussion and consensus reading were performed for papers with discrepant results. This resulted in 482 papers for which the full text needed to be retrieved (eligible and potentially eligible) and 1103 papers that could definitely be excluded.
The open source ASreview framework (https://asreview.nl/) was used for data analysis [5]. Using ASreview, we implemented a naïve Bayes classifier for classification of titles and abstracts. Naïve Bayes classifiers are standard tools for natural language processing tasks such as spam filters and provide robust, computationally simple classification [6]. Naïve Bayes classifiers offer explainable classification. The impact of each predictive variable can be analyzed by studying the conditional probability table on which the classifier is based. Diagnostic information for the variables used can also be visualized for nonstatisticians [7].
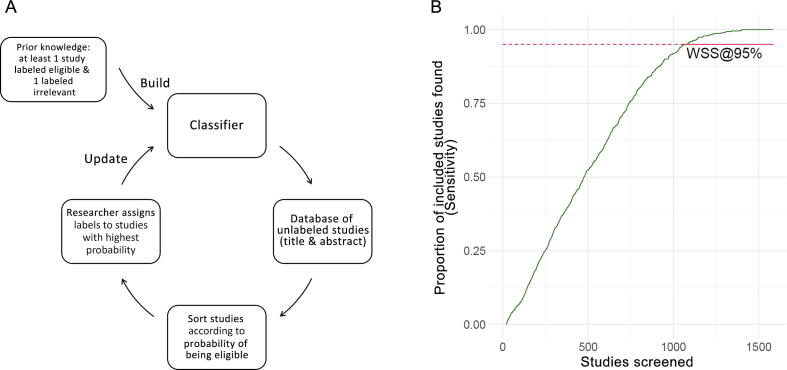
An active learning approach is applied. This means that the classification algorithm is updated after new studies have been screened and labeled by the reviewer (categorization: eligible and potentially eligible vs exclude). ASreview is run in simulation mode: the entire data set is labeled, but only a predefined count is taken for initial classifier training. Figure 1A explains the active learning pipeline.
Fig. 1.
(A) Active learning pipeline. The pipeline starts with building the classifier from prior knowledge; at least one study labeled eligible and one labeled irrelevant are needed. The classifier is then applied to the unlabeled data set and ranks all studies according to the probability of being eligible. Studies with the highest assigned probability are manually labeled. The classifier is updated after a certain number of studies have been labeled, and the cycle is repeated. (B) Performance of the naïve Bayes classifier for title and abstract screening (10 negative and positive studies are taken as prior knowledge, with a classifier update after every classification). The plot shows the sensitivity (y-axis, percentage of eligible/potentially eligible studies identified) as a function of the number of studies screened. The work saved over sampling (WSS), which is the percentage of studies that do not have to be screened, can be derived at any level of sensitivity considered sufficient. We show the WSS at 95% sensitivity (WSS@95%, solid red line). The formal definition of WSS@95% is: (n true negative studies + n false negative studies)/n all studies − 0.05, at 95% sensitivity [9].
We define the work saved over sampling at a sensitivity of 95% as our outcome variable (WSS@95%). WSS@95% is the percentage of studies that require no screening after 95% of relevant studies have been identified. To investigate the impact of the amount of prior knowledge used for initial classifier training and of the frequency of classifier updating, we systematically altered these two parameters. We also investigated how the inclusion or exclusion of studies used for initial classifier training in the calculation for WSS@95% affect this outcome. For all resulting combinations of the amount of prior knowledge and the frequency of classifier updating, we ran 20 simulations of the entire screening process and derived mean values and standard error of the mean for WSS@95%.
Figure 1B presents an example run for title and abstract screening using the active learning approach and illustrates the concept of WSS@95%. Figure 2 shows results for the review simulation runs for WSS@95%, presenting scenarios in which the initially labeled studies were and were not included for calculation of WSS@95%.
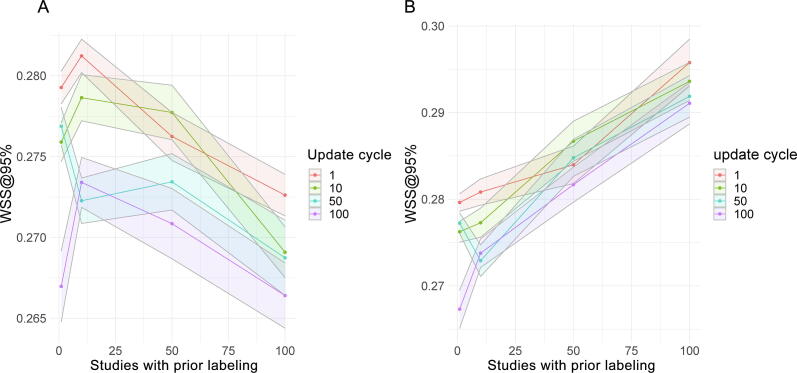
Fig. 2.
Results from simulation studies with initially labeled studies (A) included and (B) excluded. (A) WSS@95% (work saved over sampling at 95% sensitivity) for different combinations of the amount of prior knowledge (x-axis; numbers denote positive and negative studies used as prior knowledge for initial classifier training) and the frequency of classifier updating (color coded according to the number of studies after which a classifier update is performed). Studies that are used as prior knowledge are included in the WSS@95% calculation. With a high number of manually labeled instances as prior knowledge for initial classifier training, there is a slight reduction in WSS@95%. Each combination was run 20 times. The shaded areas represent the standard error of the mean for the point estimates of mean WSS@95%. (B) WSS@95% from the same simulation runs as in A but with studies used for the initial classifier training excluded for WSS@95% calculation. There is a trend for better WSS@95% results with greater initial training data.
