Abstract
In the present investigation we developed a method for the detection of Mycoplasma hyopneumoniae in bronchoalveolar lavage fluid (BALF) of pigs by PCR with a primer pair flanking a DNA fragment of 853 bp specific for M. hyopneumoniae. Several methods were tested to eliminate the amplification inhibitors present in BALFs. The best results were obtained by the extraction of the DNA from the BALFs. By the PCR performed with the extracted DNA, 102 CFU of M. hyopneumoniae could be detected in 1 ml of BALF from specific-pathogen-free swine experimentally inoculated with M. hyopneumoniae. DNA from 11 other mycoplasma species and 17 cell-walled bacterial species colonizing the respiratory tracts of pigs was not amplified. In a field study BALFs from 40 pigs from farms with a history of chronic pneumonia were tested for M. hyopneumoniae by cultivation and by PCR (i) with BALFs incubated in Friis medium and (ii) with DNA extracted from the BALFs. In addition, PCR was performed with postmortem lung washings from 19 of the 40 pigs, and immunofluorescence tests were carried out with sections of lungs from 18 of the 40 pigs. M. hyopneumoniae could not be detected in 18 of the 40 pigs by any of the five methods tested. The remaining 22 pigs showed a positive reaction by the PCR with DNA extracted from the BALFs and variable positive reactions by the other tests. A complete correspondence could be observed between the immunofluorescence test result and the result of PCR with DNA. The investigation shows that the PCR with DNA extracted from BALFs is a suitable technique for the sensitive and specific in vivo detection of M. hyopneumoniae.
Mycoplasma hyopneumoniae is the primary agent of enzootic pneumonia of pigs (14, 22). The disease has a worldwide distribution and causes considerable economic losses in swine production due to reduced growth rate and feed conversion efficiency (24). The detection of M. hyopneumoniae is usually based on the isolation of the organisms by culture or by immunofluorescence tests with lung sections (3). The cultivation of M. hyopneumoniae is difficult due to the fastidious culture requirements and the extremely slow growth of M. hyopneumoniae, often resulting in overgrowth by other mycoplasmas colonizing the respiratory tracts of pigs (12). Cross-reactions with Mycoplasma flocculare and Mycoplasma hyorhinis reduce the specificity of conventional immunological detection methods (7).
With the development of PCR an alternative diagnostic method is now available. This method is suitable for the fast, sensitive, and specific detection of fastidiously growing microorganisms (5). Different PCR procedures have been described for the detection of M. hyopneumoniae (4, 6, 16, 30, 31), but none of these has been evaluated under field conditions. Only Mattsson et al. (23) and Verdin et al. (33) tested the PCRs that they developed with samples from fattening pigs, namely, with nasal swabs or tracheobronchiolar washings, respectively. The results obtained by PCR were compared with serological results but not with the results obtained by the classical antigen detection methods.
Previous studies demonstrated M. hyopneumoniae in lavage fluids of the respiratory system including bronchoalveolar lavage fluid (BALF) from living pigs (1, 25, 33). In the present investigation we developed a method for the detection of M. hyopneumoniae in BALF by PCR. Different sample preparations were tested for their ability to remove or inactivate amplification inhibitors present in BALFs. The PCR procedure was evaluated under field conditions, and the detection of M. hyopneumoniae by PCR was compared with that by culture and the immunofluorescence test.
MATERIALS AND METHODS
Organisms and growth conditions.
The mycoplasmas and walled bacteria used in this study are listed in Table 1. Mycoplasma buccale, Acholeplasma axanthum, and Acholeplasma laidlawii were cultivated in modified Hayflick-medium (18). Mycoplasma sualvi was cultivated in the SP 4 medium described by Tully et al. (32), and the other mycoplasmas were cultivated in the medium developed by Friis (12) (Friis medium). The numbers of CFU of the mycoplasmas and the acholeplasmas were estimated by the method of Albers and Fletcher (2). Walled bacteria were grown in nutrient broth.
TABLE 1.
