Abstract
Mycobacterium tuberculosis can persist in sputum for long periods of time after the initiation of antituberculosis chemotherapy. The purpose of this study was to determine whether quantitative estimates of M. tuberculosis DNA in sputum correlate with the numbers of viable bacilli and thus measure the therapeutic response of patients during treatment. Two methods of M. tuberculosis DNA quantification were examined by using DNA isolated from sputum specimens serially collected during the course of chemotherapy. A competitive PCR assay was compared to an automated system of real-time quantification with the ABI Prism 7700 Sequence Detection System (TaqMan). The ABI 7700 system uses standard PCR in conjunction with a fluorogenic probe in which the intensity of fluorescence is proportional to the amount of target DNA present. The results showed that both PCR systems are reproducible and accurate. The amounts of M. tuberculosis DNA quantified in sputum corresponded well with the numbers of acid-fast bacilli (AFB) counted by microscopy. Before initiation of antituberculosis therapy, measures of AFB, M. tuberculosis DNA, and cultivable bacilli were similar, suggesting that quantification of DNA is a good method for measuring the initial bacillary load. However, the rate of disappearance of both AFB and M. tuberculosis DNA did not correlate with the decline in cultivable bacilli in the specimen; therefore, these tests are not appropriate for monitoring treatment efficacy.
The emergence of drug-resistant strains of Mycobacterium tuberculosis has led to the need to rapidly measure treatment efficacy. Drug susceptibility profiles can take several weeks. Additionally, in vitro drug susceptibility test results do not reflect in vivo drug absorption difficulties that can also lead to the development of drug resistance and the spread of disease. Currently, sputum smear and culture are used to monitor the response to therapy for patients with pulmonary tuberculosis. Conversion of positive results to negative results after 1 to 2 months of therapy is indicative of effective treatment (7, 30). The objective of this work was to determine whether quantitative changes in the amount of mycobacterial DNA present in sputum reflect bactericidal drug activity.
Competitive PCR is a standard method for the quantification of DNA and has been used successfully in a number of studies (6, 9, 21, 25, 29, 33). However, this technique is labor intensive and requires results from multiple reactions to be analyzed for each sample. Recently, the ABI Prism 7700 Sequence Detection System (TaqMan) has been shown to be a rapid and sensitive method for quantification of PCR and reverse transcription-PCR products (8, 10). The system uses a fluorogenic probe, and the amount of fluorescence detected is proportional to the amount of accumulated PCR product. The amount of target DNA in a sample is interpolated from a standard curve run simultaneously with the unknown samples. Quantification of PCR products occurs in real time during the amplification process, with no postamplification handling being necessary. This eliminates potential sources of carryover contamination and reduces the handling time. In this report, we compare two methods for the quantification of mycobacterial DNA in patient samples. Sputum samples were collected before and during the course of chemotherapy to evaluate whether the measure of M. tuberculosis DNA can be used as an indicator of treatment efficacy.
MATERIALS AND METHODS
Patients and specimens.
Sputa were collected from two patients with newly diagnosed, smear-positive pulmonary tuberculosis at the Universidade Federal do Espìrito Santo Hospital in Vitória, Brazil. Subjects presented with cavitary disease and were human immunodeficiency virus negative. Patients harbored drug-sensitive M. tuberculosis strains and were treated with a standard four-drug regimen of isoniazid, rifampin, ethambutol, and pyrazinamide for 2 months, followed by isoniazid and rifampin treatment for an additional 4 months. The patients responded to therapy and showed no clinical signs of relapse for up to 8 months following the cessation of treatment. Noninduced spot sputum samples were collected in triplicate before the start of chemotherapy (day 0) and in duplicate on days 2, 4, 7, 14, 30, 60, and 120. Sputa were stored at 4°C until homogenization with 2.5% N-acetyl-l-cysteine and 4-mm glass beads as described previously (2). The specimens were divided into aliquots of 0.5 ml of homogenate and were frozen at −70°C until nucleic acid extraction. Sputum was further processed by 2% NaOH-sodium citrate decontamination and centrifuge sedimentation for quantitative microscopy and culture (16).
Quantitative AFB microscopy and culture.
For quantitative AFB microscopy, 20 μl of homogenized and decontaminated sputum was fixed onto a 78-mm2 area on a slide and subjected to auramine O staining, and the number of AFB per field was counted microscopically for an average of 50 fields per sample. AFB enumeration was shown to be reproducible by repeated counting on separate days.
