Abstract
Stenotrophomonas maltophilia was isolated from the respiratory tracts of 41 (25%) of 163 children attending our pediatric cystic fibrosis unit between September 1993 and December 1995. The extents of S. maltophilia contamination of environmental sites frequented by these patients were investigated with a selective medium incorporating vancomycin, imipenem, and amphotericin B. Eighty-two isolates of S. maltophilia were cultured from 67 different environmental sites sampled between January and July 1996. The organism was widespread in the home environment, with 20 (36%) and 25 (42%) of sampled sites positive in the homes of colonized and noncolonized patients, respectively. In the nosocomial setting, it was isolated from 18 (32%) sites in the hospital ward and from 4 (17%) sites in the outpatient clinic area. The most common sites of contamination were sink drains, faucets, and other items frequently in contact with water. All environmental and clinical isolates were genotyped with enterobacterial repetitive intergenic consensus sequences as primers. A total of 33 of the 41 patients were colonized with unique strains, and four pairs of patients shared strains. Further characterization by pulsed-field gel electrophoresis after digestion with XbaI found that there was no evidence of patient-to-patient transmission; however, there was some evidence that a small number of patients may have acquired the organism from the hospital environment. Resampling of environmental sites in the hospital ward in January 1997 revealed evidence of genetic drift, complicating the accurate determination of environmental sources for clinical strains. The source of the majority of S. maltophilia strains colonizing the respiratory tracts of these patients with cystic fibrosis remained uncertain but may have represented multiple, independent acquisitions from a variety of environmental sites both within and outside the hospital.
Stenotrophomonas maltophilia is an increasingly recognized nosocomial pathogen, particularly for immunocompromised patients (29). The respiratory tract is the most common site of isolation for hospitalized patients, accounting for 56 to 69% of all isolates (6, 17, 22, 25). Risk factors for S. maltophilia colonization and infection include mechanical ventilation (22, 35), previous exposure to broad-spectrum antibiotics (11, 21, 22, 34, 35), prolonged hospitalization (11, 21, 22, 34), and the use of equipment in contact with the respiratory tract, such as nebulizers (12, 27). Epidemiological studies of S. maltophilia have employed serotyping (21, 35), pyrolysis mass spectrometry (27), ribotyping (2, 17), random amplification of polymorphic DNA (5, 6, 32, 38), enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) (5), and pulsed-field gel electrophoresis (PFGE) (22, 28, 30, 32, 38). A consistent observation of all of the genotyping studies has been that a wide diversity of strain types is isolated from patients. Although small clusters of related types have been observed, most patients harbor apparently unique strains.
S. maltophilia is found in a wide variety of aquatic, soil, and rhizosphere environments and on various contaminated materials and fomites, including faucets (21), showers (30), disinfectants (37), ice-making machines (24), nebulizers (27), dialysis machines (15, 33), and arterial-venous pressure monitoring devices (14). Several of these have been implicated as sources of nosocomial infection with S. maltophilia. Despite this, few studies have included systematic environmental sampling in the investigation of individual outbreaks, and none have sampled the home environment.
The prevalence of colonization of the respiratory tracts of patients with cystic fibrosis (CF) by S. maltophilia has also increased in recent years (8), with some clinics reporting rates in excess of 30% (1). This increase has been associated with the extensive use of antipseudomonal antibiotics for early treatment of Pseudomonas aeruginosa colonization and control of chronic P. aeruginosa respiratory tract infections (9). Although S. maltophilia has been isolated from equipment used to deliver aerosolized antibiotic therapy (18), the sources of S. maltophilia and modes of transmission between patients with CF remain to be elucidated. One small study with PFGE following DNA digestion with XbaI revealed that five patients attending the same French CF clinic were colonized with different strains of S. maltophilia (36). However, no genotyping study has analyzed large numbers of S. maltophilia strains isolated from patients with CF attending the same clinic.
In this study, the extents of S. maltophilia contamination of environmental sites in the hospital ward, outpatient clinic areas, and six homes were assessed with a selective medium for S. maltophilia incorporating vancomycin, imipenem, and amphotericin B as selective agents (VIA medium) (20). Environmental and clinical isolates of S. maltophilia were subjected to genotyping to assess the role of the environment in the colonization of the respiratory tracts of children with CF. All isolates were analyzed by ERIC-PCR, and those with profiles differing by fewer than two bands were analyzed further by PFGE.
MATERIALS AND METHODS
Setting.
The Regional Paediatric Cystic Fibrosis Unit (RPCFU) at St. James’s University Hospital in Leeds, United Kingdom, provides full-time care for children with CF from birth to 18 years of age. Inpatient care is provided in a 20-bed general pediatric ward comprising three 4-bed bays, and eight single-bed rooms. All rooms contain sinks, and there are communal bath and shower facilities. The ward also has designated play areas, a separate room for physiotherapy, and a large kitchen area shared with an adjoining pediatric surgery ward. Outpatient care is provided in a specially designed clinic used solely by patients with CF. There are four examination rooms, each with sinks, and separate rooms for respiratory function testing, physiotherapy, and weighing.
Infection control policy.
All communal respiratory function equipment has disposable mouthpieces, and all patients have their own respiratory therapy equipment. During admissions for inpatient care, patients who are colonized and/or infected with P. aeruginosa are placed into single rooms and receive all physiotherapy in isolation. Patients who are colonized and/or infected with P. aeruginosa are allowed to mix in communal areas at all other times. Patients who are colonized and/or infected with P. aeruginosa attend the outpatient clinic on days separate from those for noncolonized patients. However, those colonized with Burkholderia cepacia are seen in the outpatient clinic at separate times from all other patients and, when admitted, are kept in strict isolation. Patients colonized with S. maltophilia (solely on the basis that it is a multidrug-resistant organism) are placed in single rooms during admissions and seen in outpatient clinics at the same time as P. aeruginosa-positive patients.
