Abstract
Objective:
This study aimed to examine the expression of Histone H3.3 glycine 34 to tryptophan (G34W) mutant protein in Giant Cell Tumor of Bone (GCTB).
Methods:
This analytic observation research used a cross-sectional study design on 71 bone tumors. The cases involved 54 tissue samples diagnosed as GCBT. It was divided into GCTB primer (n=37), recurrent GCTB (n=5), GCTB with metastasis (n=9), and malignant GCTB (n=3). There were 17 samples mimics of GCTB also tested, including chondroblastoma (n=1), giant cell reparative granuloma (n=2), giant cell of tendon sheath (n=7), chondromyxoid fibroma (n=2), aneurysmal bone cyst (n=2), and giant cell-rich osteosarcoma (n=3). The Immunohistochemistry was used to evaluate the expression of G34W-mutated protein in these bone tumors.
Result:
The representation H3.3 (G34W) was expressed in the nuclei of mononuclear stromal cells but not stained on osteoclast-like giant cells. This study was analyzed by the Chi-square test, Fisher’s test, specificity test, and sensitivity test. We obtained p = 0.001 for Histone H3.3 (G34W) mutant expression in GCTB vs Non-GCTB. Statistically, there was no significant difference in the expression level of Histone H3.3 (G34W) in the GCTB and its variants p-value = 0.183. We also obtained that the specificity of Histone H3.3 expression on GCTB was 100% and the sensitivity of Histone H3.3 on GCTB was 77.8%.
Conclusion:
Histon H3.3 mutant as a mutated driver gene in an Indonesian GCTB can assist to diagnose GCTB and compare it from other bone tumors.
Key Words: Giant cell tumor of bone (GCTB), Histone H3.3, G34W, osteoclast-like giant cells
Introduction
Giant cell tumor of bone (GCTB) known as osteoclastoma is characterized by a dense distribution of mononuclear stromal cells with rounded oval nuclei, monotonous and not atypical resembling epithelioid histiocytes. There is a distribution of osteoclast-like giant cells with mononuclear cells among the stromal, the nuclei in osteoclast-like giant cells are similar to the nuclei of stromal cells. These tumor cells can infiltrate the surrounding bone trabeculae (Antonescu et al., 2020; Kim et al., 2012).
This tumor is a benign bone tumor however is locally aggressive and tends to advance into local recurrences and can transform into malignant and metastasize to the lungs (Cao et al., 2017; Chen et al., 2014; Cheng et al., 2015; Noh & Park, 2018). Interestingly, GCTB was often discovered in Southeast Asia and East Asia with an incidence of around 20% in contrast to 4-5% in Western countries (Liede et al., 2018; Siddiqui et al., 2014; Zheng et al., 2001). GCTB is an interesting thing to study, the mononuclear cell population is a major component of this tumor. It is responsible and allows it to be a true neoplastic component (Unni et al., 2010; Yamamoto et al., 2020; Zheng et al., 2001).
The histone family occurs in almost all eukaryotic cell types and is subdivided into five classes, H1, H2A, H2B, H3, and H4. The H3.3 protein is divided into two genes located in different loci: H3F3A on chromosome 1 and the other H3F3B on chromosome 17. Both encode the same sequence of amino acids but differ in the sequence of nucleotide and gene organization (Gong et al., 2021; Yamamoto et al., 2020).
Recently, a monoclonal antibody targeting the G34W mutation of the gene in chromosome 1 (H3F3A) has been developed (Gong et al., 2021). In GCTBs, only H3F3A has these mutations, changing the amino acid glycine 34 to tryptophan (G34W). It was mutated in 90% of cases, the most common mutation. This mutation is confined to the neoplastic stromal cells of GCTB (Behjati et al., 2013).
Stromal cells of this tumor has a heterozygous H3F3A somatic mutation, one of the two genes encoding the histone 3 variant H3.3 (Trovato et al., 2020; Yamamoto et al., 2020). This was proven in previous studies which reported that >90% of GCTB cases were related to mutations in the H3F3A gene (Amary et al., 2017; Yoshida et al., 2019). According to Lim et.al, the H3.3 G34W/L mutation is the only recurrent molecular change in GCT, potentially driving the devastating bone disease (Lim et al., 2017). How the process of tumor development occurs in bone is still unknown.
Previously we have reported our findings that the prevalence of GCTB is slightly higher in males than females. This is particularly interesting in our region because it contradicts some of the previous literature (Futriani et al., 2022). The Aim of this study was to prove whether there are differences in Histone H3.3 Mutations between GCTB and Non-GCTB.