Quantitative culture was performed by plating serial dilutions of homogenized-decontaminated sputum on 7H10/7H10S agar (S = 200 U of polymyxin B per ml, 10 μg of amphotericin B per ml, 50 μg of carbenicillin per ml, and 20 μg of trimethoprim per ml) (19). A series of 10-fold dilutions was made from 0.5 ml of decontaminated specimen with a 0.85% NaCl–0.25% Tween 80 solution. A total of 30 μl of each dilution was plated in duplicate onto separate quadrants of the biplate. Colonies were counted after incubation for 3 to 4 weeks at 37°C in 5% CO2, with the ideal dilution considered to be between 10 and 50 colonies per quadrant.
DNA isolation.
DNA was isolated from 0.5 ml of homogenized sputum by a combination of organic and mechanical lysis with glass beads. The entire 0.5-ml aliquot of sputum was mixed with 1 ml of TriZOL-LS (Gibco BRL Life Technologies, Grand Island, N.Y.), transferred to a 2-ml matrix tube (Bio 101, Inc., La Jolla, Calif.), and processed twice for 45 s each time in a FastPrep 120 processor (Bio 101, Inc.). The aqueous phase was removed and DNA was extracted from the interface and organic fractions. A total of 400 μl of back extraction solution (100 mM Tris [pH 8], 1 mM EDTA [pH 8], 50 mM NaCl, 1% sodium dodecyl sulfate) was added to the organic phase-interface fractions and was mixed by vortexing for 2 min. The samples were incubated at 65°C for 15 min, mixed for 2 min by vortexing, and centrifuged for 15 min at 18,000 × g. The newly formed, DNA-containing aqueous phase was transferred to a 1.5-ml microcentrifuge tube. Extraction of the organic phase-interface fractions was repeated as described above to remove any residual DNA from the organic fraction. Aqueous phases from both the first and second extractions were combined and 2 μl of linear acrylamide (Ambion, Austin, Tex.), a 1/10 volume of 3 M sodium acetate, and an equal volume of isopropanol were added, followed by precipitation of the samples overnight at −20°C. The tubes were centrifuged at 18,000 × g for 30 min, and the pellets were washed twice with 75% ethanol before drying and resuspension in 100 μl of distilled H2O.
Mycobacterial genomic DNA was isolated from cultured Mycobacterium bovis 410, which contains a single copy of the IS6110 insertion element, by the method of Beggs et al. (1). The concentration and purity of the DNA were determined spectrophotometrically, and the DNA had an A260/A280 of 1.9.
Recombinant plasmids pIS6110 and pQIS6110 were isolated from DH5α cells (GibcoBRL Life Technologies) by the PERFECTprep DNA isolation kit (5 Prime ⇒ 3 Prime, Inc., Boulder, Colo.).
Competitive PCR. (i) Internal control.
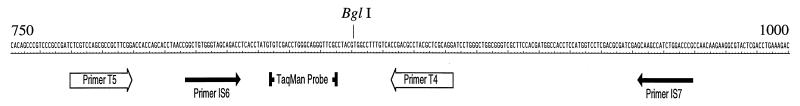
The entire sequence of IS6110 from a clone kindly provided by Ndingsa Fomukong was subcloned into pBluescript (Stratagene, La Jolla, Calif.) and was named pIS6110. An internal control plasmid, named pQIS6110, was constructed by ligating an 80-bp Mycobacterium avium DNA fragment with the same G+C content as IS6110 into a BglI site (see Fig. 1).
FIG. 1.
Sequence of the region of M. tuberculosis IS6110 amplified in both competitive and TaqMan PCRs. The primers for competitive PCR are labeled T4 and T5. The primers used in the ABI 7700 system are labeled IS6 and IS7. The BglI site is the site of insertion of the 80-bp M. avium fragment to make the internal control plasmid pQIS6110.
(ii) PCR.
The appropriate dilution of test DNA that would fall within the range of dilutions of DNA for the internal controls was initially determined empirically by PCR analysis of serial dilutions without the pQIS6110 competitor. Dilutions of the internal control pQIS6110, from 10 to 10,000 molecules/μl, were made in a 10-ng/μl yeast RNA solution (Ambion) and were coamplified with the appropriate dilution of test DNA.