Environmental sampling.
Environmental sampling was carried out in the pediatric ward in January 1996 and in the outpatient department in July 1996. Six homes were randomly selected for environmental sampling, three of which were inhabited by patients known to be colonized with S. maltophilia, and three of which were inhabited by patients not colonized with S. maltophilia. These were sampled between March and May 1996. Sites yielding S. maltophilia in the pediatric ward in 1996 were resampled in January 1997. Targeted areas were moist sites, which were found predominantly in kitchens and bathrooms. These included faucets, sink drains, showerheads, sponges, toothbrushes, waste disposal units, washing machines, dishwashers, and refrigerators. Patients’ own respiratory function-therapy equipment was sampled during home visits. All sites were sampled with sterile swabs (moistened with sterile saline when the site was dry), which were then immediately applied to blood agar and VIA medium (20). Faucets, sink drains, showerheads, and other moist sites were sampled by rubbing the swabs vigorously on all available surfaces until visible soiling was observed. Flat surfaces were examined by rubbing a swab over a 10-cm2 area as marked by a template. Plates were transferred back to the laboratory as quickly as possible and incubated in air at 37°C for 48 h.
Water samples were collected in sterile containers and examined with autoclavable filtration equipment (Sartorius SM 16510; Sartorius AG, Göttingen, Germany). Each sample was passed through a sterile 0.45-μm-pore-size cellulose acetate filter (Sartorius AG) by using a vacuum drawn through a side arm. With sterile forceps, filters were applied to the surface of VIA medium, ensuring that all of the surface had come into contact with the medium. The plates were incubated as described above. Presumptive S. maltophilia isolates were confirmed with API 20 NE (bioMerieux, Marcy l’Etoile, France). S. maltophilia NCTC 10258 was used as a control strain. Profiles generated were used to assign each strain a biotype.
Carriage of S. maltophilia on staff hands.
A point prevalence survey of the carriage of S. maltophilia on hands by members of staff with daily hands-on contact with CF patients was carried out during July 1996 by using VIA medium. Staff members were asked to place the fingertips of both hands onto the surfaces of blood agar and then of VIA medium. The plates were incubated, and presumptive S. maltophilia isolates were identified as described above.
Carriage of S. maltophilia in the gastrointestinal tracts of CF patients.
VIA agar was used to determine if the gastrointestinal tract was a reservoir for S. maltophilia in patients with CF. Patients attending the RPCFU were asked to volunteer to submit a single stool sample for analysis. On the day of receipt, a pea-size piece of stool was added to a preweighed vial containing 1 ml of 10% glycerol broth to allow calculation of the added weight of feces. The samples were stored at −70°C until they were cultured. All samples were screened for the presence of S. maltophilia by placing a sterile swab into the glycerol broth-feces mixture, which was then applied to blood agar, MacConkey agar, and VIA medium. The plates were incubated at 37°C in air for 72 h. All S. maltophilia isolates were identified as previously described.
Susceptibility testing.
Susceptibility to four antimicrobials was determined by an agar dilution breakpoint method (3). Briefly, all strains were grown overnight at 37°C in air in 10 ml of nutrient broth held in glass universals with plastic tops. Nineteen-milliliter aliquots of Iso-Sensitest agar were melted, and 1 ml of the antibiotic solution of the required concentration was added and mixed thoroughly. The molten agar was poured into sterile petri dishes and allowed to set. Each strain was diluted 1:100 with sterile saline, and 1 μl was applied to the surface of the agar by using a multipoint inoculator (Denley Instruments Ltd., Sussex, United Kingdom). The plates were incubated overnight in air at 37°C. S. maltophilia NCTC 10258, P. aeruginosa NCTC 10662, and Escherichia coli NCTC 10418 were included as control organisms. The plates were read by eye, and single colonies or a barely visible haze of growth were disregarded. The breakpoint concentrations used were based on British Society for Antimicrobial Chemotherapy guidelines (3) and were as follows: aztreonam, sensitive (<8 mg/liter); tobramycin, sensitive, (<1 mg/liter) and resistant (>4 mg/liter); ciprofloxacin, sensitive (<1 mg/liter) and resistant (>4 mg/liter); and ceftazidime, sensitive (<2 mg/liter). Antibiotics were obtained from the following sources: ceftazidime and tobramycin from Sigma Chemical Co., Dorset, United Kingdom); aztreonam from Bristol-Myers Squibb, Hounslow, United Kingdom; and ciprofloxacin from Bayer plc, Newbury, United Kingdom. The results obtained were used to formulate antibiograms for each S. maltophilia strain. Statistical analysis was performed by the chi-square test.
Molecular typing by ERIC-PCR.
