Abstract
Exogenous biomolecule delivery into plants is difficult because the plant cell wall poses a dominant transport barrier, thereby limiting the efficiency of plant genetic engineering. Traditional DNA delivery methods for plants suffer from host-species limitations, low transformation efficiencies, tissue damage, or unavoidable and uncontrolled DNA integration into the host genome. We have demonstrated efficient plasmid DNA delivery into intact plants of several species with functionalized high-aspect-ratio carbon nanotube (CNT) nanoparticles (NPs), enabling efficient DNA delivery into a variety of non-model plant species (arugula, wheat, and cotton) and resulting in high protein expression levels without transgene integration. Herein, we provide a protocol that can be implemented by plant biologists and adapted to produce functionalized single-walled CNTs (SWNTs) with surface chemistries optimized for delivery of plasmid DNA in a plant species–independent manner. This protocol describes how to prepare, construct, and optimize polyethylenimine (PEI)-functionalized SWNTs and perform plasmid DNA loading. The authors also provide guidance on material characterization, gene expression evaluation, and storage conditions. The entire protocol, from the covalent functionalization of SWNTs to expression quantification, can be completed in 5 d.
Introduction
Plant genetic transformation is an important component of agricultural engineering, small-molecule synthesis, and bioenergy efforts1–4. Maximizing the throughput of testing and generating genetically engineered plants is of core importance in both academic research and agro-industry, in which a toolset is needed that is (i) plant species independent and (ii) capable of high performance despite the physical barriers presented in intact plant tissues such as the plant cell wall. The two traditionally used plant DNA delivery tools either limit the range of plant species that can be transformed (Agrobacterium)5,6 or exhibit low transformation efficiencies and tissue damage from the use of high external force (biolistic DNA delivery)7–9. Furthermore, both methods yield uncontrolled transgene integration of the plant genome, and even if care is taken to deliver DNA with engineered Agrobacterium in a non-integrating manner, its nature as a plant pathogen elicits strict regulatory oversight of the plant product as a genetically modified organism (GMO). To our knowledge, there has yet to be a plant transformation method that enables high-efficiency DNA delivery, without transgene integration, in a plant species–independent manner.
Recently, gene and protein delivery into intact plant cells without external force or aid has been reported with nanomaterials such as CNTs10,11, mesoporous silica NPs (MSNs)12, DNA nanostructures13, fusion peptide–based nanomaterials14, and layered double hydroxide clay nanosheets15. Among these techniques, SWNTs exhibit several characteristics that are optimal for intact plant cell delivery: (i) high aspect ratio and exceptional tensile strength for efficient plant cell wall traversal, (ii) biocompatibility, and (iii) cargo biomolecule protection from cellular metabolism and degradation for optimal activity of the delivered cargoes16,17. Therefore, SWNTs are a promising candidate for plasmid DNA delivery into intact plants.
In this protocol, we describe the preparation, characterization, and optimal usage of a SWNT-based DNA delivery platform. This platform can efficiently transport plasmid DNA or linear DNA amplicons into both model and crop plants—without external mechanical aid, in a non-toxic manner, and without transgene integration—a combination of features that is not attainable with existing plant transformation approaches18. Specifically, the SWNT-based transient plant transformation approach demonstrated herein is beneficial for plant biotechnology applications in which transient protein production without transgene integration is desired. This approach may also aid high-throughput screening in mature plants so as to rapidly identify genotypes that result in desired phenotypes, mapping and optimization of plant biosynthetic pathways, and maximization of plant-mediated natural product synthesis.
Overview of the procedure
In this protocol, we detail the use of SWNTs as a tool to deliver DNA plasmids to intact plants (Fig. 1). The general method for DNA plasmid loading on modified SWNTs is through electrostatic grafting, in which carboxylated SWNTs (COOH-SWNTs) are first covalently modified with a cationic polymer (PEI) carrying a net positive charge (Steps 1–22). Next, positively charged CNTs (PEI-SWNTs) are incubated with negatively charged DNA plasmid vectors (Steps 23–25). The covalent attachment of PEI and electrostatic adsorption of DNA to SWNTs is confirmed by atomic force microscopy, X-ray photoelectron spectroscopy, and zeta potential measurements. We highlight checkpoint steps for assessing procedural quality before proceeding with applications of the protocol. After preparation of DNA plasmid-loaded PEI-SWNT conjugates, DNA-PEI-SWNTs are infiltrated into the true leaves of mature plants (Steps 26–29). Negative controls include delivery of free plasmid DNA, DNA-PEI without SWNTs, and PEI-SWNTs without plasmid DNA; positive-control studies include delivery of the same gene with Agrobacterium. Confocal microscopy, qPCR, droplet digital PCR (ddPCR), western blot, GFP-Trap Agarose, and Qubit assays are implemented to evaluate and quantify gene expression (Step 30). Throughout the procedure, we highlight the parameters that can be tuned for optimization or modification of the protocol and discuss how to troubleshoot issues that might be encountered during the procedure.
Fig. 1 |. Overview of the procedure for plant transformation with PEI-SWNTs.
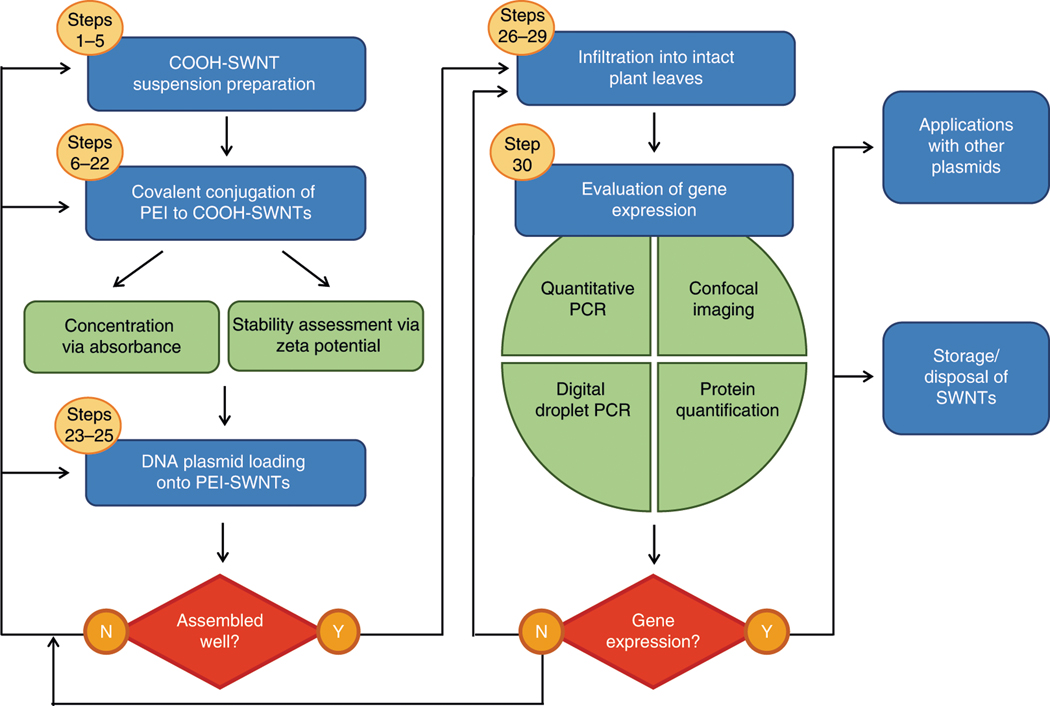
Blue boxes represent main experimental steps; green boxes represent characterization and evaluation steps; red boxes represent checkpoint steps to decide whether to continue with the procedure or to return to any previous step for optimization and re-trial (shown by arrows). Yellow circles indicate the corresponding step numbers.