PCR was performed with oligonucleotide primers T4 and T5, which amplify a 123-bp region of the IS6110 sequence (5). PCR primers were radiolabeled with 32P, and a total of 5 × 105 cpm of labeled primers, in addition to 0.4 μM unlabeled oligonucleotide primers, was added to each reaction mixture. The PCR conditions were as described previously (2). Amplification was performed with a GeneAmp PE9600 thermal cycler (Applied BioSystems/Perkin-Elmer [ABI/PE], Foster City, Calif.) with the profile of 1 cycle of 50°C for 5 min (for uracil DNA glycosylase decontamination); 1 cycle of 94°C for 5 min for denaturation; 32 cycles of 94°C for 30 s, 68°C for 1.5 min, and 72°C for 1 min; and 1 cycle of 72°C for 20 min (final extension). Target DNA concentrations were chosen so that the DNA was still in exponential amplification after 32 cycles of PCR (data not shown).
(iii) Quantification of products.
PCR products were electrophoresed on 8% nondenaturing polyacrylamide gels, dried, and exposed for 4 h to a PhosphorImager screen (Molecular Dynamics, Sunnyvale, Calif.). The intensities of the internal control bands and of authentic products were quantified with the PhosphorImager by using ImageQuant, version 3.1, software (Molecular Dynamics).
ABI 7700 quantification. (i) Oligonucleotide primers and probes.
The primers for M. tuberculosis DNA quantification for use with the ABI Prism 7700 Sequence Detection System (ABI/PE) were in the same region of the IS6110 sequence as the primers used for competitive PCR, but different sequences were selected for the oligonucleotide primers in order to optimize the PCR for a two-step profile, which is preferred by TaqMan chemistry. Selection of new primers also allowed adjustment of the melting temperatures to those optimal for the ABI 7700 system (PCR primers and fluorogenic probe) (see Fig. 1). The upstream PCR primer (IS6) corresponds to the region from base 807 to base 824 (sequence, 5′-GGCTGTGGGTAGCAGACC-3′ [bases are numbered as for the sequence with GenBank accession no. X52471]). The reverse primer (IS7) corresponds to the region of IS6110 from base 952 to base 969 (sequence, 5′-CGGGTCCAGATGGCTTGC-3′).
The internal oligonucleotide probe was labeled with the fluorescent dyes 5-carboxyfluoroscein (FAM) on the 5′ end and N,N,N′,N′-tetramethyl-6-carboxyrhodamine (TAMRA) on the 3′ end. The internal probe hybridizes within the 163-bp region amplified by the PCR primers and has the sequence 5-(FAM)-TGTCGACCTGGGCAGGGTTCG-(TAMRA)-3′. When the two dyes are in close proximity, as with an intact oligonucleotide probe, TAMRA acts as a quencher for FAM by absorbing at the FAM emission spectra. The 5′ exonuclease activity of Taq polymerase will degrade an internally hybridizing probe during the course of PCR (13). Degradation of the probe leads to separation of these two dyes in solution, with a subsequent increase in the level of fluorescence in the reaction mixture. The amount of fluorescence measured in a sample is proportional to the amount of specific PCR product generated (10).
(ii) PCR conditions.
The PCR mixture (50 μl total volume) consisted of primers IS6 and IS7 (0.8 μM each), 100 nM IS6110 TaqMan probe, dATP, dCTP, and dGTP (Pharmacia, Alameda, Calif.) each at a concentration of 200 nM, 400 nM dUTP (Pharmacia), 5 mM MgCl2, 100 ng of bovine serum albumin per μl (Pharmacia), 10 ng of yeast RNA (Ambion) per μl, 1 U of uracil DNA glycosylase (New England Biolabs, Beverly, Mass.), 1 U of Taq polymerase (Fisherbrand, Plano, Tex.), and 1× PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl).
Amplification and detection were performed with the ABI 7700 system with the following profile: 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 5 min, and 45 cycles of 94°C for 30 s and 68°C for 1 min. The TAMRA signal was used to standardize the reaction (10).
(iii) Standard curve.
The threshold is set at 10 times the standard deviation of the mean baseline emission calculated for PCR cycles 3 to 10. The fractional cycle number reflecting a positive PCR result is called the cycle threshold (Ct). The Ct values for standards and samples were usually in the range of between 18 and 32 cycles of amplification. The amount of product in a particular reaction mixture is measured by interpolation from a standard curve of Ct values generated from known starting concentrations of DNA.