All strains of S. maltophilia isolated during environmental sampling and one strain from each patient found to be colonized with S. maltophilia between September 1993 and December 1995 were subject to typing by ERIC-PCR. Strains were tested in triplicate to ensure reproducibility of the method. DNA was extracted from strains by the method of De Lamballerie et al. (7). Briefly, isolates were grown overnight on Iso-Sensitest agar. A single, well-isolated colony was emulsified by vortexing with 200 μl of Chelex extraction buffer (15% Chelex 100 resin [BioRad Laboratories, Hercules, Calif.], 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 1% Tween 20) held in a 0.6-ml Eppendorf tube. Each sample was placed into boiling water for 10 min and then vortexed again before centrifugation at 13,000 × g for 30 s. The supernatant was removed into a clean 0.6-ml Eppendorf tube and adjusted to a final concentration of 10 mM Tris-HCl and 1 mM EDTA with a 20-fold-concentrated solution of TE buffer (200 mM Tris-HCl, 20 mM EDTA [pH 8.4]). DNA extracts were kept at −70°C when there was a delay prior to use. ERIC-PCRs were carried out in 0.6-ml Eppendorf tubes containing 50 μl of PCR mix, which was made up of 10× buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 0.1% Triton X-100), 200 μM each nucleoside dATP, dTTP, dCTP, and dGTP, 5 μl of dimethyl sulfoxide, 1 mM magnesium chloride, 2 U of Taq polymerase (HT Biotechnology, Cambridge, United Kingdom), 0.4 μM ERIC-2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′) primer, and 3 μl of DNA template. Each reaction mix was overlaid with mineral oil, and PCR was performed with an air-cooled thermocycler (Quatro TC-40; Quatro Biosystems, Manchester, United Kingdom) under the following conditions: 4 min at 94°C, followed by 35 cycles each of 45 s at 94°C, 1 min at 58°C, and 2 min at 72°C, followed by a final extension of 10 min at 72°C. Amplification products were analyzed by electrophoresis through 2% agarose gels with a 123-bp ladder as a size marker and were visualized following staining with ethidium bromide. PCR product patterns were compared by eye and by computer analysis (Gelworks, UVP Inc., Upland, Calif.). PCR patterns were considered identical when the positions of all bands matched. Differences in band intensity were ignored.
Molecular typing by PFGE.
Strains were inoculated into 5 ml of nutrient broth (Unipath, Basingstoke, United Kingdom) and incubated for 6 h at 37°C with shaking to attain exponential growth. The cells were pelleted by centrifugation at 3,000 × g for 10 min, and the supernatant was discarded. The cells were then resuspended in 1 ml of SE buffer (25 mM EDTA [pH 7.4], 75 mM NaCl), mixed 1:1 (vol/vol) with 2% low-melting-point agarose dissolved in SE buffer, inserted into plug moulds (10 by 5 by 1.5 mm), and allowed to set. Each plug was placed into a 1.5-ml Eppendorf tube containing 1 ml of lysis buffer (10 mM Tris-HCl [pH 7.6], 100 mM EDTA [pH 8.0], 50 mM NaCl, 0.2% deoxycholic acid, 1% lauroylsarcosine, 2-mg/ml lysozyme, 30-μg/ml RNase), and the tubes were incubated overnight at 37°C. The lysis buffer was then replaced by 1 ml of a solution containing 0.5 M EDTA (pH 9.5)–1% lauroylsarcosine. Plugs were then incubated at 42°C overnight in a water bath. The solution was then replaced with 1 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA), and the plugs were stored at 4°C until use.
In preparation for digestion, each plug was placed in 1 ml of 1:10 dilution of TE buffer, washed for 15 min with rolling (Spiramix 5; Denley Instruments Ltd., Sussex, United Kingdom), and then placed in 750 μl of restriction digest buffer H (50 mM Tris-HCl, 10 mM MgCl2, 100 mM NaCl, 1 mM dithioerythritol [pH 7.5]) and washed again for 15 min with rolling. A 500-μl volume of fresh H buffer was used to replace the previous buffer, 25 U of XbaI restriction endonuclease (Boehringer, Mannheim, Germany) was added, and plugs were then incubated at 37°C for 6 h. Electrophoresis was performed by 1.2% PFGE agarose gels run on a contour-clamped homogeneous electric field DRII system (BioRad Laboratories) over 20 h at 14°C with 5 to 35 s of linear ramping at 6 V/cm. Lambda ladder was incorporated as a size standard. Electrophoresis products were visualized by ethidium bromide staining, and patterns were compared by eye. Interpretation was based on the criteria of Tenover et al. (31). Strains were tested in duplicate to determine reproducibility of the technique.
RESULTS
The total number of patients attending the RPCFU on a full-time basis between September 1993 and December 1995 was 163. S. maltophilia was isolated on at least one occasion during this period from 41 of these patients, an overall incidence of 25%. A total of 17 patients were male and 24 were female (ratio, 1:1.4). Some patients had been colonized with S. maltophilia for several years before the study began. The dates of the first positive respiratory tract culture for S. maltophilia were 1988 (1 patient), 1989 (2 patients), 1990 (1 patient), 1991 (2 patients), 1992 (5 patients), 1993 (7 patients), 1994 (12 patients), and 1995 (10 patients). One patient was positive for S. maltophilia on arrival from another clinic in September 1993, but it was uncertain when the first positive culture for S. maltophilia had occurred. Both the median and the mean ages at the time of the first positive sputum culture for S. maltophilia were 9 years and 8 months. Only 4 (10%) of the 41 patients were chronically colonized with S. maltophilia (i.e., >50% of sputum samples taken during the study period were positive). Twenty-four (59%) were also chronically colonized with other organisms (P. aeruginosa, Aspergillus fumigatus, Staphylococcus aureus, Haemophilus influenzae, Brevundimonas vesicularis, or atypical mycobacteria), although only 10 (24%) of these were chronically colonized with P. aeruginosa.
The results of environmental screening carried out in 1996 are shown in Table 1. The 67 positive environmental sites yielded a total of 82 isolates of S. maltophilia. Of the 16 positive sites in the ward in 1996 available for resampling (a toothbrush and a bunch of flowers were not available), 12 were again positive for S. maltophilia during 1997.
TABLE 1.
