Abstract
Phospholipase Ds (PLDs) are multifunctional and disease-relevant enzymes operating at the center of phospholipid metabolism and signaling. Physiologically, they hydrolyze abundant phospholipids into phosphatidic acid (PA), a potent lipid second messenger and central biosynthetic intermediate. Given the pleiotropic nature of PA, the multiple locations of PLD activity within single cells, and differences in PLD activities across cell types in vivo, tools with spatiotemporal precision are urgently needed to dissect the signaling functions of PLDs. Here, we describe a toolset for visualizing and quantifying cellular PLD activity with high spatial and temporal resolution. Our approach capitalizes on the ability of PLDs to catalyze transphosphatidylation reactions with exogenous alcohols to generate phosphatidyl alcohols, lipids whose location and abundance report on the extent of PLD-mediated PA synthesis. Our key innovation is to employ functionalized, “clickable,” alcohols as PLD substrates, which enables subsequent tagging of the resultant phosphatidyl alcohols with fluorophores or other functional probes for detection via highly selective click chemistry reactions. In this chapter, we describe this method, termed IMPACT (Imaging PLD Activity with Clickable Alcohols via Transphosphatidylation), which can be coupled to downstream analysis by fluorescence microscopy, flow cytometry, HPLC, or mass spectrometry. We describe two variants of IMPACT, one with greater sensitivity, for detecting PLD activity at single-cell and population levels, and one with greater spatiotemporal resolution (“real-time,” or RT-IMPACT), for accurately visualizing PLD activity at the subcellular, individual-organelle level. Together, IMPACT represents a major advance in our ability to dissect PLD-mediated PA signaling in native biological settings.
1. Introduction
Lipids are fascinating, structurally diverse, and vitally important biomolecules. They comprise the key membrane barriers between cells and the surrounding environment, serve as primary energy stores, and play numerous signaling roles. Despite this tremendous physiological importance, lipids have proven exceptionally challenging to study due to their redundant biosynthetic pathways, their nature as non-genetically encoded metabolites, and their immiscibility with aqueous environments (Bumpus & Baskin, 2018). In addition to highly abundant lipids such as cholesterol, phosphatidylcholine, and sphingomyelin, which are essential for membrane structure and cellular homeostasis, cells generate a variety of short-lived and/or low-abundance lipid second messengers in a spatiotemporally controlled manner, which precisely intersect and interact with the cell’s signaling cascades (Hannun & Obeid, 2008; Sunshine & Iruela-Arispe, 2017).
Phosphatidic acid (PA) is a pleiotropic lipid second messenger that elicits a variety of changes in cell behaviors. Aberrant PA-dependent signaling has been linked to several pathological conditions, including many cancers (Brown, Thomas, & Lindsley, 2017; Nelson & Frohman, 2015). A full understanding of the precise physiological effects of localized PA production requires a robust set of methods for visualizing and perturbing PA biosynthesis. Despite the widespread and sustained interest in the biology of PA, there remain a number of challenges associated with studying this lipid, which can be produced by three distinct biosynthetic enzyme families: lysophosphatidic acid acyltransferases, diacylglycerol kinases, and phospholipase Ds (PLD) (Selvy, Lavieri, Lindsley, & Brown, 2011; Shulga, Topham, & Epand, 2011). First, affinity-based biosensors for PA exhibit only moderate specificity and frequently rely on coincidence detection of other lipids or proteins within the membrane (Kassas et al., 2017). Critically, even in an idealized form, biosensors cannot differentiate newly synthesized PA from the total pool, nor can they distinguish between pools of PA generated by each of the three biosynthetic pathways (Bumpus & Baskin, 2018). Second, whereas fluorescently tagging specific PA-producing enzymes may mitigate the aforementioned challenges, this approach fails to reveal the active subset of enzymes, many of which exhibit dynamic localizations after stimulation (Du et al., 2003).
To address these challenges, we sought to develop tools that would enable visualization and quantification of PA production by a specific pathway. We focused our efforts on PA generated by PLD enzymes, which are important in both physiological and pathological cell signaling (Nelson & Frohman, 2015). PLDs generate PA by the hydrolysis of phosphatidylcholine, the most abundant phospholipid in cellular membranes, upon activation by several upstream factors, including many families of cell-surface receptors and intracellular GTPases and kinases (Du et al., 2000; Hammond et al., 1997; Selvy et al., 2011). Classic biochemical assays for PLD activity, described in previous volumes in this periodical (Brown, Henage, Preininger, Xiang, & Exton, 2007; Philip, Ha, Seeliger, & Frohman, 2017), rely on a second reaction that PLDs can catalyze, transphosphatidylation, wherein the water molecule that performs hydrolysis is replaced with an exogenous primary alcohol, enabling PLD-mediated generation of phosphatidyl alcohol lipids, whose abundance reports directly on PLD activity. Typically, these transphosphatidylation-based methods use radio- or stable isotope-labeling, followed by bulk biochemical measurements (e.g., thin layer chromatography or mass spectrometry) (Brown et al., 2007; Philip et al., 2017; Selvy et al., 2011). Thus, whereas the phosphatidyl alcohols are generated in cells at precise subcellular membrane locations where endogenous PLDs are activated, this spatial information is lost during sample workup steps.
We sought to develop a method based on this ability of PLDs to perform transphosphatidylation reactions that would preserve and reveal the localizations of PLD activity, at the single-cell and even subcellular levels. To accomplish this goal, we devised a two-step labeling strategy termed IMPACT (Imaging Phospholipase D Activity with Clickable Alcohols via Transphosphatidylation) (Bumpus & Baskin, 2016, 2017; Bumpus, Liang, & Baskin, 2018; Liang et al., 2019). Our method uses a functionalized primary alcohol for the transphosphatidylation reaction that enables us to subsequently install a fluorescent tag or other detection reagent, via a click chemistry reaction, to the phosphatidyl alcohol reporter, within intact cells. Thus, the two-step IMPACT protocol enables the visualization of membranes containing active PLD enzymes within intact and−in many variants of the protocol−live cells.
Here, we describe two versions of IMPACT that we have optimized for different applications for quantifying and imaging PLD activity (Fig. 1). First, we describe our original version of IMPACT, using azide-containing alcohols for transphosphatidylation and click chemistry tagging via a [3 + 2] azide-alkyne cycloaddition reaction, for measuring PLD activity at the bulk population or the single-cell levels. Second, to accurately visualize PLD activity at the subcellular, organelle-membrane level, we describe a real-time variant of IMPACT (RT-IMPACT) that uses oxo-trans-5-oxocene (oxoTCO) alcohols for transphosphatidylation and fluorescent tagging via the inverse electron-demand Diels-Alder (IEDDA) click chemistry reaction. After describing the representative experimental protocols and example results, we provide guidance on the use cases for each variant of IMPACT.