RESULTS
Competitive PCR quantification.
IS6110 is a multicopy insertion element found in M. tuberculosis complex organisms and has been the target of numerous diagnostic assays. The region targeted for amplification in these studies has been shown to be specific for the M. tuberculosis complex (11, 27) and is diagrammed in Fig. 1.
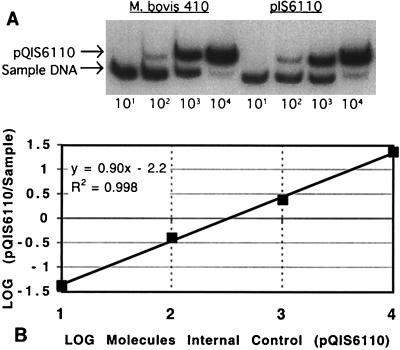
Purified DNA from M. bovis 410 and the plasmid pIS6110, each containing a single copy of IS6110, was quantified. An example of a competitive PCR gel is shown in Fig. 2A. The radioactivity in individual bands was measured with the PhosphorImager, and the values were plotted (Fig. 2B). The line has a slope of approximately 1, which indicates equivalent amplification efficiencies of test and competitor DNAs. It has been reported that a slope of 1 is required for the accurate and reproducible quantification of unknown samples by competitive PCR (24). However, other factors must be involved since we and others have observed accurate and reproducible quantification with slopes of less than 1 (23).
FIG. 2.
Competitive PCRs for M. bovis genomic DNA and pIS6110 plasmid DNA. (A) Autoradiograph of two competitive PCRs. PCR amplification of the internal control plasmid, pQIS6110, results in a 203-bp product. Amplification of test DNAs from both M. bovis and pIS6110 results in a 123-bp product. The number of pQIS6110 molecules added to each PCR mixture is listed under the lane. (B) Relative amounts of sample and internal control DNA for the data in the M. bovis gel in panel A. The equation and fit of the line are shown.
Quantitative competitive PCR analyses were performed in triplicate with an estimated 500 molecules of the target DNA described above. The calculated values for M. bovis and pIS6110 were 338 ± 33 molecules (mean ± standard deviation) and 289 ± 40, respectively. These replicate values demonstrate that the assay is reproducible, with coefficients of variation of 10% for M. bovis and 14% for pIS6110. The number of molecules measured (∼300) is lower than the expected 500 molecules of input template, perhaps due to the use of 10-fold dilutions of competitor. The use of large dilutions of competitor allows for a greater dynamic range of the assay, although some accuracy may be sacrificed (6, 33). While it is possible that an unequal amplification of the internal control versus the authentic IS6110 target occurred, further experiments suggest that this is not likely (see below). Importantly, the assay is reproducible and IS6110 molecules are quantified similarly whether they are in a genomic or a plasmid background. This suggests that in this case, the DNA structure surrounding the target does not appear to affect PCR amplification efficiency.
Automated DNA quantification.
A quantitative assay was also developed for IS6110 using the ABI Prism 7700 Sequence Detection System (TaqMan). Known amounts of M. bovis 410 genomic DNA, from 50 to 500,000 molecules per reaction mixture, resulted in Ct values ranging from 18 to 31 cycles, with coefficients of variation of less than 1% between replicate standards (Table 1).
TABLE 1.
Measurement of replicate standard curves ranging from 50 to 500,000 molecules of M. bovis 410 genomic DNA per reaction mixture
| Quantity (no. of molecules) | Ct for the following reaction:
|
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Mean | SD | % CVa | |
| 50 | 31.3 | 31 | 31 | 31.5 | 31.19 | 0.27 | 0.87 |
| 500 | 27.3 | 27.7 | 27.7 | 27.77 | 27.63 | 0.22 | 0.78 |
| 5,000 | 24.6 | 24.8 | 24.5 | 24.56 | 24.62 | 0.16 | 0.64 |
| 50,000 | 21.6 | 21.4 | 21.5 | 21.46 | 21.48 | 0.08 | 0.37 |
| 500,000 | 18.2 | 18.3 | 18.2 | 18.24 | 18.22 | 0.06 | 0.31 |
CV, coefficient of variation.