Environmental sources of S. maltophilia in the hospital and in the homes of patients with CF
| Location | Total no. of samples | No. of positive sites/no. of sites sampled from the following sources:
|
||||
|---|---|---|---|---|---|---|
| Water | Faucets | Sink drains | Othersa | Total (%) | ||
| Ward | 57 | 3/6 | 3/19 | 9/10 | 3/22 | 18 (32) |
| Clinic | 23 | 1/3 | 1/8 | 1/7 | 1/5 | 4 (17) |
| Homes (colonized) | 55 | NDb | 6/16 | 5/9 | 9/30 | 20 (36) |
| Homes (noncolonized) | 59 | ND | 6/17 | 8/9 | 11/33 | 25 (42) |
| Total | 194 | 4/9 | 16/60 | 23/35 | 17/79 | 67 (35) |
Sources were as follows for the following locations: ward, toothbrush, flowers, and kitchen work surface; clinic, waste disposal unit; homes (colonized), four sponges, potatoes, washing machine, dishcloth, washing brush, and waste disposal unit; homes (noncolonized), three sponges, two washing machines, pan scrubber, dishcloth, washing bowl, waste disposal unit, kitchen work surface, and flannel.
ND, not determined.
Twelve members of the staff (five doctors, four nurses, and three physiotherapists) were examined for S. maltophilia hand carriage during July 1996. All were found to be negative.
Twenty-nine patients volunteered to submit stool samples for analysis. Twenty-two were from noncolonized patients, and seven were from colonized patients. All 29 samples failed to yield S. maltophilia on culture.
Susceptibilities to antimicrobials were determined for 40 clinical and 80 environmental S. maltophilia isolates. Three isolates (one clinical and two environmental) failed to grow on Iso-Sensitest agar. Significant differences in resistance to ceftazidime (48 versus 16% [P < 0.0001]), ciprofloxacin (68 versus 45% [P < 0.05]), and aztreonam (73 versus 51% [P < 0.05]) were found for clinical and environmental strains of S. maltophilia. No significant differences were observed for tobramycin (93 versus 84%).
ERIC-PCR analysis of series of strains from patients with S. maltophilia isolated on more than one occasion during the study period revealed only one patient with more than one strain of S. maltophilia. He was chronically colonized with a single strain throughout the whole study period (identical ERIC-PCR profile throughout, with a difference of one band in the PFGE profile between an isolate from September 1993 and all other isolates). Four other strains were isolated during the study period, each with unique ERIC-PCR and PFGE profiles.
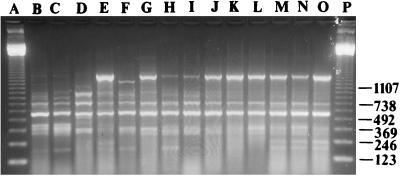
The 45 clinical isolates (1 isolate each from 40 patients and 5 isolates from 1 patient) of S. maltophilia were represented by 10 different biotypes, and 13 different antibiograms. The two most common biotypes accounted for 15 (33%) and 14 (31%) of the strains, respectively. The most common antibiogram (resistance to all four of the tested agents) accounted for 16 (36%) of the strains. The same 45 clinical isolates were represented by 41 different genotypes by ERIC-PCR. Thirty-two patients carried unique ERIC-PCR types, four pairs of patients shared the same type, and one patient was colonized with five different strains, one of them chronically. Most clinical strains had easily distinguishable ERIC-PCR profiles, but eight patients had isolates with similar profiles (Fig. 1). In this group, there were two pairs (lanes B and C and H and I) and four unique strains (lanes D to G).
FIG. 1.
ERIC-PCR profiles of S. maltophilia strains from eight different patients and from six environmental sites in the CF ward in 1996. Lanes: A and P, 123-bp ladder; B to I, clinical strains; J, toothbrush; K, sink drain 6; L, water sample, room 13; M, faucet, room 8; N, sink drain 5; and O, sink drain 4.
The 21 S. maltophilia isolates from the CF ward environment in 1996 were represented by 12 different ERIC-PCR types. Six isolates (sources for which were three sink drains, one faucet, one water sample, and one toothbrush) shared the same ERIC-PCR type (Fig. 1, lanes J to O) and were indistinguishable to one of the pairs of clinical isolates (Fig. 1, lanes H and I). Three other isolates (sources for which were one sink drain, one faucet, and one water sample) from the CF ward environment shared another ERIC-PCR type which was identical to that of another of the pairs of clinical isolates (data not shown). Two other pairs of isolates and eight unique isolates were also identified.
Three different ERIC-PCR profiles were obtained from the four outpatient clinic isolates, and none of these were found to match the profiles of clinical S. maltophilia isolates. Clusters of identical S. maltophilia strains were found in each of the six sampled homes, sometimes occurring in separate areas of the house. None of the isolates from the home environments of S. maltophilia-colonized patients had ERIC-PCR types that matched those of the clinical isolates from the patients.
Comparison of the ERIC-PCR types of S. maltophilia isolates from the inpatient ward environment in 1996 and 1997 revealed that six of the sites harbored strains with different genotypes in 1997 compared to 1996 (five sink drains and one water sample), whereas the other six had identical genotypes on both occassions (four sink drains, one faucet, and one water sample).