Materials and Methods
Sample
This research used the total sampling method using a sample of paraffin blocks. The samples collected were 54 patients diagnosed as GCTB, and 17 patients diagnosed as NON-GCTB and categorized as benign and malignant GCTB according to medical records data and histopathological review at the RS Anatomical Pathology Laboratory Hasanuddin University Makassar, between October 2021 to January 2022. Samples that met the inclusion criteria were collected including biopsy and tumor resection preparations accompanied by basic clinical data of the patient, paraffin blocks and tissue slides (of adequate size for multi-field microscopic examination) from resected tumors diagnosed as giant cell tumors (GCTB and Non-GCTB) and re-evaluated by two Anatomical Pathologists to establish a consistent diagnosis. The giant cell tumors examined included primary, recurrent, metastatic, and malignant giant cell tumors. Non-GCTB lesions are histopathological tissues characterized by the presence of giant cells resembling osteoclasts which are the differential diagnosis of giant cell tumor of bone consisting of giant cell reparative granuloma, giant cell tumor of the tendon sheath, bone cyst aneurysm, chondromyxoid fibroma, giant cell-rich osteosarcoma and chondroblastoma.
Immunohistochemical staining
Paraffin blocks are cut by 4 µm in thickness, immersed in a water bath and put on the poly-L-lysine glass object. Immunohistochemical staining procedure used Anti-Histone H3.3 G34W Rabbit Monoclonal Antibody, Clone RM263 cat.#: 31-1145-00-S, diluted 1:100. Hydrogen Peroxide 3% has been used to block Endogenous Peroxidase activity. Enzyme Label with Horseradish Peroxidase (HRP) as secondary antibody then protein visualization using 3,3’-diaminobenzidine (DAB) followed by Mayer’s Hematoxylin counterstaining.
Expression H3.3 (G34W) is the accumulation of Histone H3.3 (G34W) protein in the nuclei detected by immunohistochemical methods. Tumor cells had been marked with positive staining when it displayed brown color in the nuclei and negative staining when brown color expression was not detected.
The expression of this protein is calculated based on positive cell proportions of Histone H3.3 if it stains strongly and firmly in the nucleus of the tumor cells and negative if the expression was not detected, cytoplasmic staining or pale staining in the nucleus of the tumor cells (Amary et al., 2017; Gong et al., 2021).
Statistical Analysis
The results of this study were analyzed by the Chi-square test, Fisher’s test, specificity test, and sensitivity test.
Results
Among 54 samples of Giant Cell Tumor of the Bone (GCTB) were obtained, consisting of 37 samples of primary Giant Cell Tumor of the Bone (GCTB), 5 samples of recurrent Giant Cell Tumor of the Bone (GCTB), 9 samples of Giant Cell Tumor of the Bone (GCTB) metastases, 3 samples of malignant Giant Cell Tumor of the Bone (GCTB). This study also included 17 Non-GCTB samples for comparison. The cases included Non-GCTB included chondroblastoma 1 sample, giant cell reparative granuloma 2 samples, giant cell of tendon sheath 7 samples, chondromyxoid fibroma 2 samples, aneurysmal bone cyst 2 samples, and giant cell-rich osteosarcoma 3 samples.
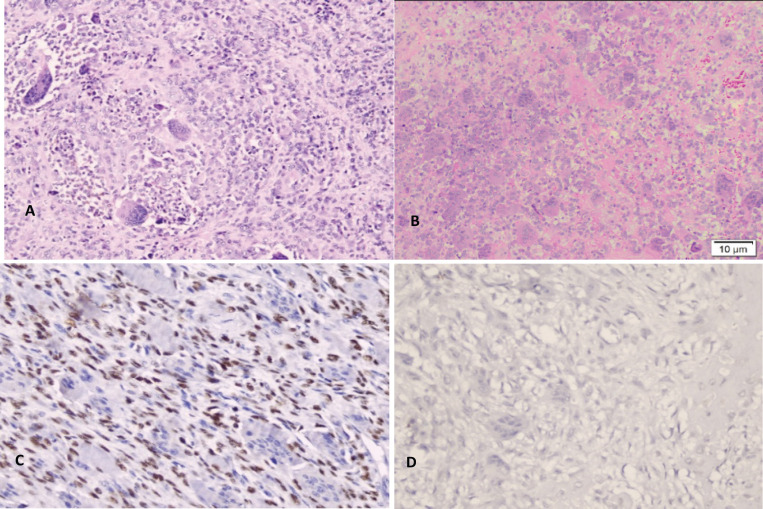
GCTB shows osteoclast-like giant cells and mononuclear cells with similar nuclear features on the hematoxylin & eosin staining (Figure 1A). H3.3 G34W was diffusely positive in the nucleus-plump mononuclear cells, but not in osteoclast-like giant cells. (Figure 1C). All cells confirmed negative expression of H3.3 G34W in Non-GCBT (Figure 1 D)
Figure 1.