Overall, WSS@95% ranged from 27% to 28% (Fig. 2). When the labeling of prior studies is considered relevant for WSS@95% calculation, the impact of the amount of prior knowledge and of the frequency of classifier updating on absolute WSS@95% is small (Fig. 2A). The maximum WSS@95% is achieved with only a few studies labeled as prior knowledge and classifier updates after every manual classification (Fig. 2A, red curve). If the initial work is not considered relevant, WSS@95% improves with classifiers initially built from larger data sets (Fig. 2B). If the classification approach is applied prospectively (ie, in update searches), we expect even higher WSS@95% in the subsequent screening process, because a large part of the present data set is used for classifier training.
We can assume that the time for screening a single study by title and abstract ranges between 30 s and 7 min, depending on reviewer experience and domain complexity [8]. Accordingly, if 444 of 1585 studies (28%) do not have to be screened, this would result in manual research time saved between 3.7 and 52.8 h per reviewer. This sums to 7.4–105.6 h for two reviewers. WSS@95% generally provides a good balance between screening sensitivity and work saved [9]. A systematic review of meta-analyses finds only a negligible effect on the overall results when a small percentage of relevant studies are missing [10], which in our view contributes to consideration of WSS@95% as a reasonable outcome. One drawback of WSS@95% is its dependence on the prevalence of eligible studies in the data set. With a prevalence of 50%, the maximum WSS@95% is 45% [9]. Our WSS@95% result of 27–28% may demonstrate the impact of this relatively high prevalence, as higher WSS@95% values have been reported for applications in other domains [5].
For transparency, we provide the python code for running the simulation experiments and our data set comprising the list of digital object identifiers and the labels assigned to them as Supplementary material. Together with our search strategies [4], this allows full reproduction of our work. We plan to report on the prospective evaluation of our classification support when the review is updated. We are optimistic that this approach will save valuable research time in the future in our project and comparable projects, and will allow researchers to focus more on data analysis.
Author contributions: Matthias Benndorf had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Nedelcu, Oerther, Engel, Benndorf.
Acquisition of data: Nedelcu, Oerther, Engel, Sigle, Benndorf.
Analysis and interpretation of data: Nedelcu, Sigle, Schmucker, Schoots, Eisenblätter, Benndorf.
Drafting of the manuscript: Nedelcu, Oerther, Benndorf.
Critical revision of the manuscript for important intellectual content: Nedelcu, Oerther, Engel, Sigle, Schmucker, Schoots, Eisenblätter, Benndorf.
Statistical analysis: Benndorf.
Obtaining funding: Benndorf.
Administrative, technical, or material support: None.
Supervision: None.
Other (statistical planning): Benndorf.
Financial disclosures: Matthias Benndorf certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: Ivo G. Schoots is a full panel member of the PI-RADS steering committee. The remaining authors have nothing to disclose.
Funding/Support and role of the sponsor: The underlying living systematic review is supported by a grant from the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, project 01KG2202). The sponsor played a role in data collection and management.
Associate Editor: Guillaume Ploussard
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.euros.2023.07.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schoots I.G., Padhani A.R. Delivering clinical impacts of the MRI diagnostic pathway in prostate cancer diagnosis. Abdom Radiol. 2020;45:4012–4022. doi: 10.1007/s00261-020-02547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American College of Radiology Committee on PI-RADS. Prostate Imaging-Reporting and Data System version 2.1. American College of Radiology; 2019. https://www.acr.org/Clinical-Resources/Reporting-and-Data-Systems/PI-RADS.
- 3.Kim K.W., Lee J., Choi S.H., Huh J., Park S.H. Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: a practical review for clinical researcherspart I. General guidance and tips. Korean J Radiol. 2015;16:1175–1187. doi: 10.3348/kjr.2015.16.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oerther B., Schmucker C., Schwarzer G., et al. Living systematic review and meta-analysis of the prostate MRI diagnostic test with Prostate Imaging Reporting and Data System (PI-RADS) assessment for the detection of prostate cancer: study protocol. BMJ Open. 2022;12 doi: 10.1136/bmjopen-2022-066327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Schoot R., de Bruin J., Schram R., et al. An open source machine learning framework for efficient and transparent systematic reviews. Nat Mach Intell. 2021;3:125–133. doi: 10.1038/s42256-020-00287-7. [DOI] [Google Scholar]
- 6.Hand D.J., Yu K. Idiot’s Bayes: not so stupid after all? Int Stat Rev. 2001;69:385–398. doi: 10.2307/1403452. [DOI] [Google Scholar]
- 7.Kulesza T., Burnett M., Wong W.K., Stumpf S. Proceedings of the 20th International Conference on Intelligent User Interfaces. Association for Computing Machinery; 2015. Principles of explanatory debugging to personalize interactive machine learning; pp. 126–133. [DOI] [Google Scholar]
- 8.Wallace B.C., Trikalinos T.A., Lau J., Brodley C., Schmid C.H. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinform. 2010;11:55. doi: 10.1186/1471-2105-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusa W., Lipani A., Knoth P., Hanbury A. An analysis of work saved over sampling in the evaluation of automated citation screening in systematic literature reviews. Intell Syst Appl. 2023;18 doi: 10.1016/j.iswa.2023.200193. [DOI] [Google Scholar]
- 10.Waffenschmidt S., Knelangen M., Sieben W., Bühn S., Pieper D. Single screening versus conventional double screening for study selection in systematic reviews: a methodological systematic review. BMC Med Res Methodol. 2019;19:132. doi: 10.1186/s12874-019-0782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.