Abstract
We have developed a PCR assay for one-step differentiation of the three complexes of New World Leishmania (Leishmania braziliensis, Leishmania mexicana, and Leishmania donovani). This multiplex assay is targeted to the spliced leader RNA (mini-exon) gene repeats of these organisms and can detect all three complexes simultaneously, generating differently sized products for each complex. The assay is specific to the Leishmania genus and does not recognize related kinetoplastid protozoa, such as Trypanosoma cruzi, Trypanosoma brucei, and Crithidia fasciculata. It correctly identified Leishmania species with a broad geographic distribution in Central and South America. The sensitivity of the PCR amplification ranged from 1 fg to 10 pg of DNA (0.01 to 100 parasites), depending on the complex detected. Crude extracts of cultured parasites, prepared simply by boiling diluted cultures, served as excellent templates for amplification. Crude preparations of clinical material were also tested. The assay detected L. braziliensis in dermal scrapings from cutaneous leishmanial lesions, Leishmania chagasi in dermal scrapings of atypical cutaneous leishmaniasis, and L. mexicana from lesion aspirates from infected hamsters. We have minimized the material requirements and maximized the simplicity, rapidity, and informative content of this assay to render it suitable for use in laboratories in countries where leishmaniasis is endemic. This assay should be useful for rapid in-country identification of Leishmania parasites, particularly where different Leishmania complexes are found in the same geographical area.
Parasitic protozoa of the genus Leishmania cause a broad spectrum of disease in humans throughout tropical and subtropical regions worldwide and are considered a major public health problem (41). Leishmaniasis is associated with a variety of clinical manifestations, depending on the species of the parasite, the host immune response, and factors in the saliva of the sand fly vector (19). In the Americas, 15 species of New World Leishmania are grouped into three complexes, which are responsible for (i) a self-curing ulcerative disease and diffuse cutaneous leishmaniasis (typically caused by the Leishmania mexicana complex), (ii) persistent cutaneous and often disfiguring mucocutaneous lesions (Leishmania braziliensis complex), and (iii) the potentially fatal visceral disease attributed to Leishmania chagasi (Leishmania donovani complex) (19). In addition, there have been reports of atypical infections which constitute exceptions to these rules (2, 3, 24, 44).
Parasitological confirmation of diagnosis is critical because the wide spectrum of symptoms can be caused by a number of other etiological agents, leading to potential misdiagnosis. In addition, the treatment for leishmaniasis is expensive, lengthy, and associated with toxic side effects (41). Further, species identification is important because different species can require distinct treatment regimens (19, 32, 35, 36). Characterization of Leishmania parasites is also necessary for epidemiological objectives, such as documenting the distribution of Leishmania species and designing appropriate control measures.
Microscopic examination of clinical material or cultured parasites is not adequate for species identification due to the morphological similarity of the different species (36). Alternative methods for identifying the complex and species of Leishmania include biochemical techniques such as isoenzyme (zymodeme) analysis (27), immunological approaches with monoclonal antibodies (serodeme analysis) (18), and classical molecular techniques involving restriction analysis of kinetoplast DNA (kDNA) (schizodeme analysis) (52). Zymodeme and schizodeme analysis involve lengthy, complicated, and expensive procedures that require large-scale cultivation of parasites and a sophisticated laboratory setting. While certain monoclonal antibodies appear promising for species identification by serodeme analysis, the reactivity patterns of some species vary significantly depending on the geographical origin of the parasites (17).
PCR can offer a rapid, sensitive, specific, and low-cost alternative. A number of PCR assays for identification of Leishmania at the genus level (8, 30, 48) or for characterization of individual complexes of L. braziliensis (14, 20, 28, 33), L. mexicana (15, 33), or L. donovani (13, 23, 33, 43, 47, 51) have been described. However, none of these PCR protocols identifies all three complexes simultaneously. Randomly amplified polymorphic DNA (RAPD) analysis has been reported for species-level identification of Leishmania, but it requires purified DNA and highly standardized conditions and results in a sensitivity much lower than that of standard PCR (38). Other molecular approaches involve identification of the Leishmania complexes by PCR coupled with DNA probe hybridization (10, 39, 45, 46, 53), restriction enzyme analysis, or single-strand conformation polymorphism (54). DNA probes have been developed for speciation of Leishmania, but they are substantially more time-consuming and less sensitive than PCR amplification (56).