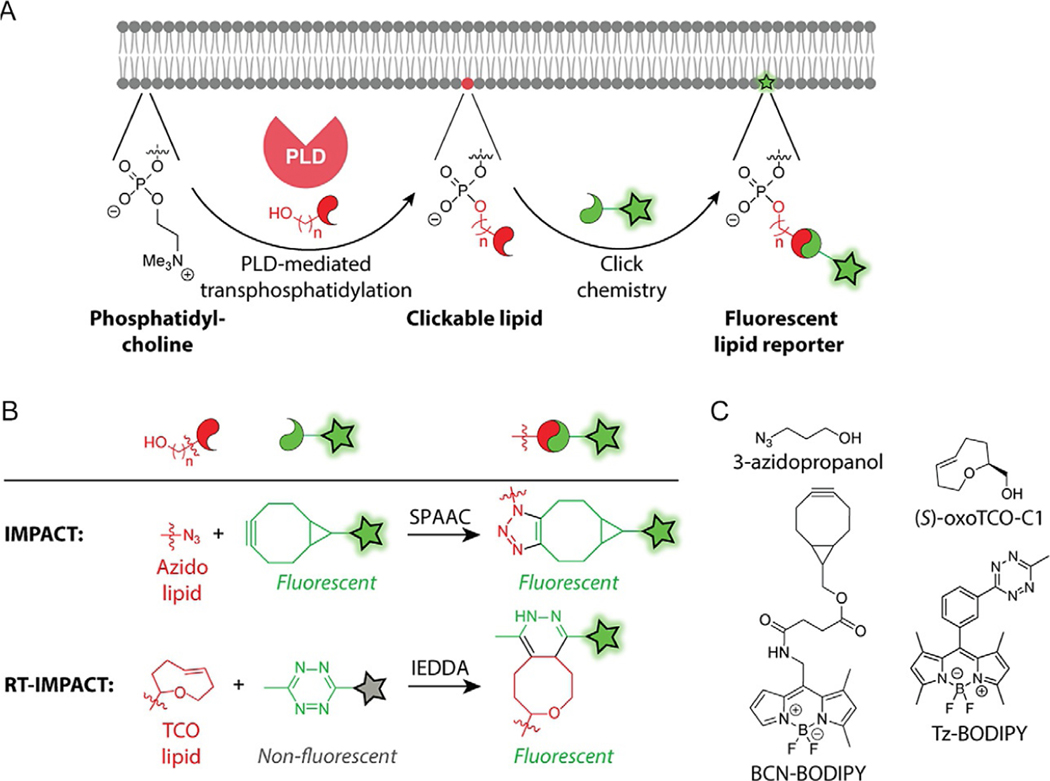
Fig. 1.
(A) Schematic of the IMPACT methods for detecting cellular PLD activity. PLD catalyzes the transphosphatidylation of phosphatidylcholine with an exogenous alcohol, producing a clickable lipid product that can then be detected following the introduction of a fluorophore using a click chemistry tagging reaction. (B and C) Structures of reagents, reactions, and reaction products utilized in IMPACT and RT-IMPACT. See text for abbreviations.
2. IMPACT for bulk and single-cell measurement of PLD activity
This section focuses on IMPACT techniques with relatively lower temporal resolution, but whose reagents are either commercially available or easily synthesized and exhibit excellent aqueous stabilities, making them better suited to large-scale and bulk analysis such as flow cytometry, HPLC, and mass spectrometry (Fig. 2). Our recommendation is to use 3-azidopropanol (Bumpus & Baskin, 2017) as the alcohol for transphosphatidylation and detection via the appropriate [3 + 2] azide-alkyne cycloaddition reaction; however, some reagents require moderate organic synthetic capabilities. If these are not available, an alternate protocol using 5-hexyn-1-ol (Bumpus & Baskin, 2016) is provided, which has some minor drawbacks but relies entirely on commercially available reagents.
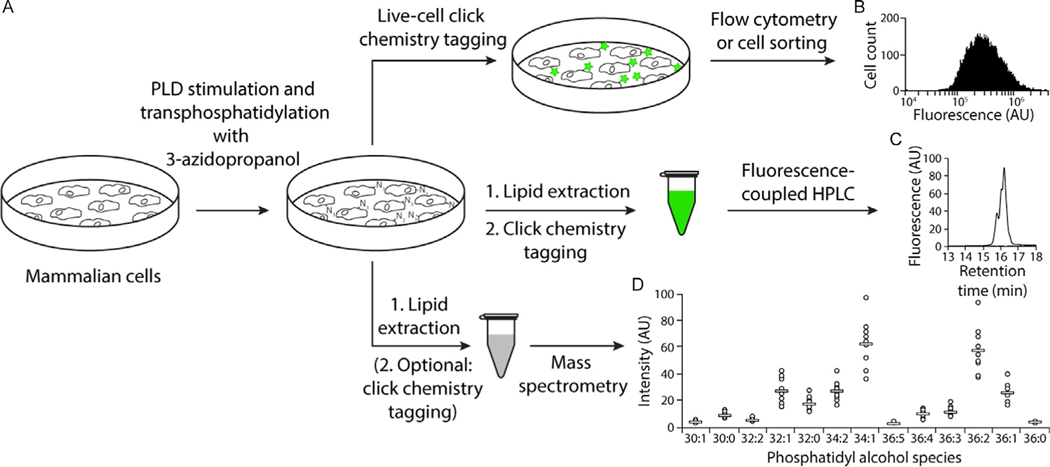
Fig. 2.
Overview of IMPACT with 3-azidopropanol and SPAAC detection. (A) Workflow for IMPACT labeling and detection of PLD activity by flow cytometry, HPLC with fluorescence detection, and mass spectrometry. (B) Flow cytometry histogram of HeLa cells labeled using 3-azidopropanol and BCN-BODIPY in IMPACT. (C) HPLC chromatograph of a lipid extract of HeLa cells after IMPACT labeling with 3-azidopropanol and click chemistry tagging of lipid extracts with BCN-BODIPY. (D) Quantification of the phosphatidyl alcohol products generated when HeLa cells were treated with 3-azidopropanol and PLD activity was stimulated by PMA. Lipid extracts were reacted via click chemistry with Alk-QA to increase detection sensitivity. This figure was adapted with permission from ACS Central Science (https://pubs.acs.org/doi/abs/10.1021/acscentsci.7b00222). Further permissions requests for this material should be directed to the ACS.