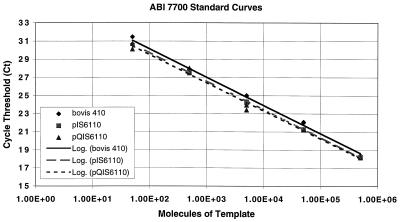
Standard curves generated with DNA from M. bovis 410, plasmid pIS6110, and the internal control pQIS6110 plasmid were assayed in parallel. All standard curves were linear over 6 orders of magnitude, with the R2 values of the lines being greater than 0.99 in each case (Fig. 3). The slopes of the curves were similar for all three sources of control DNA, with no appreciable differences between genomic and plasmid DNA. These data suggest that amplification efficiencies were similar for mycobacterial genomic DNA, plasmid DNA, and the engineered plasmid DNA used as an internal control in the competitive assay.
FIG. 3.
Standard curves generated by analysis of known amounts of template DNA with the ABI 7700 system. Purified DNA from M. bovis 410, plasmid pIS6110, and internal control plasmid pQIS6110, each containing one copy of the IS6110 insertion element, were assayed in parallel. The regression lines calculated for the datum points are shown. The R2 values for each of the lines was >0.99.
Measurements of M. tuberculosis DNA in sputum.
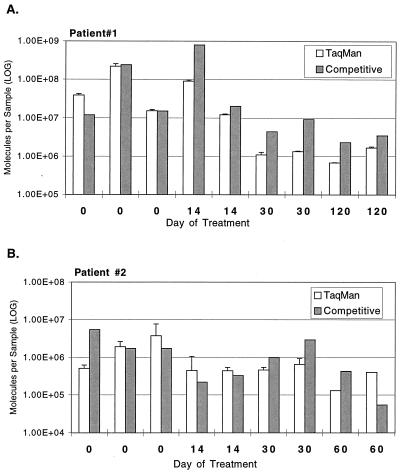
The number of IS6110 molecules present in DNA isolated from sputum specimens was quantified by both the competitive and the ABI 7700 PCR systems. The results for multiple samples collected from two patients during the first 2 to 3 months of antituberculosis therapy are presented in Fig. 4. Both DNA quantification assays resulted in the measurement of similar levels of IS6110 DNA.
FIG. 4.
Comparison of TaqMan and competitive PCR assays for two patients receiving tuberculosis chemotherapy. IS6110 DNA levels were determined with both assay systems for DNA isolated from sputum collected during the course of chemotherapy. The same DNA sample was tested in both systems. Specimens were collected in triplicate on day 0, before the onset of antimicrobial chemotherapy, and duplicate samples were collected thereafter. The competitive assay was performed once per sample. Error bars show the standard deviations for duplicate TaqMan measurements.
Duplicate quantifications of IS6110 DNA with the ABI 7700 system showed a coefficient of variation of less than 10% for all samples, demonstrating the reproducibility of this assay. Due to the laborious nature of the competitive assay, only two patient samples were tested in duplicate by this technique, resulting in higher coefficients of variation (98 and 74%, respectively) than those observed with the ABI 7700 system.
The variation between replicate spot sputum specimens collected sequentially before and after the initiation of chemotherapy can also be seen in Fig. 4. In most cases, replicate spot samples resulted in similar numbers of IS6110 molecules; however, differences of up to 10-fold were observed.
Comparison of M. tuberculosis DNA to quantitative microscopy and culture.
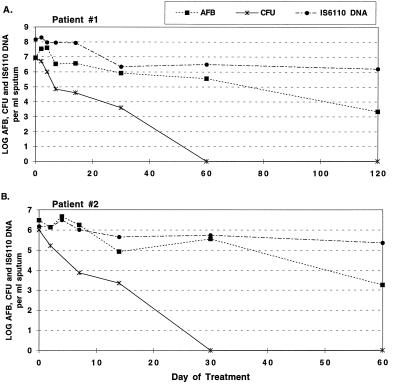
The levels of M. tuberculosis DNA, the numbers of AFB found on microscopy, and the numbers of CFU were measured in sputum collected at each time point, and relative levels were compared during the course of chemotherapy for two patients. For the first patient, 108 IS6110 DNA molecules per sample were found on day 0 and more than 106 IS6110 DNA molecules per sample were measured after 120 days of treatment (Fig. 5A). Quantitative microscopy for AFB revealed a similar trend, but with values about 10-fold lower than those for IS6110 at each time point. The higher levels of IS6110 DNA may be due to the fact that IS6110 is a multicopy element, with up to 25 copies per M. tuberculosis genome (28). At pretreatment (day 0), the numbers of CFU were similar to the total numbers of AFB present and within 60 days declined to below the limit of detection (∼102 CFU), a decrease of at least 106-fold. In contrast, AFB and IS6110 DNA levels decreased by less than 100-fold during the same time period.
