Abstract
Salmonella is one of the most important zoonotic pathogens and a major cause of foodborne illnesses, posing a serious global public health hazard. The emergence of plasmid-mediated mcr genes in Salmonella has greatly reduced the clinical choice of salmonellosis treatment. The aim of this study was to investigate the plasmid characteristics of mcr-positive Salmonella identified from patients in Sichuan, China during 2014 to 2017 by whole genomes sequencing. In this study, a total of 12 mcr-positive isolates (1.15%, ; mcr-1, n=10; mcr-3, n=2) were identified from 1046 Salmonella isolates using PCR. Further characterization of these isolates was performed through antimicrobial susceptibility testing, conjugation assays, whole genome sequencing, and bioinformatics analysis. The mcr-1 gene in these isolates were carried by three types of typical mcr-1-bearing plasmids widely distributed in Enterobacteriaceae (IncX4, IncI2 and IncHI2). Of note, two mcr-1-harboring IncHI2 plasmids were integrated into chromosomes by insertion sequences. Two mcr-3-bearing plasmids were IncC and IncFIB broad-host-range plasmids respectively. Genetic context analysis found that mcr-1 was mainly located in Tn6330 or truncated Tn6300, and mcr-3 shared a common genetic structure tnpA-mcr-3-dgkA-ISKpn40. Overall, we found that mcr gene in clinical Salmonella were commonly carried by broad-host plasmids and have potential to transfer into other bacteria by these plasmids. Continuous surveillance of MDR Salmonella in humans and investigation the underlying transmission mechanisms of ARGs are vital to curb the current severe AMR concern.
Keywords: Salmonella enterica, colistin resistance, mcr, plasmid, genomics
1. Introduction
Colistin is a polypeptide antimicrobial with a narrow antibacterial spectrum that is largely effective against Gram-negative bacteria and has long been used in clinical medicine and animal husbandry (Landman et al., 2008; Biswas et al., 2012). In livestock industry, it was allowed and widely used for non-therapeutic usage, such as growth promotion, in many countries (Li et al., 2006). In clinical practice, it was commonly used as a non-preferred treatment for multidrug-resistant (MDR) Gram-negative bacteria due to its toxicity to human previously (Conway et al., 1997). However, the emergence of carbapenem-resistant Enterobacteriaceae (CRE) had limited the clinical treatment options. The World Health Organization (WHO) listed colistin as the highest priority critically important antibiotic fighting against CRE. The emergence of plasmid-medicated colistin resistance gene, mcr-1, in animal Escherichia coli in 2015 fundamentally changed the administration practice of colistin (Liu et al., 2016). Several countries have banned the use of colistin as an animal growth promoter (Wang et al., 2020). However, the current global prevalence of the mobile colistin resistance gene mcr-1 and its variants are still a serious concern (Wang et al., 2017; Sun et al., 2018). To date, ten different mcr variants (mcr-1 to mcr-10) have been identified in different bacteria isolated from animals, foods, humans, and the environment (Hussein et al., 2021). The majority of them were found in plasmids, suggesting that mcr variants could be transmitted across intraspecific and interspecific bacteria. The mcr-1 gene was shown to be more prevalent in bacteria from animal sources than that from patients with nosocomial illnesses (Huang et al., 2017; Tian et al., 2017). This phenomenon may be related to the different exposure of colistin in different settings.
Salmonella is a group of gram-negative bacteria found in the intestinal tract of humans and animals as well as in the environment that can cause gastrointestinal diseases and fever called salmonellosis (Lin et al., 1995; Holschbach and Peek, 2018). Recently, a global systematic meta-analysis estimated that approximately 15% of patients with non-typhoidal Salmonella invasive disease die (Marchello et al., 2022). In some developing countries, infections caused by Salmonella are endemic and represent a serious public health hazard (Abd El-Ghany, 2020). Most healthy people infected by Salmonella can recover without specific treatment and do not require treatments of antimicrobials. However, young children and people with weakened immune systems may have an increased risk of developing bacteremia, meningitis and osteomyelitis (Marchello et al., 2022). Although mcr genes have seriously spread in Enterobacteriaceae, colistin resistance Salmonella were significantly less than E. coli and Klebsiella pneumoniae. Nonetheless, colistin resistance has emerged in clinical Salmonella in many countries, and mcr-1 and mcr-3 were the most common variants (Portes et al., 2022). The public health risk caused by colistin-resistant Salmonella has increasingly arisen. However, there are few systematic investigations of the plasmid characteristics in mcr-positive clinical Salmonella (Shen et al., 2021; Portes et al., 2022). In this study, we comprehensively investigated the characteristics of mcr-bearing plasmids in clinical Salmonella isolates isolated from Sichuan province during 2014 to 2017.
2. Materials and methods
2.1. Bacterial isolates
The clinical Salmonella isolates were collected from different hospitals in Sichuan province, China between 2014 and 2017. The Salmonella isolates were recovered from patients following the previous methods (Xia et al., 2009). The mcr-positive isolates were screened by the multiplex PCR method and confirmed by Sanger sequencing (Rebelo et al., 2018). All mcr-positive isolates were stored at -80°C in LB broth containing 20% glycerol for further investigation.
2.2. Antimicrobial susceptibility testing and conjugation assay
The MICs (minimum inhibitory concentrations) of mcr-positive isolates against different antimicrobials, including kanamycin, ciprofloxacin, meropenem, ampicillin, colistin, enrofloxacin, tetracycline and ceftiofur, were tested using broth microdilution as per Clinical and Laboratory Standards Institute (CLSI) standards (CLSI, 2020). E. coli ATCC25922 was used for quality control. In order to test the transferability of mcr genes, conjugation experiments were performed using E. coli C600 (RifR) as recipients. Briefly, the donor and recipient strains were grown in LB broth until they reached the logarithmic growth phase, then mixed at the ratio of 1:1 and cultured overnight on LB agar plates. The transconjugants were screened on LB agar plates with rifampin (300 mg/L) and colistin (2 mg/L), and validated using PCR targeting the mcr genes and 16S rDNA gene sequencing.