Bacterial strains used in the PCR specificity tests
| Species | Strain | PCR results |
|---|---|---|
| Mycoplasma hyopneumoniae | J | + |
| Mycoplasma hyopneumoniae | 241-4 | + |
| Mycoplasma hyopneumoniae | 257 | + |
| Mycoplasma hyopneumoniae | 724-2-3 | + |
| Mycoplasma arginini | G230 | − |
| Mycoplasma buccale | GH20274 | − |
| Mycoplasma flocculare | Ms42 | − |
| Mycoplasma hyopharyngis | H3-6BF | − |
| Mycoplasma hyorhinis | BTS7 | − |
| Mycoplasma hyosynoviae | S16 | − |
| Mycoplasma sualvi | Mayfield B | − |
| Acholeplasma axanthum | H86N | − |
| Acholeplasma granularum | BTS39 | − |
| Acholeplasma laidlawii | PG8 | − |
| Acinetobacter calcoaticus | Field strain | − |
| Actinobacillus pleuropneumoniae | Field strain | − |
| Actinomyces pyogenes | Field strain | − |
| Bacillus subtilis | Field strain | − |
| Bordetella bronchiseptica | Field strain | − |
| Enterococcus faecalis | Field strain | − |
| Escherichia coli | Field strain | − |
| Haemophilus parasuis | Field strain | − |
| Klebsiella spp. | Field strain | − |
| Neisseria spp. | Field strain | − |
| Pasteurella multocida | Field strain | − |
| Proteus spp. | Field strain | − |
| Pseudomonas spp. | Field strain | − |
| Staphylococcus aureus | Field strain | − |
| Staphylococcus hyicus | Field strain | − |
| Streptococcus dysgalactiae bv. equisimilis | Field strain | − |
| Streptococcus suis type 1 | Field strain | − |
+, amplification; −, no amplification.
DNA extraction.
The DNAs of the walled bacteria and of M. hyopneumoniae were obtained after incubation with lysis buffer (0.1 M NaCl, 10 mM Tris HCl [pH 8.0], 1 mM EDTA, 5% [vol/vol] Triton X-100, 2.8 mg of lysozyme per ml) and isopropanol precipitation as described by Sambrook et al. (27).
BALF.
BALF was taken by fiberoptic bronchoscopy as described by Hensel et al. (17) and Ganter and Hensel (13). In short, the pigs were anesthetized with 2 mg of azaperone kg of body weight−1 intramuscularly and 15 mg of metomidate kg−1 intra-abdominally or with 10 mg of tiletamine-zolazepam (Tilest) kg−1 intramuscularly and were positioned in a sling in sternal recumbency. After intubation the tip of the endoscope was placed in the bronchus trachealis supplying the right anterior lung lobus. Five portions of 20 ml of 0.15 M phosphate-buffered saline (PBS) were instilled and aspirated immediately.
PCR.
An 853-bp fragment specific for the M. hyopneumoniae genome was amplified with the oligonucleotides H1 (5′-TAGAAATGACTGGCAGACAA-3′) and H2 (5′-GAGGCTTTGATTTTGGAGTC-3), which were used as primers. Amplification was carried out in a 50-μl reaction mixture containing 10 mM Tris HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 0.01% (wt/vol) gelatin, 1.25% (vol/vol) formamide, each deoxynucleoside triphosphate at a concentration of 200 μM, 7.5 pmol of each primer, and 2.5 U of Taq DNA polymerase (Stratagene, Heidelberg, Germany). A hot start technique was used with Chill-out 14 liquid wax (Biozym, Hessisch Oldendorf, Germany) according to the manufacturer’s instructions. After an initial denaturation step of 5 min at 94°C 50 cycles were performed in an automated thermal cycler (Perkin-Elmer Cetus, Langen, Germany). The samples were denatured at 93°C for 30 s. The primers were annealed at 55°C for 30 s and extended at 72°C for 90 s. The extension step of the last cycle was 10 min. Aliquots of the amplified sample (10 μl) were analyzed by electrophoresis in a 1% (wt/vol) agarose gel containing ethidium bromide (0.5 μg/ml).
To determine the specificity the PCR was performed with 10-μl cultures of the mycoplasmas and 1 μg of DNA of the walled bacteria listed in Table 1. The sensitivity of the PCR was determined by investigation of 10-fold dilutions of M. hyopneumoniae cultures or DNA.
Preparation of BALFs for PCR.
Several methods were tested with BALFs from M. hyopneumoniae-free, specific-pathogen-free (SPF) swine inoculated with 101 to 105 CFU of M. hyopneumoniae per ml and mixed with 0.1 mg of dithiothreitol per ml. The mycoplasmas were harvested from 2-ml aliquots of the BALFs by centrifugation at 12,000 × g for 30 min. For the following preparations the pellets were suspended in 10 μl of distilled water.
(i) Boiling and centrifugation.
The samples were inoculated into 100 μl of distilled water, boiled for 10 min, and centrifuged at 12,000 × g for 30 min. A total of 10 μl of the supernatant was used for PCR.