FIG. 5.
Amounts of AFB, numbers of CFU, and IS6110 DNA levels in sputum collected during the first 2 to 3 months of chemotherapy. Data for two patients are shown. Values represent the highest measurement for replicate specimens collected on a given day.
A similar analysis for the second patient was performed with samples collected over a course of 60 days from the initiation of chemotherapy (Fig. 5B). Bacterial load estimates for day 0 were similar by AFB microscopy, quantitative culture, and IS6110 PCR, the numbers being approximately 106 per sample. AFB and IS6110 DNA levels decreased by approximately 10- to 100-fold over the first 30 days of treatment, while the numbers of CFU fell to undetectable levels at this same point in treatment, a decrease of at least 105-fold.
DISCUSSION
The most common methods used to monitor response to therapy for patients with pulmonary tuberculosis are conversion of positive results of smears for AFB or culture of sputum specimens to negative results. Culture can take up to 4 weeks for a result, and AFB staining lacks sensitivity and specificity. Although some reports indicate that PCR of M. tuberculosis DNA in sputum is an effective means of measuring treatment efficacy (15, 18, 31, 32), our work showed that a qualitative assessment of M. tuberculosis DNA in sputum does not correspond to the stage or severity of disease in patients receiving standard chemotherapy (12). It has been suggested that quantitative levels of DNA might reflect bacterial load and thus indicate the efficacy of chemotherapy (12, 14, 22). To test this idea, we used a new PCR system, the ABI 7700 system (TaqMan), to determine DNA levels in a real-time format. This system measures the amount of target DNA produced during each cycle of an amplification reaction; thus, all samples are quantified during the exponential phase of amplification, the phase considered to produce the most accurate results. The system also allows for a large dynamic range, and we have achieved linearity over 6 logs of input DNA. There are also advantages of speed and ease of use with this system. In our laboratory, it would require a person a minimum of 1 week to analyze a set of serially collected samples from a single patient by the standard competitive method, whereas less than a day is required with the automated system.
When either the competitive PCR or the automated assay is performed with purified control DNA, the coefficients of variation are typically 10% or less. Quantification of DNA isolated from sputa showed a similar reproducibility with the ABI 7700 system, although by the competitive PCR sample DNA tended to have a larger variation than control DNA. This has been noted in the past for competitive PCR (6, 26). A possible reason for the larger sample variation obtained by competitive PCR is the increased handling necessary to assay multiple dilutions of samples. It is also possible that when dealing with relatively impure DNA, such as that isolated from sputum, the presence of contaminants in these preparations may interfere with uniform and consistent dilution and amplification of target DNA. This is supported by the uniformity of replicate measurements with purified DNA compared to that with sample DNA (Table 1). The competitive assay is also more labor intensive than the assay with the ABI 7700 system, so replicate measurements of a single sample are not often performed, potentially leading to errors in quantification.
The data show that measurement of IS6110 DNA before and during the course of chemotherapy corresponds to the number of bacilli enumerated by staining for AFB. For one patient, the numbers of AFB and the amount of IS6110 DNA were nearly the same. For the other patient, the amount of IS6110 DNA was consistently 10-fold higher than the number of AFB, perhaps because the insertion element was present in multiple copies in the chromosome of this strain of M. tuberculosis. Importantly, neither the AFB count nor the IS6110 DNA level corresponded to the number of cultivable bacilli (number of CFU) after the initiation of chemotherapy, and thus, these measures do not reflect the bactericidal activities of chemotherapeutic agents. These data conflict with observations by others who have shown a strong correlation between AFB and culture positivity, in which the AFB result becomes negative at the same time as or before the sputum becomes culture negative (20, 32). However, patients who have advanced cavitary disease and who are treated with a rifampin-containing regimen often become culture negative but remain smear positive (17), as was the case in this investigation. Although only two patients were initially examined, we are testing an additional 18 patients and thus far have observed similar results (3). The result that neither AFB counts nor M. tuberculosis DNA levels correlated with the numbers of viable organisms suggests that neither is a good indicator of bactericidal effect. Similar data have been observed in mice, in which quantitative M. tuberculosis DNA levels did not correspond to the numbers of bacilli cultured from either the spleens or the lungs of animals treated with antituberculosis drugs (4).