2.3. Genomic DNA extraction and whole genome sequencing
In order to decipher the genetic structure features of mcr genes in these Salmonella isolates. We first extracted their genomic DNA using FastPure Bacteria DNA Isolation Mini Kit (Vazyme™, China) according to the instructions of the manufacturer. The purity and concentration of genomic DNA were evaluated using the Titertek-Berthold Colibri (Berthold™, Germany) and the QubitR Fluorometer (Thermo Fisher™, US) respectively. Then, extracted genomic DNA was sequenced using both short-read and long-read sequencing methods. Short-read DNA sequencing was conducted on the BGISEQ-50 platform with the SE50 strategy. The long-read sequencing was performed on ONT (Oxford Nanopore Technologies) MinION platform using the SQK-LSK109 library preparation kit in R9.4 flow cells.
2.4. Genome assembly and genomic feature analysis
The complete genomic sequences of mcr-positive Salmonella isolates were obtained using Unicycler (Wick et al., 2017) with hybrid assemble strategy based on short- and long-read data. An online fully-automated service, RAST was used to annotate the complete genomes (https://rast.nmpdr.org/). Antimicrobial resistance genes (ARGs), insertion sequences (ISs) and plasmid replicon genes were detected using abricate tool (https://github.com/tseemann/abricate) based on NCBI AMRFinderPlus, ISFinder and PlasmidFinder databases. The plasmid data were downloaded from NCBI RefSeq database (https://ftp.ncbi.nlm.nih.gov/genomes/refseq/plasmid/). The Salmonella serovars were identified using SISTR (Yoshida et al., 2016). The multi-locus sequence type (MLST) was analyzed using an mlst tool (https://github.com/tseemann/mlst). BRIG v.0.95 and Easyfig v.2.2.3 were used to generate plasmid comparison maps (Alikhan et al., 2011; Sullivan et al., 2011).
3. Results
3.1. Identification of mcr-positive S. enterica and antimicrobial susceptibility testing
In total, 12 mcr-positive isolates (ten mcr-1 and two mcr-3) were detected from 1046 clinical Salmonella isolates using PCR. The prevalence and basic genomic features have been identified in our previous study (Li et al., 2022). For the 12 mcr-positive S. enterica, five isolates each year were detected in 2016 and 2017. The remaining two isolates were found in 2014. No mcr-positive isolates were detected in 2015. Antimicrobial susceptibility testing showed that the 12 mcr-positive S. enterica isolates were all resistant to colistin but sensitive to meropenem. In addition, the majority of them were resistant to ampicillin, tetracycline, aztreonam and kanamycin, whereas they were almost all sensitive to ciprofloxacin and enrofloxacin ( Table 1 ). The majority of 12 mcr-positive S. enterica isolates showed multidrug resistance against three or more types of antimicrobials, designated as MDR isolates. According to the conjugation assay, the colistin resistance phenotype of the 12 S. enterica isolates could be transferred into E. coli C600. Antimicrobial susceptibility testing showed that all transconjugants were resistant to colistin. In addition, majority of transconjugants displayed resistance to other antimicrobials ( Table 1 ). This indicated that the resistance genes in donor S. enterica were probable carried by conjugative MDR plasmids.
Table 1.
The basic information and MICs (mg/L) of mcr-positive isolates and their transconjugants in this study.
| Isolates | Gene | Serovar | STs a | Antimicrobials b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KAN | CIP | MEM | AMP | CL | ENR | TET | CFF | ||||
| SC2014107 | mcr-1 | Orion | 684 | 8 | ≤0.25 | ≤0.25 | 8 | 8 | ≤0.25 | 4 | ≤0.25 |
| SC2014238 | mcr-1 | Meleagridis | 463 | 4 | ≤0.25 | ≤0.25 | >128 | 4 | ≤0.25 | 8 | >128 |
| SC2016025 | mcr-1 | I 4,[5],12:i:- | 34 | 128 | 4 | ≤0.25 | >128 | 8 | 2 | 128 | >128 |
| SC2016042 | mcr-1 | I 4,[5],12:i:- | 34 | 8 | ≤0.25 | ≤0.25 | >128 | 8 | ≤0.25 | 128 | 8 |
| SC2016090 | mcr-1 | I 4,[5],12:i:- | 34 | 8 | ≤0.25 | ≤0.25 | >128 | 4 | 2 | 64 | >128 |