A. H&E staining GCTB, B. H&E staining NON-GCTB (giant cell rich osteosarcoma), C. Histone H3.3 (G34W) positive expression, D. Histone H3.3 (G34W) Negative expression. x20
Table 1 shows a total of 54 GCT samples, 83,8% of which are Primary GCTB samples. There were 31 (83,8%) samples that expressed positive Histone H3.3 (G34W) and only 6 (16,2%) samples were negative expressions. Recurrent GCTB had positive expression of Histone H3.3 (G34W) in 4 samples (80%) and 1 sample was negative expression (20%). Metastatic GCTB, there were 6 samples (66,7%) and 3 samples (33,3%) with negative expression. But, the reverse condition for malignant GCTB, only 1 sample (33,3%) showed positive expression. In the Chi-Square test, the p-value was >0.005 (P=0.183).
Table 1.
Expression of Histone H3.3 (G34W) in GCTB
| GCTB and its variant group | Expression of Histone H3.3 (G34W) | |
|---|---|---|
| Positive | Negative | |
| Primary GCTB (n=37) | 31 (83,8%) | 6 (16,2%) |
| Recurrent GCTB (n=5) | 4 (80%) | 1 (20%) |
| Metastatic GCTB (n=9) | 6 (66,7%) | 3 (33,3%) |
| Malignant GCTB (n=3) | 1 (33,3%) | 2 (66,7%) |
| Total | 42 (77,8%) | 12 (22,2%) |
*Chi-Square test, P= 0,183; GCTB, Giant Cell Tumor of the Bone
Table 2 shows that of the 71 samples studied, 42 (77.8%) GCTB samples were positively expressed by Histone H3.3 (G34W) staining, and 12 (22.2%) samples were not expressed by Histone staining H3.3 (G34W). Whereas in the 17 Non-GCTB samples, none of them were expressed by Histone H3.3 (G34W) staining. By comparing the expression of Histone H3.3 (G34W) in GCTB and Non-GCTB, the p-value on Chi-Square test and Fisher’s test was P = 0.001 (P <0.05).
Table 2.
Expression of Histone H3.3 (G34W) in GCTB and Non-GCTB
| Tumor type | Score expression | Total | |
|---|---|---|---|
| Positive | Negative | ||
| GCTB | 42 (77,8%) | 12 (22,2%) | 54 (100%) |
| Non-GCTB | 0 (0 %) | 17 (100%) | 17 (100%) |
*Fisher test P= 0,001
Table 2 shows the results of diagnostic tests to determine sensitivity, specificity, positive predictive value, and negative predictive value. Based on the table above, it can be calculated: Sensitivity = 42 / 54 = 0.778 = 77.8%; Specificity = 17 / 17 = 1 = 100%; Positive predictive value = 42 / 42 = 1 = 100%; Negative predictive value = 17 / 29 = 0.586 = 58.6%.
Discussion
This research stated that the clone RM263, anti-histone H3.3 G34W monoclonal antibody is highly specific and even quite sensitive for tumors which has H3.3 mutation and is an effective marker for GCTB, greatly improves diagnostic accuracy in biopsy specimens. This is consistent with previous studies which detected strongly stained Histone H3.3 (G34W) antibodies in mononuclear cell populations that had H3F3A mutations in GCTB and Unstained in osteoclastic giant cells. Lüke et al., (2017) demonstrated that strong and consistent nuclear staining was present in nuclei of stromal cells in approximately 10-90%. In all GCTB classification groups, primary GTCB is the most common. Generally, it shows a positive expression of Histone H.3.3 (G34W) immunohistochemistry. There were 2 cases of malignant GCTB that failed to show immunoreactivity which is probably to be defined by tissue fixation and decalcification, the significance of setting up procedures for tissue handling and needs to perform with a larger population of the tumor.