2.1. Single-cell quantification of PLD activity by flow cytometry using IMPACT
2.1.1 The day before the experiment, 125,000 cells are seeded per well of a 24-well plate. On the day of the experiment, negative control samples are treated for 30 min prior to the start of the labeling with a specific PLD inhibitor (PLD1/2 dual inhibitor, FIPI (Su et al., 2009), 750 nM; PLD1 isoform-selective inhibitor, VU0359595 (Lewis et al., 2009), 250 nM; PLD2 isoform-selective inhibitor, VU0364739 (Lavieri et al., 2010), 350 nM). Cells are then incubated in the presence or absence of an agonist to stimulate endogenous PLD enzymes (e.g., phorbol 12-myristate 13-acetate, PMA, 100 nM, for 20 min as a positive control) and 3-azidopropanol (1 mM) with or without the inhibitor as suitable. The cells are then rinsed three times with PBS to remove excess alcohol and subjected to a strain-promoted azide-alkyne cycloaddition (SPAAC) click chemistry tagging reaction with a fluorescent cyclooctyne reagent (BCN-BODIPY (Alamudi et al., 2016), 1 μM) in Tyrode’s-HEPES buffer (135 mM NaCl, 5 mM KCl, 1.8 mM CaCl2,1 mM MgCl2, 1 mg/mL glucose, 1 mg/mL bovine serum albumin, 20 mM HEPES, pH7.4) for 10 min at 37 °C. Following the SPAAC reaction, cells are rinsed two times with PBS and incubated at 37 °C in Tyrode’s-HEPES buffer for 10–15 min. (Note: this rinse-out step, both the time and temperature, is absolutely critical to remove excess BCN-BODIPY, and its duration may have to be further optimized depending on cell type.) The cells are lifted from the plate with trypsin, transferred to a 96-well plate, rinsed twice with cold PBS containing FBS (0.5% v/v) and finally resuspended in 50 μL cold PBS containing FBS (0.5% v/v) for analysis (BCN-BODIPY-derived fluorescence is detected in the GFP channel). Comparison of mean fluorescence intensities of a population of at least 10,000 live cells, as determined by forward/side scattering, with those of negative controls, enables an accurate quantification of the extent of PLD activity in the population and therefore serves as a suitable alternative for the bulk, extractive analytic techniques described in Section 2.2.
2.1.2 If flow cytometry facilities are unavailable, a similar, though smaller scale, quantification can be performed using fluorescence microscopy. Cells should be prepared as described above, but rather than seeding cells into a 24-well plate, approximately 200,000 cells should be seeded on a 35-mm glass-bottomed imaging dish, and rather than adding trypsin to lift cells after rinsing of fluorophore, 1 mL of Tyrode’s-HEPES buffer should be added to the dish prior to imaging.
2.1.3 Larger samples (generally more than 1,000,000 cells) prepared for flow cytometry as in Section 2.1, can be analyzed and subpopulations collected using a fluorescence-activated cell sorter. If the number of cells is such that sorting the population will take place over several hours, we recommend including a final fixation step in which cells are treated with paraformaldehyde (4% (v/v) in PBS) for 10 min followed by an additional rinse with PBS, after the fluorophore labeling and subsequent rinses. This order of operations (i.e., performing BCN-BODIPY labeling and rinse-out prior to fixation) helps to minimize background incorporation of the fluorophore into the sample and maximize the signal-to-noise ratio.
2.1.4 The above flow cytometry and microscopy protocols can be adapted to utilize the oxoTCO reagents described below in Section 3 to validate new PLD agonists for detection via RT-IMPACT. (Please see Section 3 for a detailed description of these reagents and this approach.) OxoTCO is dosed at 3 mM in conjunction with the PLD agonist for 5 min in DMEM or relevant buffers at 37 °C. Subsequent click chemistry tagging with Tz-BODIPY (0.5 μM) requires only 1 min and, because fluorescence is activated upon click chemistry tagging in this instance, this protocol does not require the additional extensive BODIPY rinse-out step prior to cells being lifted by trypsin.
2.2. Bulk measurement of PLD activity via transphosphatidylation using IMPACT reagents
One day prior to the experiment, 500,000 cells are seeded on a 60-mm dish. On the day of the experiment, cells are incubated in the presence of a PLD agonist and 3-azidopropanol (1 mM) for varying amounts of time (typically 20 min). The cells are then placed on ice, the media aspirated, and rinsed with cold PBS to remove excess alcohol. The labeling is stopped by the addition of a solution containing 100 μL cold PBS, 125 μL acetic acid (20 mM), and 250 μL methanol (Bligh & Dyer, 1959). The cells are scraped from the dish and transferred to a 1.5 mL Eppendorf tube. To the tube is then added 250 μL of chloroform. The mixture is then vortexed for 1 min and centrifuged at maximum speed (13,300 × g). The bottom organic layer is then transferred to a new tube, a further 250 μL chloroform is then added to the original mixture, which is again vortexed and centrifuged. The organic layers are combined and dried under a stream of nitrogen gas.
2.2.1 For detection of total PLD activity, we recommend normal-phase HPLC separation of the final reaction mixture with fluorescence detection. The SPAAC reaction is performed by dissolving the dried lipids in 37 μL of a mixture containing chloroform:methanol:water (73:23:3 v/v) and BCN-BODIPY (2.5 μM) and incubating at 42 °C for 16 h in the dark. After the SPAAC reaction step, the sample is diluted with 113 μL chloroform:methanol:water (73:23:3 v/v) and filtered (0.45 μm) prior to analysis.
2.2.2 For mass spectrometry analysis, click chemistry tagging is performed using an alternate reagent (an alkyne–quaternary ammonium salt (N,N,N-trimethylpropargylamine bromide, or Alk-QA), via copper-catalyzed azide-alkyne cycloaddition (CuAAC)) to increase the sensitivity of detection by LC–MS (though in many cases the initial transphosphatidylation products could also be detected). To the dried lipid residue in a 1.5 mL Eppendorf tube is added 7 μL of degassed chloroform followed by 30 μL of a reaction master mix (e.g., for 10 reactions, master mix contains 1 mg of Alk-QA (representing a substantial molar excess), 60 μL of 10 mM [acetonitrile]4CuBF4 in degassed acetonitrile, and 240 μL of degassed ethanol). The tube is briefly flushed with argon gas (nitrogen gas is a suitable substitute if argon is not available) and placed in a 42 °C water bath, such that the top half of the tube is not submerged. After 5 h, the reactions are diluted with 113 μL of a chloroform:methanol:water (73:23:3) mixture and filtered (0.45 μm) for analysis.