| SC2016091 | mcr-3 | I 4,[5],12:i:- | 34 | 8 | ≤0.25 | ≤0.25 | >128 | 8 | ≤0.25 | 128 | >128 |
| SC2016290 | mcr-3 | I 4,[5],12:i:- | 34 | 4 | ≤0.25 | ≤0.25 | >128 | 8 | ≤0.25 | 32 | >128 |
| SC2017030 | mcr-1 | I 4,[5],12:i:- | 34 | >128 | ≤0.25 | ≤0.25 | >128 | 8 | ≤0.25 | 64 | >128 |
| SC2017057 | mcr-1 | I 4,[5],12:i:- | 34 | >128 | ≤0.25 | ≤0.25 | >128 | 1 | ≤0.25 | 16 | 128 |
| SC2017100 | mcr-1 | I 4,[5],12:i:- | 34 | 2 | 0.5 | ≤0.25 | >128 | 4 | ≤0.25 | 32 | >128 |
| SC2017167 | mcr-1 | I 4,[5],12:i:- | 34 | >128 | ≤0.25 | ≤0.25 | >128 | 4 | ≤0.25 | 64 | >128 |
| SC2017297 | mcr-1 | I 4,[5],12:i:- | 34 | >128 | ≤0.25 | ≤0.25 | >128 | 8 | 1 | 32 | >128 |
| cSC2014107 | mcr-1 | / | / | 1 | ≤0.25 | ≤0.25 | 8 | 2 | 1 | 0.5 | ≤0.25 |
| cSC2014238 | mcr-1 | / | / | 1 | ≤0.25 | ≤0.25 | 32 | 2 | 0.5 | 0.5 | 0.5 |
| cSC2016025 | mcr-1 | / | / | 4 | 0.5 | ≤0.25 | >128 | 2 | 1 | 32 | 128 |
| cSC2016042 | mcr-1 | / | / | 2 | ≤0.25 | ≤0.25 | 32 | 2 | 0.5 | 0.5 | 0.5 |
| cSC2016090 | mcr-1 | / | / | 4 | ≤0.25 | ≤0.25 | >128 | 2 | 1 | 0.5 | 16 |
| cSC2016091 | mcr-3 | / | / | 1 | ≤0.25 | ≤0.25 | >128 | 2 | 1 | ≤0.25 | >128 |
| cSC2016290 | mcr-3 | / | / | 1 | ≤0.25 | ≤0.25 | >128 | 2 | 0.5 | 4 | >128 |
| cSC2017030 | mcr-1 | / | / | >128 | ≤0.25 | ≤0.25 | >128 | 2 | 0.5 | 32 | 32 |
| cSC2017057 | mcr-1 | / | / | 8 | ≤0.25 | ≤0.25 | >128 | 2 | 0.5 | 32 | 64 |
| cSC2017100 | mcr-1 | / | / | 2 | ≤0.25 | ≤0.25 | >128 | 2 | 0.5 | 0.5 | 16 |
| cSC2017167 | mcr-1 | / | / | >128 | ≤0.25 | ≤0.25 | >128 | 2 | 0.5 | 0.5 | 16 |
| cSC2017297 | mcr-1 | / | / | >128 | ≤0.25 | ≤0.25 | >128 | 1 | 0.5 | 16 | 64 |
STs indicate sequence types.
KAN, kanamycin; CIP, ciprofloxacin; MEM, meropenem; AMP, ampicillin; CL, colistin; ENR, enrofloxacin; TET, tetracycline; CFF, ceftiofur.
3.2. Genetic context of mcr-1 and mcr-3
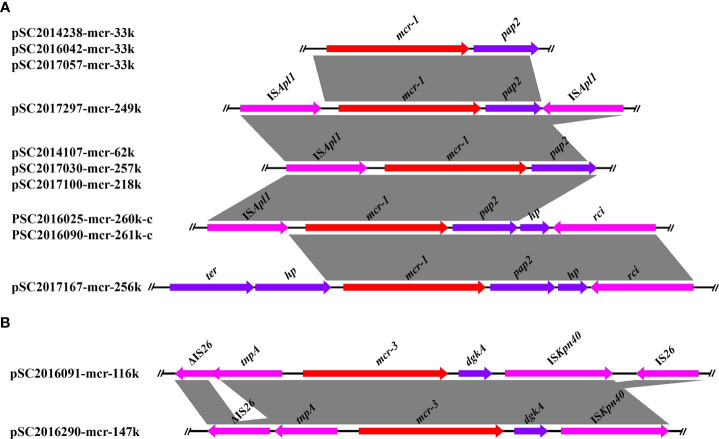
The genetic contexts of mcr-1 in the ten mcr-1 positive Salmonella isolates were classified as five types ( Figure 1 ). The genetic structures of mcr-1 in IncX4 plasmids were identical. No mobile elements were detected around the mcr-1 gene in IncX4 plasmids. In IncHI2 plasmids, four different mcr-1 genetic structures were found. Apart from plasmid pSC2017167-mcr-256k, we found an ISApl1 in the upstream of mcr-1 in the other IncHI2 plasmids. In addition, we detected a reversed ISApl1 in the downstream of mcr-1 in plasmid pSC2017297-mcr-249k. ISApl1 was an important element associated with the rapid dissemination of mcr-1, which is usually located in both upstream and downstream of mcr-1 forming an ISApl1-mcr-1-pap2-ISApl1 (Tn6330) transposon (Snesrud et al., 2018). Another study demonstrated that mcr-1 genetic structures with only one ISApl1 or with ISApl1 absent were formed by deletion of ISApl1 from the ancestral Tn6330 (Snesrud et al., 2018). We detected truncated Tn6330 in many mcr-1-bearing plasmids, suggesting that the genetic structures of mcr-1 in these plasmids have evolved for a long time.
Figure 1.
Linear comparison of the core genetic contexts of mcr genes investigated in this study. (A) The genetic context of mcr-1 in plasmids and chromosomal plasmids. PSC2016025-mcr-260k-c and PSC2016090-mcr-261k-c were two chromosomally integrated plasmids. (B) The genetic structure of mcr-3 in plasmids pSC2016091-mcr-116k and pSC2016290-mcr-147.
The genetic structures of mcr-3 in plasmids pSC2016091-mcr-116k and pSC2016290-mcr-147k were similar and shared a common core structure tnpA-mcr-3-dgkA-ISKpn40 ( Figure 1 ). In plasmid pSC2016091-mcr-116k, we found a truncated IS26 present upstream and an intact IS26 present downstream of mcr-3. Only an intact IS26 present upstream of mcr-3 was detected in plasmid pSC2016290-mcr-147k. Similar to the mobilization of mcr-1 medicated by Tn6330, the mobility of mcr-3 is usually associated with a composite transposon structure ISKpn40-mcr-3.1-dgkA-ISKpn40 (He et al., 2020). Apart from ISKpn40, the highly mobilized IS26 around mcr-3 deserves our attention, as it may facilitate the horizontal translocation of mcr-3.