Development of the approach
SWNTs were first reported to passively penetrate the cell wall and cell membrane of intact plant cells in 2009 by Liu et al.10. Since then, SWNTs have been used in exploratory studies of biocompatibility and sensing of small molecules in plant tissues by introducing SWNTs complexed to synthetic fluorescent dyes or oligonucleotides16,19,20. Motivated by these previous studies, we aimed to develop a SWNT-based platform that can, for the first time (to our knowledge), deliver functional plasmids into intact plant cells for gene expression. For biosensing or chirality separation purposes, DNA (or RNA) oligonucleotides are conventionally loaded on SWNTs via non-covalent π–π interactions using direct probe-tip sonication of SWNTs with nucleotides21. However, this approach cannot be used to load large double-stranded DNA plasmids, because probe-tip sonication shears the double-stranded DNA, and double-stranded DNA is largely unstable on SWNT owing to base-paired interactions that decrease π–π interactions between the DNA and the SWNT.
Therefore, we developed a new platform, in which we covalently modified the surface of SWNTs with a cationic polymer (PEI) to electrostatically load and deliver plasmid DNA into intact plant cells. In our paper18, we performed the covalent conjugation of PEI to COOH-SWNTs with a heat-driven reaction, in which the mixture was heated to high temperatures to favor the chemical bonding between the COOH groups on SWNTs and the primary amines on PEI. We have recently further optimized and increased the reaction efficiency by using carboxylic acid activators (N-(3-dimethylaminopropyl)-Nʹ-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide sodium salt (NHS)) rather than heat. We showed that in both the heat and activator-driven methods, the covalent conjugation, DNA loading, and transgene expression took place (Supplementary Fig. 1), but higher reaction yields and transgene expression efficiencies were achieved with the EDC–NHS-driven reaction as compared to the heat reaction. Published in tandem with our study, Kwak et al.22 used a different cationic polymer (chitosan) covalently functionalized to SWNTs for gene delivery specifically targeted to intact plant chloroplasts.
Applications and impact of the method
We have verified that our SWNT-based transformation platform yields high-efficiency expression of gene vectors delivered without external force to leaves of a variety of model and crop plant species: Nicotiana benthamiana, Eruca sativa (arugula), Triticum aestivum (wheat), and Gossypium hirsutum (cotton)18, and is thus useful for a diverse range of plant genetic engineering applications. SWNT-based DNA delivery is amenable to multiplexing, whereby multiple gene vectors are delivered and screened rapidly in a combinatorial manner and in parallel. The ability to perform transient transformations in a high-throughput and combinatorial manner for target genes offers the capability to map important plant biosynthetic pathways rapidly, which can then be used to augment in planta production of valuable small molecules, recombinant proteins, and human therapeutics. Therefore, this protocol may be of interest to plant biologists and crop scientists who wish to utilize this tool for fast and reliable transient genetic manipulation of non-model plants and for the discovery and engineering of plant metabolic pathways to synthesize molecules endogenously in a plant host that can improve agriculture and human health.
This protocol describes the delivery of plasmids encoding fluorescent proteins such as GFP, because fluorescent proteins provide an easily scorable phenotype and enable expression quantification at the level of (i) fluorescence (via confocal microscopy), (ii) mRNA (via qPCR), and (iii) protein (via western blotting and Qubit). However, the protocol is fundamentally generic for plasmid loading to SWNTs; therefore, users can substitute the vector used herein by their own plasmid of interest. For instance, owing to the non-integrating nature of PEI-SWNT-based DNA delivery, one of the most useful applications will be to implement SWNTs for delivery of plasmids coding for nuclease-based genome-editing proteins such as zinc finger nucleases, transcription activator-like effector nucleases, and Cas (CRISPR-associated) nucleases. When leveraged to deliver nuclease-encoding plasmids, SWNTs could endow transient expression of these nucleases, which may generate permanent (stable) edits. As such, SWNT-based nuclease plasmid delivery could enable high-efficiency genome modification without transgene integration and is particularly beneficial for heterogeneous plant species such as cassava, cacao, and sugarcane, in which crossing cannot be used to remove integrated transgenes. In addition, delivery of CRISPR genome-editing cargoes to achieve permanent editing without the integration and stable expression of the nuclease proteins has been shown to result in lower off-target editing and toxicity23. Delivery of CRISPR genome-editing cargoes with SWNT may also be a way to circumvent GMO labeling, owing to the non-integrating nature of the transgene and non-pathogenic nature of the delivery tool.
This protocol describes delivery of plasmids for expression in the plant cell nucleus. Moreover, SWNTs can also be used to deliver plasmids to plant chloroplasts18,22, which house genes coding for many photosynthetic proteins. Genome transformation of plastids such as the chloroplast offers orthogonal advantages over nuclear transformations, including lack of gene silencing, which enables high efficiency and stable expression of transgenes. This feature makes chloroplasts an attractive target for biosynthetic production of commodity chemicals, drugs, and even larger protein products such as antibodies and biologics. Notably, the chloroplast is not transformable with Agrobacterium; thus PEI-SWNTs present an opportunity to perform genetic transformation of chloroplasts through the current protocol with a promoter and codon-optimized plasmid that express only in the chloroplast.
Last, SWNTs can also be used to deliver RNA (siRNA and single-guide RNA) directly (i.e., without an RNA-encoding DNA vector) but with a different process altogether that does not involve covalent modification of the pristine SWNT surface17.
It has been shown that polymer-functionalized SWNTs (at the SWNT working concentrations used for gene delivery and expression studies, <10 mg L−1) do not cause cytotoxicity or tissue damage to plants18,22. Regarding genetic transformation of edible plants with SWNTs, we note that the transformed plant material constitutes the experimental generation To. Plants genetically edited using SWNT-based delivery would undergo several generations of progeny production before their seeds are brought to market, and thus edible plants would constitute generations that have never undergone direct exposure to nanomaterials.
Comparison with other methods
Despite the utility of plant biotechnology, many plant species and tissue types remain difficult to transform genetically2 and are subject to species- or tissue-dependent heuristic optimizations of genetic transformation procedures. Limitations in plant genetic transformation are due to (i) the difficulty of delivering DNA, RNA, or protein across the rigid cell wall, which has a small (~20-nm) size exclusion limit24; and (ii) challenges in efficient plant regeneration technologies. Currently, two well-established DNA delivery platforms (Agrobacterium and biolistics) exist to transfer biomolecules into plant cells, but these techniques suffer from host-species limitations, low transformation efficiencies, tissue damage, or unavoidable and uncontrolled DNA integration into the host genome (Table 1). The most commonly used tool today for plant genetic transformations—Agrobacterium-mediated transformation technology—is unable to perform species-independent transgene-free editing, cannot be used for chloroplast transformation, and yields random DNA integration. Similarly, DNA delivery methods that utilize a gene gun25 or other external forces, such as ultrasound26 or vortexing27, can cause cell damage, which leads to increased rates of transgene integration and lower efficiency of edited plant regeneration.
Table 1 |.