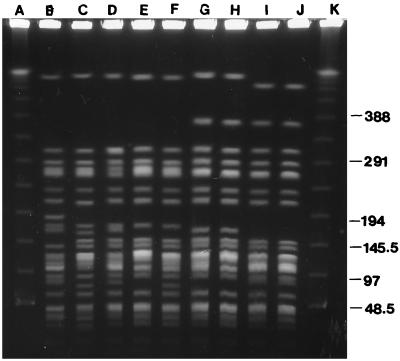
Of the four pairs of clinical isolates identified by ERIC-PCR, all could be differentiated by PFGE. One pair of isolates with similar (but distinguishable) ERIC-PCR profiles (Fig. 1, lanes C and G) differed by only one band by PFGE (Fig. 2, lanes E and F). The other clinical S. maltophilia strains with similar ERIC-PCR genotypes were all distinguishable from one another by PFGE. PFGE of the six environmental isolates with the indistinguishable ERIC-PCR type (Fig. 1, lanes J to O) identified three different PFGE types (Fig. 2). The first type was represented by a water sample isolate and two others (isolated from a toothbrush and a sink drain) which differed from the first by only three bands (Fig. 2, lanes H to J). The water sample strain was identical to one of the clinical strains (Fig. 2, lanes G and H). The second type, represented by an isolate from a sink drain, differed by only two bands from two of the clinical isolates (Fig. 2, lanes D to F). The third type was represented by two CF ward environmental isolates (isolated from a faucet and a sink drain), differing from each other by only three bands (Fig. 2, lanes B and C). Three other environmental strains with identical ERIC-PCR types were differentiated by two different PFGE patterns. One pattern was shared by isolates from a water faucet and a water sample taken through it. Both of these PFGE patterns were different from those of the two clinical isolates with the same ERIC-PCR type, which themselves had clearly distinguishable patterns.
FIG. 2.
PFGE profiles of selected strains of S. maltophilia shown in Fig. 1. Lanes: A and K, multimers of phage lambda DNA (48.5 kb) as molecular mass markers; B, faucet, room 8; C, sink drain 4; D, sink drain 5; E to G, clinical strains; H, water sample, room 13; I, toothbrush; and J, sink drain 6.
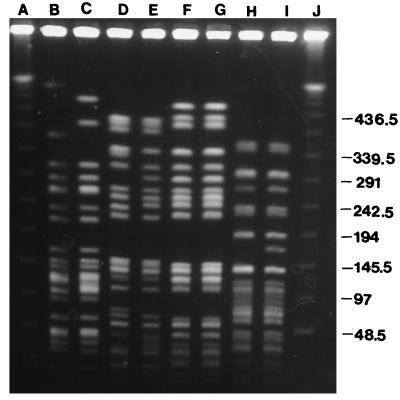
PFGE analysis of the six CF ward environment strains with identical ERIC-PCR patterns in 1996 and 1997 revealed that four strains differed from the previous year’s PFGE pattern by one to three bands. Three pairs of these strains are shown in Fig. 3 (lanes B and C, D and E, and H and I). The other two strains had identical PFGE profiles on both occasions.
FIG. 3.
PFGE profiles of S. maltophilia strains isolated from the same environmental sites in 1996 and 1997. Lanes: A and J, multimers of phage lambda DNA (48.5 kb) as molecular mass markers; B, water sample, room 13, 1996; C, water sample, room 13, 1997; D, water sample, nursing station, 1996; E, water sample, nursing station, 1997; F, shower drain, 1996; G, shower drain, 1997; H, sink drain 7, 1996; and I, sink drain 7, 1997.
DISCUSSION
S. maltophilia was relatively easy to find in the environment of patients with CF. This is in direct contrast to attempts to locate environmental sources of clinical strains of B. cepacia (4, 26). Interestingly, for an organism primarily thought of as a nosocomially acquired pathogen, it was more widespread in the home (39% of samples) than it was in the hospital (28%). Colonized patients did not appear to contaminate their surroundings with S. maltophilia. There were no significant differences in the prevalence of the organism in the homes of colonized and noncolonized patients, and none of the home environment strains of colonized patients matched their clinical strains by genotyping.
The diversity of types seen with ERIC-PCR in this study is consistent with the findings of other genotyping studies of S. maltophilia outbreaks in which the majority of patients have had unique types and only occasional small clusters of indistinguishable strains have been identified (6, 17, 22, 28, 30, 32, 38). It is also consistent with the results of one small study (five patients) of S. maltophilia strains isolated from patients with CF (36). This suggests that patient-to-patient spread of a highly transmissible strain was not a major route of spread in the RPCFU. In the absence of any convincing evidence that patients were contaminating their environment, the finding of clusters of strains with identical or very similar ERIC-PCR profiles from clinical and environmental sources suggested that the source of some of the clinical S. maltophilia strains may have been the hospital ward environment. However, the PFGE and epidemiological data were inconclusive. Only one patient had a clinical isolate with ERIC-PCR and PFGE profiles indistinguishable from those of an environmental isolate (Fig. 1, lanes I and L; Fig. 2, lanes G and H) but this patient had never been admitted to the inpatient ward prior to her first isolation of S. maltophilia, nor had she visited the ward or utilized any ward respiratory equipment. There were no other links between this patient and the others with ERIC-PCR profiles shown in Fig. 1, other than attendance of the same outpatient clinic with one other patient (Fig. 1, lane F) on a single occasion 5 months prior to the first isolation of S. maltophilia. Their respective S. maltophilia strains were easily distinguishable by PFGE. Although the other seven patients featured in Fig. 1 had been admitted to the inpatient ward at some time prior to first isolation of S. maltophilia (range, 0 to 30 months), there were very few links among any of them. One patient (Fig. 1, lane G; Fig. 2, lane E) had been admitted to the ward 3 months prior to his first isolation of S. maltophilia at a time when another patient (Fig. 1, lane I) was also present. Their strains were also distinguishable by PFGE. There were no other inpatient overlaps for the other patients. Four of these seven patients had attended the same outpatient clinics in the year prior to first isolation, but it was not known if they had had any contact during these visits. None of these patients were known to socialize together outside the hospital. The other pair of patients whose S. maltophilia strains had ERIC-PCR profiles identical to those of three ward environmental strains were easily distinguishable by PFGE. They also had inconclusive epidemiological links. One of them had not been previously admitted to the inpatient ward, but both did have the same clinic appointment date 11 months prior to the second patient’s becoming S. maltophilia positive. However, this patient did become positive during a hospital admission, although it was not known what contact, if any, this patient had with sources of the environmental strains.