3.3. Distribution of mcr-bearing plasmids in S. enterica isolates
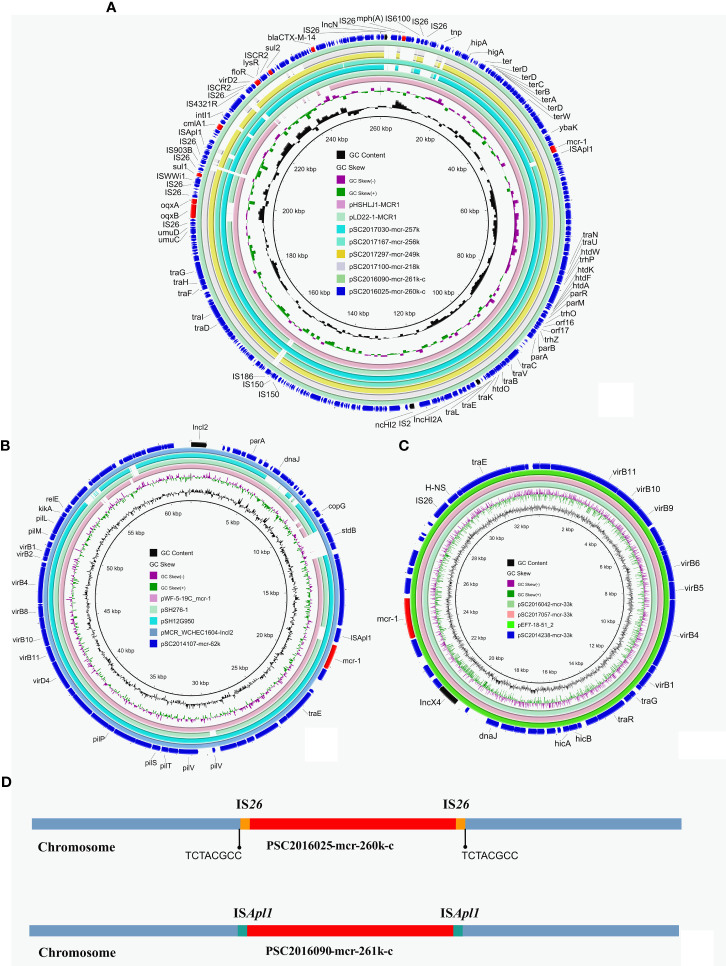
Long-read nanopore sequencing and short-read Illumina sequencing were performed on the 12 S. enterica isolates. The complete chromosomes and plasmids of these isolates were obtained using a hybrid assembly strategy and single-read analysis, as described in our previous study (Li et al., 2018). According to the assembly results, we found two chromosomally integrated IncHI2 plasmids from isolates SC2016025 and SC2016090 as a similar phenomenon previously reported ( Figures 2A, D ) (Chang et al., 2022). Similar mcr-1-bearing IncHI2/HI2A plasmids were found in other mcr-1 positive isolates from our investigation including SC2017030, SC2017100, SC2017297 and SC2017167. Moreover, these mcr-1-bearing IncHI2/HI2A plasmids were also found in NCBI nr database by BLASTn analysis ( Figure 2A ). Almost identical mcr-1 bearing plasmids were simultaneously detected in different genus, different geographical locations, different sources of isolates, implying that such plasmids play a crucial role in the horizontal transmission of mcr-1.
Figure 2.
Structure analysis of mcr-1-bearing plasmids in Salmonella isolates. (A) Comparative analysis of mcr-1-carrying IncHI2 plasmids and chromosomally integrated plasmids in this study with other similar plasmids including pLD22-1-MCR1 (CP047877.1) and pHSHLJ1-MCR1 (KX856066.1). (B) Four identical mcr-1-bearing IncX4 plasmids including three identified in this study and plasmid pEF7-18-51_2 (CP063489.1) from NCBI database. (C) Circular comparative analysis of pSC2014107-mcr-62k with four closely related plasmids including pMCR_WCHEC1604-IncI2 (KY829117.1), pSH12G950 (MH522410.1), pSH276-1 (MG299140.1) and pWF-5-19C_mcr-1 (KX505142.1). Resistance genes and plasmid replicon genes in these plasmids were highlighted in red arrows and black arrows respectively. (D) The integration sites of chromosomal plasmids PSC2016025-mcr-260k-c and PSC2016090-mcr-261k-c.
Comparative analysis found that there are some structure differences in these mcr-1-bearing IncHI2/HI2A plasmids from different isolates. A replicon gene of IncN plasmids was absent in plasmids pSC2017100-mcr-218k, pSC2017297-mcr-249k and pSC2017030-mcr-251k. In plasmid pSC2017100-mcr-218k, a region encoding multiple resistance genes containing cmlA1, floR and sul2 was lost ( Figure 2A ). The evolution of this type of plasmids in different isolates may result in their structural changing. IncHI2/HI2A plasmids with mcr-1 are commonly detected in E. coli (Lu et al., 2020). This type of mcr-1 positive plasmid has recently been found frequently in Salmonella (Vazquez et al., 2021), indicating that the mcr-1 gene has the ability to transfer to other species of bacteria with their assistance. We should pay more attention to monitor the spread of such mcr-1-bearing plasmids.
We identified three mcr-1-bearing IncX4 plasmids in isolates SC2014238, SC2016042 and SC2017057. The three IncX4 plasmids shared 100% identity and 100% coverage with each other. In addition, there are many IncX4 plasmids from other genus of bacteria that are identical to the three plasmids ( Figure 2B ). Like IncHI2/HI2A plasmids, IncX4 plasmids were also a common plasmid type carrying mcr-1, because they are self-transferable with high conjugation frequencies (Sun et al., 2017). Apart from IncHI2/HI2A and IncX4 plasmids, an IncI2 plasmid harboring mcr-1 with 62kb in length was identified in isolate SC2014107 ( Figure 2C ). The IncI2 plasmids have recently been noticed due to they are usually associated with the transmission of ESBLs genes and mcr-1 (Wong et al., 2015). A previous investigation showed that IncI2 plasmids were the most common type of mcr-1-carrying plasmids (Ricker et al., 2022). In addition, we found many mcr-1-bearing IncI2 plasmids from different bacteria in nr database ( Figure 2C ), indicating that the IncI2 plasmids were broad-host-range plasmids.