2.2.3 We have found that the 3-azidopropanol (Bumpus & Baskin, 2017) IMPACT protocol described here affords better chromatographic separation of the lipid product from the excess unreacted BCN-BODIPY reagent and results in higher signal intensity than the originally published protocol using 5-hexyn-1-ol (Bumpus & Baskin, 2016) as the transphosphatidylation substrate and azido fluorophores for tagging via CuAAC. However, if synthetic capabilities are not available, all of the reagents for the 5-hexyn-1-ol/CuAAC-based protocol are commercially available and will produce more than satisfactory results. We do not in general recommend performing these bulk, ex vivo measurements using the oxoTCO alcohol described in Section 3 due to (1) the complexity of its synthesis and (2) its lower efficacy as a PLD substrate compared to 3-azidopropanol and 5-hexyn-1-ol. However, we did utilize these HPLC and LC–MS-based assays with various oxoTCO alcohols while validating them as substrates for RT-IMPACT, and detailed protocols are reported elsewhere (Liang et al., 2019).
3. Real-time (RT)-IMPACT for visualization of subcellular localization of PLD activity
The variants of IMPACT described in Section 2 are most useful for quantifying the extent of PLD-mediated PA synthesis within individual cells or across a population of cells. At the subcellular level, the two PLD isoforms that produce PA via PC hydrolysis, PLD1 and PLD2, exhibit differential and dynamic subcellular localizations on several organelle membranes (Du et al., 2003; Du, Huang, Liang, & Frohman, 2004). Because production of PA at different subcellular locations has a profound effect on its downstream signaling consequences, methods to directly visualize the subset of PA produced only by PLDs, with organelle-level precision, can greatly enhance our ability to dissect PLD signaling pathways.
IMPACT with 3-azidopropanol requires a 10-min click chemistry tagging with BCN-BODIPY and subsequent 10–15 min rinse-out of excess BCN-BODIPY prior to imaging (see Section 2.1,). On this timescale, BODIPY-tagged lipids are transported from their site of synthesis to an equilibrium distribution that is mostly in the endoplasmic reticulum (ER) and Golgi complex (e.g., Fig. 3B, far right), and whereas their total fluorescence intensities report on intracellular PLD activity, their subcellular localizations do not (Liang et al., 2019). To overcome the temporal limitation imposed by this rapid intracellular lipid transport, we developed a real-time variant of IMPACT (RT-IMPACT) using a faster click chemistry detection step to ensure that the observed localization of IMPACT-derived fluorescent lipids would accurately report on the localizations of endogenous PLD enzymes in real-time (Liang et al., 2019). For RT-IMPACT, we selected the inverse electron-demand Diels-Alder (IEDDA) click chemistry reaction between trans-cyclooctene (TCO) and tetrazine (Tz) reagents (Blackman, Royzen, & Fox, 2008) as a substitute for SPAAC-based click chemistry. The IEDDA reaction not only has a much faster second-order rate constant (up to 106 M−1 s−1), but the tetrazine moiety can quench fluorescence, enabling development of fluorogenic, or “turn-on” reagents, which do not require a post-click chemistry rinse-out step (Oliveira, Guo, & Bernardes, 2017). Together, these two properties (shorter click chemistry reaction times and no requisite BODIPY rinse-out step) enable real-time imaging of PLD activities on the second timescale.
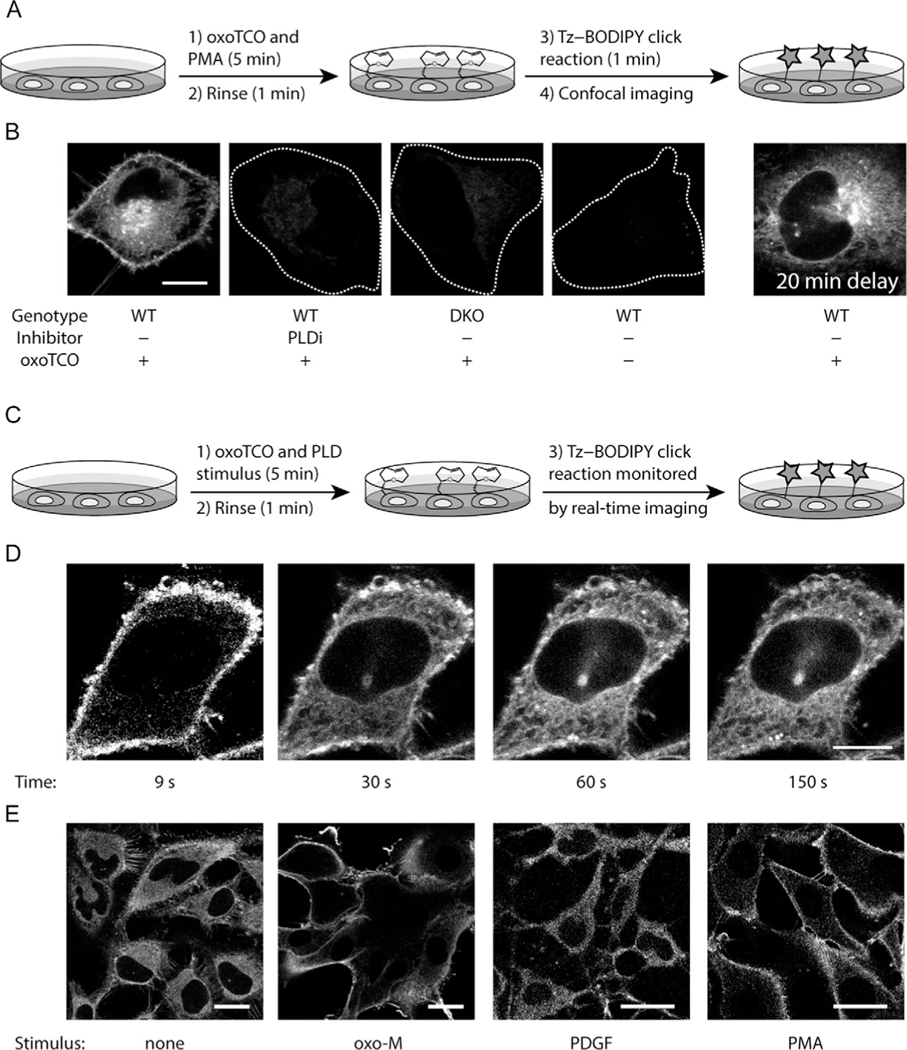
Fig. 3.