The fourth pair of patients with identical clinical ERIC-PCR profiles had no contact with each other during hospital admissions but had attended the same outpatient clinic 7 months prior to the second patient’s becoming S. maltophilia positive. These strains were distinguishable by PFGE, and they did not match any environmental isolates.
Although circumstantial evidence from the results of ERIC-PCR typing suggest that some of the clinical S. maltophilia strains may have been acquired from the ward environment, it does not account for the origin of the majority of strains. The outpatient clinic was unlikely to be a source for these strains. Although all of the S. maltophilia-positive patients had attended the clinic prior to the first isolation, the clinic environment yielded the lowest number of positive sites for the organism. These had been located in areas of the clinic not usually frequented by patients, and none of the ERIC-PCR types matched those of patient strains.
S. maltophilia was isolated from all of the sampled homes, and no significant differences were noted in the distribution between the homes of colonized and noncolonized individuals. However, none of the environmental strains isolated from the homes of colonized patients matched any of their clinical strains, and, as such, the origins of the majority of S. maltophilia clinical strains remain unclear. Evidence from resampling experiments carried out in the inpatient ward suggests that this may be difficult to clarify. Four of the 16 sites positive in 1996 were no longer positive in 1997, and 6 of the 12 positive sites in 1997 harbored strains with ERIC-PCR types different from those of the previous year. This suggests that the distribution of S. maltophilia in the environment is continually changing, and point prevalence surveys are therefore unlikely to detect all genotypes of S. maltophilia prevalent in a given location over time. This problem is compounded by the finite sensitivity of the sampling technique used and limitations on the number of potential sites of exposure that can be examined. PFGE of all of the ward environmental strains found to be indistinguishable in 1996 and 1997 by ERIC-PCR revealed that some of those isolated in 1997 differed by one to three bands from those in 1996 (Fig. 3). This suggested that these were the same strains which had undergone genetic events resulting in minor changes in PFGE profile, a problem highlighted by Tenover et al. (31). Over longer periods of time, this process may result in strains with PFGE profiles significantly different from that of the parent strain, making exact identification of environmental sources for clinical strains of S. maltophilia acquired months or years previously extremely difficult.
Further studies would be required to ascertain the precise mode of acquisition of S. maltophilia from the environment. S. maltophilia was isolated primarily from moist sites, particularly in relation to plumbing systems, such as faucets and sink drains. Aerosols containing P. aeruginosa can be generated from such sites (10); however, further experiments would be required to ascertain if this is also the case with S. maltophilia. Counts of S. maltophilia in water samples ranged from 0 to more than 2 × 103 CFU/ml. Levels also varied markedly between the two sampling times. It is also possible that levels will vary at different times of the day, with higher levels occurring after prolonged periods of faucet nonuse. Systematic sampling at different times of the day would be required to confirm if this is the case. However, the level at which water contamination with S. maltophilia becomes a significant threat to patients is unknown and may depend on the clinical status of the patient and the coadministration of antibiotics. The absence of S. maltophilia from the hands of staff members suggests that transmission via this route is not a major factor.
The decrease in antibiotic susceptibility of clinical strains of S. maltophilia relative to environmental strains does not necessarily imply that the more resistant environmental strains are most likely to colonize patients. Because S. maltophilia is noted for its ability to become increasingly resistant to a variety of antimicrobial agents during therapy (16, 23), many clinical strains may have been more susceptible at the time of initial acquisition.
Most environmental strains of S. maltophilia isolated in this study were resistant to tobramycin. The use of aerosolized aminoglycosides has been significantly associated with S. maltophilia colonization of patients with CF in a previous study (9) and S. maltophilia is also known to contaminate equipment used to deliver aerosolized antibiotics (18). Since solutions of tobramycin used in nebulizers come ready-made in sealed sterile vials, any contamination of nebulizer equipment by S. maltophilia is likely to result from postuse washing. This is often performed by using water taken from faucets. If drying is incomplete, the equipment may become contaminated with S. maltophilia, particularly since the organism is known to adhere to plastics (19). It is not known if S. maltophilia would be capable of surviving exposure to the extremely high concentrations of tobramycin that occur in this setting, but it is interesting to note that a strain of S. maltophilia has been reported that utilized streptomycin as an energy source (13). Further studies would be needed to assess the level of contamination of nebulizers with S. maltophilia.
This study has revealed the widespread distribution of S. maltophilia in the homes and hospital environments of patients with CF, particularly in water and plumbing systems. The majority of patients possessed strains with unique genotypes. Although circumstantial evidence suggested that some patients may have acquired the organism from the hospital ward environment, the origins of most of the isolates of S. maltophilia colonizing patients remained uncertain. The ease of isolation from the home environment suggests that acquisition of S. maltophilia outside the nosocomial setting may be much more common than previously anticipated. Although PFGE was more discriminatory than ERIC-PCR for typing S. maltophilia in this study, its usefulness for investigating potential environmental sources of strains acquired months or years previously was weakened by the evolution of different band patterns within clones. The findings of this study have wider implications for the epidemiology of S. maltophilia in other patient groups.
ACKNOWLEDGMENTS
We thank Sue Kitchen and Sharryn McLaughlin, CF Nurse Specialists, for their assistance in arranging the home visits and all patients and their families who kindly agreed to take part. We also thank all of the Biomedical Scientists working in the Microbiology Department at St. James’s University Hospital who saved isolates from clinical samples for the study. We also thank Val Keer for her assistance in producing the PFGE gels for publication.
This work was supported by the Cystic Fibrosis Trust (grant no. PJ398).