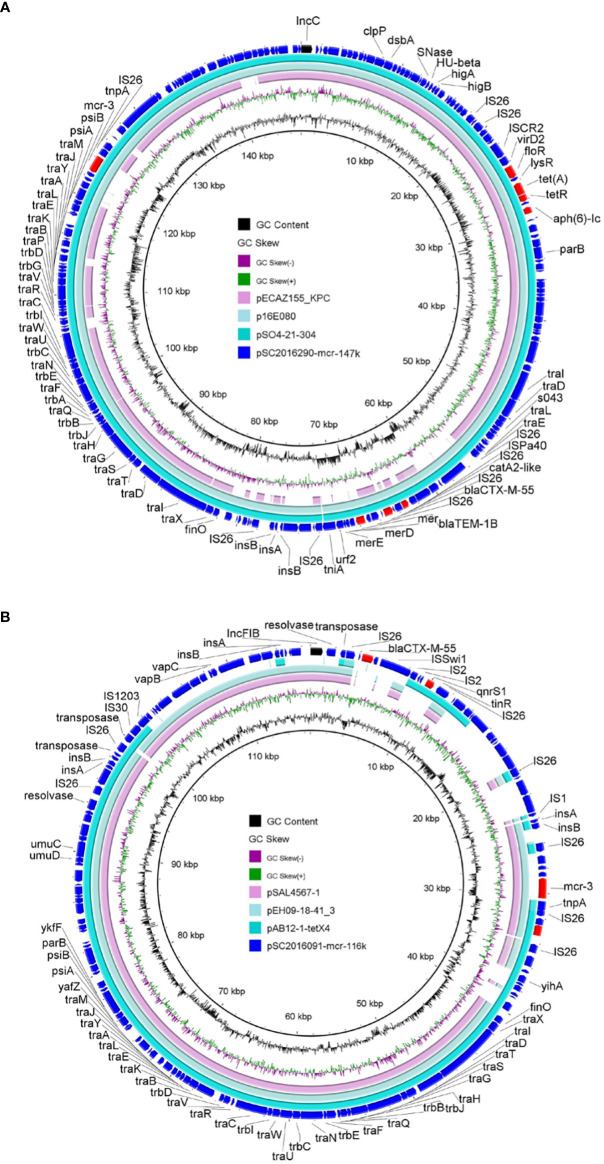
Apart from mcr-1, another mcr variant mcr-3 was detected in isolates SC2016090 and SC2016091. Whole genome analysis of the two isolates found that the mcr-3 gene was located on plasmids. In isolate SC2016090, mcr-3 was carried by an IncC plasmid pSC2016290-mcr-147k with a size 147kb. Besides, the plasmid also carried bla CTX-M-55, bla TEM-1B, catA2, aph(6)-Id, tet(A) and floR. We noticed that a few of such mcr-3-bearing plasmids have been detected in other Salmonella isolates ( Figure 3A ). Of note, the plasmid pSC2016290-mcr-147k showed a partial backbone similar to an E. coli plasmid pECAZ155_KPC carrying the carbapenemase gene bla KPC-2. This phenomenon implied that the plasmid pSC2016290-mcr-147k could integrate into other plasmids, causing the mcr-3 gene to spread to other bacteria. Another mcr-3-bearing plasmid found in isolate SC2016091 was an IncFIB type plasmid pSC2019091-mcr3-116k with 116kb. Coincidently, the plasmid also carried ESBL gene bla CTX-M-55 ( Figure 3B ). Furthermore, the IncFIB plasmids belonged to a type of broad host plasmids, and there are numerous plasmids from various bacteria sharing a backbone comparable to the IncFIB plasmid.
Figure 3.
Structure analysis of mcr-3-bearing plasmids. (A) Comparative analysis of plasmid pSC2016290-mcr-147k with plasmids pSO4-21-304 (AP014634.1), p16E080 (MN647788.1) and pECAZ155_KPC (CP019001.1). (B) Structure analysis of plasmid pSC2016091-mcr-116k with other similar plasmids including pAB12-1-tetX4 (MZ054177.1), pEH09-18-41_3 (CP063506.1) and pSAL4567-1 (AP023307.1).
3.4. Prevalence characteristics and structures of mcr-positive plasmids
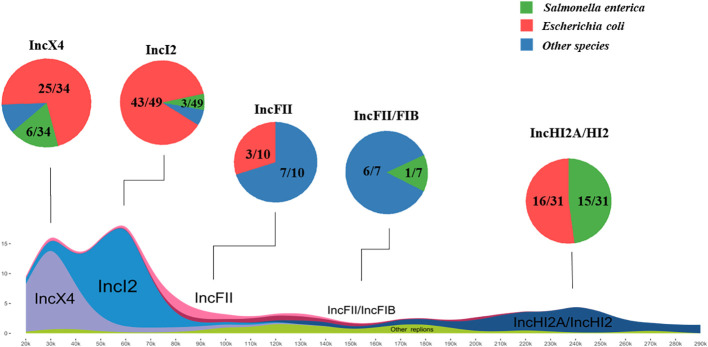
In order to further investigate the prevalence and plasmid backbone structures of mcr-positive plasmids, all plasmid data in the RefSeq database were downloaded and utilized for comparative analysis of plasmid backbone structures. Through BLASTn analysis, a total of 136 complete plasmids encoding mcr were identified among over twenty-seven thousand plasmids in database. The plasmid replicon types of plasmids were further determined using the plasmidfinder database. The major of mcr-1 variants identified in both the database and this study was found in plasmids of replicon types IncX4 (34/122), IncI2 (49/122), and IncHI2/HI2A (31/122), and all plasmids of those types carried only mcr-1, suggesting those plasmids has a strong correlation with mcr-1 horizontal transfer. Additionally, eleven plasmids encoding mcr-3 identified in the database and this study, belonging to four different replicon types, including IncFII (5/11), IncC (4/11), IncI1 (1/11), IncFII/FIB (1/11). Furthermore, the plasmids carrying mcr-8, with replicon types IncFII (1/4), IncFII/FIA (2/4) and IncFII/FIB (1/4), and eight additional plasmids carrying mcr-10 was determined to be of replicon type IncFII/FIB (5/8), IncFII (1/8), IncFII/FIA (1/8) and IncFII (1/8).