We are currently testing other microbial markers which might more appropriately reflect the bactericidal effect of antituberculosis treatment and find an excellent correlation between cell viability and M. tuberculosis mRNA levels (reference 3 and unpublished data). Levels of specific mRNA targets can be readily measured by reverse transcription-PCR, with quantification performed with the ABI 7700 system. We have observed a precipitous reduction in specific M. tuberculosis mRNA levels within as few as 2 days after the start of antituberculosis agents. Furthermore, ratios of M. tuberculosis DNA levels, as measured here, to specific mRNA levels may provide the most meaningful assessment of the effects of antituberculosis agents.
Although the quantities of M. tuberculosis DNA in sputum may not be an appropriate measure of bactericidal effect, the excellent correlation between the numbers of CFU and DNA levels in patients prior to treatment indicates that quantitative PCR could be used to estimate the initial bacterial load. Since the test for IS6110 DNA is M. tuberculosis complex specific, it would also remove the ambiguities of a positive AFB smear. The speed and sensitivity of this technique would support its use for routine tuberculosis diagnostics, in particular, when the numbers of bacilli are limiting.
ACKNOWLEDGMENTS
We thank Maria Winters, Shirley Haun, Barbie Demchuck, and Robert Pruss for expert technical assistance; Ndingsa Fomukong for the IS6110-containing plasmid; and Joseph Bates, Tobin Hellyer, Jerrold Ellner, Shirley Haun, and Gery Hehman for critical reading of the manuscript and helpful discussions.
This work was funded by the Tuberculosis Research Unit (NIH contract no. NO-AI-45244).
REFERENCES
- 1.Beggs, M. L., M. D. Cave, and K. D. Eisenach. Unpublished data.
- 2.DesJardin L E, Perkins M D, Teixiera L, Cave M D, Eisenach K D. Alkaline decontamination of sputum specimens adversely affects stability of mycobacterial mRNA. J Clin Microbiol. 1996;34:2435–2439. doi: 10.1128/jcm.34.10.2435-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DesJardin L E, Chen Y, Perkins M D, Teixiera L, Cave M D, Eisenach K D. Microbial markers as surrogates for response to chemotherapy of tuberculosis. Am J Respir Crit Care Med. 1996;155(4 part 2):A255. doi: 10.1164/ajrccm.160.1.9811006. [DOI] [PubMed] [Google Scholar]
- 4.deWit D, Wootton M, Dhillon J, Mitchison D A. The bacterial DNA content of mouse organs in the Cornell model of dormant tuberculosis. Tubercle Lung Dis. 1995;76:555–562. doi: 10.1016/0962-8479(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 5.Eisenach K D, Sifford M D, Cave M D, Bates J H, Crawford J T. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Respir Dis. 1991;144:1160–1163. doi: 10.1164/ajrccm/144.5.1160. [DOI] [PubMed] [Google Scholar]
- 6.Ferre F. Quantitative or semi-quantitative PCR: reality versus myth. PCR Methods Appl. 1992;2:1–9. doi: 10.1101/gr.2.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Gangadharam P R. The role of the laboratory in the management of tuberculosis patients. Semin Respir Med. 1981;2:182–195. [Google Scholar]
- 8.Gibson U E M, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 9.Gilliland G, Perrin S, Blanchard K, Bunn H F. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:2725–2729. doi: 10.1073/pnas.87.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 11.Hellyer T, DesJardin L E, Assaf M K, Bates J H, Cave M D, Eisenach K D. Specificity of IS6110-based amplification assays for Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:2843–2846. doi: 10.1128/jcm.34.11.2843-2846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellyer T J, Fletcher T W, Bates J H, Stead W W, Templeton G L, Cave M D, Eisenach K D. Strand displacement amplification and the polymerase chain reaction for monitoring response to treatment in patients with pulmonary tuberculosis. J Infect Dis. 1996;173:934–941. doi: 10.1093/infdis/173.4.934. [DOI] [PubMed] [Google Scholar]