(ii) Proteinase K digestion.
Samples were inoculated into 100 μl of proteinase K lysis buffer (100 μg of proteinase K [Pharmacia, Freiburg, Germany] per ml, 0.5% [vol/vol] Tween 20 [Merck, Darmstadt, Germany], 0.5% [vol/vol] Igpal [Sigma, Deisenhofen, Germany]) and were kept for 1 h at 60°C, according to the method of Campbell (8). The proteinase K was inactivated by boiling for 10 min. A total of 10 μl of the suspension was used for PCR.
(iii) DNA extraction with the QIAamp blood kit.
The DNA was extracted according to the manufacturer’s instructions (Qiagen GmbH, Hilden, Germany) and was precipitated with ethanol at 4°C overnight. DNA pellets were washed in 70% (vol/vol) ethanol and were resuspended in 20 μl of distilled water. A total of 1 μg of DNA was used for PCR.
(iv) DNA extraction with the Nucleon II DNA extraction kit.
The DNA extraction with the Nucleon II DNA extraction kit (Scotlab, Wiesloch, Germany) was performed according to the manufacturer’s instructions. In short, the samples were incubated in a solution containing 340 μl of reagent B (0.4 M Tris, 0.06 M EDTA, 0.15 M NaCl, 1% [wt/vol] sodium dodecyl sulfate) and 2.4 μl of RNase A (10 mg/ml) for 30 min at 37°C. After incubation with 100 μl of 5 M sodium perchlorate for 15 min at room temperature and for 25 min at 65°C, the DNA was extracted with chloroform and precipitated with ethanol for 2 h at 4°C. The DNA pellets were washed with 70% (vol/vol) ethanol and were resuspended in 20 μl of distilled water. A total of 1 μg DNA was subjected to PCR.
(v) DNA extraction by the CTAB method.
DNA was extracted by the cetyltrimethylammonium bromide (CTAB) method described by Maass and Dalhoff (21). The samples were incubated in 600 μl of proteinase K lysis buffer (10 mM Tris-EDTA [pH 7.5] 0.5% [vol/vol] sodium dodecyl sulfate, 100 μg of proteinase K per ml) for 1 h at 60°C. After adding 100 μl of 5 M NaCl and 80 μl of CTAB (10% [wt/vol] in 0.7 M NaCl), the samples were incubated for 10 min at 65°C. The DNA was extracted with chloroform-isoamyl alcohol (24:1) and phenol-chloroform-isoamyl alcohol (25:24:1) and precipitated with isopropanol at 4°C overnight. The DNA pellets were washed in 70% (vol/vol) ethanol and were resuspended in 20 μl distilled water. A total of 1 μg of DNA was used for PCR.
Clinical specimens.
To evaluate the PCR procedure under field conditions, BALFs were taken from 40 pigs on 18 farms with problems of chronic pneumonia immediately before they were killed. All except one of the pigs showed macroscopic bronchopneumonic alterations in the lungs at necropsy. BALFs of pigs were investigated by cultivation and PCR. Lung samples from 19 randomly selected pigs were taken at necropsy and were investigated by the indirect immunofluorescence test for M. hyopneumoniae antigen. In addition, washings of the bronchus trachealis (bronchial washings) were obtained at necropsy from 18 of the 40 pigs. The tip of a catheter was placed in the bronchus trachealis supplying the right anterior lung lobus. Two portions of 1 ml of PBS (2.7 mM KCl, 1.5 mM KH2PO4, 137 mM NaCl, 8 mM Na2HPO4 [pH 7.2]) were instilled into the bronchus and aspirated immediately. The two suspensions were combined and investigated by PCR.
Cultivation of BALFs.
Immediately after collection, 1 ml of BALF was inoculated into 2 ml of Friis medium. For cultivation each 0.2 ml of this transport mixture were transferred to each of 1.8 ml of Friis medium and 1.8 ml of Friis medium containing 5% (vol/vol) rabbit anti-M. hyorhinis serum. Further dilutions (10−2, 10−3, 10−4) were produced by transferring 0.2 to 1.8 ml of medium. The tubes were incubated on a roller at 37°C up to 4 weeks and were investigated twice a week for a color change without turbidity. Cultures showing a color shift to yellow were subcultivated in liquid medium and on solid medium. The solid media were incubated at 37°C in a humid atmosphere with 5% CO2 for at least 30 days and investigated twice a week for mycoplasma colonies. Differentiation was done by the immunobinding assay according to the method of Kotani and McGarrity (20) with rabbit antisera against M. hyopneumoniae, M. hyorhinis, M. flocculare, Mycoplasma hyosynoviae, and A. laidlawii.