The main hosts of mcr-positive plasmids in the RefSeq database were identified as Salmonella enterica (18/136) and Escherichia coli (96/136).
The plasmids from both the database and this study were merged for further investigations. Among these plasmids, those belonging to replicon types IncX4 and IncI2 were most abundant, often in the size range of approximately 30kb and 60kb, respectively, followed by IncHI2A/HI2 type plasmids mainly distributed between 220kb to 270kb in length ( Figure 4 ). Alignment of plasmid backbone structures revealed that, aside from IncFII/FIB replicon type mcr-positive plasmids, those with the same plasmid replicon types displayed similar backbone structures ( Supplement Figure 1 ). These plasmids with similar structures were found in different host bacteria especially Escherichia coli, indicating the horizontal transmission of mcr-positive plasmids carrying different mcr variants among different hosts.
Figure 4.
Host distribution and structural characteristics of mcr-positive plasmids The investigation of mcr-plasmids from RefSeq database via BLASTn comparison. A total of 136 complete plasmids encoding mcr were identified. Salmonella enterica and Escherichia coli are the Major hosts of positive plasmids. IncX4 and IncI2 plasmids as most abundant (approx. 30kb or 60kb), followed by IncHI2A/HI2 (220kb-270kb). The frequent detection of mcr in various plasmid backbones and bacterial isolates highlights the widespread distribution and prevalence of this resistance mechanism.
3.5. Genetic characteristics of multidrug resistant chromosomally integrated plasmid
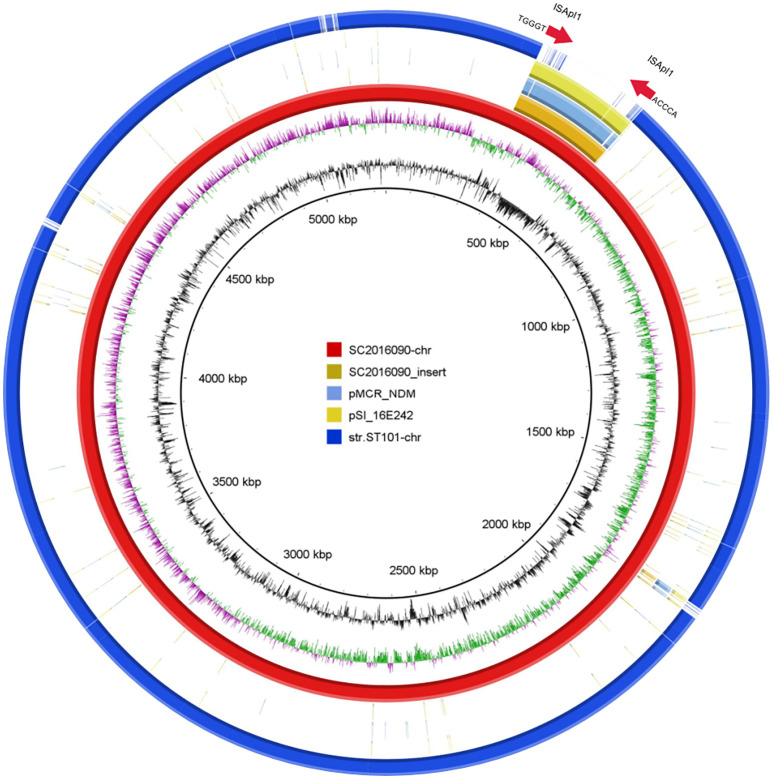
To investigate the formation mechanism of the integrated plasmid, we employed third-generation DNA sequencing to obtain accurate genome structures of strain SC2016090. The chromosome of SC2016090 was 5,247,576-bp in length, exhibited a GC content of 51.9% and comprised 4,927 predicted coding sequences (CDSs). Comparative analysis using BLASTn revealed that the chromosome of SC2016090 showed remarkable similarity (93% coverage, 99.97% identity) to the chromosome of Salmonella enterica strain ST101 (CP050731.1), isolated from a diarrheal patient in Shanghai. However, the SC2016090 chromosome contained an extra 261,335 bp DNA fragment with 46.8% GC content ( Figure 5 ) that was not present in the ST101 chromosome.
Figure 5.
The presence of the ISApl1 gene and structure analysis of multidrug-resistant chromosomally integrated plasmid. BLASTn revealed high homology (93% coverage and 99.97% identity) between SC2016090 and Salmonella enterica strain ST101 chromosomes (CP050731.1). The extra fragment showed 99% identity with plasmids pMCR_NDM (CP049111.1) and pSI_16E242 (ON960347.1), with respective coverage of 95% and 99%. The presence of the ISApl1 transposase and reverse repeating base sequence was detected at both ends of the fragment.
Further investigation revealed that this extra fragment encompassed 249 CDSs and displayed 99% identity with plasmids pMCR_NDM (CP049111.1) and pSI_16E242 (ON960347.1), with coverage of 95% and 99%, respectively. Notably, within the extra DNA fragment of strain SC2016090, several others resistance genes, including bla CTX-M , sul3, aadA2, and fosA were detected, IncHI2 replicon was also coding in the extra DNA fragment. Furthermore, a comprehensive analysis of the extra DNA fragment unveiled the presence of numerous mosaic genetic elements, including the clustered Tn26. Interestingly, we found the CDS for a D-serine transporter (DsdX, QIU08742.1) was truncated by insertion of the extra DNA fragment. Additionally, the presence of the ISApl1 in both areas of the fragment’s ends and the reverse repeating base sequence located on both sides of the extra plasmid segment, suggested the involvement of ISApl1 in the insertion of the integrated plasmid ( Figure 5 ).