Overview of RT-IMPACT with oxoTCO and IEDDA detection. (A) Schematic of steady-state imaging of PLD activities using oxoTCO as a substrate and Tz-BODIPY as the fluorogenic reporter (see Section 3.2). HeLa cells are stimulated (e.g., with PMA (100 nM)) in the presence of oxoTCO (3 mM) for 5 min, rinsed (1 min), and click chemistry tagging is performed via IEDDA with Tz-BODIPY (0.5 μM for 1 min). (B) Representative images using the protocol in (A), with indicated negative controls. White dotted lines represent cell outline in negative control cells. WT: Wild-type; DKO: PLD1/2 double knockout cells. PLDi: PLD inhibitor (FIPI, 750 nM pre-incubation for 30 min prior to and during oxoTCO/PMA stimulation step). 20 min delay refers to cells labeled as in the left-most panel but imaged after a 20-min incubation at 37 °C. Note redistribution of fluorescent lipids from PM to intracellular membranes on this timescale. Scale bar: 10 μm. (C) Schematic of real-time imaging of PLD activity using RT-IMPACT (see Section 3.3). Cells are first stimulated with a PLD stimulus either alone (optional) or together with oxoTCO (3 mM) for 5 min at 37 °C, rinsed (1 min), and imaged in real-time (snapshots taken every 3 s for a 3-min period) before and after addition of Tz-BODIPY (0.5 μM final concentration). (D) RT-IMPACT reveals lipid-trafficking events of fluorophore-tagged lipid reporters. Images shown are snapshots acquired at the indicated timepoints post-addition of Tz-BODIPY using the protocol in (C) and are adjusted to a similar brightness to facilitate viewing the localizations of fluorescent signals. Scale bar: 10 μm. (E) RT-IMPACT reveals stimulus-specific subcellular localizations of PLD activities. Representative images of indicated stimuli were acquired using the protocol described in (C). Images shown are snapshots at 9 s timepoints after addition of Tz-BODIPY and adjusted to similar brightness to facilitate visual assessment. HeLa cells stably expressing M1 muscarinic receptor (M1R) were used for the no stimulus (none) and oxotremorine-M (oxo-M, an M1R agonist) conditions, whereas NIH 3T3 cells were used for stimulation with platelet-derived growth factor (PDGF) and PMA. Scale bars: 20 μm. Note that for some stimuli, the most elevated PLD activity requires a stimulation with agonist prior to the agonist+ oxoTCO step. I.e., here, oxo-M (10 μM) was added to the cells simultaneously with oxoTCO, but PDGF (50 ng/mL) and PMA (100 nM) were added to cells 10 min before incubation together with oxoTCO.
The optimal reagents for RT-IMPACT are (S)-oxoTCO-C1 (Liang et al., 2019), referred to here as “oxoTCO,” a TCO-containing alcohol which acts as the transphosphatidylation substrate, and Tz-BODIPY (Carlson, Meimetis, Hilderbrand, & Weissleder, 2013), a cell-permeable detection reagent that is non-fluorescent until after the IEDDA reaction (Fig. 1).
3.1. General considerations for RT-IMPACT
The most compelling reason to use RT-IMPACT (Liang et al., 2019) over the conventional IMPACT method described in Section 2 (Bumpus & Baskin, 2017) is to investigate the precise subcellular localizations of PLD activities as elicited by a variety of stimuli. To date, we have used RT-IMPACT to visualize localizations of PLD activity following protein kinase C activation by phorbol esters, muscarinic M1 receptor (a prototypical G protein-coupled receptor that signals via the Gαq/phospholipase C pathway) activation by its agonist oxotremorine-M (oxo-M), and platelet-derived growth factor (PDGF) receptor (a prototypical receptor tyrosine kinase) activation by PDGF (Fig. 3). Additionally, RT-IMPACT in combination with a red ER-tracker dye can be used to examine trafficking events of fluorescently tagged lipids from the plasma membrane (PM) to ER membranes.
The multistep preparations of oxoTCO (Lambert et al., 2017; Liang et al., 2019) and Tz-BODIPY (Carlson et al., 2013) have been previously reported, and currently, neither of them is commercially available. The final oxoTCO product need only be purified by silica gel column chromatography, whereas the Tz-BODIPY should be purified by HPLC to remove any fluorescent impurities. The compounds should be stored neat at −80 °C to prevent decomposition. For ease of use, working stock solutions may be used (oxoTCO: 3 M in DMSO, corresponding to roughly 5:7 volume-to-volume ratio of oxoTCO to DMSO, stored at −20 °C; Tz-BODIPY: 1 mM in DMSO in 3 μL aliquots at −80 °C to avoid repeated freeze-thaws, where the concentration of Tz-BODIPY can be calculated by absorbance measurements at 493 nm, with extinction coefficient of BODIPY~79,000 M−1 cm−1 in methanol). Working concentrations for these reagents are 3 mM in DMEM or relevant buffers for oxoTCO and 0.5 μM in PBS for Tz-BODIPY. Due to their limited stabilities in aqueous solutions, working solutions of oxoTCO and Tz-BODIPY should be used within 30 min and 4 h of preparation, respectively. Typically, 200 μL of oxoTCO working solution is prepared in DMEM or relevant buffer and used, 100 μL each, for two consecutive dishes of imaging (covering only the center glass well of a 35-mm glass-bottom imaging dish). For flow cytometry, a master mix of oxoTCO working solution is prepared and dispensed to wells on 24-well plate (100 μL per well). A reasonable starting point, prior to optimization, for the oxoTCO-based transphosphatidylation reaction in cells is to use 3 mM concentration of oxoTCO in the presence of the stimulus of interest for 5 min at 37 °C. In some cases, incubation of agonist alone prior to alcohol treatment may be necessary to enhance the signal-to-noise ratio.
3.2. Preliminary flow cytometry and steady-state imaging to test new agonists
For applications involving unknown or novel stimuli of PLDs, we recommend performing IMPACT using oxoTCO/Tz-BODIPY and analysis by flow cytometry, to optimize concentrations and the time window for the stimulus of interest and to maximize signal-to-noise ratio for future real-time, microscopy-based imaging experiments. Such screens will also inform on whether a particular stimulus can upregulate PLD activities sufficiently above background control for detection by this method (either by comparison to parallel treatment with PLD inhibitors or in the absence of the relevant stimulus). The protocols for fluorescence-based flow cytometry are discussed in Section 2.1. For subsequent real-time imaging applications, we recommend that the signal-to-noise ratio (ratio of mean fluorescence intensities between samples of interest and negative controls) be at least 2.