PCR with BALFs and postmortem lung washings.
PCR was performed with the BALFs incubated in Friis medium and with DNA extracted from the BALFs from the 40 pigs with a history of chronic pneumonia. To investigate the BALFs incubated in Friis medium by PCR, 10 μl was taken from the tubes prepared for cultivation (containing 1.8 ml of Friis medium and 0.2 ml of the transport mixture) after 3 days of incubation. For the PCR with the DNA of the BALFs, DNA was extracted from BALFs with the Nucleon kit and by the CTAB method. To ensure that the amplification inhibitors were eliminated by the preparation procedure, simulated positive samples from all BALFs collected in the field were prepared by adding 104 CFU of M. hyopneumoniae/ml of BALF and were investigated. In addition, PCR was performed with bronchial washings taken at necropsy from 18 of the 40 pigs. PCR was performed with DNA extracted from the washings by the CTAB method.
Immunofluorescence test with lung sections.
Cryostat sections (3 to 4 μm) were obtained from two locations of pneumonic areas of the left apical lobe. In case M. hyopneumoniae was not detected at these two locations samples from two other locations were investigated. The sections were dried on the slides for 10 min at 37°C, fixed with acetone (20°C) for 10 min, incubated with rabbit anti-M. hyopneumoniae or rabbit anti-Mycoplasma bovirhinis serum (negative control), and diluted 1:300 with PBS containing 1% (vol/vol) swine serum for 30 min. After washing with PBS containing 0.05% (vol/vol) Tween 20 (PBS-Tween) for 10 min, the sections were incubated with fluorescein isothiocyanate-conjugated swine anti-rabbit immunoglobuline (DAKO, Glostrup, Denmark) diluted 1:50 with PBS containing 1% (vol/vol) swine serum for 30 min. All incubation steps were performed at room temperature. The sections were then washed in PBS-Tween, covered with 90% (vol/vol) glycerol in 10% (wt/vol) barbital buffer, mounted with a cover glass, and investigated under UV light. Lung sections from pigs artificially infected with M. hyopneumoniae served as positive controls.
Statistical analysis.
The sensitivities and specificities of the different methods of detection of M. hyopneumoniae were calculated in comparison with a “gold standard” (see Tables 3 and 4) as described by Fletcher et al. (11). The 95% confidence interval was estimated as described by Sachs (26). The Fisher exact test was performed with the software SAS (SAS, Heidelberg, Germany) to examine if the sensitivities or specificities of the detection methods differ significantly.
TABLE 3.
Sensitivities and specificities of detection of M. hyopneumoniae by PCR with different samples in comparison to those of detection by PCR with DNA extracted from BALFsa
| Sample used for PCR | Sensitivity (% [95% confi- dence interval]) | Specificity (% [95% confi- dence interval]) |
|---|---|---|
| DNA extracted from BALFs (CTAB method) (n = 40)b | 100 | 100 B |
| Cultures of BALFs (n = 40) | 27 A (11–50) | 100 B (81–100) |
| DNA extracted from postmortem bronchial washings (CTAB method) (n = 18) | 60 A (26–88) | 100 B (63–100) |
Groups sharing letters are not significantly different by Fisher’s exact test (P < 0.05).
This sample and its results represent the gold standard.
TABLE 4.
Sensitivities and specificities of detection of M. hyopneumoniae by culture and PCR in comparison to those of detection by the immunofluorescence testa
| Examination method | Sensitivity (% [95% confidence interval]) | Specificity (% [95% confidence interval]) |
|---|---|---|
| Immunofluorescence test (n = 19)b | 100 A | 100 B |
| Culture (n = 19) | 9 (0.2–41) | 100 B (63–100) |
| PCR with DNA extracted from BALFs (n = 19) | 100 A (72–100) | 100 B (63–100) |
Groups sharing letters are not significantly different by Fisher’s exact test (P < 0.05).
This test and its results represent the gold standard.
RESULTS
Specificity of the PCR.
A specific PCR amplification product of 853 bp was obtained from the type strain M. hyopneumoniae J and from the M. hyopneumoniae field strains. No amplification products appeared in the PCR with the other mycoplasmas and the walled bacteria investigated (Table 1). A minimum of 103 CFU of M. hyopneumoniae J/ml of culture or 10 fg of M. hyopneumoniae DNA could be detected.