4. Discussion
Although colistin was not the first-line antibiotic for the treatment of Salmonella infections, emergence of colistin resistant Salmonella in clinical posed a great threaten to human health. To date, many studies have proved that colistin resistance genes mcr have spread in Salmonella of animal and human sources (Lu et al., 2019; Yang et al., 2022). This alarmed that colistin resistant Salmonella should not be neglected. Plasmids play an important role in the dissemination of mcr in many bacteria, including Salmonella. Understanding the mcr-bearing plasmids characteristics of Salmonella is important to prevent the diffusion of mcr in Salmonella. In this study, we comprehensively investigated the characteristics of mcr-bearing plasmids in 1046 clinical S. enterica isolates collected in Sichuan province from 2014 to 2017. We found that mcr-1 was predominated mcr variant in clinical Salmonella. Three different types of mcr-1-bearing plasmids (IncHI2/IncHI2A, IncX4 and IncI2) were found in the 10 mcr-1 positive Salmonella isolates. Of note, the three types of plasmids were commonly detected in E. coli (Sun et al., 2017; Lu et al., 2020), implying that mcr-1 in Salmonella was probably came from E. coli. Apart from mcr-1, we also detected two mcr-3 positive Salmonella isolates. The mcr-3 gene in the two Salmonella isolates were carried by two different types of plasmids (IncC and IncFIB). It is worth noting that the mcr-3-bearing plasmids detected in Salmonella were also broad-host plasmids. In addition, both mcr-1- and mcr-3-harboring plasmids in our study could be transferred into E. coli by conjugation assay, indicating that mcr genes in clinical Salmonella isolates might spread into other bacteria.
According to our investigation, we found majority of mcr-positive Salmonella isolates belonged to S. 4 [5],,12:i:-. Previous study showed that MDR phenotype of S. 4,[5],12:i:- was determined by IncHI2 plasmids they carried (Mu et al., 2022). We also found a high detection rate of IncHI2 plasmids in mcr-1 positive clinical S. 4,[5],12:i:-. This alerted us to pay more attention to monitor the spread of IncHI plasmids in different ecological niche.
In summary, the prevalence of mcr genes was low in clinical Salmonella in Sichuan, China. The mcr-1 was more prevalence than mcr-3. The propagation of mcr genes in Salmonella was mainly mediated by plasmids. The mcr-bearing plasmids detected in clinical Salmonella could transfer among different Enterobacteriaceae bacteria. Continuous surveillance of mcr-bearing Salmonella in different settings as the One Health approach should be performed to curb the potential risk caused by MDR Salmonella.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XY, RL, and ZW contributed to conception and design of the study. XS, LZ, JM performed the experiments and organized the data. KP and WH conducted the sequencing and bioinformatics analysis. WH and GL did data curation and visualization. XS and KP wrote the first draft of the manuscript. RL and XY reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the Key Research and Development Program of Sichuan Province (Major Science and Technology Projects) (2022ZDZX0017) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1240580/full#supplementary-material
References
- Abd El-Ghany W. A. (2020). Salmonellosis: a food borne zoonotic and public health disease in Egypt. J. Infect. Dev. Ctries 14, 674–678. doi: 10.3855/jidc.12739 [DOI] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S., Brunel J. M., Dubus J. C., Reynaud-Gaubert M., Rolain J. M. (2012). Colistin: an update on the antibiotic of the 21st century. Expert Rev. Anti Infect. Ther. 10, 917–934. doi: 10.1586/eri.12.78 [DOI] [PubMed] [Google Scholar]
- Chang M. X., Zhang J., Zhang J. F., Ding X. M., Lu Y., Zhang J., et al. (2022). Formation, transmission, and dynamic evolution of a multidrug-resistant chromosomally integrated plasmid in salmonella spp. Front. Microbiol. 13, 846954. doi: 10.3389/fmicb.2022.846954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2020). “Performance Standards for antimicrobial susceptibility Testing,” in CLSI supplement M100, 30th ed (Wayne, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Conway S. P., Pond M. N., Watson A., Etherington C., Robey H. L., Goldman M. H. (1997). Intravenous colistin sulphomethate in acute respiratory exacerbations in adult patients with cystic fibrosis. Thorax 52, 987–993. doi: 10.1136/thx.52.11.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. Z., Long T. F., Chen C. P., He B., Li X. P., Schuyler J., et al. (2020). ISKpn40-Mediated Mobilization of the Colistin Resistance Gene mcr-3.11 in Escherichia coli. Antimicrob. Agents Chemother. 64. doi: 10.1128/AAC.00851-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holschbach C. L., Peek S. F. (2018). Salmonella in dairy cattle. Vet. Clin. North Am. Food Anim. Pract. 34, 133–154. doi: 10.1016/j.cvfa.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Yu L., Chen X., Zhi C., Yao X., Liu Y., et al. (2017). High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in china. Front. Microbiol. 8, 562. doi: 10.3389/fmicb.2017.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein N. H., Al-Kadmy I. M. S., Taha B. M., Hussein J. D. (2021). Mobilized colistin resistance (mcr) genes from 1 to 10: a comprehensive review. Mol. Biol. Rep. 48, 2897–2907. doi: 10.1007/s11033-021-06307-y [DOI] [PubMed] [Google Scholar]
- Landman D., Georgescu C., Martin D. A., Quale J. (2008). Polymyxins revisited. Clin. Microbiol. Rev. 21, 449–465. doi: 10.1128/CMR.00006-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nation R. L., Turnidge J. D., Milne R. W., Coulthard K., Rayner C. R., et al. (2006). Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6, 589–601. doi: 10.1016/S1473-3099(06)70580-1 [DOI] [PubMed] [Google Scholar]