Here we present a simple, sensitive, and specific one-step PCR assay for differentiating the three complexes of New World Leishmania, using the multicopy spliced leader (SL) RNA (mini-exon) gene as a target. This assay generates complex-specific products of different sizes for L. braziliensis, L. mexicana, and L. donovani and is suitable for use in nonsophisticated laboratories in countries where leishmaniasis is endemic.
MATERIALS AND METHODS
Parasite strains and clinical samples.
The strains of Leishmania used in this study are listed in Table 1 and include nine World Health Organization reference strains and seven characterized isolates from Nicaragua, Honduras, Venezuela, and Peru. Related kinetoplastid parasites were obtained from the American Type Culture Collection, International Laboratory for Research on Animal Disease, and Nicaragua, as indicated in the table. Clinical samples from the following patients were analyzed in this study: (i) 30 patients from areas where cutaneous leishmaniasis is endemic who were referred to the Centro Nacional de Diagnóstico y Referencia, Managua, Nicaragua, for diagnosis of leishmaniasis, presenting with ulcerated cutaneous lesions, and 11 patients presenting with papular, nonulcerated lesions (from Leon and Apompua, Nicaragua), (ii) 1 patient referred to the Instituto de Servicios de Laboratorio de Diagnóstico y Investigación en Salud (SELADIS), Universidad Mayor de San Andrés, La Paz, Bolivia, for diagnosis of leishmaniasis; and (iii) 1 patient who presented at the California Pacific Medical Center (CPMC) in San Francisco with multiple lesions of presumptive cutaneous leishmaniasis and who had visited an area in Bolivia where leishmaniasis is endemic. Dermal scrapings from lesions were examined by microscopic observation for the presence of amastigotes on Giemsa-stained smears of lesion scrapings carried out according to World Health Organization recommendations (57). All samples from Nicaragua were taken as part of routine diagnostic procedures for leishmaniasis, which include PCR assays. Informed consent was obtained from the patients at CPMC and the Instituto SELADIS. Fine-needle aspirates from lesions of four hamsters (Mesocricetus auratus) infected with Leishmania isolates from Bolivia were also analyzed. These isolates had been previously characterized as L. mexicana by multilocus enzyme electrophoresis at the Instituto Boliviano de Biología de las Alturas (29). The following non-Leishmania-infected specimens were examined as negative controls: (i) normal buccal scrapings from three uninfected individuals and (ii) dermal scrapings from three Nicaraguan patients presenting with cutaneous lesions with confirmed diagnosis for other diseases (leprosy, candidiasis, and varicose ulcer) that were demonstrated to be negative for Leishmania by microscopy, kDNA-specific PCR, and serological tests.
TABLE 1.