Summary and comparison of the DNA delivery methods in intact plants
| Delivery platform | Target range | Genetic fate of target | Advantages | Platform limitations | Time line leading up to delivery to intact plants | |
|---|---|---|---|---|---|---|
| Agrobacterium-mediated delivery | Mature plants, immature tissue (calli, meristems, embryos), protoplasts | Gene integration into genome, transient or stable expression, transformation of germ-line cells | Amenable to large sizes of DNA, high efficiency, stable transformation, no specialized equipment needed | High host specificity, time-consuming protocol, pathogenic and therefore regulated as GMO | Days 1–3: Agrobacterium-competent cell preparation Days 4 and 5: Agrobacterium transformation Days 6 and 7: preparation of Agrobacterium inoculum Day 8: Agrobacterium activation and infiltration |
|
| Biolistic delivery | Mature plant tissue explants (leaves, petioles), immature tissue | Gene integration into genome; multiple or partial copy insertions, typically requires regeneration and selection (species dependent) | Amenable to large sizes of DNA, rapid protocol pre-delivery, can deliver proteins | Low transformation efficiency, specialized equipment needed, large amount of DNA needed, tissue/cell damage | Day 1: sterilization of macrocarriers, holders Coating of carriers with DNA Particle bombardment25 |
|
| NP-mediated passive delivery | MSNs12 | Mature tissue (roots), protoplasts | Fate of delivered DNA (genome integration versus transient expression) remains uninvestigated | High DNA-loading capacity | Unknown target species range (validated only in Arabidopsis) | Days 1 and 2: synthesis of MSN Day 3: functionalization of MSN Preparation of DNA-MSN complexes Plant incubation with DNA-MSN |
| Peptide carriers28,34 | Mature tissue (leaves), immature tissue (embryos), targeted delivery to mitochondria35,36 | Fate of delivered DNA (genome integration versus transient expression) remains uninvestigated | Rapid protocol, no specialized equipment needed, no external force required | Unknown target species range (validated only in Nicotiana benthamiana and Arabidopsis thaliana) | Day 1: preparation of peptide-DNA constructs Infiltration into target tissue |
|
| SWNTs18,22 | Mature tissue (leaves18,22, roots and callus (data not shown)), protoplasts18,22, for nuclear18,22 or chloroplast22 expression | Transient expression of gene18,22, no integration into genome18 | No transgene integration, scalable, no external force required | Unknown target species range (validated in tobacco, arugula, wheat, cotton18, Arabidopsis thaliana22, sorghum (data not shown) | Day 1: COOH-SWNT suspension PEI-SWNT reaction Day 2: PEI-SWNT washing Preparation of PEI-SWNT + DNA Infiltration into/exposure to target tissue |
|
Nanomaterials have emerged as a promising candidate for delivery of genetic cargoes to intact plant cells. Recently, several reports have described passive uptake of nanomaterials by intact plant cells: MSNs12, DNA nanostructures13, and peptide carriers28 have demonstrated the possibility of nanoscale internalization into walled plant cells without strong mechanical aid to deliver functional biological cargoes (Table 1). These methods have achieved DNA delivery and gene expression across various tissues (leaves and roots) in intact plants, as well as immature tissues and protoplasts. Compared to Agrobacterium-mediated gene delivery, DNA delivery for all NP methods can be executed in less time, expediting gene expression studies. Because NP-based delivery methods are nascent to the field of plant genetic engineering, most successful DNA delivery has been demonstrated only in model plants; more testing must be done to ascertain the target species limitations of NP-based methods.
The SWNT-based delivery platform we present here enables DNA plasmid delivery without transgene integration in both model and crop plants—and in both dicot and monocot plants—with high efficiency and without toxicity or tissue damage. The SWNT-mediated delivery platform described here is well suited for transient gene-editing applications because it is easy, fast, cost effective, non-destructive, species independent, and scalable. SWNTs also protect DNA cargo against nuclease degradation in cells18, a feature of SWNT-based delivery that may be extended to the protection of other biological cargoes of interest. Furthermore, SWNTs for gene delivery and their associated chemistries cost <US$3 per transformation (including labor costs) and circumvents the high cost of gene gun equipment and gold particles commonly used in biolistic delivery or the molecular biology tools needed for use of Agrobacterium. Although their optimal storage conditions are at 4 °C, SWNTs used for genetic transformation of plants remain stable at room temperature (20–21 °C) for a month post synthesis, and are thus amenable to distribution and use in locations devoid of standard laboratory equipment.
Limitations
This SWNT-based plant gene delivery platform may be of interest for use in diverse applications in plant biology. Although this protocol demonstrates DNA plasmid delivery in plant leaves, certain parameters may need to be optimized (e.g., DNA-SWNT ratio, exposure time, buffer conditions, and sterility) to apply SWNT-based delivery to different plant tissues, such as embryonic tissue, germ-line cells, or callus. The protocol described herein is optimized for the delivery of an ~5-kb DNA plasmid. Although we have also obtained successful expression of proteins from ~12-kb plasmids, optimization of DNA loading for larger plasmids may be needed, and it is possible that there is an as-yet-undetermined upper limit on the maximum plasmid size for delivery with PEI-SWNTs.
The aforementioned optimizations might also be needed for plant species that are different in size or morphology from the plant tissues assayed herein. In addition, although SWNTs could be used for delivery of biomolecules other than DNA vectors, such as proteins and RNA17, surface chemistry optimization of the SWNT would probably be needed for other cargo attachments and their efficient delivery.
Materials
Biological materials
In the example described in this protocol, we use wild-type N. benthamiana plants (seeds obtained from the Staskawicz lab at UC Berkeley).
We have also successfully used our approach with wild-type E. sativa (seeds from Renee’s Garden, cat. no. 5450), wheat (seeds from Botanical Interests, cat. no. 3037), and cotton plants (seedlings from Cottonman, cat. no. 6WS). We expect that our protocol will be compatible with many monocot and dicot plant species; however, the infiltration process becomes more challenging for plant species with very thick and hydrophobic leaves (such as maize and kale). We recommend testing a range of DNA-PEI concentrations and volumes for transformations in different tissue types.
Reagents
Milli-Q water (Millipore Sigma, cat. no. ZR0Q008WW)
Nuclease-free water (Qiagen, cat. no. 129114)
10× PBS (Corning, cat. no. 46-013-CM)
Carbon nanotubes (single-walled, carboxylic acid functionalized; Sigma-Aldrich, cat. no. 652490) !CAUTION Because dry nanomaterials are easily dispersed into the air, they pose a respiratory health hazard. All dry nanomaterials should be handled in a fume hood or nanomaterial enclosure. Any solid or liquid waste resulting from nanomaterial handling should be disposed of as hazardous chemical waste.
MES hydrate (Sigma-Aldrich, cat. no. M5287)
Polyethylenimine (PEI, branched, molecular weight (MW) 25,000; Sigma-Aldrich, cat. no. 408727)
Hydrochloric acid (HCl, 37% (vol/vol); Sigma-Aldrich, cat. no. 320331)
Sodium hydroxide (NaOH; Sigma-Aldrich, cat. no. S8045)
Magnesium chloride hexahydrate (MgCl2.6H20; Sigma-Aldrich, cat. no. M2670)
N-(3-dimethylaminopropyl)-Nʹ-ethylcarbodiimide hydrochloride (EDC; Sigma-Aldrich, cat. no. E1769)
N-hydroxysulfosuccinimide sodium salt (NHS; Sigma-Aldrich, cat. no. 106627-54-7)
Pierce 660nm Protein Assay (Thermo Fisher Scientific, cat. no. 22660)
GFP-Trap Agarose (ChromoTek, cat. no. GTA-10)
iScript cDNA synthesis kit (Bio-Rad, cat. no. 1708891)
PowerUp SYBR Green Master Mix (Applied Biosystems, cat. no. A25742)
Qubit Protein Assay (Thermo Fisher Scientific, cat. no. Q33211)
RNeasy Plant Mini Kit (Qiagen, cat. no. 74904)
Plasmid DNA containing the gene of interest: in this example, we use HBT95::sGFP(S65T)-NOS29
GFP primers: forward 5′-CGCCGAGGTGAAGTT-3′, reverse 5′-GTGGCTGTTGTAGTTGTAC-3′ (IDT, custom order)
EF1 primers: forward 5′-TGGTGTCCTCAAGCCTGGTATGGTTGT-3′, reverse 5′-ACGCTTGAGATCCTTAACCGCAACATTCTT-3′ (IDT, custom order)
Liquid nitrogen !CAUTION Handle liquid nitrogen carefully. Protect hands at all times with cryo gloves. Protect your eyes with safety goggles and wear appropriate personal protective equipment (lab coat, closed-toe shoes, and long pants).