With optimal concentrations and the experimental time window established for a stimulus of interest, one can then proceed to “steady-state imaging” of PLD activity using oxoTCO/Tz-BODIPY, to optimize settings for fluorescence microscopy and verify the signal-to-noise ratio in this context. Steady-state imaging here refers to IMPACT labeling with fluorescence microscopy readout, which can be performed either by epifluorescence or confocal microscopy depending on the available setup. The key difference between it and the real-time imaging IMPACT protocol described in Section 3.3 is that the goal is to evaluate overall cellular fluorescence rather than subcellular localization, and as a result these experiments are not as time sensitive. For steady-state imaging, the same concentrations of oxoTCO and PLD stimulus should be used as determined by flow cytometry results. For example, for stimulation of PLDs by phorbol 12-myristate 13-acetate (PMA), cells are treated with 3 mM oxoTCO and 100 nM PMA for 5 min at 37 °C in DMEM, followed by aspiration and a one-minute rinse with DMEM at 37 °C. Depending on the signal-to-noise ratio for a particular stimulus, this rinse step may be lengthened or shortened accordingly. For the click chemistry tagging step, cells are treated with Tz-BODIPY (0.5 μM for 1 min). Following the reaction, the BODIPY solution is aspirated and replaced with 1–2 mL Tyrode’s-HEPES buffer, and the dish can be imaged immediately without further rinsing. Typically, only 100 μL volumes of oxoTCO or Tz reagents are used to cover the center glass well in 35-mm glass-bottom imaging dishes to conserve reagents for transphosphatidylation and click chemistry steps. For rinsing with DMEM or other longer incubations, however, such as incubation with PLD inhibitors or with stimulus alone without the RT-IMPACT reagents, it is recommended to cover the entire imaging dish with relevant buffers to minimize effects of evaporation.
3.3. RT-IMPACT for real-time imaging of PLD activity
The parameters established in Section 3.2 serve as a starting point for using oxoTCO and Tz-BODIPY to accurately visualize the precise subcellular localization of PLD activity, with the following modifications. First, cells grown on 35-mm glass-bottom dishes are incubated with the PLD stimulus and oxoTCO for the appropriate amount of time (e.g., for PMA stimulation of PLD activity in HeLa cells: 100 nM PMA and 3 mM oxoTCO for 5 min at 37 °C in 100 μL DMEM in the center of the dish). After aspiration and a one-minute rinse, 100 μL of Tyrode’s-HEPES buffer is added to the center glass well of the dish, and the dish is then mounted on the microscope and the cells are located. Working quickly to minimize the trafficking of the oxoTCO-containing but not-yet-fluorescently-tagged lipid product, a region of interest with 10–20 cells is selected for imaging, and acquisition of a time-lapse movie is begun, using the microscope settings optimized during the steady-state imaging described in Section 3.2, ideally utilizing the instrument’s focus control (i.e., “Definite Focus” for Zeiss instruments or similar). For laser-scanning confocal microscopy, single-channel time-lapse movies typically use an interval of 3 s and a total duration of 3 min for localization (with optimal localization observable at the 9-s timepoint after Tz-BODIPY addition, after sufficient IEDDA tagging but prior to lipid trafficking). For PM-to-ER lipid trafficking studies, a two-channel time-lapse movie is acquired, intervals of 8 s and a duration of 4 min, with addition of a red-fluorescent ER-tracker (Thermo Fisher) during the oxoTCO/stimulus incubation step. Once the time-series has started, 100 μL Tz-BODIPY (1 μM, or 2 ×, in PBS) is carefully added, dropwise but rapidly, to the center glass well and acquisition continued for 3 min.
3.4. Blinded image analysis and statistical methods to determine accurate subcellular localizations of PLD activity using RT-IMPACT
To quantitatively analyze the localizations of PLD activities, all cells in frame at 9 s timepoints after Tz-BODIPY addition should be categorized, or identified, as falling into one of three categories, based on locations of the cellular IMPACT-derived fluorescence: (i) predominantly PM, (ii) predominantly intracellular, or, (iii) a mixture of both PM and intracellular (Liang et al., 2019). This categorization should be performed by a different individual (analyzer) other than the experimenter who obtained the images, and it should be done in a blinded manner. We recommend that, prior to categorization, the analyzer be given examples of exclusively PM and ER-labeled cells for visual calibration of appearances of PM and ER for each cell line. For statistical analyses, the entire dish is treated as a biological replicate and each condition should be repeated with at least 3 dishes with 10–20 cells each in frame. The mean fractions of each categories for each experimental condition can then be represented with stacked bar graphs as previously described (Liang et al., 2019). Chi-square test of independence is an appropriate statistical test, as it determines whether the distribution of fluorescence localization (denoted by fractions in three categories) is independent from a particular experimental condition (e.g., in the presence/absence of a stimulus, time course of a stimulus, etc.). For the use of RT-IMPACT to quantify the rate of fluorescent lipid trafficking from PM to ER, a plot of Pearson’s colocalization coefficient between green (RT-IMPACT) and red (ER-tracker) channel over time will give curves that best fit to an inverse exponential decay curve (curve fitting can be performed using Origin Pro 8 or other similar software). This analysis then enables the calculation of rate of trafficking of fluorescent lipids as quantified by half-life of the fluorescent lipid at the PM.
4. Guidance for selecting which variant of IMPACT to use
This final section provides a guide for selecting between IMPACT and RT-IMPACT. This analysis and decision-making process takes into account technical considerations, including reagent availability and stability (Table 1). As well, we highlight the different applications that each method is best suited to, based on the types of information that it yields (Table 2). We also provide a flow chart to determine which variant of IMPACT is best suited to a given application (Fig. 4). We note that there may be some applications where these two related methodologies may overlap.
Table 1.