- 13.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jamil S, Keer J T, Lucas S B, Dockrell H M, Chiang T J, Hussain R, Stoker N G. Use of polymerase chain reaction to assess efficacy of leprosy chemotherapy. Lancet. 1993;342:264–268. doi: 10.1016/0140-6736(93)91816-5. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy N, Gillespie S H, Saruni A O S, Kisyombe G, McNerney R, Ngowi F I, Wilson S. Polymerase chain reaction for assessing treatment response in patients with pulmonary tuberculosis. J Infect Dis. 1994;170:713–716. doi: 10.1093/infdis/170.3.713. [DOI] [PubMed] [Google Scholar]
- 16.Kent P T, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. Atlanta, Ga: Centers for Disease Control and Prevention; 1985. Isolation procedures; pp. 31–70. [Google Scholar]
- 17.Kim T C, Blackman R S, Heatwole K M, Kim T, Rochester D F. Acid-fast bacilli in sputum smears of patients with pulmonary tuberculosis. Am Rev Respir Dis. 1984;129:264–268. [PubMed] [Google Scholar]
- 18.Levèe G, Glaziou P, Giquel B, Chantreau S. Follow-up of tuberculosis patients undergoing standard anti-tuberculosis chemotherapy by using a polymerase chain reaction. Res Microbiol. 1994;145:5–8. doi: 10.1016/0923-2508(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 19.Mitchison D A, Allen B W, Carol L, Dickinson J M, Aber V R. A selective oleic acid albumin agar medium for tubercle bacilli. J Med Microbiol. 1972;5:165–175. doi: 10.1099/00222615-5-2-165. [DOI] [PubMed] [Google Scholar]
- 20.Moore D F, Curry J I, Knott C A, Jonas V. Amplification of rRNA for assessment of treatment response of pulmonary tuberculosis patients during antimicrobial therapy. J Clin Microbiol. 1996;34:1745–1749. doi: 10.1128/jcm.34.7.1745-1749.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piatek M, Jr, Saag M S, Yang L C, Clark S J, Kappes J C, Luk K-C, Hahn B H, Shaw G M, Lifson J D. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 22.Pfaller M A. Application of new technology to the detection, identification, and antimicrobial susceptibility testing of mycobacteria. Am J Clin Pathol. 1994;101:329–337. doi: 10.1093/ajcp/101.3.329. [DOI] [PubMed] [Google Scholar]
- 23.Piatak M, Jr, Luk K C, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–77. [PubMed] [Google Scholar]
- 24.Raeymaekers L. Quantitative PCR: theoretical considerations with practical implications. Anal Biochem. 1993;214:582–585. doi: 10.1006/abio.1993.1542. [DOI] [PubMed] [Google Scholar]
- 25.Scadden D T, Wang Z, Groopman J E. Quantitation of plasma human immunodeficiency virus type 1 RNA by competitive polymerase chain reaction. J Infect Dis. 1992;165:1119–1123. doi: 10.1093/infdis/165.6.1119. [DOI] [PubMed] [Google Scholar]
- 26.Souaze F, Ntodou-Thomi A, Tran C Y, Rostene W, Forgez P. Quantitative RT-PCR: limits and accuracy. BioTechniques. 1996;21:280–285. doi: 10.2144/96212rr01. [DOI] [PubMed] [Google Scholar]
- 27.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Soolinger D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang A M, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeager H, Jr, Lacy J, Smith L R, LaMaistre C A. Quantitative studies of mycobacterial populations in sputum and saliva. Am Rev Respir Dis. 1967;95:1004. doi: 10.1164/arrd.1967.95.6.998. [DOI] [PubMed] [Google Scholar]
- 31.Yuen K Y, Chan K S, Chan C M, Ho B S W, Dai L K, Chau P Y, Ng M H. Use of PCR in routine diagnosis of treated and untreated pulmonary tuberculosis. J Clin Pathol. 1993;46:318–322. doi: 10.1136/jcp.46.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuen K-Y, Chan K-S, Chan C-M, Ho P-L, Ng M-H. Monitoring the therapy of pulmonary tuberculosis by nested polymerase chain reaction assay. J Infect. 1997;34:29–33. doi: 10.1016/s0163-4453(97)80006-0. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann K, Mannhalter J W. Technical aspects of quantitative competitive PCR. BioTechniques. 1996;21:268–279. doi: 10.2144/96212rv01. [DOI] [PubMed] [Google Scholar]