Sensitivity of the PCR.
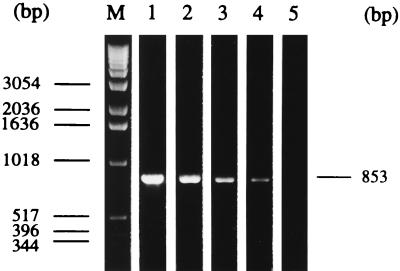
Of the several BALF preparation methods tested, the best results were achieved by DNA extraction with the Nucleon II DNA extraction kit and by the CTAB method, allowing the detection of 102 CFU of M. hyopneumoniae/ml of BALF from SPF pigs (experimentally inoculated with M. hyopneumoniae) by PCR (Fig. 1). By the PCR with DNA extracted with the QIAamp blood kit, an amplification product was obtained only in the presence of at least 103 CFU of M. hyopneumoniae/ml of BALF. After boiling and centrifugation as well as after proteinase K digestion, the detection limit of the PCR was only 105 CFU of M. hyopneumoniae/ml of BALF.
FIG. 1.
Limit of detection of M. hyopneumoniae by PCR with DNA extracted from BALFs from SPF pigs inoculated with M. hyopneumoniae. BALFs from SPF pigs were inoculated with 101 to 105 CFU of M. hyopneumoniae/ml. DNA was extracted from 2-ml aliquots of these BALFs by the CTAB method. A total of 1 μg of the DNA was used in the PCR. The amplification products were separated by agarose gel electrophoresis (1% agarose; 10 μl sample per vial) and were visualized by ethidium bromide staining and with UV light. Lane M, molecular size marker (1-kb DNA ladder; Gibco BRL, Eggenstein, Germany); lane 1, 105 CFU of M. hyopneumoniae/ml of BALF from SPF pigs; lane 2, 104 CFU; lane 3, 103 CFU; lane 4, 102 CFU; lane 5, 10 CFU. The specific amplification product of 853 bp appears in lanes 1 to 4.
Detection of M. hyopneumoniae in BALFs of pigs with chronic pneumonia.
The Nucleon kit and the CTAB method were used for the preparation of the BALFs obtained from the pigs with chronic pneumonia for PCR because of the highest sensitivity achieved by these two preparation methods in the PCR with BALFs from SPF pigs. In the treatment of the BALFs from pigs with pneumonia, these two preparation methods, however, had different effects on the PCR. These effects appeared in the PCR with the simulated positive samples. The removal of the amplification inhibitors succeeded by the CTAB method for 39 of the field-collected BALFs and with the Nucleon II DNA extraction kit for only 34 of the 40 field-collected BALFs.
The rates of detection of M. hyopneumoniae in pigs with chronic pneumonia obtained by cultivation, immunofluorescence tests, and PCR (i) with BALFs incubated in Friis medium, (ii) with DNA extracted from BALFs, and (iii) with washings from lungs are summarized in Table 2. M. hyopneumoniae could not be detected by any of the five methods tested in 18 of the 40 pigs investigated. All remaining 22 pigs showed a positive reaction in the PCR with DNA extracted from BALFs. The lungs of 11 of these 22 pigs were also examined by the immunofluorescence test. M. hyopneumoniae could be detected in all of them, indicating a complete correspondence of the results of these two methods. Of the 22 pigs showing M. hyopneumoniae by PCR with the DNA extracted from BALF, only 6 had a positive result by the PCR with BALFs incubated in Friis medium. The PCR with postmortem lung washings from 10 of these 22 pigs was positive for 6 pigs. The lowest detection rate was obtained by cultivation of the BALFs, which was positive for only 1 of the 40 pigs investigated. M. hyorhinis was isolated from 15 of the 40 pigs. The cultures of the BALFs from 13 pigs could not be evaluated because of the high level of contamination by walled bacteria. Mycoplasmas could not be cultivated from the BALFs of the remaining 11 pigs.
TABLE 2.
Detection of M. hyopneumoniae in pigs with chronic pneumonia by cultivation, immunofluorescence test, and PCR
| No. of pigs | Cultivation result | Immunofluorescence test result | Result of PCR with the following samples:
|
||
|---|---|---|---|---|---|
| BALF incubated in Friis medium | DNA extracted from BALFs | DNA extracted from bronchial washingsa | |||
| 1 | +b | + | − | + | + |
| 5 | −c | + | − | + | + |
| 4 | − | + | − | + | − |
| 1 | − | + | + | + | NT |
| 5 | − | NTd | + | + | NT |
| 6 | − | NT | − | + | NT |
| 10 | − | NT | − | − | NT |
| 7 | − | − | − | − | − |
| 1 | − | − | − | ?e | − |
| No. positive/no. investigated (%) | 1/40 (2.5) | 11/19 (58) | 6/40 (15) | 22/40 (55) | 6/18 (30) |
Washings from lungs obtained at necropsy.