REFERENCES
- 1.Ballestero S, Virseda I, Escobar H, Suarez L, Baquero F. Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1995;14:728–729. doi: 10.1007/BF01690887. [DOI] [PubMed] [Google Scholar]
- 2.Bingen E H, Denamur E, Lambert-Zechovsky N Y, Bourdois A, Mariani-Kurkdjian P, Cezard J-P, Navarro J, Elion J. DNA restriction fragment length polymorphism differentiates crossed from independent infections in nosocomial Xanthomonas maltophilia bacteremia. J Clin Microbiol. 1991;29:1348–1350. doi: 10.1128/jcm.29.7.1348-1350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.British Society for Antimicrobial Chemotherapy. 1991. A guide to sensitivity testing. J. Antimicrob. Chemother. 27(Suppl. D):1–50. [PubMed]
- 4.Butler S L, Doherty C J, Hughes J E, Nelson J W, Govan J R W. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard? J Clin Microbiol. 1995;33:1001–1004. doi: 10.1128/jcm.33.4.1001-1004.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatelut M, Dournes J L, Chabanon G, Marty N. Epidemiological typing of Stenotrophomonas (Xanthomonas) maltophilia by PCR. J Clin Microbiol. 1995;33:912–914. doi: 10.1128/jcm.33.4.912-914.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davin-Regli A, Bollet C, Auffray J P, Saux P, De Micco P. Use of random amplified polymorphic DNA for epidemiological typing of Stenotrophomonas maltophilia. J Hosp Infect. 1996;32:39–50. doi: 10.1016/s0195-6701(96)90163-2. [DOI] [PubMed] [Google Scholar]
- 7.De Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one step microbial DNA extraction method using “Chelex 100” suitable for gene amplification. Res Microbiol. 1992;143:785–790. doi: 10.1016/0923-2508(92)90107-y. [DOI] [PubMed] [Google Scholar]
- 8.Denton M. Stenotrophomonas maltophilia: an emerging problem in cystic fibrosis patients. Rev Med Microbiol. 1997;8:15–19. [Google Scholar]
- 9.Denton M, Todd N J, Littlewood J M. Role of antibiotics in the emergence of Stenotrophomonas maltophilia in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 1996;15:402–405. doi: 10.1007/BF01690098. [DOI] [PubMed] [Google Scholar]
- 10.Doring G, Ulrich M, Muller W, Bitzer J, Schmidt-Koenig L, Munst L, Grupp H, Wolz C, Stern M, Botzenhardt K. Generation of Pseudomonas aeruginosa aerosols during handwashing from contaminated sink drains, transmission to hands of hospital personnel, and its prevention by use of a new heating device. Zentralbl Hyg. 1991;191:494–505. [PubMed] [Google Scholar]
- 11.Elting L S, Khardori N, Bodey G P, Fanstein V. Nosocomial infection caused by Xanthomonas maltophilia: a case-control study of predisposing factors. Infect Control Hosp Epidemiol. 1990;11:134–138. doi: 10.1086/646136. [DOI] [PubMed] [Google Scholar]
- 12.Feeley T W, du Moulin G C, Hedley-White J, Bushnell L S, Gilbert J P, Feingold D S. Aerosol polymyxin and pneumonia in seriously ill patients. N Engl J Med. 1975;293:471–475. doi: 10.1056/NEJM197509042931003. [DOI] [PubMed] [Google Scholar]
- 13.Fenton J J, Harsch H H, Klein D. Production of volatile nitrogenous compounds from the degradation of streptomycin by Pseudomonas maltophilia. J Bacteriol. 1973;116:1267–1272. doi: 10.1128/jb.116.3.1267-1272.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher M C, Long S S, Roberts E M, Dunn J M, Balsara R K. Pseudomonas maltophilia bacteremia in children undergoing open heart surgery. J Am Med Assoc. 1981;246:1571–1574. [PubMed] [Google Scholar]
- 15.Flaherty J P, Garcia-Houchins S, Chudy R, Arnow P M. An outbreak of Gram-negative bacteremia traced to contaminated O-rings in reprocessed dialyzers. Ann Intern Med. 1993;119:1072–1078. doi: 10.7326/0003-4819-119-11-199312010-00003. [DOI] [PubMed] [Google Scholar]
- 16.Garrison M W, Anderson D E, Campbell D M, Carroll K C, Malone C L, Anderson J D, Hollis R J, Pfaller M A. Stenotrophomonas maltophilia: emergence of multidrug-resistant strains during therapy and an in vitro pharmacodynamic chamber model. Antimicrob Agents Chemother. 1996;40:2859–2864. doi: 10.1128/aac.40.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerner-Smidt P, Bruun B, Arpi M, Schmidt J. Diversity of nosocomial Xanthomonas maltophilia (Stenotrophomonas maltophilia) as determined by ribotyping. Eur J Clin Microbiol Infect Dis. 1995;14:137–140. doi: 10.1007/BF02111874. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson G R, Parker S, Pryor J A, Duncan-Skingle F, Hoffman P N, Hodson M E, Kaufmann M E, Pitt T L. Home-use nebulizers: a potential primary source of Burkholderia cepacia and other colistin-resistant, gram-negative bacteria in patients with cystic fibrosis. J Clin Microbiol. 1996;34:584–587. doi: 10.1128/jcm.34.3.584-587.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr K G, Anson J, Hawkey P M. Abstracts of the 94th General Meeting of the American Society for Microbiology, Las Vegas, Nev. Washington, D.C: American Society for Microbiology; 1994. Adherence of clinical and environmental strains of Xanthomonas maltophilia to plastic material, abstr. B-339; p. 89. [Google Scholar]