Strains of Leishmania used in this study
| Species | International reference no. | Origin | Sourcea |
|---|---|---|---|
| L. (Viannia) braziliensis | MHOM/NI/89/XD37 | Nicaragua | 1 |
| MHOM/BR/84/LTB300 | Brazil | 2 | |
| L. (Viannia) panamensis | MHOM/NI/91/ZF11 | Nicaragua | 1 |
| MHOM/NI/89/XD26 | Nicaragua | 1 | |
| MHOM/PA/71/LS94 | Panama | 2 | |
| L. (V.) braziliensis-L. (V.) panamensis | MHOM/NI/88/XD42 | Nicaragua | 1 |
| L. (V.) braziliensis-L. (Viannia) guyanensis | MHOM/VE/92/P-2 | Venezuela | 3 |
| L. (V.) guyanensis | MHOM/BR/75/M4147 | Brazil | 2 |
| L. (Viannia) peruviana | MHOM/PE/84/LC26 | Peru | 4 |
| L. (Viannia) lainsoni | MHOM/BR/81/M16426 | Brazil | 5 |
| L. amazonensis | MHOM/BR/67/PH8 | Brazil | 2 |
| MHOM/BR/73/M2269 | Brazil | 2 | |
| L. mexicana | MNYC/BZ/62/M379 | Belize | 2 |
| L. chagasi | MHOM/BR/74/PP75 | Brazil | 2 |
| MHOM/HN/87/029 | Honduras | 6 | |
| L. donovani | MHOM/IN/80/DD8 | India | 2 |
| T. cruzi | TCN (human, 1988) | Nicaragua | 1 |
| T. brucei gambiense | ILRAD 1325 | Zaire | 7 |
| C. fasciculata | ATCC 11745 | United States | 7 |
Sources: 1, B. Rodriguez and A. Belli, Centro Nacional de Diagnóstico y Referencia, Ministerio de Salud, Managua, Nicaragua; 2, London School of Hygiene and Tropical Medicine, London, United Kingdom; 3, R. Bonfante-Garrido, Escuela de Medicina, Universidad Centro Occidental “Lisandro Alvarado,” Barquisimeto, Lara, Venezuela; 4, M. López, Instituto de Medicina Tropical “Alexander von Humboldt,” Universidad Peruana Cayetano Heredia, Lima, Peru; 5, S. Revollo, Instituto SELADIS, Universidad Mayor de San Andrés, La Paz, Bolivia; 6, C. Ponce, Laboratorio Central, Ministerio de Salud Pública, Tegucigalpa, Honduras; 7, N. Agabian, Program in Molecular Pathogenesis, University of California, San Francisco.
Preparation of samples for PCR amplification. (i) DNA purification of parasites in culture.
Leishmania spp. were grown at 26°C in RPMI 1640 medium (Gibco BRL, Grand Island, N.Y.) modified as described by Belli et al. (7). For DNA extraction, 1 × 109 to 5 × 109 promastigotes were harvested, washed in cold phosphate-buffered saline (pH 7.2), and resuspended in lysis buffer (50 mM Tris [pH 8.0], 50 mM NaCl, 50 mM EDTA, 1% sodium dodecyl sulfate). Proteinase K (Promega Corp., Madison, Wis.) was added to a final concentration of 100 μg/ml, and the suspension was incubated overnight at 37°C. After sequential extraction with phenol, phenol-chloroform, and chloroform, the DNA was precipitated by the addition of 100 mM magnesium chloride and 2 volumes of ethanol. The pellet was washed with 70% ethanol, air dried, resuspended in TE (10 mM Tris HCl [pH 7.2], 1 mM EDTA), and then treated with 100 μg of RNase (Sigma Chemical Co., St. Louis, Mo.) per ml.
(ii) Crude extraction of parasites in culture.
Approximately 35 μl (2 drops from a Pasteur pipette) of Leishmania parasites grown in culture was diluted in 200 μl of sterile distilled water and heated at 100°C for 10 min. After centrifugation at 13,600 × g for 1 min, 1 μl of the supernatant was used for PCR amplification.
(iii) Scrapings from cutaneous lesions.
Two skin scrapings were taken from the border of each cutaneous lesion with a sterile wooden toothpick, placed in 100 or 200 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) or 5% Chelex 100 (Bio-Rad Laboratories, Richmond, Calif.), heated for 10 min in a boiling-water bath, and frozen at −20°C until use (5). After centrifugation at 13,600 × g for 1 min, 5 μl of the supernatant was used for PCR amplification. Normal human buccal scrapings and dermal scrapings from nonleishmanial lesions were resuspended in 5% Chelex 100 and boiled for 10 min as described above.
(iv) Fine-needle aspirates.
Fine-needle aspirates were taken from lesions of Leishmania-infected hamsters with 26-gauge needles on 3-ml syringes containing 0.3 ml of sterile normal saline solution (0.85% NaCl). Each aspirate sample (300 μl) was placed in a microcentrifuge tube containing 100 μl of 4× TE, heated for 10 min in a boiling-water bath, and frozen at −20°C until use. After centrifugation at 13,600 × g for 1 min, 5 μl of the supernatant was used for PCR amplification.