+, positive reaction.
−, negative reaction.
NT, not tested.
Not evaluable due to the presence of remaining amplification inhibitors.
Table 3 presents the sensitivities and specificities, with confidence intervals, estimated for PCR with different samples, with PCR with DNA extracted from BALFs used as the apparent gold standard. PCR with cultures of BALFs and PCR with DNA extracted from bronchial washings showed by Fisher’s exact test significantly lower sensitivities than that of the apparent gold standard.
The sensitivities and specificities of detection of M. hyopneumoniae by culture and PCR in comparison to those of detection by the immunofluorescence test are presented in Table 4.
DISCUSSION
In the present investigation a PCR procedure and a sample preparation method for the detection of M. hyopneumoniae in BALFs have been developed, and the results were compared with those of the detection of M. hyopneumoniae by culture and an immunofluorescence test. The PCR procedure described here proved to be specific and sensitive for the detection of M. hyopneumoniae.
BALFs contain large amounts of substances inhibiting the PCR, and these substances must be removed before PCR. Several methods were tested for their suitability at removing the amplification inhibitors from BALFs from SPF pigs inoculated with different amounts of M. hyopneumoniae. In these investigations boiling and centrifugation as well as digestion with proteinase K appeared to be unsuitable methods since the sensitivity of detection of M. hyopneumoniae in the subsequent PCR was very low. This is in agreement with the results of other studies, which showed that lysis of BALFs by boiling and detergent treatment and subsequent centrifugation cannot remove all inhibitory substances (9) and that polysaccharides and lipids which are not degraded by proteinase K can still act as amplification inhibitors (15, 19, 21, 28).
In the investigation of BALFs from SPF swine, the highest sensitivity was observed with the PCR with DNA extracted with the Nucleon kit and by the CTAB method. Less sensitive was the PCR with DNA from BALFs extracted with the QIAamp blood kit, probably due to unsufficient removal of the amplification inhibitors. The limit of detection of M. hyopneumoniae by PCR in field samples may be somewhat lower than the sensitivity determined with BALFs from SPF pigs due to a larger amount of foreign DNA originating from purulent inflammations and the possible presence of additional inhibitory substances in BALFs from clinically affected swine (14, 28). Although identical regarding the sensitivity of the PCR with BALFs from SPF swine, the Nucleon kit and the CTAB method differed in their abilities to remove amplification inhibitors from the BALFs from pigs with respiratory problems. The present investigations showed a clear superiority of the CTAB method regarding the removal of amplification inhibitors from the BALFs from the clinically affected swine.
In the field study the highest rate of detection of M. hyopneumoniae was achieved by the PCR with DNA extracted from the BALFs from the pigs. PCR with DNA extracted from bronchial washings from the lungs at autopsy yielded fewer positive results. This may be the result of contamination with blood, which could act as an additional inhibitor (10, 29) and which could increase the concentration of nonmycoplasmal DNA. The lowest sensitivity was obtained by the PCR with BALFs cultivated in Friis medium. Apparently, the field strains of M. hyopneumoniae did not multiply in these cultures and the amplification inhibitors in the medium (for example, the serum content) as well as the fact that the target DNA was not enriched by centrifugation (as was done with BALFs investigated by PCR with extracted DNA) prevented amplification of the small amount of the DNA of the organisms present. M. hyopneumoniae did not multiply under the cultivation conditions used, and this also occurred in the cultivation of the BALFs, in which M. hyopneumoniae could be detected in only 1 of the 22 BALFs that were M. hyopneumoniae positive by the PCR. Because of its fastidious cultivation requirements M. hyopneumoniae is generally difficult to cultivate, and it appears that it is particularly hard to cultivate it from BALFs. This was also reported by Mattsson et al. (23), who could cultivate M. hyopneumoniae from lungs but not from BALFs.
A complete correlation between the results obtained by PCR and the results obtained by the immunofluorescence test with lung sections was observed in the present investigations, indicating the similar sensitivities of both of these methods. While the immunofluorescence test, however, is restricted to the investigation of dead animals, the PCR with BALF can also be performed with samples from living pigs.