Tris–HCl (Sigma, cat. no. T5941-100G)
NaCl (Sigma, cat. no. S7653-1KG)
EDTA (RPI, cat. no. E57040-100)
NP-40 (Fisher Scientific, cat. no. T01164-500g)
Glycerol (Fisher Scientific, cat. no. G33-500)
Protease inhibitor cocktail (Sigma, cat. no. P9599-1ML)
Equipment
Delicate task wipes (Kimberly-Clark, cat. no. 06-666)
Sterile syringe filter (0.45 μm; VWR, cat. no. 28145-481)
Microcentrifuge tubes (1.5 mL; VWR, cat. no. 89000-028)
Conical tubes (50 mL; Olympus, cat. no. 28-106)
Pipette tips (low-retention 10, 200, and 1,000-μL filter tips; USA Scientific, cat. nos. 1181-3710, 1180-8710, and 1182-1730)
Extended-length pipette tips (1,000 μL; Eppendorf, cat. no. 0030073614)
Analytical balance (Radwag, model no. AS 60/220.R2)
Ultrasonic bath (Branson, cat. no. 15-336-100)
Ultrasonic homogenizer with 6-mm tip (Cole-Parmer, cat. nos. UX-04711-70 and UX-04712-14)
Vortex mixer (Fisher Scientific, cat. no. 02-215-365)
pH meter (Spectrum, cat. no. 242-97839)
Orbital shaker (Waverly, cat. no. S1CE)
100,000-MWCO filter units (Amicon, cat. no. UFC910024)
NanoVue Plus spectrophotometer (GE Life Sciences, cat. no. 28-9569-61)
Visible spectrophotometer (Thermo Fisher Scientific, cat. no. 14-385-445)
Tabletop centrifuge (Eppendorf, cat. no. 5418000017)
Dynamic light-scattering instrument (Zetasizer Nano ZS; Malvern Instruments)
Folded capillary zeta cell (Malvern Instruments, cat. no. DTS1070)
Microscope cover glass (no. 1; Fisher Scientific, cat. no. 12-542B)
Microscope slides (VWR, cat. no. 16004-422)
Tweezers (VWR, cat. no. 63042-518)
Scissors (VWR, cat. no. 82027-582)
Mortar and pestle (Cole-Parmer, cat. no. EW-63100-54)
Syringe (1 mL; BD, cat. no. 14-823-434)
Plastic beakers (Nalgene, cat. no. 1201-0600)
Cryo gloves (US Solid, cat. no. JFLNT00008)
Sharpie marker (Sharpie, cat. no. 37001)
Software
ZEN Blue v.2.6 (https://www.zeiss.com/microscopy/us/downloads.html)
GraphPad Prism v.7.0a (https://www.graphpad.com/scientific-software/prism/)
Fiji ImageJ v.2.0.0 (https://imagej.net/Fiji/Downloads)
Reagent setup
5 M HCl solution
▲ CRITICAL Milli-Q water should be used wherever water is mentioned, unless otherwise specified. 5 M HCl solution
Add 4.2 mL of 37% (vol/vol) HCl dropwise to 40 mL of Milli-Q water. Bring the volume to 50 mL with water. Store the solution at room temperature for up to 3 months. Sterilization is not necessary !CAUTION Always add acid to water and not vice versa.
10 N NaOH solution
To 80 mL of H2O, slowly add 40 g of NaOH pellets, stirring continuously. As an added precaution, place the beaker on ice after the addition of NaOH pellets. When the pellets have dissolved completely, adjust the volume to 100 mL with H2O. Store the solution in a plastic container at room temperature for up to 3 months. Sterilization is not necessary !CAUTION The preparation of 10 N NaOH is highly exothermic, which can cause breakage of glass containers. Prepare this solution with extreme care in plastic beakers.
100 mM MgCl2 solution
To prepare 100 mL of 0.1 M MgCl2 solution, dissolve 2.03 g of MgCl2.6H20 in 80 mL of nuclease-free water. Adjust the volume to 100 mL using H2O. Sterilize with a 0.45-μm sterile syringe, filter, and store the solution at room temperature for up to 1 month !CAUTION MgCl2 is extremely hygroscopic. Buy small bottles and do not store opened bottles for long periods of time. Once the crystals become saturated with H2O, dispose of the chemical properly or release the water under vacuum via heat treatment at <50 °C.
500 mM MES buffer solution (pH = 4.5–5.0)
Prepare 80 mL of nuclease-free water in a suitable container. Add 9.762 g of MES hydrate to the solution. The starting pH for 0.5 M MES solution is ~3.2. Add 10 N NaOH to adjust the pH to 4.5–5.0. Add nuclease-free water until the volume is 100 mL. Sterilize with a 0.45-μm sterile syringe filter and store the solution at room temperature for up to 1 month.
MES delivery buffer: 25 mM MES, 15 mM MgCl2 at pH 6
Mix 5 mL of 500 mM MES buffer solution and 15 mL of 100 mM MgCl2 solution in a suitable container. Bring the volume to 80 mL with nuclease-free water and fix the pH to 6 with 10 N NaOH. Add nuclease-free water until volume is 100 mL. Sterilize with a 0.45-μm sterile syringe filter and store the solution at room temperature for up to 1 month.
Procedure
!CAUTION All SWNT waste (SWNT solutions, leaves infiltrated with SWNTs, flow-through solutions from 100,000-MWCO filters) should be discarded as hazardous chemical waste and should be disposed of per your institution’s guidelines.
COOH-SWNT suspension preparation ● Timing 2 h 30 min
-
1
Weigh out 30 mg of dry COOH-SWNTs into a 50-mL conical tube inside a chemical fume hood and add 30 mL of nuclease-free water.
!CAUTION All dry CNTs must be handled in a chemical fume hood and their waste should be discarded as hazardous chemical waste.
-
2
Bath-sonicate for 10 min at room temperature. COOH-SWNTs should be visibly suspended in water to form a dark black solution.
-
3
Probe tip–sonicate continuously for 30 min at 10% amplitude (with a 6-mm probe tip) in an ice bath using an ultrasonic homogenizer. The typical power we observe for these sonication parameters is ~30–40 W. After probe-tip sonication, the COOH-SWNT suspension should still be dark black.
▲ CRITICAL STEP Make sure that the ice does not completely melt during the course of sonication.If the ice is melting, replace the ice bath and continue sonication.
▲ CRITICAL STEP Ensure that the conical tube is either clamped or affixed so that the probe tip is in contact with only the COOH-SWNT suspension, not the walls of the conical tube (Fig. 2).
-
4
Remove the tube from the ice bath and allow it to rest at room temperature for 10 min.
-
5
Centrifuge at maximum speed (~18,000g) for 1 h at room temperature. Next, collect the supernatant, which contains individually suspended COOH-SWNTs and determine the COOH-SWNTs concentration.
▲ CRITICAL STEP Collecting supernatant is challenging because the concentrated nature of SWNTs makes it difficult to differentiate supernatant from pellet. Begin pipetting slowly and carefully; recover all the supernatant except the last ~2–3 mL to ensure minimal disruption of pellet.
▲ CRITICAL STEP Measure concentration using absorbance at 632 nm with an extinction coefficient of 0.036 L mg−1 cm−1. Typical COOH-SWNT solution yields are ~100–150 mg L−1 after centrifugation. Because this solution has very high optical density, dilute a small quantity of it ~10 times for absorbance measurements. See Troubleshooting for yields with substantially lower concentrations.
? TROUBLESHOOTING
■ PAUSE POINT The COOH-SWNT suspension can be stored for a month at room temperature or 4 °C for further reactions. Bath-sonicate for 30 min before the reaction, if the COOH-SWNTs have been stored for more than a week.
Fig. 2 |. Probe-tip sonication setup.
a, Use a beaker and copper wire to fix the conical tube to the beaker. b, Fill the beaker with ice and place the probe tip into the solution, making sure that the tip does not touch the tube bottom or walls and is submerged in the solution (for Steps 3 and 19). For maximum energy dispersal, position the edge of the probe tip in the middle of the solution.