Primer design.
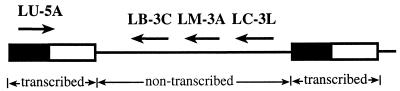
Sequences for the SL RNA region of the New World Leishmania species were aligned with GeneJockey II software (Biosoft, Ferguson, Mo.) to identify potential sites for genus- as well as complex-specific PCR priming. Candidate amplification primers were then analyzed with Oligo version 4.0 software (National Biosciences, Inc., Plymouth, Minn.) for false priming regions and internal stability. The sequences of the primers were examined for homology to the sequences of related parasites (Trypanosoma cruzi, Trypanosoma brucei, Trypanosoma rangeli, and Crithidia fasciculata) and to other sequences in the GenBank database. We chose oligonucleotide primers which were either conserved in all Leishmania species (LU-5A) or specific to each New World complex (LB-3C, LM-3A, and LC-3L). The primers and their sequences are as follows: LU-5A, 5′-TTT ATT GGT ATG CGA AAC TTC-3′; LB-3C, 5′-CGT (C/G)CC GAA CCC CGT GTC-3′; LM-3A, 5′-GCA CCG CAC CGG (A/G)CC AC-3′; and LC-3L, 5′-GCC CGC G(C/T)G TCA CCA CCA T-3′. The locations and directions of the primers are shown in Fig. 1. Oligonucleotides were synthesized by Operon Technologies, Inc. (Alameda, Calif.), or Ransom Hill Bioscience, Inc. (Ramona, Calif.).
FIG. 1.
Schematic diagram of the SL RNA gene repeat region and location of the primers. Filled box, conserved SL exon (39 nt); open box, SL intron (55 to 101 nucleotides); solid line, variable nontranscribed spacer.
PCR amplification.
We prepared a 50-μl reaction mixture containing 50 mM KCl, 10 mM Tris (pH 8.3), 200 μM each deoxynucleotide triphosphate, 1.5 mM MgCl2, 10.5% dimethyl sulfoxide, 50 mM tetramethylammonium chloride (11), 0.6 M betaine (34), 1 mM dithiothreitol, 0.4 μM 5′ primer LU-5A, 0.2 μM each 3′ primer (LB-3C, LM-3A, and LC-3L), and 0.04 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer, Foster City, Calif.) per μl. An initial denaturation step of 95°C for 5 min was followed by 35 cycles of 95°C for 30 s, 54°C for 45 s, and 72°C for 30 s, with a final extension at 72°C for 5 min.
Amplification was conducted in 0.6-ml tubes (Robbins Scientific Corp., Sunnyvale, Calif.) with a model 480 thermal cycler (Perkin-Elmer, Norwalk, Conn.). It was important that the tubes fit snugly in the wells of the thermocylcer block; if not, it was necessary to add 1 to 2 drops of mineral oil to the wells. Ten microliters of the product was analyzed by electrophoresis on 1.5% agarose gels in 1× TBE (89 mM Tris borate, 2 mM EDTA [pH 8.3]) or 8% polyacrylamide gels in 0.5× TBE. No difference in sensitivity was observed between the two gel systems. The expected product sizes were as follows: 351 to 397 bp for the L. donovani complex, 218 to 240 bp for the L. mexicana complex, and 146 to 149 bp for the L. braziliensis complex.
RESULTS
Primer design.
To develop an assay that would amplify products of different sizes for each New World complex, certain criteria were chosen for primer design, namely, (i) elevated copy number, (ii) the presence of sequences conserved among all Leishmania species, and (iii) the presence of variable sequences which differed between New World complexes but were conserved among species within each complex. Potential targets in the Leishmania genome included the kDNA minicircle (50), the small-subunit (SSU) ribosomal DNA (rDNA) (54), and the SL RNA (mini-exon) gene repeat (16). The kDNA minicircles are present at a very high copy number (∼10,000 per cell), and they contain both sequences which are conserved and sequences which vary among species. However, due to the locations of the variable and conserved regions, it was not possible to identify sites for complex-specific primers that would generate products of the appropriate size for each complex. In the highly conserved SSU rDNA region, distinct Leishmania complexes differed only by point mutations; thus, we could not find divergent regions large enough to design highly variable complex-specific primers that resulted in differently sized products for each complex. The SL RNA gene repeat region best satisfied the necessary criteria. It is present in the nuclear genome as a tandem array of approximately 200 copies, where each copy contains both a transcribed conserved region and a nontranscribed variable region that differs among the Leishmania complexes in both sequence and size (16, 22).
Four primers were used in the multiplex assay: one 5′ primer conserved in all Leishmania species and three distinct 3′ primers, specific to the variable region of each of the three New World Leishmania complexes (Fig. 1). Amplification with these primers yielded products of different sizes for each complex (L. braziliensis, L. mexicana, and L. donovani) based on the sequence specificities of the primers.
Species specificity of the PCR assay.
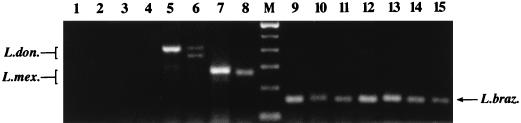
The species specificity of the multiplex assay was evaluated with a number of Leishmania species from geographically disparate regions of Central and South America, in addition to several related trypanosomatids. As shown in Fig. 2, the multiplex assay resulted in products in the expected size range (351 to 397 bp) for species of the L. donovani complex; namely, L. chagasi (lane 5) and L. donovani (lane 6). Similar results were obtained for the L. mexicana complex (218 to 240 bp) from representative samples of Leishmania amazonensis (lane 7) and L. mexicana (lane 8). A number of species belonging to the L. braziliensis complex were tested, including L. braziliensis strains from different geographical regions (lanes 9 to 10), a putative L. braziliensis-Leishmania guyanensis hybrid (9) (lane 11), Leishmania panamensis (lane 12), a putative L. braziliensis-L. panamensis hybrid (7) (lane 13), L. guyanensis (lane 14), and Leishmania peruviana (lane 15). All yielded products in the expected size range (146 to 149 bp), as did Leishmania lainsoni (data not shown). No product was obtained from the related kinetoplastids T. brucei gambiense (lane 2), T. brucei rhodesiense (data not shown), T. cruzi (lane 3), and C. fasciculata (lane 4) or from the negative amplification control (water; lane 1). Negative controls consisting of normal human tissue and specimens from nonleishmanial lesions reproducibly yielded either no amplified products (see Fig. 5, lane 2) or minor nonspecific products that formed a distinctive pattern distinguishable from those of specific Leishmania-derived PCR products (see Fig. 5, lane 3).
FIG. 2.
Species specificity of the multiplex PCR assay. One-nanogram samples of DNAs from the organisms indicated below were amplified in the multiplex assay. Lane 1, water (negative control); lane 2, T. brucei gambiense (ILRAD 1325), lane 3, T. cruzi (TCN); lane 4, C. fasciculata (ATCC 11745); lane 5, L. chagasi (MHOM/BR/74/PP75); lane 6, L. donovani (MHOM/IN/80/DD8); lane 7, L. amazonensis (MHOM/BR/67/PH8); lane 8, L. mexicana (MNYC/BZ/62/M379); lane M, 100-bp ladder (Gibco BRL; the lowest band shown is 100 bp); lane 9, L. braziliensis (MHOM/NI/89/XD37); lane 10, L. braziliensis (MHOM/BR/84/LTB300), lane 11, L. braziliensis-L. guyanensis hybrid (MHOM/VE/92/P-2); lane 12, L. panamensis (MHOM/NI/89/XD26); lane 13, L. braziliensis-L. panamensis hybrid (MHOM/NI/88/XD42); lane 14, L. guyanensis (MHOM/BR/75/M4147); lane 15, L. peruviana (MHOM/PE/84/LC26). Expected product sizes were 146 to 149 bp for L. braziliensis (L. braz.), 218 to 240 bp for L. mexicana (L. mex.), and 351 to 397 bp for L. donovani (L. don.).
FIG. 5.