The finding of H3.3 mutations in both benign and malignant stromal components support the concept that these H3.3 mutations are mutation drivers. It is thought that there may be additional, undetermined genetic alterations (Amary et al., 2017). Furthermore, 17 tumors other than GCTB were negative or undetectable as immunoreactive for this clone antibody including chondroblastomas, osteoblastoma, aneurysmal bone cyst, osteosarcoma giant cell-rich, giant cell reparative granuloma, chondromyxoid fibroma GCT of tendon sheath. Cleven et al., (2015) also reported that other tumors (Non-GCTB) containing giant cells did not has the H3F3A mutation so it did not express Histone H3.3 (G34W) (Cleven et al., 2015).
This study shows that the value of the diagnostic test obtained a specificity of Histone H3.3 expression is 100% and the sensitivity of Histone H3.3 expression is 77.8% so that in this study it can be concluded that Histone H3.3 expression can be used as a diagnostic marker for GCTB. The specificity of the antibody for the mutation was confirmed by the absence of H3.3 G34W expression in the cases. The investigation of the histopathological features and H3F3A mutation status of GCTBs can also be used as a basis for selecting therapeutic targets (Kato et al., 2018; Lübbehüsen et al., 2019).
Histone H3.3 is highly specific for GCTB differentiation from other giant cell tumors (Cleven et al., 2015; van der Heijden et al., 2020). We found Histone H3.3 (G34W) is a highly specific and sensitive marker useful for the differential diagnosis of GCTB that has a similar histological appearance. Immunohistochemistry should serve as a good substitute for molecular testing (Ogura et al., 2017).
There were some boundaries to this study since our study consisted of only a small sample size. In future studies, it is crucial to increase the number of GCTB samples by extending the research time so that the prognosis of the Giant Cell Tumor of Bone can be assessed as well.
In conclusion, Histone H3.3 (G34W) expression assessment can be used as a diagnostic marker for GCTB so that it can be considered by clinicians in ruling out a differential diagnosis but there was no significant difference with Histone H3.3 (G34W) staining based on the variant of the Giant Cell Tumor of Bone group.
Author Contribution Statement
The methodology was planned and designed by UM and F; DA, F, NK and MJ were involved in data gathering, processing, and reporting. UM, DA and HD were conducted a comprehensive conceptual, writing manuscript and editorial evaluation; the finalization of the article was amended and approved by all the contributors..
Acknowledgements
The Anatomical Pathology Laboratory at Hasanuddin University Hospital contributed to the feasibility of this study especially for the technicians (Mardiati and Juniarsih) who helped preparation of Immunohistochemistry.
Study and ethical Approval
The research committee of the Faculty of Medicine at Hasanuddin University approved this project.
The Ethics Committee of the Faculty of Medicine granted it this study (Protocol #UH21080529, Archive No. 585/UN4.6.4.5.31/PP36- 2021KOMETIK).
Availability of Data
On reasonable request, the associated author will release the datasets used in this work.
Conflict of Interest
All contributors report having no competing interests.
References
- Amary F, Berisha F, Ye H, et al. H3F3A (Histone 3 3) G34W Immunohistochemistry. Am J Surg Pathol. 2017;41:1059–68. doi: 10.1097/PAS.0000000000000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR. WHO Classification of Tumours Editorial Board, World Health Organization (2020). WHO Classification of Tumours: Soft tissue and bone tumours (5th ed.) [Online] France: International Agency for Research on Cancer; 2020. [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–82. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Lin F, Hu Y, et al. Epidemiological and Clinical Features of Primary Giant Cell Tumors of the Distal Radium: A Multicenter Retrospective Study in China. Sci Rep. 2017;7:1–6. doi: 10.1038/s41598-017-09486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Li C, Wu B, et al. Identification of differentially expressed genes and their subpathways in recurrent versus primary bone giant cell tumors. Int J Oncol. 2014;45:1133–42. doi: 10.3892/ijo.2014.2501. [DOI] [PubMed] [Google Scholar]