PEI reaction with COOH-SWNTs ● Timing 3 h + overnight
▲ CRITICAL The scale of the reaction should be tuned on the basis of the quantity of the product needed. In this section, we will present a reaction protocol that starts with 2 mg of suspended COOH-SWNT solution and results in sufficient product for 4,000 infiltrations at the quantities suggested below. Be aware that the lower amount of SWNT will wash faster with the filters; hence, adjust the centrifugation time accordingly.
-
6
Prepare a 500 mM MES buffer solution in nuclease-free water at pH 4.5–5.0 (see ‘Reagent setup’ section).
-
7
Calculate the volume of COOH-SWNTs needed to react 2 mg. Add the appropriate volume of 500 mM MES buffer solution to 2 mg of suspended COOH-SWNTs to have a final MES concentration of 100 mM. For example, if the final COOH-SWNT concentration is 100 mg L−1, use 20 mL of COOH-SWNTs and add 5 mL of 500 mM MES to it. This will result in 25 mL of COOH-SWNT suspension that has a 100 mM MES final concentration.
▲ CRITICAL STEP Make sure the pH of this solution is between 4.5 and 6, because EDC–NHS activation reaction is most efficient in this pH range30.
-
8
Prepare a fresh EDC–NHS solution in 100 mM MES (pH 4.5–5.0). Add 10 mg EDC and 10 mg NHS to 2.5 mL of 100 mM MES solution. Make sure the EDC and NHS powders are completely dissolved and mixed well.
▲ CRITICAL STEP EDC–NHS solution must be prepared fresh for each reaction immediately before the experiment, because the activity of these chemicals in aqueous solutions will decrease substantially over time.
-
9
Add the complete EDC–NHS solution slowly dropwise to the COOH-SWNT suspension from Step 7 under stirring. Bath-sonicate for 15 min at room temperature. Let the reaction continue for 45 min on an orbital shaker at ~180 r.p.m. at room temperature.
-
10
Pre-wash two 100,000-MWCO filters once with 15 mL of 0.1× PBS to wet the membranes by centrifuging at maximum speed for 2 min at room temperature.
-
11
Wash the activated COOH-SWNT solution from Step 9 with 0.1× PBS (pH 7.4) three times using (pre-washed) 100,000-MWCO filters to remove free EDC, NHS, and by-products as follows: split the activated COOH-SWNT solution from Step 9 into two and pour into two filter units. Fill the volume up to 50 mL line in each filter unit with 0.1× PBS (pH 7.4). Centrifuge at 300g for 8 min at 21 °C, discard the flow-through and repeat the wash two more times with 0.1× PBS (pH 7.4), following the same protocol (Fig. 3a,b). Vortex the activated COOH-SWNT solution briefly after each wash step.
▲ CRITICAL STEP If the reaction is scaled up to use a higher amount of SWNTs, it may be necessary to use more than two filter units for efficient washing.
? TROUBLESHOOTING
-
12
After the last wash step, bath-sonicate the two 100,000-MWCO filters (only the top parts) containing the activated COOH-SWNTs for 1 min while pipetting the solution up and down. Place the filters back into the collection tubes, close the tubes with their caps, and vortex briefly. Recover the activated COOH-SWNTs with a 1,000-μL extended-length pipette tip (Fig. 3c). A typical recovery volume from each filter should be ~4–5 mL. Merge the activated COOH-SWNT solutions in one tube after recovery from filters.
-
13
Add 0.1× PBS (pH 7.4) to the activated COOH-SWNT solution until reaching the initial COOH-SWNT solution volume used in Step 7 before the addition of MES buffer (20 mL in this example). Resuspend the washed and activated COOH-SWNTs via bath sonication for 15 min.
-
14
Add 40 mg of PEI (25,000 MW, branched) to 5 mL of 0.1× PBS and dissolve fully. Fix the pH of this PEI solution to 7.4–7.6 with 5 M HCl.
▲ CRITICAL STEP PEI dissolution in PBS can take a long time. Using an inoculation loop and shaking on the orbital shaker for 10–15 min expedites the process. Make sure the PEI is fully dissolved before fixing the pH.
-
15
Add the activated COOH-SWNTs (from Step 13) to the PEI solution dropwise. Run the reaction overnight (~16 h) at room temperature on an orbital shaker at ~180 r.p.m. It is okay if SWNTs agglomerate slightly after mixing with PEI solution and after the reaction is performed overnight.
▲ CRITICAL STEP For less agglomeration, add activated COOH-SWNTs to PEI solution, and not the other way around.
▲ CRITICAL STEP This amination reaction is most efficient at a pH range of 7–8. Make sure the solution—after mixing in the PEI and COOH-SWNTs—is still in this pH range; make adjustments with 5 M HCl or 10 N NaOH if necessary.
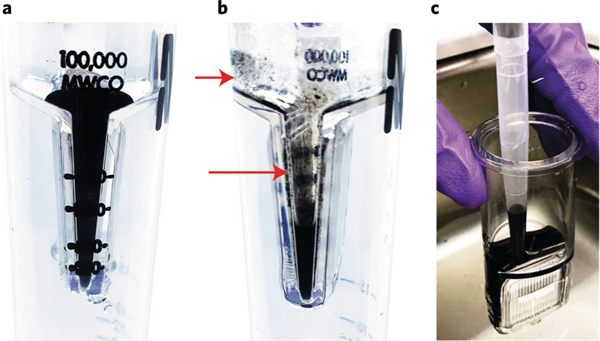
Fig. 3 |. Suggested usage of Amicon 100,000-MWCO filters for washing.
a, Correct amount of solution passing through the membrane leaves enough liquid to cover the membrane surface. b, Excessive centrifugation times or speeds cause too much solution to pass through the membrane, which leaves the membrane surface exposed and aggregates the SWNTs on the membrane (shown by red arrows). c, Recover the solution from the spin filter using a 1,000-μL extended-length pipette tip after quick bath sonication and mixing. Descriptions here apply to Steps 11, 12, 17, and 18.
Washing and suspending the PEI-SWNT product ● Timing 5 h
-
16
Pre-wash two 100,000-MWCO filters once with 15 mL of nuclease-free water to wet the membranes by centrifuging at maximum speed for 2 min at room temperature.
-
17
Split the PEI-SWNT reaction solution from Step 15 into two and transfer each solution half into one of the (prewashed) 100,000-MWCO filters. Wash the PEI-SWNT solutions with nuclease-free water six times as follows: fill the volume up to 50 mL line in each filter unit with nuclease-free water. Centrifuge at 1,000g for 15–20 min at 21 °C, discard the flow-through, and repeat the wash five more times with nuclease-free water, following the same protocol. Vortex the PEI-SWNT solution briefly after each wash step.
▲ CRITICAL STEP It is important not to let the PEI-SWNT liquid level go below the membrane level when washing, because this will contribute to PEI-SWNT aggregation and sample loss (Fig. 3b). Adjust the centrifugation speed and time accordingly. Lower centrifugation speeds reduce sample loss on membranes.
▲ CRITICAL STEP If the reaction is scaled up to use higher amount of SWNTs, it may be necessary to use more than two filter units for efficient washing.
-
18
After the last wash step, bath-sonicate the two 100,000-MWCO filters (only the top parts) containing the PEI-SWNT solution for 1 min while pipetting up and down. Place the filters back into the collection tubes, close the tubes with their caps, and vortex briefly. Recover the PEI-SWNTs with a 1,000-μL extended-length pipette tip (Fig. 3c) and merge them in one tube.
-
19
Add nuclease-free water to the PEI-SWNT solution until reaching the initial COOH-SWNT solution volume used in Step 7 before the addition of MES buffer (20 mL in this example). Resuspend the washed PEI-SWNTs through bath and probe-tip sonication. Bath-sonicate the PEI-SWNTs for 15 min (the PEI-SWNT solution should be well dispersed and dark in color after the bath sonication step), and then probe tip–sonicate the PEI-SWNTs at 10% amplitude for 15 min with a 6-mm probe tip in an ice bath using an ultrasonic homogenizer. The typical power we observe for this sonication is ~30–40 W. The final PEI-SWNT solution after probe-tip sonication should still have high optical density (dark black in color).
▲ CRITICAL STEP Replace the ice bath with fresh ice after the first half of the probe-tip sonication to prevent overheating of the solution and continue sonication. Ensure that the conical tube is either clamped or affixed so that the probe tip is in contact with only the PEI-SWNT suspension and not the walls of the conical tube.
? TROUBLESHOOTING
-
20
Centrifuge at 16,000g for 1 h at room temperature and collect the supernatant.
-
21
Measure concentration of PEI-SWNTs via absorbance at 632 nm with an extinction coefficient of 0.036 L mg−1 cm−1.
It is normal for some SWNT pelleting to occur during the centrifugation step. We typically obtain 40–70 mg L−1 of PEI-SWNTs after centrifugation and observe a reaction efficiency of ~40 ± 10%, with efficiency calculated using Eq. (1).(1) -
22
Measure the zeta potential of the PEI-SWNTs using the Zetasizer Nano ZS to assess their colloidal stability and to confirm PEI attachment. We typically obtain zeta potential values between +50 and +70 mV.
■ PAUSE POINT The PEI-SWNT suspension can be stored at 4 °C for a month for further experiments. Bath-sonicate for 30 min before cargo loading, if PEI-SWNTs have been stored for more than a week.
DNA loading onto PEI-SWNTs ● Timing 1 h
▲ CRITICAL For DNA delivery to plants, optimization may be necessary for different plant species, plant tissues, and DNA constructs. Parameters that can be varied are total DNA amount and SWNT/DNA ratio in the infiltration mix. The protocol here is optimized for delivery of ~4.2 kb of plasmid DNA into 1-month-old N. benthamiana plant leaves.
-
23
Prepare MES delivery buffer (see ‘Reagent setup’ section).
-
24
To load 500 ng PEI-SWNTs with 167 ng plasmid DNA in a 3:1 PEI-SWNT:DNA mass ratio, first dilute 500 ng of PEI-SWNTs in 100 μL of MES delivery buffer per infiltration (account for the volume that DNA will be occupying when diluting the PEI-SWNTs in MES buffer). Add the diluted PEI-SWNTs drop by drop to the 167 ng of plasmid solution. Pipette up and down 10 times to mix.
▲ CRITICAL STEP For example, if the starting PEI-SWNT and plasmid DNA concentrations are 40 ng μL−1, add 12.5 μL of PEI-SWNTs to 83.3 μL of MES delivery buffer and mix well. Pipette 4.2 μL of DNA into a separate microcentrifuge tube and add the diluted PEI-SWNT solution to the DNA solution drop by drop.
▲ CRITICAL STEP The 100-μL infiltration volume and its constituents can be scaled up, if desired, to infiltrate a larger leaf area or an entire leaf for western blot analysis.
-
25
Incubate at room temperature for 30 min for the formation of the DNA-PEI-SWNT complex (Fig. 4).
▲ CRITICAL STEP If agglomeration of SWNTs is visible (it will look like tiny black specs in the solution), do not continue with the infiltration, because the agglomerated SWNTs will not be able to internalize into cells.
? TROUBLESHOOTING
Fig. 4 |. Stability of the DNA-loaded PEI-SWNT suspension.
a, Representative image of stable DNA loading on PEI-SWNTs, showing no SWNT agglomeration (Step 25). b, Representative image of unstable DNA loading on PEI-SWNTs, showing substantial SWNT agglomeration (black specks).
Infiltration of DNA-PEI-SWNTs into leaves ● Timing 1 min per infiltration
-
26
After a 30-min incubation, put the DNA-PEI-SWNT solution into a 1-mL needleless syringe. Depending on the plant species, it may help to induce a tiny puncture with a pipette tip on the leaf abaxial surface; thicker and more hydrophobic leaf tissue may require this puncture for infiltration. Center the syringe tip at puncture area and gently push the syringe plunger until all the liquid is infiltrated. Rest the leaf between your thumb and index finger while infiltrating (Fig. 5).
▲ CRITICAL STEP Ensure there are no air bubbles inside the syringe when infiltrating. If air is infiltrated into the leaves, it may cause tissue deformation or damage. To remove air bubbles interspersed with fluid, in a ventilated chemical hood, orient syringe outlet toward the ceiling and gently flick the syringe tip.
▲ CRITICAL STEP For protein quantification in Step 30C, we suggest infiltrating the entire leaf area (or multiple leaves) to collect enough leaf sample for quantification.
-
27
Remove the excess solution from leaf surfaces after the infiltration with a Kimwipe. Gently dab the leaf surface.
▲ CRITICAL STEP When delivering DNA encoding for fluorescent proteins, placing the infiltrated plant back in the growth chamber may cause underestimation of expression efficiency. High-intensity growth chamber light may cause photobleaching of the expressed fluorescent protein (depending on the protein photostability and light conditions in the chamber) over 2–3 d.
-
28
Gently mark the area of infiltration with a Sharpie marker on the leaf to keep note of which areas received the gene of interest.
-
29
Wait 1 d for plasmid DNA transcription to mRNA, and 2–3 d for mRNA translation into protein.
? TROUBLESHOOTING
Fig. 5 |. Infiltration of leaves with DNA-loaded PEI-SWNTs.
a, A tiny puncture induced with a pipette tip on the leaf abaxial surface. b, Infiltration through puncture area by gently pushing the syringe plunger until all the liquid is infiltrated. c, Remove excess solution with a Kimwipe. Area of infiltration can be increased by infiltrating a larger volume of DNA-PEI-SWNT solution (Step 26).
Evaluation of gene expression
-
30Evaluate transgene expression. The choice of the preferred method of evaluating transgene expression largely depends on the properties of the expressed protein. For reporter fluorescent proteins, we suggest performing both qPCR analysis (option A) and confocal microscopy imaging (option B). For evaluating the expression of non-fluorescent proteins, we suggest qPCR (option A) and our modified western blot analysis (option C), if antibodies are available for the given protein. If desired, a ddPCR method can also be used on genomic DNA to test whether the delivered plasmid has been integrated into the plant nuclear genome31,32.
- qPCR analysis ● Timing 5 h
-
24 h after infiltration, cut the infiltrated leaf area (maximum of 100 mg of leaf tissue) and extract total RNA with an RNeasy Plant Mini Kit. After cutting the leaf, immediately proceed with the first step of the RNeasy RNA extraction protocol (i.e., grinding the tissue in liquid nitrogen) to make sure gene expression levels do not change after cutting.!CAUTION Handle liquid nitrogen carefully. Protect hands at all times with cryo gloves. Protect your eyes with safety goggles and wear appropriate personal protective equipment (lab coat, closed-toe shoes, and long pants).
-
Following RNA extraction, measure the concentration and reverse-transcribe 1 μg of total RNA into cDNA with an iScript cDNA synthesis kit.■ PAUSE POINT The cDNA can be stored at 4 or −20 °C overnight.
-
Use PowerUp SYBR Green Master Mix for the qPCR step with 2 μL of cDNA from Step 30A(ii) and specific primers for the target and housekeeping genes.▲ CRITICAL STEP It is necessary to run a housekeeping gene (such as EF1) in addition to the target gene to account for differences in plant/leaf conditions and possibly slightly different amounts of starting leaf tissue mass and total mRNA extracted. We suggest checking the literature for appropriate housekeeping genes, because these may differ for different plant species.
-
Analyze the qPCR data using the delta–delta cycle threshold (ddCt) method33.? TROUBLESHOOTING
-
- Confocal microscopy imaging ● Timing 30 min per sample
- Wait for 48–72 h after DNA-PEI-SWNT infiltration to allow protein expression and accumulation to take place.