Technical considerations for the use of IMPACT and RT-IMPACT for measurement of cellular PLD activities.
| Technical considerations | IMPACT | RT-IMPACT |
|---|---|---|
| Recommended types of applications (see Table 2 for details) | Determination of PLD activity at the single-cell or bulk population levels, amenable to either short- or long-term labeling timescales | Determination of organelle-level, subcellular localizations ofPLD activity, amenable to short-term labeling timescales |
| Detection methods (see Table 2 for details) | Flow cytometry or fluorescence microscopy (single-cell measurements); HPLC or mass spectrometry (bulk population measurements) | Fluorescence microscopy for real-time imaging; flow cytometry recommended to establish optimal labeling conditions depending on PLD stimulus |
| Reagents required for transphosphatidylation and click chemistry steps | (1) Recommended protocol: 3-azidopropanol and BCN-BODIPY (λex: 495; λem: 510) (2) Alternative protocol: 5-hexyn-1-ol and azido-fluorophores |
(S)-oxoTCO-C1 and Tz-BODIPY (λex: 490; λem 510) |
| Storage conditions for reagents | −20 °C (neat or in DMSO stocks; stable indefinitely) | −80°C (neat; stable indefinitely); —20°C (DMSO stock; use within a few months) |
| Stability of reagents in aqueous buffer/cell media | Stable indefinitely | Limited stability; use oxoTCO within 30 min and Tz-BODIPY within 4 h of dilution into aqueous buffer |
| Availability of reagents | (1) Recommended protocol: 3-azidopropanol is commercially available and moderate organic synthesis required to prepare BCN-BODIPY. (2) Alternative protocol: 5-hexyn-1-ol and azido-fluorophores are commercially available |
Complex, multistep organic synthetic sequences (involving photochemistry, flow chemistry, and pyrophoric reagents) are required to prepare oxoTCO and Tz-BODIPY |
Table 2.
Recommended applications and detection methods for IMPACT and RT-IMPACT.
| Detection method | IMPACT | RT-IMPACT |
|---|---|---|
| Flow cytometry (analytical) | Profiling of PLD activity within and across different cell lines under various stimulants | Similar to IMPACT |
| Fluorescence-activated cell sorting (preparative) | Enrichment of cells with high or low PLD activity within a population; availability and stability of reagents make this a well-suited application | Not recommended for this application, due to high value and limited aqueous stability of reagents |
| Fluorescence microscopy | Overall cellular fluorescence reflects PLD activity; however, observed subcellular localizations of IMPACT-derived fluorescence may not reflect accurate localizations of PLD activity due to fluorescent lipid trafficking | Rapid, real-time imaging protocol can reveal both extent and accurate subcellular localizations of PLD activities, with minimal interference from lipid-trafficking events due to the rapid timescale of the RT-IMPACT protocol. N.B.: Extended time-lapse movies of RT-IMPACT may be used to visualize intracellular trafficking of fluorescent lipids to study lipid transport mechanisms |
| Fluorescence-coupled HPLC | Determination of PLD activity at the bulk population level; availability and stability of reagents make this a well-suited application | Not recommended for this application, due to high value and limited aqueous stability of reagents |
| Mass spectrometry | Determination of precise lipid species (defined by acyl tail composition) produced by active PLDs, reflecting substrate preferences | Similar to IMPACT but with lower signal-to-noise ratio |
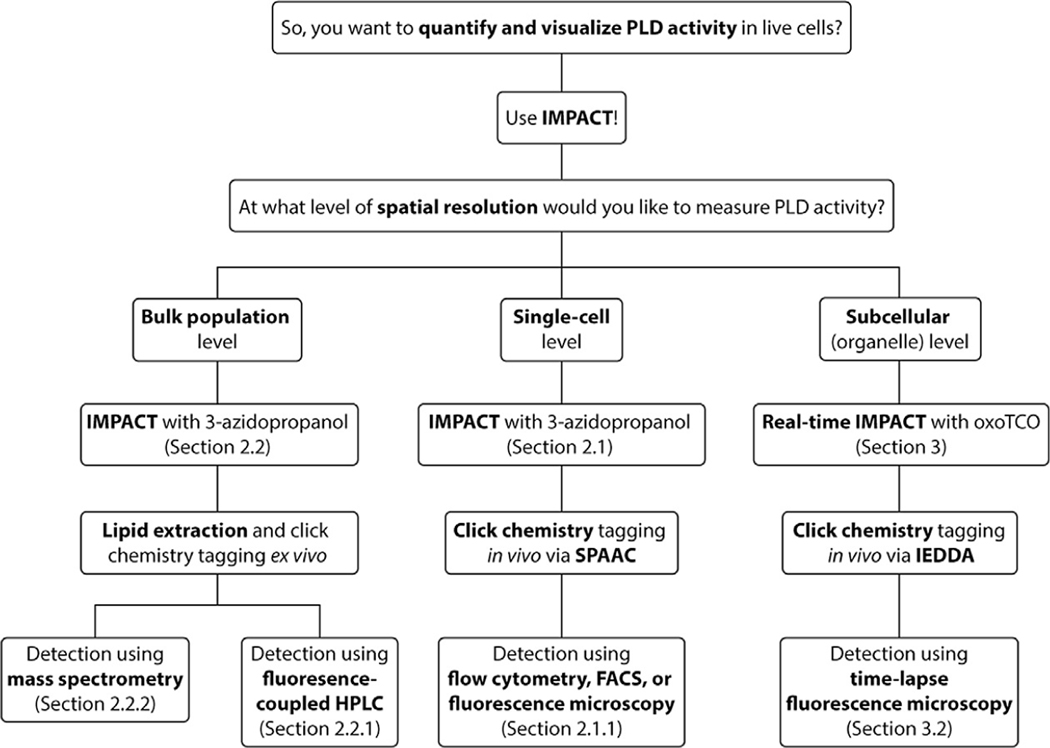
Fig. 4.
Flow chart to aid in determination of which IMPACT variant to use for a desired application.
IMPACT with 3-azidopropanol or 5-hexyn-1-ol (Section 2) allows relatively simple access to live-cell based assays of PLD activity. The reagents are chemically accessible, and the protocols are straightforward. The readouts provide robust signal above background and faithfully report on PLD activities with minimal perturbations over long periods of time. From a practical perspective, the excellent aqueous stability of 3-azidopropanol makes IMPACT ideal for applications such as single-cell measurements, cell sorting, bulk biochemical measurements, pulse-chase labeling, and (potentially, in principle) in vivo labeling.
As a complement to IMPACT, RT-IMPACT enables accurate visualization of active subpopulations of PLD enzymes at the subcellular, organelle-level. This unique capability is enabled by the rapid kinetics and fluorogenic nature of the IEDDA reaction, and both of these aspects are indispensable with respect to real-time, background-free applications. Because RT-IMPACT operates on roughly the same timescale as phospholipid trafficking (~tens of seconds), it is useful not only for visualizing the sites of PLD activity at early timepoints in the RT-IMPACT time-lapse movie, but its rapid temporal resolution enables quantification of the intracellular transport of the fluorescent, RT-IMPACT-derived lipids. The preparation of both oxoTCO and Tz-BODIPY by multistep organic synthetic procedures, however, present challenging obstacles, and their limited aqueous stability allows for only transient incubation in cellular media. Collectively, these two IMPACT variants complement one another, and together they represent an exciting and significant advancement in our ability to understand PLD- and PA-based signaling.