- Cheng DD, Hu T, Zhang HZ, Huang J, Yang QC. Factors Affecting the Recurrence of Giant Cell Tumor of Bone after Surgery: A Clinicopathological Study of 80 Cases from a Single Center. Cell Physiol Biochem. 2015;36:1961–70. doi: 10.1159/000430164. [DOI] [PubMed] [Google Scholar]
- Cleven AHG, Höcker S, Briaire-de Bruijn I, et al. Mutation Analysis of H3F3A and H3F3B as a Diagnostic Tool for Giant Cell Tumor of Bone and Chondroblastoma. Am J Surg Pathol. 2015;39:1576–83. doi: 10.1097/PAS.0000000000000512. [DOI] [PubMed] [Google Scholar]
- Futriani, Johan MP, Ketut N, Zainuddin AA, Achmad D, Miskad UA. Clinical Characteristics of Giant Cell Tumour of Bone in Makassar , Indonesia. Int J Sci Basic Appl Res. 2022;62:177–81. [Google Scholar]
- Gong L, Bui MM, Zhang W, et al. H3F3A G34 mutation DNA sequencing and G34W immunohistochemistry analysis in 366 cases of giant cell tumors of bone and other bone tumors. Histol Histopathol. 2021;36:61–8. doi: 10.14670/HH-18-264. [DOI] [PubMed] [Google Scholar]
- Kato I, Furuya M, Matsuo K, et al. Giant cell tumours of bone treated with denosumab: histological, immunohistochemical and H3F3A mutation analyses. Histopathology. 2018;72:914–22. doi: 10.1111/his.13448. [DOI] [PubMed] [Google Scholar]
- Kim Y, Nizami S, Goto H, Lee FY. Modern interpretation of giant cell tumor of bone: Predominantly osteoclastogenic stromal tumor. Clin Orthop Surg. 2012;4:107–16. doi: 10.4055/cios.2012.4.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liede A, Hernandez RK, Tang ET, et al. Epidemiology of benign giant cell tumor of bone in the Chinese population. J Bone Oncol. 2018;12:96–100. doi: 10.1016/j.jbo.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Park JH, Baude A, et al. The histone variant H3 3 G34W substitution in giant cell tumor of the bone link chromatin and RNA processing. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-13887-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübbehüsen C, Lüke J, Seeling C, et al. Characterization of Three Novel H3F3A-mutated Giant Cell Tumor Cell Lines and Targeting of Their Wee1 Pathway. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-42611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke J, von Baer A, Schreiber J, et al. H3F3A mutation in giant cell tumour of the bone is detected by immunohistochemistry using a monoclonal antibody against the G34W mutated site of the histone H3 3 variant. Histopathology. 2017;71:125–33. doi: 10.1111/his.13190. [DOI] [PubMed] [Google Scholar]
- Noh BJ, Park YK. Giant cell tumor of bone: updated molecular pathogenesis and tumor biology. Hum Pathol. 2018;81:1–8. doi: 10.1016/j.humpath.2018.06.017. [DOI] [PubMed] [Google Scholar]
- Ogura K, Hosoda F, Nakamura H, et al. Highly recurrent H3F3A mutations with additional epigenetic regulator alterations in giant cell tumor of bone. Genes Chromosom Cancer. 2017;56:711–8. doi: 10.1002/gcc.22469. [DOI] [PubMed] [Google Scholar]
- Siddiqui MA, Seng C, Tan MH. Risk factors for recurrence of giant cell tumours of bone. J Orthop Surg. 2014;22:108–10. doi: 10.1177/230949901402200127. [DOI] [PubMed] [Google Scholar]
- Trovato M, Patil V, Gehre M, Noh KM. Histone Variant H3 3 Mutations in Defining the Chromatin Function in Mammals. Cells. 2020:9. doi: 10.3390/cells9122716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni KK, Inwards CY, Research MFME. Dahlin’s Bone Tumors. Philadelphia: Lippincott Williams & Wilkins; 2010. [Google Scholar]
- Van der Heijden L, Dijkstra S, van de Sande M, Gelderblom H. Current concepts in the treatment of giant cell tumour of bone. Curr Opin Oncol. 2020;32:332–8. doi: 10.1097/CCO.0000000000000645. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Ishihara S, Toda Y, Oda Y. Histone H3 3 mutation in giant cell tumor of bone: an update in pathology. Med Mol Morphol. 2020;53:1–6. doi: 10.1007/s00795-019-00238-1. [DOI] [PubMed] [Google Scholar]
- Yoshida KI, Nakano Y, Honda-Kitahara M, et al. Absence of H3F3A mutation in a subset of malignant giant cell tumor of bone. Mod Pathol. 2019;32:1751–61. doi: 10.1038/s41379-019-0318-5. [DOI] [PubMed] [Google Scholar]
- Zheng MH, Robbins P, Xu J, et al. The histogenesis of giant cell tumour of bone: A model of interaction between neoplastic cells and osteoclasts. Histol Histopathol. 2001;16:297–307. doi: 10.14670/HH-16.297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
On reasonable request, the associated author will release the datasets used in this work.