- Cut a small area of the infiltrated leaf within the infiltration area and put the tissue on a glass slide with the abaxial leaf surface facing upward. Cover with a thickness no. 1 coverslip and add water between the slide and coverslip. Make sure there are no water bubbles and no air bubbles between the leaf and coverslip. If bubbles are present, tap on the coverslip several times to remove air and water bubbles. Image through the abaxial side of the leaf.
-
Image the sample with the appropriate filter sets depending on the fluorescent protein of interest.? TROUBLESHOOTING
- Protein quantification ● Timing 3 h
- Wait for 48–72 h after DNA-PEI-SWNT infiltration for protein expression and accumulation to take place.
- Extract total proteins from the infiltrated leaves as described in ref. 18. Briefly, grind infiltrated plant leaves in liquid nitrogen to recover dry frozen powders. Transfer the frozen powders to microcentrifuge tubes with pre-prepared lysis buffer containing 10 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% (vol/vol) NP-40, 5% (vol/vol) glycerol, and 1% (vol/vol) protease inhibitor cocktail. After lysis at 4 °C overnight, centrifuge the tubes at 10,000 r.p.m. for 15 min at 21 °C and collect the supernatants containing whole proteins into a new tube. Quantify the total extracted proteins with a Pierce 660nm Protein Assay.
- A GFP-Trap Agarose (GFP-Trap A) or similar available kits for the protein of interest canbe used to purify and concentrate the protein extracted in Step 30C(ii).
-
Quantify the protein amount with a Qubit Protein Assay.? TROUBLESHOOTING
Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2 |.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | Low concentration of COOH-SWNT suspension | Not enough dry COOH-SWNT added | It is hard to accurately measure small amounts of dry CNTs; use more COOH-SWNT powder for making the suspension |
| Probe tip was touching the sides of the conical tube or was not optimally placed | Make sure the probe tip is not touching the conical tube and is submerged fully in the solution, resting at a distance halfway from the fluid meniscus | ||
| The solution overheated during the probe-tip sonication | Change the ice bath every 10 min to prevent overheating. If none of these solutions work, the resulting COOH-SWNT suspension can be concentrated using 100,000-MWCO centrifugal filters | ||
| 11 | Excessive agglomeration or pelleting of activated COOH-SWNTs during washes with PBS (some degree of SWNT destabilization can happen, and it will not affect the next steps, if the SWNTs can be suspended again with bath sonication) | PBS salt concentration is too high | Make sure to use 0.1× PBS for washes, as 1x PBS will affect the colloidal stability of SWNTs adversely because of the high salt concentration |
| The starting COOH-SWNT solution was not well suspended or was too old | Suspend the COOH-SWNTs well in water and use only the supernatant of the centrifuged COOH-SWNTs. If using COOH-SWNTs from a previously suspended batch, bath-sonicate for at least 30 min or make a new COOH-SWNT suspension | ||
| Too much EDC and/or NHS was used | EDC-NHS is used in excess for this reaction, but too much of it can agglomerate the SWNTs. Decrease the amount of EDC-NHS until no agglomeration is observed | ||
| Too much solution was passed through the filtration membrane during washing | Always keep the membranes submerged in the reaction solution to prevent pelleting of SWNTs on membranes. If more liquid than desired is passing through and the membrane surface is exposed, decrease the centrifugation speed and/or time | ||
| 19 | PEI-SWNT reaction product is not suspending in water or has a low final concentration (<20 mg L−1) | The PEI was not well-dissolved in solution before addition to the activated COOH-SWNTs | After making the PEI suspension, place it on an orbital shaker at maximum speed overnight |
| Overheating of the solution during probe-tip sonication | Make sure to change the ice bath at least once or twice during the probe-tip sonication. Alternatively, use the pulse feature of the ultrasonic homogenizer with a ‘1 s on and 2 s off’ setting | ||
| Addition of not enough or too much PEI polymer for the reaction (not enough PEI may cause multiple SWNTs to be covalently bonded together by one PEI polymer, and too much PEI may cause formation of larger particles through non-specific adsorption, both of which will prevent the successful suspension of individual SWNTs) | Use the optimized amount suggested in this paper and, if scaling the reaction up or down, also scale the amount of PEI accordingly. Here, we suggest using 40 mg of PEI (25,000 MW, branched) for 2 mg of COOH-SWNTs | ||
| 25 | PEI-SWNT agglomeration upon plasmid DNA addition | PEI-SWNT positive charge density is not enough to carry added plasmid negative charge | Either increase the PEI-SWNT amount or decrease the plasmid DNA amount. Alternatively, redo PEI-SWNT synthesis to achieve particles with higher zeta potential values |
| 29 | Observed toxicity or damage in the infiltrated leaves | Too much PEI-SWNT was used for infiltration | For the reaction detailed here, we suggest using 500–1,000 ng of PEI-SWNTs per infiltration, because more may cause damage. Use less PEI-SWNTs if damage is observed with 500 ng |
| PEI-SWNT particles contain too much free PEI | If the washing is not performed efficiently, there may be too much free PEI left in the solution. Perform more wash cycles after the PEI reaction | ||
| SWNTs were functionalized with too much PEI | Instead of using 40 mg of PEI for 2 mg of COOH-SWNTs, repeat the reaction with less PEI. We suggest testing PEI amounts between 20 and 40 mg per 2 mg of COOH-SWNTs to troubleshoot | ||
| 30A(iv), B(iii), C(iv) | No gene expression was observed with any method | Plants or leaves used for delivery were too old or unhealthy | Use 1-month-old healthy plants and young leaves to test the constructs |
| Need to optimize the DNA-loading protocol | For DNA-loading optimization, we suggest testing between 500 and 1,000 ng of PEI-SWNTs in a total of 100 μL of solution per leaf infiltration, with the following SWNT/DNA mass ratios: 6:1, 3:1,1:1, 1:3, and 1:6 | ||
| PEI-SWNTs may be toxic to cells even though no visible leaf damage is observed | Follow the troubleshooting suggestions for Step 29 | ||
| Plasmid DNA construct is not working | Make sure the plasmid DNA is fully sequenced and induces expression when delivered via Agrobacterium to leaves or via PEG transfection in protoplasts | ||
| 30B(iii) | No fluorescent protein expression is detected with confocal microscopy | Low laser power | Use high laser power (80–100% of maximum) and a 20× objective. Scan the entire area carefully for cells that are expressing the fluorescent protein |
| Gene expression was not strong and uniform as with Agrobacterium-mediated delivery | Unlike Agrobacterium-mediated expression, the expression via SWNTs is less strong, and not all cells in the infiltration area will express the protein of interest |
Timing
Steps 1–5, COOH-SWNT suspension preparation: 2 h 30 min
Steps 6–15, PEI reaction with COOH-SWNTs: 3 h + overnight
Steps 16–22, washing and suspension of the PEI-SWNT product: 5 h
Steps 23–25, DNA loading onto PEI-SWNTs: 1 h
Steps 26–29, infiltration of DNA-PEI-SWNTs into leaves: 1 min per infiltration
Step 30A, qPCR analysis: 5 h
Step 30B, confocal microscopy imaging: 30 min per sample
Step 30C, protein quantification: 3 h
Anticipated results
Using this protocol, we anticipate researchers will be able to generate and characterize PEI-modified SWNTs to deliver plasmid DNA into intact cells of mature model and crop plants for genetic transformations. Using a reporter gene (35S-GFP), we have demonstrated SWNT-based gene expression in N. benthamiana, E. sativa (arugula), T. aestivum (wheat), and G. hirsutum (cotton) leaves with confocal microscopy imaging (Fig. 6a) and with qPCR analysis (Fig. 6b) at 3 d post infiltration18. We have also shown that the expression is transient and disappears at 10 d post infiltration, with confocal imaging and qPCR (Fig. 6b)18. Furthermore, we have quantified the GFP protein amount that is transiently produced in plant leaf cells via GFP protein extraction, purification, and concentration using a GFP-Trap Agarose kit and a Qubit Protein Assay, which revealed that 13.6 μg of GFP was obtained per gram of fresh leaf18.