In conclusion, the PCR with DNA extracted from BALFs by the CTAB method by the procedure described here appears to be a suitable method for the detection of M. hyopneumoniae in living pigs. Due to its high sensitivity, even small amounts of organisms, characteristic of early stages of infection and subclinical infection, can be detected by this procedure, and importantly, noncultivable strains of M. hyopneumoniae can also be detected by this method. BALFs can be collected from living pigs, allowing random sampling of a representative number of diseased as well as healthy animals of a herd. With the PCR with the BALFs, reliable and fast information about the occurrence of M. hyopneumoniae in a pig herd can be obtained.
ACKNOWLEDGMENTS
We thank P. Ahrens for providing the oligonucleotide sequences and multiple valuable suggestions. We thank also N. F. Friis for providing the field strains of M. hyopneumoniae and helpful advice concerning the cultivation of M. hyopneumoniae.
REFERENCES
- 1.Abiven P, Pommier P. Technique de lavage tracheobronchique par la voie transnasal pour la détection de Mycoplasma hyopneumoniae chez le porc vivant non anesthésie. Vet Res. 1993;24:515–522. [PubMed] [Google Scholar]
- 2.Albers A C, Fletcher R D. Simple method for quantitation of viable mycoplasmas. Appl Environ Microbiol. 1982;43:958–960. doi: 10.1128/aem.43.4.958-960.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong C H, Scheidt A B, Thacker H L, Runnels L J, Freeman M J. Evaluation of criteria of the postmortem diagnosis of mycoplasmal pneumonia of swine. Can J Comp Med. 1984;48:278–281. [PMC free article] [PubMed] [Google Scholar]
- 4.Artiushin S, Stipkovits L, Minion F C. Development of polymerase chain reaction primers to detect Mycoplasma hyopneumoniae. Mol Cell Probes. 1993;7:381–385. doi: 10.1006/mcpr.1993.1056. [DOI] [PubMed] [Google Scholar]
- 5.Bej A K, Mahbubani M H, Atlas R M. Amplification of nucleic acids by polymerase chain reaction (PCR) and other methods and their applications. Crit Rev Biochem Mol Biol. 1991;26:301–334. doi: 10.3109/10409239109114071. [DOI] [PubMed] [Google Scholar]
- 6.Blanchard B, Abiven P, Saillard C, Kobisch M, Bové J M. Proceedings of the 6th International Symposium of ADILA and O.I.E. 1992. Specific detection of Mycoplasma hyopneumoniae in clinical samples; p. 12. [Google Scholar]
- 7.Bölske G, Strandberg M-L, Bergström K, Johansson K-E. Species-specific antigen of Mycoplasma hyopneumoniae and cross-reaction with other porcine mycoplasmas. Curr Microbiol. 1987;15:233–239. [Google Scholar]
- 8.Campbell L A. PCR detection of Chlamydia pneumoniae. In: Persing D H, Smith T S, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 247–252. [Google Scholar]
- 9.Clarridge J E, Shawar R M, Shinnick T M, Plikaytis B B. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J Clin Microbiol. 1993;31:2049–2056. doi: 10.1128/jcm.31.8.2049-2056.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Francis R, Cross N C P, Foulkes N S, Cox T M. A potent inhibitor of Taq polymerase copurifies with human genomic DNA. Nucleic Acids Res. 1988;16:10355. doi: 10.1093/nar/16.21.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher R H, Fletcher S W, Wagner E W. Clinical epidemiology. The essentials. Baltimore, Md: The Williams & Wilkins Co.; 1982. Diagnosis; pp. 42–75. [Google Scholar]
- 12.Friis N F. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare. Nord Veterinaermed. 1975;27:337–339. [PubMed] [Google Scholar]
- 13.Ganter M, Hensel A. Cellular variables in bronchoalveolar lavage fluids (BALF) in selected healthy pigs. Res Vet Sci. 1997;63:215–217. doi: 10.1016/s0034-5288(97)90023-0. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin R F W, Pomeroy A P, Whittlestone P. Production of enzootic pneumonia in pigs with a mycoplasma. Vet Rec. 1965;77:1247–1249. [Google Scholar]
- 15.Greenfield L, White T J. Sample preparation methods. In: Persing D H, Smith T S, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 122–137. [Google Scholar]
- 16.Harasawa R, Koshimizu K, Takeda O, Uemori T, Asada K, Kato I. Detection of Mycoplasma hyopneumoniae DNA by the polymerase chain reaction. Mol Cell Probes. 1991;5:103–109. doi: 10.1016/0890-8508(91)90003-3. [DOI] [PubMed] [Google Scholar]
- 17.Hensel A, Ganter M, Kipper S, Krehon S, Wittenbrink M M, Petzoldt K. Prevalence of aerobic bacteria in bronchoalveolar lavage fluid from healthy pigs. Am J Vet Res. 1994;55:1697–1702. [PubMed] [Google Scholar]
- 18.Kirchhoff H, Rosengarten R. Isolation of a motile mycoplasma from fish. J Gen Microbiol. 1984;130:2439–2445. doi: 10.1099/00221287-130-9-2439. [DOI] [PubMed] [Google Scholar]
- 19.Kolk A H J, Schuitema A R J, Kuijper S, van Leeuwen J, Hermans P W M, van Embden J D A, Hartskeerl R A. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a nonradioactive detection system. J Clin Microbiol. 1992;30:2567–2575. doi: 10.1128/jcm.30.10.2567-2575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotani H, McGarrity G J. Identification of mycoplasma colonies by immunobinding. J Clin Microbiol. 1985;23:783–785. doi: 10.1128/jcm.23.4.783-785.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maass M, Dalhoff K. Comparison of sample preparation methods for the detection of Chlamydia pneumoniae in bronchoalveolar lavage fluid by PCR. J Clin Microbiol. 1994;32:2616–2619. doi: 10.1128/jcm.32.10.2616-2619.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maré C J, Switzer W P. New species: Mycoplasma hyopneumoniae—a causative agent of virus pig pneumonia. Vet Med. 1965;60:841–846. [PubMed] [Google Scholar]
- 23.Mattsson J G, Bergström K, Wallgren P, Johansson K-E. Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene. J Clin Microbiol. 1995;33:893–897. doi: 10.1128/jcm.33.4.893-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross R F. Mycoplasmal diseases. In: Leman A D, Click R D, Mengerling W L, Penny R H C, Scholl E, Straw B, editors. Diseases of swine. Ames: Iowa State University Press; 1992. pp. 537–551. [Google Scholar]
- 25.Runge M, Ganter M, Delbeck F, Hartwick W, Ruffer A, Franz B, Amtsberg U G. Etiological diagnosis of pneumonia in fattening pigs: investigations of bronchoalveolar lavage (BAL) by culture methods and immunofluorescence microscopy as well as serological findings. Berl Münch Tierärztl Wochenschr. 1996;109:101–107. [PubMed] [Google Scholar]
- 26.Sachs L. Angewandte Statistik. Berlin, Germany: Springer Verlag; 1992. Die Häufigkeit von Ereignissen; pp. 432–443. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Skakni L, Sardet A, Just J, Landman-Parker J, Costil J, Moniot-Ville N, Bricout F, Garbarg-Chenon A. Detection of Mycoplasma pneumoniae in clinical samples from pediatric patients by polymerase chain reaction. J Clin Microbiol. 1992;30:2638–2643. doi: 10.1128/jcm.30.10.2638-2643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soini H, Skurnik M, Lippo K, Tala E, Viljanen M K. Detection and identification of mycobacteria by amplification of a segment of the gene coding the 32-kilodalton protein. J Clin Microbiol. 1992;30:2025–2028. doi: 10.1128/jcm.30.8.2025-2028.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sørensen V, Ahrens P, Barford K, Feenstra A A, Feld N C, Friis N F, Bille-Hansen V, Jensen N E, Pedersen M W. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and evaluation of four different assays. Vet Microbiol. 1997;54:23–34. doi: 10.1016/s0378-1135(96)01266-7. [DOI] [PubMed] [Google Scholar]
- 31.Stemke G W, Phan R, Young T F, Ross R F. Differentiation of Mycoplasma hyopneumoniae, M. flocculare, and M. hyorhinis on the basis of amplification of a 16S rRNA gene sequence. Am J Vet Res. 1994;55:81–84. [PubMed] [Google Scholar]
- 32.Tully J G, Whitcomb R F, Clark H F, Williamson D L. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science. 1977;195:892–894. doi: 10.1126/science.841314. [DOI] [PubMed] [Google Scholar]
- 33.Verdin E, Blanchard B, Kobisch M, Bové J M, Saillard C. Use of nested PCR diagnosis test to detect Mycoplasma hyopneumoniae under field conditions. IOM Lett. 1996;4:101–102. [Google Scholar]