Acknowledgments
Work in the Baskin Laboratory is supported by a Beckman Young Investigator award from the Arnold and Mabel Beckman Foundation, a Sloan Research Fellowship from the Alfred P. Sloan Foundation, and grants from the National Science Foundation (CAREER CHE-1749919) and the National Institutes of Health (R01GM131101). T.W.B. was supported by a National Science Foundation Graduate Research Fellowship (DGE-1650441), and D.L. was supported by a Cornell Fellowship.
References
- Alamudi SH, Satapathy R, Kim J, Su D, Ren H, Das R, et al. (2016). Development of background-free tame fluorescent probes for intracellular live cell imaging. Nature Communications, 7(May), 11964. 10.1017/CBO9781107415324.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman ML, Royzen M, & Fox JM. (2008). Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. Journal of the American Chemical Society, 130(41), 13518–13519. 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, & Dyer WJ. (1959). A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology, 37(8), 911–917. [DOI] [PubMed] [Google Scholar]
- Brown HA, Henage LG, Preininger AM, Xiang Y, & Exton JH. (2007). Biochemical analysis of phospholipase D. Methods in Enzymology, 434, 49–87. 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- Brown HA, Thomas PG, & Lindsley CW. (2017). Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nature Reviews Drug Discovery, 16, 351–367. 10.1038/nrd.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus TW, & Baskin JM. (2016). A chemoenzymatic strategy for imaging cellular phosphatidic acid synthesis. Angewandte Chemie (International Ed. in English), 55(42), 13155–13158. 10.1002/anie.201607443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus TW, & Baskin JM. (2017). Clickable substrate mimics enable imaging of phospholipase D activity. ACS Central Science, 3(10), 1070–1077. 10.1021/acscentsci.7b00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus TW, & Baskin JM. (2018). Greasing the wheels of lipid biology with chemical tools. Trends in Biochemical Sciences, 43(12), 970–983. 10.1016/j.tibs.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumpus TW, Liang FJ, & Baskin JM. (2018). Ex Uno Plura: Differential labeling of phospholipid biosynthetic pathways with a single bioorthogonal alcohol. Biochemistry, 57, 226–230. 10.1021/acs.biochem.7b01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JCT, Meimetis LG, Hilderbrand SA, & Weissleder R. (2013). BODIPY-tetrazine derivatives as superbright bioorthogonal turn-on probes. Angewandte Chemie, International Edition, 52(27), 6917–6920. 10.1002/anie.201301100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du GW, Altshuller YM, Kim Y, Han JM, Ryu SH, Morris AJ, et al. (2000). Dual requirement for Rho and PKC in direct activation of phospholipase D1 through G protein-coupled receptor signaling. Molecular Biology of the Cell, 11(December), 4359–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Altshuller YM, Vitale N, Huang P, Chasserot-Golaz S, Morris AJ, et al. (2003). Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. Journal of Cell Biology, 162(2), 305–315. 10.1083/jcb.200302033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Huang P, Liang BT, & Frohman MA. (2004). Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Molecular Biology of the Cell, 15(March), 1024–1030. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/14742705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, et al. (1997). Characterization of two alternately spliced forms of phospholipase D1. Journal of Biological Chemistry, 272(6), 3860–3868. 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- Hannun YA, & Obeid LM. (2008). Principles of bioactive lipid signalling: Lessons from sphingolipids. Nature Reviews Molecular Cell Biology, 9(2), 139–150. 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Kassas N, Tanguy E, Thahouly T, Fouillen L, Heintz D, Chasserot-Golaz S, et al. (2017). Comparative characterization of phosphatidic acid sensors and their localization during frustrated phagocytosis. Journal of Biological Chemistry, 292, 4266–4279. 10.1074/jbc.M116.742346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert WD, Scinto SL, Dmitrenko O, Boyd SJ, Magboo R, Mehl RA, et al. (2017). Computationally guided discovery of a reactive, hydrophilic: Trans-5-oxocene dienophile for bioorthogonal labeling. Organic and Biomolecular Chemistry, 15(31), 6640–6644. 10.1039/c7ob01707c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavieri RR, Scott SA, Selvy PE, Kim K, Jadhav S, Morrison RD, et al. (2010). Design, synthesis, and biological evaluation of halogenated N-(2-(4-Oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: Discovery of an isoform-selective small molecule phospholipase D2 inhibitor. Journal of Medicinal Chemistry, 53(18), 6706–6719. 10.1021/jm100814g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Scott SA, Lavieri R, Buck JR, Selvy PE, Stoops SL, et al. (2009). Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorganic & Medicinal Chemistry Letters, 19(7), 1916–1920. 10.1016/j.bmcl.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wu K, Tei R, Bumpus TW, Ye J, & Baskin JM. (2019). A real-time, click chemistry imaging approach reveals stimulus-specific subcellular locations of phospholipase D activity. Proceedings of the National Academy of Sciences of the United States of America, 116(31), 15453–15462. 10.1073/pnas.1903949116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RK, & Frohman MA. (2015). Physiological and pathophysiological roles for phospholipase D. Journal of Lipid Research, 56, 2229–2237, jlr.R059220. 10.1194/jlr.R059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira BL, Guo Z, & Bernardes GJL. (2017). Inverse electron demand Diels-Alder reactions in chemical biology. Chemical Society Reviews, 46(16), 4895–4950. 10.1039/c7cs00184c. [DOI] [PubMed] [Google Scholar]
- Philip F, Ha EE, Seeliger MA, & Frohman MA. (2017). Measuring phospholipase D enzymatic activity through biochemical and imaging methods. Methods in Enzymology, 583, 309–325. 10.1016/bs.mie.2016.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvy PE, Lavieri RR, Lindsley CW, & Brown HA. (2011). Phospholipase D: Enzymology, functionality, and chemical modulation. Chemical Reviews, 111(10), 6064–6119. 10.1021/cr200296t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga YV, Topham MK, & Epand RM. (2011). Regulation and functions of diacylglycerol kinases. Chemical Reviews, 111(10), 6186–6208. 10.1021/cr1004106. [DOI] [PubMed] [Google Scholar]
- Su W, Yeku O, Olepu S, Genna A, Park J, Ren H, et al. (2009). 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Molecular Pharmacology, 75(3), 437–446. 10.1124/mol.108.053298.have. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine H, & Iruela-Arispe ML. (2017). Membrane lipids and cell signaling. Current Opinion in Lipidology, 28(5), 408–413. 10.1097/MOL.0000000000000443. [DOI] [PMC free article] [PubMed] [Google Scholar]