Fig. 6 |. Anticipated results.
a, Wild-type N. benthamiana (Nb), arugula, wheat, and cotton leaves infiltrated with DNA-PEI-SWNTs are imaged with confocal microscopy to determine GFP expression levels in the leaf lamina of each plant species (Step 30B). b, qPCR analysis of GFP mRNA expression levels at days 3 and 10 in DNA-PEI-SWNT-treated Nb leaves (top; Step 30A). **P = 0.0003 in two-way ANOVA. Quantitative fluorescence intensity analysis of confocal images at 3 and 10 d post infiltration (bottom). ***P = 0.0001 in two-way ANOVA. Scale bars, 50 μm. Error bars indicate s.e.m. (n = 3). Image adapted with permission from ref. 18, Springer Nature Limited.
Supplementary Material
Acknowledgements
We acknowledge support from a Burroughs Wellcome Fund Career Award at the Scientific Interface (CASI), a Stanley Fahn PDF Junior Faculty Grant under award no. PF-JFA-1760, a Beckman Foundation Young Investigator Award, a USDA AFRI award, a grant from the Gordon and Betty Moore Foundation, a USDA NIFA award, a USDA-BBT EAGER award, support from the Chan-Zuckerberg Foundation, and an FFAR New Innovator Award (to M.P.L.). G.S.D. was supported by a Schlumberger Foundation Faculty for the Future Fellowship. H.Z. acknowledges the support of the National Natural Science Foundation of China (21605153). We also acknowledge support from the UC Berkeley Molecular Imaging Center (supported by the Gordon and Betty Moore Foundation), the QB3 Shared Stem Cell Facility, and the Innovative Genomics Institute (IGI). We are grateful to the Staskawicz lab at UC Berkeley for the N. benthamiana seeds.
Footnotes
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41596-019-0208-9.
Peer review information Nature Protocols thanks Ardemis Boghossian, Mohammad Ramezani and other anonymous reviewer(s) for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Data availability
All materials are available from commercial sources or can be derived using methods described in this study. All primary data underlying the figures reported in the article can be obtained from the corresponding author upon reasonable request.
References
- 1.Goswami R, Dasgupta P, Saha S, Venkatapuram P. & Nandi S. Resource integration in smallholder farmsfor sustainable livelihoods in developing countries. Cogent Food Agric. 2, 10.1080/23311932.2016.1272151(2016). [DOI] [Google Scholar]
- 2.Altpeter F. et al. Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Himmel ME et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Baltes NJ, Gil-Humanes J. & Voytas DF Genome engineering and agriculture: opportunities andchallenges. Prog. Mol. Biol. Transl 149, 1–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herreraestrella L, Depicker A, Vanmontagu M. & Schell J. Expression of chimaeric genes transferred intoplant-cells using a Ti-plasmid-derived vector. Nature 303, 209–213 (1983). [Google Scholar]
- 6.Binns AN Agrobacterium-mediated gene delivery and the biology of host range limitations. Physiol. Plant 79, 135–139 (1990). [Google Scholar]
- 7.Klein TM, Wolf ED, Wu R. & Sanford JC High-velocity microprojectiles for delivering nucleic-acidsinto living cells. Nature 327, 70–73 (1987). [PubMed] [Google Scholar]
- 8.Cunningham FJ, Goh NS, Demirer GS, Matos JL & Landry MP Nanoparticle-mediated deliverytowards advancing plant genetic engineering. Trends Biotechnol. 36, 882–897 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russell JA, Roy MK & Sanford JC Physical trauma and tungsten toxicity reduce the efficiency of biolistic transformation. Plant Physiol. 98, 1050–1056 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q. et al. Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 9, 1007–1010 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Serag MF et al. Trafficking and subcellular localization of multiwalled carbon nanotubes in plant cells. ACS Nano 5, 493–499 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Chang F-P et al. A simple plant gene delivery system using mesoporous silica nanoparticles as carriers. J. Mater. Chem B 1, 5279–5287 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Zhang H. et al. DNA nanostructures coordinate gene silencing in mature plants. Proc. Natl Acad. Sci. USA 116, 7543–7548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng KK et al. Intracellular delivery of proteins via fusion peptides in intact plants. PLoS One 11, e0154081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitter N. et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses.Nat. Plants 3, 16207 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Giraldo JP et al. Plant nanobionics approach to augment photosynthesis and biochemical sensing.Nat. Mater 13, 400–408 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Demirer GS, Zhang H, Goh NS, Chang R. & Landry MP Nanotubes effectively deliver siRNA tointact plant cells and protect siRNA against nuclease degradation. Preprint at bioRxiv, 10.1101/564427 (2019). [DOI]
- 18.Demirer GS et al. High aspect ratio nanomaterials enable delivery of functional genetic material withoutDNA integration in mature plants. Nat. Nanotechnol 14, 456–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak S-Y et al. Nanosensor technology applied to living plant systems. Annu. Rev. Anal. Chem 10, 113–140 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Wong MH et al. Nitroaromatic detection and infrared communication from wild-type plants using plantnanobionics. Nat. Mater 16, 264 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Del Bonis-O’Donnell JT et al. Engineering molecular recognition with bio-mimetic polymers on single walled carbon nanotubes. J. Vis. Exp 2017, e55030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak S-Y et al. Chloroplast-selective gene delivery and expression in planta using chitosan-complexedsingle-walled carbon nanotube carriers. Nat. Nanotechnol 14, 447–455 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun 7, 12617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Lombi E, Zhao FJ & Kopittke PM Nanotechnology: a new opportunity in plant sciences.Trends Plant Sci. 21, 699–712 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Kikkert JR, Vidal JR & Reisch BI in Transgenic Plants: Methods and Protocols 61–78 (Springer, 2005). [Google Scholar]
- 26.Liu Y, Yang H. & Sakanishi A. Ultrasound: mechanical gene transfer into plant cells by sonoporation. Biotechnol. Adv 24, 1–16 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Asad S. & Arshad M. Silicon carbide whisker-mediated plant transformation. in Properties and Applications of Silicon Carbide, 345–358 (InTech Open, 2011).
- 28.Lakshmanan M, Kodama Y, Yoshizumi T, Sudesh K. & Numata K. Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules 14, 10–16 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Yoo S-D, Cho Y-H & Sheen JJ Np Arabidopsis mesophyll protoplasts: a versatile cell system fortransient gene expression analysis. Nat. Protoc 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Vashist SK Comparison of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide based strategies to crosslinkantibodies on amine-functionalized platforms for immunodiagnostic applications. Diagnostics 2, (23–33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobnik D, Stebih D, Blejec A, Morisset D. & Zel J. Multiplex quantification of four DNA targets in one reaction with Bio-Rad droplet digital PCR system for GMO detection. Sci. Rep 6, 35451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collier R. et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR.Plant J. 90, 1014–1025 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Zhang JD, Ruschhaupt M. & Biczok R. ddCt method for qRT–PCR data analysis http://bioconductor.jp/packages/2.14/bioc/vignettes/ddCt/inst/doc/rtPCR.pdf (2010).
- 34.Zheng N, Song Z, Liu Y, Yin L. & Cheng J. Gene delivery into isolated Arabidopsis thaliana protoplasts and intact leaves using cationic, α-helical polypeptide. Front. Chem. Sci. Eng 11, 521–528 (2017). [Google Scholar]
- 35.Chugh A. & Eudes F. Study of uptake of cell penetrating peptides and their cargoes in permeabilized wheatimmature embryos. FEBS J. 275, 2403–2414 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Chuah J-A, Yoshizumi T, Kodama Y. & Numata KJ Sr Gene introduction into the mitochondria of Arabidopsis thaliana via peptide-based carriers. Sci. Rep 5, 7751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials are available from commercial sources or can be derived using methods described in this study. All primary data underlying the figures reported in the article can be obtained from the corresponding author upon reasonable request.