- 20.Kerr K G, Denton M, Todd N J, Corps C M, Kumari P, Hawkey P M. A novel selective culture medium for the isolation of Stenotrophomonas maltophilia. Eur J Clin Microbiol Infect Dis. 1996;15:607–610. doi: 10.1007/BF01709373. [DOI] [PubMed] [Google Scholar]
- 21.Khardori N, Elting L, Wong E, Schable B, Bodey G P. Nosocomial infections due to Xanthomonas maltophilia (Pseudomonas maltophilia) in patients with cancer. Rev Infect Dis. 1990;12:997–1003. doi: 10.1093/clinids/12.6.997. [DOI] [PubMed] [Google Scholar]
- 22.Laing F P Y, Ramotar K, Read R R, Alfieri N, Kureishi A, Henderson E A, Louie T J. Molecular epidemiology of Xanthomonas maltophilia colonization and infection in the hospital environment. J Clin Microbiol. 1995;33:513–518. doi: 10.1128/jcm.33.3.513-518.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manian F A, Meyer L, Jenne J, Owen A, Taff T. Loss of antimicrobial susceptibility in aerobic gram negative bacilli repeatedly isolated from intensive care patients. Infect Control Hosp Epidemiol. 1996;17:222–226. doi: 10.1086/647284. [DOI] [PubMed] [Google Scholar]
- 24.Medical Services Directorate. Ice cubes: infection caused by Xanthomonas maltophilia. London, United Kingdom: Department of Health; 1993. . (Hazard 93:42.) [Google Scholar]
- 25.Morrison A J, Jr, Hoffman K K, Wenzel R P. Associated mortality and clinical characteristics of nosocomial Pseudomonas maltophilia in a university hospital. J Clin Microbiol. 1986;24:52–55. doi: 10.1128/jcm.24.1.52-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortensen J E, Fisher M C, LiPuma J J. Recovery of Pseudomonas cepacia and other Pseudomonas species from the environment. Infect Control Hosp Epidemiol. 1995;56:152–162. doi: 10.1086/646999. [DOI] [PubMed] [Google Scholar]
- 27.Orr K, Gould F K, Sisson P R, Lighfoot N F, Freeman R, Burdess D. Rapid inter-strain comparison by pyrolysis mass spectrometry in nosocomial infection with Xanthomonas maltophilia. J Hosp Infect. 1991;17:187–195. doi: 10.1016/0195-6701(91)90230-6. [DOI] [PubMed] [Google Scholar]
- 28.Sader H S, Pignatari A C, Frei R, Hollis R J, Jones R N. Pulsed-field gel electrophoresis of restriction-digested genomic DNA and antimicrobial susceptibility of Xanthomonas maltophilia strains from Brazil, Switzerland, and the USA. J Antimicrob Chemother. 1994;33:615–618. doi: 10.1093/jac/33.3.615. [DOI] [PubMed] [Google Scholar]
- 29.Spencer, R. C. 1995. The emergence of epidemic, multiple-antibiotic-resistant Stenotrophomonas (Xanthomonas) maltophilia and Burkholderia (Pseudomonas) cepacia. J. Hosp. Infect. 30(Suppl.):453–464. [DOI] [PubMed]
- 30.Talon D, Bailly P, Leprat R, Godard C, Deconnink E, Cahn J-Y, Michel-Briand Y. Typing of hospital strains of Xanthomonas maltophilia by pulsed-field gel electrophoresis. J Hosp Infect. 1994;27:209–217. doi: 10.1016/0195-6701(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 31.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Couwenberghe C J, Cohen S H, Tang Y J, Gumerlock P H, Silva J., Jr Genomic fingerprinting of epidemic and endemic strains of Stenotrophomonas maltophilia (formerly Xanthomonas maltophilia) by arbitrarily primed PCR. J Clin Microbiol. 1995;33:1289–1291. doi: 10.1128/jcm.33.5.1289-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanholder R, Vanhaecke E, Ringoir S. Pseudomonas septicemia due to deficient disinfectant mixing during reuse. Int J Artific Org. 1992;15:19–24. [PubMed] [Google Scholar]
- 34.Victor M A, Arpi M, Bruun B, Jonsson V, Hansen M M. Xanthomonas maltophilia bacteremia in immunocompromised hematological patients. Scand J Infect Dis. 1994;26:163–170. doi: 10.3109/00365549409011780. [DOI] [PubMed] [Google Scholar]
- 35.Villarino M E, Stevens L E, Schable B, Mayers G, Miller J M, Burke J P, Jarvis W R. Risk factors for epidemic Xanthomonas maltophilia infection/colonisation in intensive care unit patients. Infect Control Hosp Epidemiol. 1992;13:201–206. doi: 10.1086/646510. [DOI] [PubMed] [Google Scholar]
- 36.Vu-Thien H, Moissenet D, Valcin M, Dulot C, Tournier G, Garbarg-Chenon A. Molecular epidemiology of Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans in a cystic fibrosis center. Eur J Clin Microbiol Infect Dis. 1996;15:876–879. doi: 10.1007/BF01691221. [DOI] [PubMed] [Google Scholar]
- 37.Wishart M M, Riley T V. Infection with Pseudomonas maltophilia: hospital outbreak due to contaminated disinfectant. Med J Aust. 1976;2:710–712. doi: 10.5694/j.1326-5377.1976.tb128238.x. [DOI] [PubMed] [Google Scholar]
- 38.Yao J D C, Conly J M, Krajden M. Molecular typing of Stenotrophomonas (Xanthomonas) maltophilia by DNA macrorestriction analysis and random amplified polymorphic DNA analysis. J Clin Microbiol. 1995;33:2195–2198. doi: 10.1128/jcm.33.8.2195-2198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]