- Li R., Peng K., Huang W., Sun X., Huang Y., Lei G., et al. (2022). The genomic epidemiology of mcr-positive Salmonella enterica in clinical patients from 2014 to 2017 in Sichuan, China and global epidemiological features. J. Infect 85(6), 702–769. doi: 10.1016/j.jinf.2022.08.042 [DOI] [PubMed] [Google Scholar]
- Li R., Xie M., Dong N., Lin D., Yang X., Wong M. H. Y., et al. (2018). Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7, 1–9. doi: 10.1093/gigascience/gix132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Lee I. S., Frey J., Slonczewski J. L., Foster J. W. (1995). Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol 177, 4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Lu X., Xiao X., Liu Y., Huang S., Li R., Wang Z. (2020). Widespread Prevalence of Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli from Pere David's Deer in China. mSphere 5(6), e01221-20. doi: 10.1128/mSphere.01221-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Zeng M., Xu J., Zhou H., Gu B., Li Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. Ebiomedicine 42, 133–144. doi: 10.1016/j.ebiom.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchello C. S., Birkhold M., Crump J. A., iN.T.S.c.c. Vacc-i (2022). Complications and mortality of non-typhoidal salmonella invasive disease: a global systematic review and meta-analysis. Lancet Infect. Dis. 22, 692–705. doi: 10.1016/S1473-3099(21)00615-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y., Li R., Du P., Zhang P., Li Y., Cui S., et al. (2022). Genomic epidemiology of ST34 monophasic salmonella enterica serovar typhimurium from clinical patients from 2008 to 2017 in Henan, China. Engineering 15(8), 34–44. doi: 10.1016/j.eng.2022.05.006 [DOI] [Google Scholar]
- Portes A. B., Rodrigues G., Leitao M. P., Ferrari R., Conte Junior C. A., Panzenhagen P. (2022). Global distribution of plasmid-mediated colistin resistance mcr gene in Salmonella: a systematic review. J. Appl. Microbiol. 132, 872–889. doi: 10.1111/jam.15282 [DOI] [PubMed] [Google Scholar]
- Rebelo A. R., Bortolaia V., Kjeldgaard J. S., Pedersen S. K., Leekitcharoenphon P., Hansen I. M., et al. (2018). Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 23(6), 17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker N., Chalmers G., Whalen E., Allen H. K., Meinersmann R. J. (2022). Genomic changes within a subset of IncI2 plasmids associated with dissemination of mcr-1 genes and other important antimicrobial resistance determinants. Antibiotics (Basel) 11. doi: 10.3390/antibiotics11020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Ma F., Deng S., Zhong L. L., El-Sayed Ahmed M. A. E., Zhang G., et al. (2021). Prevalence, genomic characteristics, and transmission dynamics of mcr-1-positive Salmonella enterica Typhimurium from patients with infectious diarrhea. Int. J. Med. Microbiol. 311, 151501. doi: 10.1016/j.ijmm.2021.151501 [DOI] [PubMed] [Google Scholar]
- Snesrud E., McGann P., Chandler M. (2018). The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9. doi: 10.1128/mBio.02381-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. J., Petty N. K., Beatson S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Fang L. X., Wu Z., Deng H., Yang R. S., Li X. P., et al. (2017). Genetic analysis of the incX4 plasmids: Implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7, 424. doi: 10.1038/s41598-017-00095-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhang H., Liu Y. H., Feng Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/j.tim.2018.02.006 [DOI] [PubMed] [Google Scholar]
- Tian G. B., Doi Y., Shen J., Walsh T. R., Wang Y., Zhang R., et al. (2017). MCR-1-producing Klebsiella pneumoniae outbreak in China. Lancet Infect. Dis. 17, 577. doi: 10.1016/S1473-3099(17)30266-9 [DOI] [PubMed] [Google Scholar]
- Vazquez X., Garcia V., Fernandez J., Bances M., de Toro M., Ladero V., et al. (2021). Colistin resistance in monophasic isolates of salmonella enterica ST34 collected from meat-derived products in Spain, with or without CMY-2 co-production. Front. Microbiol. 12, 735364. doi: 10.3389/fmicb.2021.735364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tian G. B., Zhang R., Shen Y., Tyrrell J. M., Huang X., et al. (2017). Prevalence, risk factors, outcomes, and molecular epidemiology of mcr-1-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect. Dis. 17, 390–399. doi: 10.1016/S1473-3099(16)30527-8 [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu C., Zhang R., Chen Y., Shen Y., Hu F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3 [DOI] [PubMed] [Google Scholar]
- Wick R. R., Judd L. M., Gorrie C. L., Holt K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. H., Liu L., Yan M., Chan E. W., Chen S. (2015). Dissemination of IncI2 Plasmids That Harbor the blaCTX-M Element among Clinical Salmonella Isolates. Antimicrob. Agents Chemother. 59, 5026–5028. doi: 10.1128/AAC.00775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Hendriksen R. S., Xie Z., Huang L., Zhang J., Guo W., et al. (2009). Molecular characterization and antimicrobial susceptibility of Salmonella isolates from infections in humans in Henan Province, China. J. Clin. Microbiol. 47, 401–409. doi: 10.1128/JCM.01099-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Chen K., Ye L., Heng H., Chan E. W. C., Chen S. (2022). Genetic and drug susceptibility profiles of mcr-1-bearing foodborne Salmonella strains collected in Shenzhen, China during the period 2014-2017. Microbiol. Res. 265, 127211. doi: 10.1016/j.micres.2022.127211 [DOI] [PubMed] [Google Scholar]
- Yoshida C. E., Kruczkiewicz P., Laing C. R., Lingohr E. J., Gannon V. P., Nash J. H., et al. (2016). The salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft salmonella genome assemblies. PLoS One 11, e0147101. doi: 10.1371/journal.pone.0147101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .