Abstract
Background
Cervical cancer is a common gynecological malignancy. Gene microarray found that TCP11 gene was highly expressed in cervical cancer. However, the effect of TCP11 gene on the proliferation, apoptosis and migration of cervical cancer cells and its underlying molecular mechanisms are unclear.
Methods
GEPIA database, tissue microarray, western blot and qRT-PCR were used to analyze the expression of TCP11 gene in cervical cancer tissues and cells and its relationship with patients’ survival rate. The cell cycle and apoptosis were detected by flow cytometry, and the expressions of cell cycle and apoptosis related molecules and EMT-related molecules were detected by Western blot and qRT-PCR.
Results
The results showed that TCP11 gene was highly expressed in cervical cancer tissues and cells compared with normal cervical tissues and cells, and its expression was positively correlated with patients’ survival rate. The results of proliferation and migration assays showed that TCP11 overexpression inhibited the proliferation and migration of HeLa and SiHa cells. The results showed that TCP11 overexpression blocked the cell cycle of HeLa and SiHa cells, decreased the expression of CDK1 and Cyclin B1, and increased the apoptosis and the expression of caspase-3, cleaved-caspase-3 and cleaved-PARP. TCP11 overexpression increased the protein and mRNA expression of EMT-related molecules ZO-1 and E-cadherin. Conversely, TCP11 knockdown promoted the proliferation of HeLa and SiHa cells and the migration of HeLa cells.
Conclusions
TCP11 overexpression significantly inhibited the occurrence and development of cervical cancer cells, it may be a potentially beneficial biomarker for cervical cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-023-11129-1.
Keywords: Cervical cancer, TCP11 gene, Cell cycle, Apoptosis, Migration and proliferation
Background
Cervical cancer is a common gynecological malignant disease in women [1–4]. The morbidity and mortality of cervical cancer have gradually increased in China, and it has gradually become younger [5]. In 2020, there were about 604,000 newly diagnosed cases and 342,000 deaths of cervical cancer worldwide [6]. Human papilloma virus (HPV) infection is a common sexually transmitted disease [7], and more than 90% of cervical cancer is caused by persistent high-risk HPV (HR-HPV) infection [8, 9]. Cervical cancer has no obvious symptoms in the early stage of infection, which is easy to be ignored and misdiagnosed. Moreover, cervical cancer has a high degree of malignancy and is prone to metastasis and invasion [10]. Although the diagnosis and treatment of cervical cancer has been improved, and more than 90% of patients with cervical cancer in the early stage of infection can be cured, the survival rate of patients with advanced cervical cancer is still very low, especially for patients with metastatic cervical cancer [11]. Tumor invasion and metastasis are still important obstacles to the treatment of cervical cancer [12].
According to PSORT (https://psort.hgc.jp/) analysis, TCP11 protein may be mainly distributed in the cytoplasm. TCP11 is evolutionarily conserved in most metazoans and contains an uncharacterized protein domain, the TCP11 domain, that comprises most of the protein [13]. TCP11 gene is a human homologue of mouse Tcp11 gene, which can encode fertilization-promoting peptide (FPP) receptor, and there are three splicing products named TCP11a, TCP11b and TCP11c [14]. It has been reported that TCP11 protein plays a role in sperm capacitation and acrosome reaction, and can regulate the activity of adenylate cyclase cAMP pathway [15]. In addition, changes in DNA methylation of TCP11 gene are a characteristic of the sarcoma component of Uterine Carcinosarcoma (UCS) [16]. TCP11 is overexpressed in bilateral varicocele patients, which can be used as a potential biomarker for bilateral varicocele [17].
At present, there is no article report on TCP11 in cervical cancer. Interestingly, GEPIA database analysis showed that TCP11 gene was highly expressed in cervical cancer, and its high expression was beneficial to the prognosis of cervical cancer patients. But, the effect of TCP11 gene on the development of cervical cancer remains unclear. The purpose of this study was to evaluate the effect of TCP11 on proliferation, apoptosis and migration of cervical cancer cells. The study revealed that TCP11 may be a potential biomarker of cervical cancer, playing a role in the prevention, treatment and analysis of cervical cancer prognosis.
Materials and methods
The 22 K human genome array of CapitalBio
Four cases of cervical cancer and corresponding normal tissues adjacent to cancer were selected to extract tissue RNA by Trizol (Invitrogen, Gaithersburg, MD, USA) one-step method, and the total RNA was further purified by the NucleoSpin® RNA clean up kit (MACHEREY-NAGEL, Germany), quantified by spectrophotometer, and examined by formaldehyde denaturing gel electrophoresis. Fluorescent labeling of sample RNA (crystal core® CRNA amplification labeling kit). Hybridize and wash, and the labeled DNA is dissolved in 80 µL in the hybridization solution (3×SSC, 0.2% SDS, 5×Denhart’s, 25% formamide), hybridization at 42 ℃, overnight. After hybridization, 0.2% SDS, 2×Wash in liquid of SSC for 5 min, and then wash in 0.2×Wash in SSC at room temperature for 5 min. The slides can be used for scanning after being dried. The chip is scanned by LuxScan 10KA dual channel laser scanner (CapitalBio).
Gene expression profiling interactive analysis (GEPIA) database
GEPIA database (http://gepia.cancer-pku.cn/index.html), developed by Peking University team, was used for gene expression analysis of 9736 tumors and 8587 normal samples from TCGA and GTEx databases [18]. In this paper, GEPIA database was used to analyze the expression of TCP11 gene in cervical cancer and its relationship with survival rate.
Sample Collection
The tissue wax blocks, Hematoxylin-Eosin (HE) sections and relevant medical history information of patients in the First Affiliated Hospital of School of Medicine, Shihezi University, Xinjiang from 2017 to 2019 were collected, including 31 normal cervical tissues and 35 cervical cancer tissues. The clinical information can be seen in supplementary Table S1. All specimens were diagnosed by two experienced pathologists, and all patients provided written informed consent. It has been approved by the Medical Ethics Committee of the First Affiliated Hospital of Shihezi University Medical School (Approval Number: KJ2020-051-01).
Tissue microarray and immunohistochemistry
According to the HE sections of collected cervical tissue wax block, pathological experts marked the site of the cervical epithelium on the sections. Sampling was carried out according to the marker, and tissue microarray were made. The sections were deparaffinized with xylene and hydrated with alcohol, and then treated with citrate buffer for antigen-retrieval. After incubation in the dark in 3% H2O2, they were incubated with TCP11 antibody (Proteintech, China, 1:200) overnight. The next day, the sections were incubated with rabbit/mouse general secondary antibody (ZSGB-BIO, China) for 30 min. After DAB staining, the sections were stained with hematoxylin, dried and sealed, and the results were interpreted by pathological experts.
Cell origin and cell culture
Cervical cancer SiHa cell line and HeLa cell line were purchased from Wuhan Boster Biological Technology Co., Ltd. in 2015. Cervical cancer C33A cell line and immortalized normal epithelial HaCaT cell line were purchased from Wuhan Procell Life Science&Technology Co., Ltd. in 2019. HaCaT cells were cultured on Dulbecco’s Modified Eagle Medium (DMEM) containing 15% Fetal Bovine Serum (FBS) and 1% penicillomycin, and cervical cancer cells were cultured on DMEM containing 10% FBS and 1% penicillomycin. The cells were cultured in a 37 °C incubator with 5% CO2, and passaged at intervals of 2–3 days.
Cell transfection
The lentivirus overexpressing TCP11 gene was designed and constructed by Shanghai Genechem Co., LTD. Lentiviruses were transfected in cervical cancer HeLa and SiHa cells. After 72 h of infection, puromycin (Solarbio, China) was used to select the stably transfected cells. The lentvirus that overexpresses TCP11 gene is built with the transcript NM-001370687 (containing 503 amino acids) as the template. The selected GV341 vector is added with 3 Flag tags (each containing 8 amino acids), and the number of amino acids of TCP11 protein is 527. Therefore, after infection with lentivirus in cervical cancer cells, the molecular weight of TCP11 protein will increase, and the molecular weight size is about 58 kDa. The siRNA that interfered with TCP11 gene was designed and constructed by Shanghai GenePharma Co., Ltd. Cervical cancer HeLa cells were transfected with siRNA using Lipofectamine 2000 (Invitrogen, USA) reagent. TCP11 siRNA sequence is as follows:
siRNA-TCP11-1: GCCUGAGAAUUGAGAUUGATT;
siRNA-TCP11-2: GCAGCCUAGUCUCCUUAAUTT;
siRNA-TCP11-3: GCUCUAAGCAGUGAUAAUATT;
siRNA-Negative Control: UUCUCCGAACGUGUCACGUTT.
Western blot
Total cell protein was extracted by using cell lysate buffer (RIPA/ PMSF, Solarbio, China). The protein concentration was detected using BCA kit (Beyotime, China). Proteins were separated by SDS-PAGE electrophoresis, and transfered to PVDF membrane (Immobilon-P, USA). Then, the membrane strips were blocked with blocking solution (5% nonfat dry milk), cut according to different molecular weights of the target strips, and the primary antibody was incubated overnight at 4℃. After washing the membrane strips, the rabbit/mouse secondary antibody (ZSGB-BIO, China) was incubated at room temperature for 2 h. Finally, a chemiluminescence reagent (Thermo, USA) was added dropwise to the membrane strips, and a chemiluminescence instrument (Tanon, China) was used for exposure. Anti-GAPDH (TA-08) and anti-β-actin (TA-09) were purchased from ZSGB-BIO (China). Anti-TCP11 (14606-1-AP) and anti-caspase-3 (8193T) were purchased from Proteintech (China). Anti-cleaved-PARP (BSM-33,138 M), anti-CDK1 (BS-0542R), and anti-CyclinB1 (BS-0572R) were purchased from Bioss (China). Anti-ZO-1 (66470-2-IG), anti-Snail (C15D3), anti-Vimentin (D21H3), anti-β-catenin (D10A8), anti-Claudin-1 (D5H1D) and anti-E-cadherin (3195T) were purchased from Cell Signaling Technology (USA).
Real-time quantitative PCR (qRT-PCR)
Trizol lysate buffer (Ambion, USA) was used to extract total cell RNA. Using the reverse transcription kit (TaKaRa, Japan), 1ug of each group of RNA was reversely transcribed into cDNA, and then PCR amplification was performed with GAPDH as internal reference. The amplification conditions were 95℃ for 30s, 95℃ for 3s, 60℃ for 30s, 40 cycles. The mRNA relative expression of target gene was calculated by 2−ΔΔCt method. See Supplementary Table S2 for primer sequences.
Thiazole blue (MTT) assay
Cervical cancer cells transfected with lentivirus or siRNA for 24 h were digested with trypsin and counted. Each well of 96-well plates was seeded with 1000 cells, and 5 replicates were set up for each group. MTT reagent (Solarbio, China) was added into wells, and incubated in the incubator for 3 h. The liquid in the wells was carefully discarded, and added with DMSO (MACKLIN, China). The 96-well plate was incubated on the shaker for 15 min in darkness. Cell proliferation was measured at 490 nm with BIO-RAD (USA) at 0, 24, 48, 72 and 96 h, respectively.
Colony formation assay
Cervical cancer cells transfected with lentivirus or siRNA for 24 h were digested with trypsin and counted. Six-well plates were seeded with 500 cells/well. The culture was continued for about 2 weeks, and the medium was changed every 3–4 days. When the number of colony cells was > 50, or the cell colonies were visible to the naked eye, the culture was stopped. After washing the cells with PBS, they were fixed with 4% paraformaldehyde (Biosharp, China) for 30 min and stained with 0.1% crystal violet (Solarbio, China) for 30 min. Finally, the difference of colony number was analyzed by photographing and counting.
Cell drop slice
Cervical cancer cells with stable infection of lentivirus were collected, fixed with 4% paraformaldehyde for 20 min and then incubated with 0.1% Triton X-100 (Solarbio, China) for 15 min to increase cell permeability. After the cells were washed with PBS, an appropriate amount of PBS was added for resuspension, and the trace suspension was absorbed and added to the 12-well slides soaked in poly-lysine, so that the cells presented a monolayer and a density of about 70%. After drying, 12-well slides were stored at -20℃ for using in subsequent experiment. After the slides were immunochemically stained (Ki67, ZSGB-BIO, 1:400), 4 areas were randomly selected, and Image Pro Plus software was used to measure the integrated optical density (IOD) value and area value. Mean density = IOD/area was calculated, which could reflect the protein expression level.
Transwell transfer assay
Cell migration was measured using a 24-well plate with Transwell chambers (Corning Costar, USA). The cells were suspended in serum-free medium, and 200 µL cell suspension (100 cells/ml) was added into the upper chamber, and 600 µL DMEM medium containing 10% FBS was added into the lower chamber. After incubating for 36 h in an incubator at 37℃ and 5% CO2, they were fixed with 4% paraformaldehyde, stained with crystal violet, washed with water and dried. The number of transmembrane cells was counted in 9 fields of view (×400) randomly taken from each chamber membrane under a high magnification microscope.
Cell scratch test
4 × 105 cells transfected with lentivirus were collected and inoculated in 6-well plates for culture. When the cells have grown to about 80% of the cell density, the cells were scratched in a straight using 200 µL pipette tip perpendicular to the bottom. The inverted microscope was used to take pictures at 0, 24 and 48 h, trying to keep the same position.
Cell cycle assay
Cervical cancer cells stably infected with lentivirus were collected and immobilized with pre-cooled 70% ethanol according to the instructions of the Cell Cycle Detection Kit (LIANKE, China). On the day of detection, 1mL DNA Staining Solution was added and let stand for 30 min avoiding light at room temperature. The cell cycle distribution was detected and analyzed by flow cytometry, and > 10,000 cells were collected per sample.
Cell apoptosis assay
Cervical cancer cells stably infected with lentivirus were collected. According to the instructions of apoptosis Detection Kit (LIANKE, China), apoptosis was detected by flow cytometry using Annexin V-FITC and PI double staining.
Statistical analysis
All data input was organized by Excel software, processed and analyzed by SPSS 22.0. Statistics of measurement data were expressed as  ± s, differences between groups were tested by t test, and tissue microarray results were tested by χ2 test. P < 0.05 was considered to be statistically significant.
± s, differences between groups were tested by t test, and tissue microarray results were tested by χ2 test. P < 0.05 was considered to be statistically significant.
Results
TCP11 is highly expressed in cervical cancer tissues and cells, which is closely related to the survival rate of cervical cancer patients
In the early stage, the research group collected 4 cases of cervical cancer and corresponding adjacent normal tissues to make a gene microarray (The 22 K Human Genome Array of CapitalBio) to screen differentially expressed genes. The previous gene microarray results found that the expression of TCP11 gene in cervical cancer tissue was significantly higher than that in normal cervical tissue (Table 1). However, TCP11 gene was not reported in cervical cancer, which aroused our research interest. To initially explore the role of TCP11 in cervical cancer, we used GEPIA database for predictive analysis. The results showed that the mRNA expression of TCP11 in cervical cancer tissues was higher than that in normal cervical tissues (Fig. 1A). Immunohistochemical staining was performed on the tissue to confirm the predicted results. The expression of TCP11 protein in cervical cancer tissues was higher than that in normal cervical tissues. In 31 cases of normal cervical tissue, the weak positive rate of TCP11 expression was 48.39%, and the negative rate was 51.61%; in 35 cases of cervical cancer tissue, the positive rate of TCP11 expression was 14.29%, the weak positive rate was 57.14%, and the negative rate was 28.57% (Fig. 1B; Table 2). In addition, western blot and qRT-PCR were used to detect the expression of TCP11 protein and mRNA in immortalized epithelial cells HaCaT and cervical cancer HeLa, SiHa and C33A cells, respectively. Compared with immortalized epithelial HaCaT cells, TCP11 protein and mRNA were highly expressed in three cervical cancer cell lines (Fig. 1C and D). Interestingly, based on the GEPIA database, we found that cervical cancer patients with high expression of TCP11 had significantly higher overall survival than those with low expression (Fig. 1E). These results indicate that TCP11 gene may play an important role in the development of cervical cancer.
Table 1.
The statistical results of TCP11 gene expression were analyzed by gene expression chip
| Name | Mean | 1 + 2 | 3 + 4 | 3CA + 3CA side | 5CA + 5CA side |
|---|---|---|---|---|---|
| TCP11 | 6.3800 | 3.6360 | 5.9840 | 3.0817 | 12.8181 |
The “Mean” value greater than 2.0 indicates that the gene is highly expressed in cancer tissues. “1” represents cervical cancer tissue, and “2” represents corresponding adjacent tissue of cervical cancer; “3” represents cervical cancer tissue, and “4” represents cervical cancer corresponding adjacent tissue; “3CA” and “3CA side” represent cervical cancer and its corresponding adjacent tissues respectively; “5CA” and “5CA side” represent cervical cancer and its corresponding adjacent tissues respectively.
Fig. 1.
TCP11 gene is highly expressed in cervical cancer tissues and cells, and is closely associated with patients’ survival rate. A: The mRNA expression of TCP11 in normal cervical tissues and cervical cancer tissues were analyzed by GEPIA database. B: Immunohistochemical results of TCP11 protein expression in normal cervical tissues (n = 31) and cervical cancer tissues (n = 35) microarray. “-” indicates that the expression of TCP11 is negative; “+” indicates that the expression of TCP11 is weakly positive; “++” indicates that the expression of TCP11 is medium positive. Magnification: ×100 (top) and ×400 (bottom). C: Western blot was used to detect the expression of TCP11 protein in immortalized epithelial cells HaCaT and three cervical cancer cells. Full-length blots/gels are presented in Supplementary Figure S1. D: qRT-PCR was used to detect the expression of TCP11 mRNA in immortalized epithelial cells HaCaT and three cervical cancer cells. E: GEPIA database was used to analyze the relationship between the expression of TCP11 and survival rate of cervical cancer patients. Data are presented as mean ± SD of at least 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Table 2.
Statistical table of immunohistochemical results of TCP11 in normal cervix and cervical cancer tissues
| Sample | - | + | ++ | Total | P value |
|---|---|---|---|---|---|
| Normal cervical tissue | 16 | 15 | 0 | 31 | 0.019 |
| Cervical cancer tissue | 10 | 20 | 5 | 35 |
“-” indicates that the expression of TCP11 is negative; “+” indicates that the expression of TCP11 is weakly positive; “++” indicates that the expression of TCP11 is medium positive
TCP11 overexpression inhibits the proliferation of cervical cancer cells
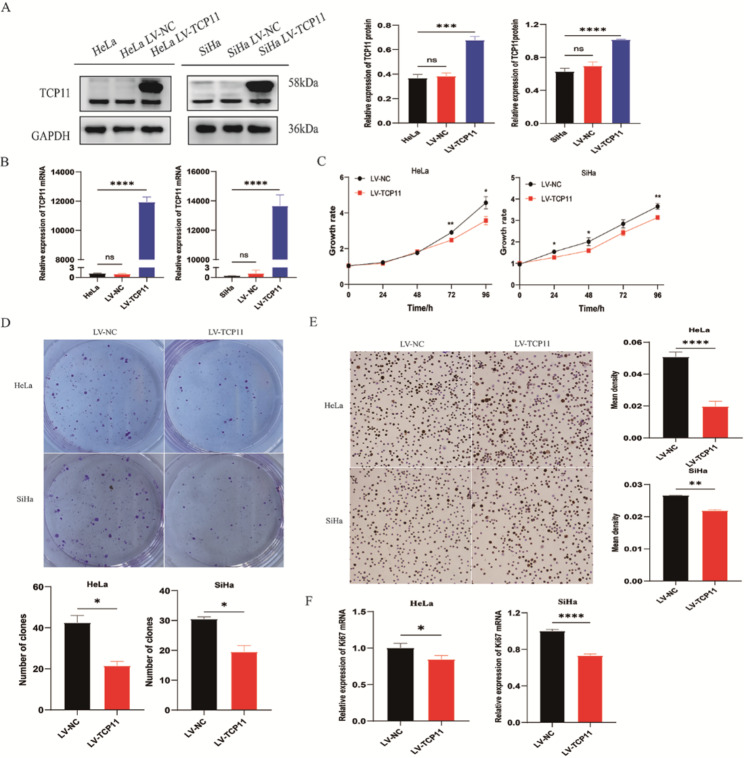
To confirm the role of TCP11 in cervical cancer, we first investigated whether overexpression of TCP11 by lentivirus infection had an effect on the proliferation of cervical cancer cells. After lentivirus infection with cervical cancer HeLa and SiHa cells, the expression of TCP11 protein and mRNA was detected to confirm the successful selection of stably transfected cells overexpressing TCP11 (Fig. 2A and B). MTT assay showed that TCP11 overexpression significantly inhibited the cell viability of HeLa and SiHa cells (Fig. 2C). This result was further confirmed by the colony formation assay. Compared with the NC group, the number of colony formation in the overexpression group was significantly reduced (Fig. 2D). In addition, we analyzed the protein and mRNA expression of the proliferation marker Ki67 by using cellular immunohistochemistry and qRT-PCR. TCP11 overexpression significantly inhibited the expression of Ki67 in cervical cancer cells (Fig. 2E and F). These results suggest that TCP11 plays an important role in inhibiting the proliferation of cervical cancer cells.
Fig. 2.
TCP11 overexpression inhibits the proliferation of HeLa and SiHa cells. A: Western blot was used to detect the overexpression of TCP11 protein in HeLa and SiHa cells after lentivirus infection. Full-length blots/gels are presented in Supplementary Figure S2. B: qRT-PCR was used to detect the overexpression of TCP11 mRNA in HeLa and SiHa cells after lentivirus infection. C and D: The cell viability of HeLa and SiHa cells was detected by MTT and colony formation assays. E: The protein expression of Ki67 in HeLa and SiHa cells was detected by cellular immunohistochemistry. Magnification: ×100. The results were analyzed by Image Pro Plus software, and the mean density was calculated (mean density = IOD/area). F: Ki67 mRNA expression in HeLa and SiHa cells were detected by qRT-PCR. Data are presented as mean ± SD of at least 3 independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001
TCP11 overexpression blocks the cell cycle progression of cervical cancer cells
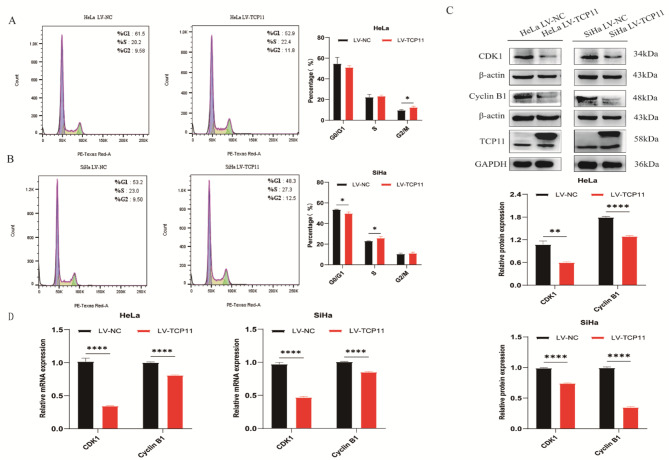
To investigate the reason why TCP11 inhibits the proliferation of cervical cancer cells, flow cytometry was used to analyze the effect of TCP11 overexpression on cell cycle progression. The results showed that the number of HeLa cells in G2/M phase in the overexpressed group increased compared with the NC group (Fig. 3A). The number of SiHa cells in S phase increased, and the number of SiHa cells in G2/M phase also increased, but there was no statistical significance (Fig. 3B).
Fig. 3.
TCP11 overexpression blocks cell cycle progression in HeLa and SiHa cells. A and B: Cell cycle distribution of HeLa and SiHa cells was determined by flow cytometry. C: Western blot was used to detect the protein expression of CDK1 and Cyclin B1 in HeLa and SiHa cells infected with overexpressing TCP11 lentivirus. Full-length blots/gels are presented in Supplementary Figure S3. D: qRT-PCR was used to detect the mRNA expression of CDK1 and Cyclin B1 in HeLa and SiHa cells infected with overexpressing TCP11 lentivirus. Data are presented as mean ± SD of at least 3 independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001
To further explore the potential reasons of TCP11 blocking cell cycle, western blot and qRT-PCR were used to detect the expression changes of cell cycle-related molecules CDK1 and Cyclin B1. After overexpression of TCP11, the protein and mRNA expressions of CDK1 and Cyclin B1 were decreased in HeLa and SiHa cells (Fig. 3C and D). The above results indicate that TCP11 may block the cell cycle by inhibiting the expression of CDK1/Cyclin B1, thereby inhibiting the proliferation of cervical cancer cells.
TCP11 overexpression promotes apoptosis of cervical cancer cells
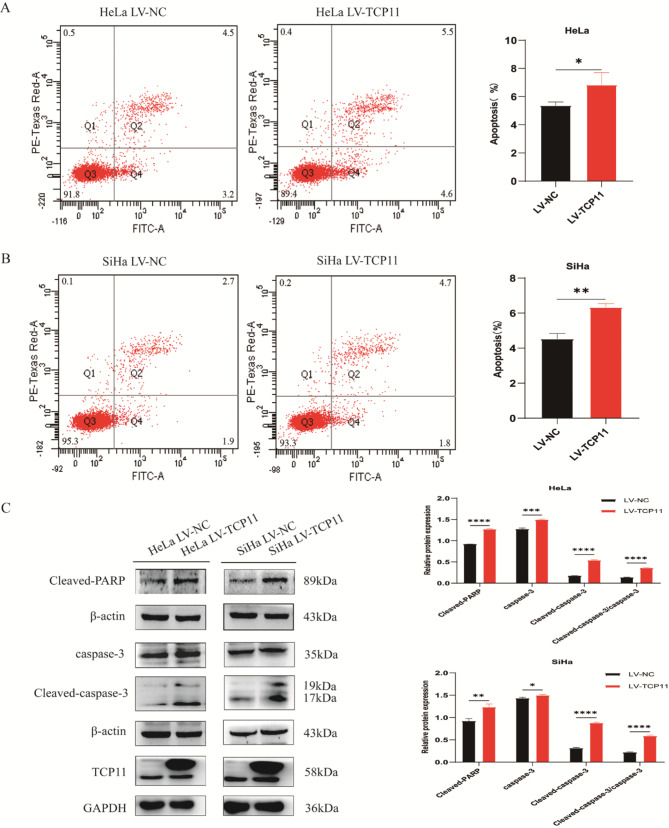
Flow cytometry was used to analyze whether the expression of TCP11 has an effect on the apoptosis of cervical cancer cells. The results showed that the apoptosis of HeLa and SiHa cells increased in the overexpressed group compared with the NC group (Fig. 4A and B). In order to better understand the potential reasons of TCP11 promoting apoptosis of cervical cancer cells, we used western blot to analyze the protein expression of apoptosis-related molecules caspase-3, cleaved-caspase-3 and cleaved-PARP. The results showed that TCP11 overexpression significantly increased the expression of caspase-3, cleaved-caspase-3 and cleaved-PARP (Fig. 4C).
Fig. 4.
TCP11 overexpression induces the apoptosis of HeLa and SiHa cells. A and B: Apoptosis of HeLa and SiHa cells were detected by flow cytometry. C: Western blot was used to detect the protein expression of caspase-3, cleaved-caspase-3 and cleaved-PARP in HeLa and SiHa cells infected with overexpressing TCP11 lentivirus. Full-length blots/gels are presented in Supplementary Figure S4. Data are presented as mean ± SD of at least 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
TCP11 overexpression inhibits cervical cancer cell migration by inhibiting EMT of cervical cancer cells
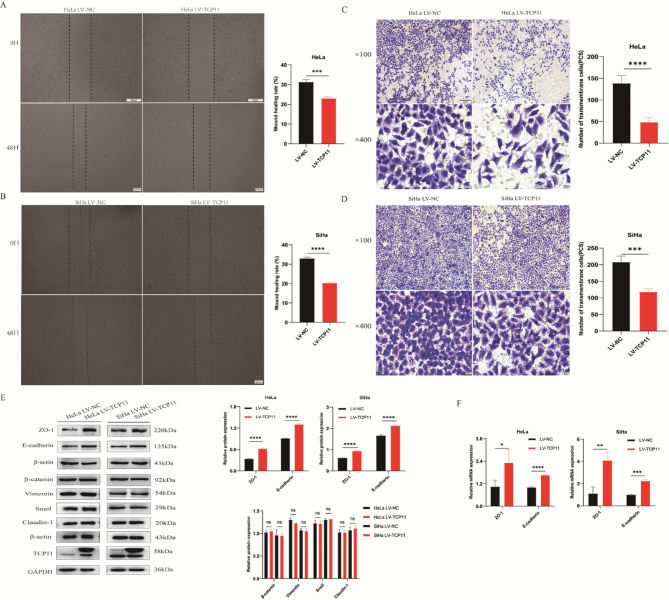
Migration is one of the key steps in tumor cells metastasis. Inhibition of cell migration can effectively prevent tumor progression. Cell scratch assay showed that TCP11 overexpression significantly inhibited the scratch healing rate of HeLa and SiHa cells (Fig. 5A and B). In addition, transwell migration assay showed that the number of transmembrane cells of HeLa and SiHa cells in the overexpressed group was significantly lower than that in the NC group (Fig. 5C and D). These results indicate that TCP11 plays an important role in inhibiting cell migration.
Fig. 5.
TCP11 overexpression inhibits cervical cancer cell migration by inhibiting EMT of HeLa and SiHa cells. A and B: Cell scratch assay was used to detect the migration ability of HeLa and SiHa cells. C and D: Transwell migration assay was used to detect the migration ability of HeLa and SiHa cells. E: Western blot was used to detect the protein expression of EMT-related molecules ZO-1, E-cadherin, Claudin-1, Snail, Vimentin and β-catenin in HeLa and SiHa cells infected with overexpressing TCP11 lentivirus. Full-length blots/gels are presented in Supplementary Figure S5. F: qRT-PCR was used to detect the mRNA expression of EMT-related molecules ZO-1 and E-cadherin in HeLa and SiHa cells infected with overexpressing TCP11 lentivirus. Data are presented as mean ± SD of at least 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
EMT is the key to the migration and invasion of malignant tumor cells, and the process of EMT plays an important role in the development of cervical cancer. Analysis of the expression changes of EMT-related molecules could reveal the potential reasons of TCP11 gene affecting cervical cancer cell migration. Our results showed that the protein and mRNA expressions of ZO-1 and E-cadherin were significantly increased after overexpression of TCP11 in HeLa and SiHa cells, but Claudin-1, Snail, Vimentin and β-catenin showed no significant difference (Fig. 5E and F). This suggests that TCP11 may inhibit the migration of cervical cancer cells by increasing tight junctions and adhesion between cells.
TCP11 knockdown promotes the proliferation and migration of cervical cancer HeLa cells
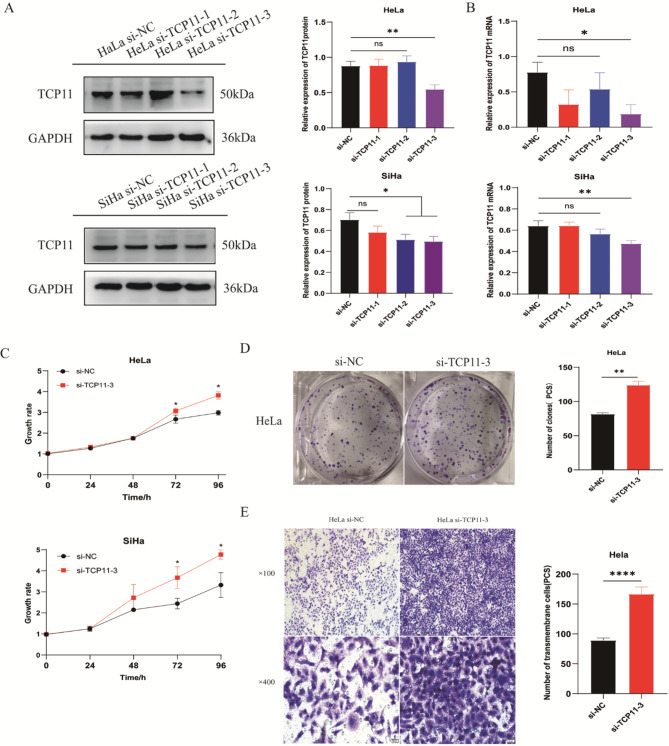
To further confirm the inhibitory effect of TCP11 on the proliferation and migration of cervical cancer cells, three siRNA fragments targeting different positions were transfected into cervical cancer HeLa and SiHa cells, and it was found that si-TCP11-3 successfully knocked down the expression of TCP11 in HeLa and SiHa cells (Fig. 6A and B). MTT assay showed that the cell viability of HeLa and SiHa cells in the knockdown group was significantly higher than that in the NC group (Fig. 6C). Colony formation assay also showed that TCP11 knockdown increased the colony number of HeLa cells (Fig. 6D). Transwell migration assay showed that TCP11 knockdown significantly increased the mobility of HeLa cells (Fig. 6E).
Fig. 6.
TCP11 knockdown promotes the proliferation of HeLa and SiHa cells and the migration of HeLa cells. A: Western blot was used to detect the knockdown effect of three designed and constructed siRNA fragments on TCP11 in HeLa and SiHa cells. Full-length blots/gels are presented in Supplementary Figure S6. B: qRT-PCR was used to detect the knockdown effect of three siRNA fragments on TCP11 in HeLa and SiHa cells. C: MTT assay was used to detect the effect of TCP11 knockdown on the proliferation of HeLa and SiHa cells. D: Colony formation assay was used to detect the effect of TCP11 knockdown on the proliferation of HeLa cells. E: Transwell migration assay was used to detect the effect of TCP11 knockdown on the migration of HeLa cells. Data are presented as mean ± SD of at least 3 independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001
Discussion
According to Uniprot database (https://www.uniprot.org/), TCP11 gene is related to cell differentiation, signal transduction, multicellular biological development and protein kinase A signal transduction. In this study, through GEPIA database and tissue microarray assay, we demonstrated that the expression of TCP11 gene was higher in cervical cancer tissues than in normal cervical tissues. It is worth noting that the TCP11 had low expression in testicular germ cell tumor in GEPIA database, which was contrary to the high expression in cervical cancer. This may be because TCP11 gene is a testicle-specific gene product and enriched in normal testicle, but the expression of TCP11 is reduced when the testis becomes cancerous, for reasons that are not clear. Interestingly, cervical cancer patients with high expression of TCP11 had higher survival rates. This suggests that TCP11 may be a prognostic indicator for cervical cancer patients. Furthermore, we found that TCP11 overexpression significantly inhibited the proliferation and migration of cervical cancer HeLa and SiHa cells. Conversely, the low expression of TCP11 promoted the cell proliferation in HeLa and SiHa cells. Knocking down the expression level of TCP11 gene can promote the cell migration in HeLa cells. TCP11 gene can affect the proliferation and migration of cervical cancer cells, possibly by affecting the expression of cell cycle related molecules and EMT related molecules. Therefore, we next analyzed the characteristics of the TCP11 gene acting on these molecules.
It has been shown that the disorder of cell cycle regulation is one of the main factors leading to the proliferation of malignant cells [19]. We found that overexpression of TCP11 arrested the cell cycle of cervical cancer cells, HeLa cells were arrested in G2/M phase, SiHa cells were arrested in S phase. Cyclin B1 is a cyclin-related protein of the M phase, which can bind to CDK1 and activate CDK1 to realize the cell cycle transition from G2 phase to M phase [20, 21]. The results showed that TCP11 overexpression inhibited the protein and mRNA expressions of CDK1 and Cyclin B1. We speculate that TCP11 gene may block the cell cycle by regulating the expression of CDK1/Cyclin B1, thereby inhibiting the proliferation of cervical cancer cells. In addition, we also explored the effect of TCP11 on apoptosis of cervical cancer cells. TCP11 gene may induce cell apoptosis by activating caspase-3, further activating its substrate PARP. EMT is the key to migration and invasion of malignant tumor cells [22, 23]. To further investigate whether TCP11 inhibits the migration of cervical cancer cells by regulating EMT, we detected the protein and mRNA expressions of EMT-related molecules ZO-1 and E-cadherin. The results showed that TCP11 overexpression can inhibit the EMT process by increasing the protein and mRNA expressions of ZO-1 and E-cadherin, thereby inhibiting the migration of cervical cancer cells. Further research is needed on the proliferation and migration effects of TCP11 gene on cervical cancer cells.
According to UniProt database analysis, TCP11 gene can regulate the activity of cAMP pathway of adenylyl cyclase. cAMP as a second messenger can activate cAMP-dependent protein kinase (PKA) [24, 25]. cAMP-PKA signaling pathway can regulate many cellular responses, including proliferation, apoptosis, migration and metabolism [26–28]. It has been shown that cAMP/PKA signaling pathway can inhibit the migration of cervical cancer HeLa cells [29]. TCP11 has been reported to be correlated with PKA signal transduction [30]. Therefore, we speculated whether TCP11 gene could inhibit the progression of cervical cancer cells by regulating the activity of cAMP/PKA signaling pathway. However, further studies are needed to verify this hypothesis.
Conclusions
In summary, our results suggest that TCP11 plays an important role in the development of cervical cancer. TCP11 overexpression can inhibit the proliferation and migration of cervical cancer cells, block cell cycle and induce apoptosis. In addition, TCP11 overexpression inhibited cell migration by inhibiting EMT-related molecules. TCP11 may be a potentially beneficial biomarker for cervical cancer. However, we still need to further study which specific signaling pathway can regulate the inhibitory effect of TCP11 on cervical cancer progression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We acknowledge that this work was supported in part by the National Natural Science Foundation of China (grant number: 82060518, U1503125); International Science and Technology Collaboration Projector of Xinjiang Production and Construction Corps (grant number: 2019BC007).
Abbreviations
- GEPIA
Gene Expression Profiling Interactive Analysis
- HE
Hematoxylin-Eosin
- DMEM
Dulbecco’s Modified Eagle Medium
- FBS
Fetal Bovine Serum
- qRT-PCR
Real-time fluorescent quantitative polymerase chain reaction
- ICC
Immunocytochemistry
- HPV
Human papilloma virus
- UCS
Uterine Carcinosarcoma
- PARP
Poly ADP ribose polymerase
- EMT
Epithelial-Mesenchymal Transition
- ZO-1
Zonula occludens-1
- CDK1
Cyclin-dependent kinases
- cAMP
Cyclic adenosine monophosphate
- PKA
Protein kinase A
Authors’ contributions
FW, SYS and BXG made main contributions to the document, designed the research, and participated in specimen collection, sorting data, data analysis and drafting the manuscript; YYL and HJW assisted in specimen collection; SWF and LYW assisted in processing the resulting images; HTL and DML were mainly responsible for guiding data analysis; RFS, XYZ and ZMP provided substantial guidance, supported help in modifying article and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China (grant number: 82060518, U1503125); International Science and Technology Collaboration Projector of Xinjiang Production and Construction Corps (grant number: 2019BC007).
Data Availability
GEPIA (http://gepia.cancer-pku.cn/index.html) and Uniprot database(https://www.uniprot.org/) were used to predict the expression and function of TCP11. TCP11 protein distribution results from PSORT (https://psort.hgc.jp/). All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Declarations
Ethics approval and consent to participate
The sample collection for this study was approved by the Ethical Committee of the First Affiliated Hospital of the Medical College of Shihezi University (Approval Number: KJ2020-051-01), and informed consent was obtained from all of the participants. All methods were carried out in accordance with relevant institutional guidelines and regulations. All methods were carried out in accordance with declaration of Helsinki.
Consent for publication
No applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fang Wang, Shuyan Song and Bingxuan Guo contributed equally to this work.
Contributor Information
Xiangyi Zhe, Email: 342480821@qq.com.
Zemin Pan, Email: panteacher89@sina.com.
References
- 1.Liu C, Wang X, Zhang Y. The roles of HK2 on Tumorigenesis of Cervical Cancer. Technol Cancer Res Treat. 2019;18:1533033819871306. doi: 10.1177/1533033819871306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu D, Dong P, Xiong Y, Yue J, Konno Y, Ihira K, et al. MicroRNA-361-Mediated inhibition of HSP90 expression and EMT in Cervical Cancer is counteracted by oncogenic lncRNA NEAT1. Cells. 2020;9:632. doi: 10.3390/cells9030632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Buskwofie A, David-West G, Clare CA. A review of Cervical Cancer: incidence and disparities. J Natl Med Assoc. 2020;112(2):229–32. doi: 10.1016/j.jnma.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Hu Z, Ma D. The precision prevention and therapy of HPV-related cervical cancer: new concepts and clinical implications. Cancer Med. 2018;7(10):5217–36. doi: 10.1002/cam4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 7.Trottier H, Franco EL. Human papillomavirus and cervical cancer: burden of illness and basis for prevention. Am J Manag Care. 2006;12(17 Suppl):462–72. [PubMed] [Google Scholar]
- 8.Arbyn M, Weiderpass E, Bruni L, de Sanjosé S, Saraiya M, Ferlay J, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health. 2020;8(2):e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Xu Y, Jiang W, Ji H, Wang ZW, Zhu X. Discovery of key genes as novel biomarkers specifically associated with HPV-negative cervical cancer. Mol Ther Methods Clin Dev. 2021;21:492–506. doi: 10.1016/j.omtm.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JW, Hanson V, Johnson GD, Royalty JE, Richardson LC. From cancer screening to treatment: service delivery and referral in the national breast and cervical Cancer Early Detection Program. Cancer. 2014;120(0 16):2549–56. doi: 10.1002/cncr.28823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Ye X, Li D, Yao Q, Li Y. Incidence, clinical risk and prognostic factors for liver metastasis in patients with cervical cancer: a population-based retrospective study. BMC Cancer. 2021;21(1):421. doi: 10.1186/s12885-021-08127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Wu X, Cheng X. Advances in diagnosis and treatment of metastatic cervical cancer. J Gynecol Oncol. 2016;27(4):e43. doi: 10.3802/jgo.2016.27.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castaneda JM, Miyata H, Archambeault DR, Satouh Y, Yu Z, Ikawa M, et al. Mouse t-complex protein 11 is important for progressive motility in sperm†. Biol Reprod. 2020;102(4):852–62. doi: 10.1093/biolre/ioz226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Jiang M, Li C, Yang P, Sun H, Tao D, et al. Human t-complex protein 11 (TCP11), a testis-specific gene product, is a potential determinant of the sperm morphology. Tohoku J Exp Med. 2011;224(2):111–7. doi: 10.1620/tjem.224.111. [DOI] [PubMed] [Google Scholar]
- 15.Adeoya-Osiguwa SA, Dudley RK, Hosseini R, Fraser LR. FPP modulates mammalian sperm function via TCP-11 and the adenylyl cyclase/cAMP pathway. Mol Reprod Dev. 1998;51(4):468–76. doi: 10.1002/(SICI)1098-2795(199812)51:4<468::AID-MRD14>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Xing X, Li D, Zhang B, Mutch DG, Hagemann IS, et al. Whole-genome DNA methylation profiling identifies epigenetic signatures of Uterine Carcinosarcoma. Neoplasia. 2017;19(2):100–11. doi: 10.1016/j.neo.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal A, Sharma R, Durairajanayagam D, Cui Z, Ayaz A, Gupta S, et al. Spermatozoa protein alterations in infertile men with bilateral varicocele. Asian J Androl. 2016;18(1):43–53. doi: 10.4103/1008-682X.153848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leal-Esteban LC, Fajas L. Cell cycle regulators in cancer cell metabolism. Biochim Biophys Acta Mol Basis Dis. 2020;1866(5):165715. doi: 10.1016/j.bbadis.2020.165715. [DOI] [PubMed] [Google Scholar]
- 20.Farshadi E, Yan J, Leclere P, Goldbeter A, Chaves I, van der Horst GTJ. The positive circadian regulators CLOCK and BMAL1 control G2/M cell cycle transition through cyclin B1. Cell Cycle. 2019;18(1):16–33. doi: 10.1080/15384101.2018.1558638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Łukasik P, Załuski M, Gutowska I. Cyclin-dependent kinases (CDK) and their role in Diseases Development-Review. Int J Mol Sci. 2021;22(6):2935. doi: 10.3390/ijms22062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amankwah EK, Lin HY, Tyrer JP, Lawrenson K, Dennis J, Chornokur G, et al. Epithelial-mesenchymal transition (EMT) gene variants and epithelial ovarian Cancer (EOC) risk. Genet Epidemiol. 2015;39(8):689–97. doi: 10.1002/gepi.21921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otani T, Furuse M. Tight Junction structure and function revisited. Trends Cell Biol. 2020;30(10):805–17. doi: 10.1016/j.tcb.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Sassone-Corsi P. The cyclic AMP pathway. Cold Spring Harb Perspect Biol. 2012;4(12). 10.1101/cshperspect.a011148. [DOI] [PMC free article] [PubMed]
- 25.Naviglio S, Caraglia M, Abbruzzese A, Chiosi E, Di Gesto D, Marra M, et al. Protein kinase A as a biological target in cancer therapy. Expert Opin Ther Targets. 2009;13(1):83–92. doi: 10.1517/14728220802602349. [DOI] [PubMed] [Google Scholar]
- 26.Chin KV, Yang WL, Ravatn R, Kita T, Reitman E, Vettori D, et al. Reinventing the wheel of cyclic AMP: novel mechanisms of cAMP signaling. Ann N Y Acad Sci. 2002;968:49–64. doi: 10.1111/j.1749-6632.2002.tb04326.x. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed MB, Alghamdi AAA, Islam SU, Lee JS, Lee YS. cAMP signaling in Cancer: a PKA-CREB and EPAC-Centric Approach. Cells. 2022;11(13):2020. doi: 10.3390/cells11132020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapio L, Di Maiolo F, Illiano M, Esposito A, Chiosi E, Spina A, Naviglio S. Targeting protein kinase A in cancer therapy: an update. EXCLI J. 2014;13:843–55. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JW, Lee J, Moon EY. HeLa human cervical cancer cell migration is inhibited by treatment with dibutyryl-cAMP. Anticancer Res. 2014;34(7):3447–55. [PubMed] [Google Scholar]
- 30.Stanger SJ, Law EA, Jamsai D, O’Bryan MK, Nixon B, McLaughlin EA et al. (2016). A novel germ cell protein, SPIF (sperm PKA interacting factor), is essential for the formation of a PKA/TCP11 complex that undergoes conformational and phosphorylation changes upon capacitation. FASEB J. 2016;30(8):2777-91. doi: 10.1096/fj.201500136R. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GEPIA (http://gepia.cancer-pku.cn/index.html) and Uniprot database(https://www.uniprot.org/) were used to predict the expression and function of TCP11. TCP11 protein distribution results from PSORT (https://psort.hgc.jp/). All data generated or analyzed during this study are included in this published article (and its supplementary information files).