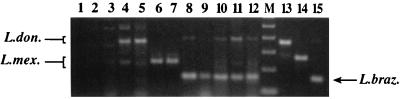
PCR detection of Leishmania in clinical material. Five microliters of the supernatant of boiled dermal scrapings or lesion aspirates (prepared as described in Materials and Methods) was amplified by the multiplex assay. Lane 1, water (negative control); lane 2, dermal scraping from a nonleishmanial cutaneous lesion (Nicaragua); lane 3, normal human buccal scraping (Nicaragua); lane 4, dermal scraping from an atypical cutaneous lesion from patient LC9853 (Leon, Nicaragua); lane 5, dermal scraping from an atypical cutaneous lesion from patient LC9786 (Apompua, Nicaragua); lane 6, lesion aspirate from a hamster infected with L. mexicana (Bolivia); lane 7, lesion aspirate from a hamster infected with L. mexicana (Bolivia); lane 8, dermal scraping from a cutaneous lesion from patient LC9708 (Nicaragua); lane 9, dermal scraping from a cutaneous lesion from patient LC11 (Nicaragua); lane 10, dermal scraping from a cutaneous lesion from patient LC9767 (Nicaragua); lane 11, dermal scraping from a cutaneous lesion from patient RG10 (Nicaragua); lane 12, dermal scraping from a cutaneous lesion from a patient from CPMC; lane M, 100-bp ladder (Gibco BRL; the lowest band shown is 100 bp); lane 13, L. chagasi DNA (MHOM/HN/87/029); lane 14, L. mexicana DNA (MNYC/BZ/62/M379); lane 15, L. braziliensis DNA (MHOM/BR/84/LTB300). L. don., L. donovani; L. mex., L. mexicana; L. braz., L. braziliensis.
Sensitivity of the PCR assay.
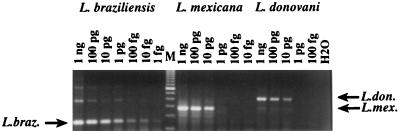
The sensitivity of the assay was determined by amplifying 10-fold serial dilutions of DNA from a representative Leishmania strain from each of the three complexes. The amount of DNA tested ranged from 1 ng (9,000 parasites) to 1 fg (0.009 parasites), with 108 bp being used as the size of the diploid Leishmania genome to calculate the number of parasites (19). As shown in Fig. 3, the multiplex assay detected 1 fg (∼0.01 parasite) of the L. braziliensis complex (L. panamensis), 1 to 10 pg (∼10 to 100 parasites) of the L. mexicana complex (L. amazonensis), and 10 pg (∼100 parasites) of the L. donovani complex (L. chagasi). The higher sensitivity obtained with the L. braziliensis complex can be attributed in part to the smaller size of the L. braziliensis amplicon, which is amplified more efficiently than larger fragments.
FIG. 3.
Sensitivity of the multiplex PCR assay. Tenfold dilutions of Leishmania DNA were amplified by the multiplex PCR to determine the sensitivity of the assay. The amounts of DNA and the Leishmania complexes are indicated. The L. braziliensis (L. braz.) complex was L. panamensis (MHOM/PA/71/LS94), the L. donovani (L. don.) complex was L. chagasi (MHOM/BR/74/PP75), and the L. mexicana (L. mex.) complex was L. amazonensis (MHOM/BR/67/PH8). Lane M, 100-bp DNA ladder (Gibco BRL); the lowest band shown is 100 bp.
Detection of cultured Leishmania and mixtures of parasites.
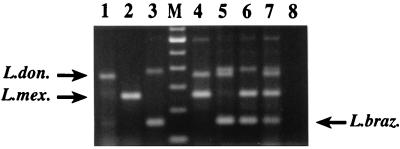
Crude extracts were prepared from cultures of Leishmania parasites by diluting the culture in sterile water and boiling. Figure 4 shows that fragments of the expected sizes were obtained from cultured L. chagasi (lane 1), L. mexicana (lane 2), and L. braziliensis (lane 3). When mixtures of cultures containing combinations of two or all three Leishmania complexes were amplified in the multiplex assay, fragments corresponding to each complex present in the mixture were obtained (Fig. 4, lanes 4 to 7).
FIG. 4.
PCR detection of Leishmania in cultures. Two drops of cultured parasites were diluted in 200 μl of sterile water and boiled for 10 min. One microliter of the boiled culture was amplified by the multiplex assay. Lane 1, L. chagasi (MHOM/BR/74/PP75); lane 2, L. mexicana (MNYC/BZ/62/M379); lane 3, L. panamensis (MHOM/NI/91/ZF11); lane M, 100-bp ladder (Gibco BRL; the lowest band shown is 100 bp); lane 4, L. chagasi (MHOM/BR/74/PP75) and L. mexicana (MNYC/BZ/62/M379); lane 5, L. chagasi (MHOM/BR/74/PP75) and L. braziliensis (MHOM/BR/84/LTB300); lane 6, L. mexicana (MNYC/BZ/62/M379) and L. braziliensis (MHOM/BR/84/LTB300), lane 7, L. braziliensis (MHOM/BR/84/LTB300), L. mexicana (MNYC/BZ/62/M379), and L. chagasi (MHOM/BR/74/PP75); lane 8, water (negative control). L. don., L. donovani; L. mex., L. mexicana; L. braz., L. braziliensis.
At higher concentrations of template, the amplification of dimers of tandem copies of the SL RNA gene repeat was observed, though in yields lower than that of the preferentially amplified smaller monomer (Fig. 3 and 4). At first glance, these dimers may appear to interfere with the interpretation of amplification results; however, the pattern generated can be easily distinguished from that obtained from a mixture of parasites. For example, in addition to the usual monomeric amplicon of 146 to 149 bp, L. braziliensis can generate a 375-bp dimer, extending from the 5′ primer of one L. braziliensis monomer to the 3′ primer of the next. This size falls in the range of the L. donovani complex; however, the L. donovani amplification is never associated with smaller products. Therefore, after a single multiplex assay, a product of ∼350 to ∼400 bp alone is indicative of the L. donovani complex, whereas a fragment of ∼375 bp accompanied by a fragment of ∼150 bp is indicative of L. braziliensis. If a mixed infection is suspected, a second assay can be performed, either by repeating the amplification separately with L. donovani- or L. braziliensis-specific primers or by hybridizing the product with L. donovani- or L. braziliensis-specific probes containing the variable region of the SL RNA gene (46). To demonstrate that the secondary band in the L. braziliensis amplification in Fig. 3 was specific to L. braziliensis, the same sample was amplified with only the L. braziliensis-specific primer (in conjunction with the conserved 5′ primer), resulting in the ∼150-bp product characteristic of L. braziliensis. When the same sample was amplified with only the L. donovani-specific primer, no product was obtained (data not shown).
Detection of Leishmania in clinical material.
Crude extracts of dermal scrapings prepared from ulcerated lesions of confirmed cutaneous leishmaniasis from 20 patients in Nicaragua, Bolivia, and San Francisco, Calif., yielded a L. braziliensis-sized fragment in the multiplex assay. Representative amplifications from five patients are shown in Fig. 5, lanes 8 to 12. These samples were also amplified with primers specific to L. braziliensis kDNA (28), and products of the expected sizes were obtained (data not shown). Dermal scrapings from nonulcerated lesions from three Nicaraguan patients with atypical cutaneous leishmaniasis (ACL) analyzed by the multiplex assay resulted in PCR products characteristic of the L. donovani complex, which in the Americas is represented by L. chagasi (Fig. 5, lanes 4 and 5). Cultured parasites isolated from these lesions were confirmed to be members of the L. donovani complex by an L. donovani-specific PCR assay (43) (data not shown). When crude preparations of lesion aspirates from L. mexicana-infected hamsters were amplified, fragments of the size expected for L. mexicana were obtained (Fig. 5, lanes 6 and 7). Results of the amplification of tissue specimens from non-Leishmania-infected negative controls are shown in Fig. 5, lanes 2 and 3, as mentioned previously.
DISCUSSION
Detection of Leishmania parasites and identification of Leishmania complexes can be accomplished by PCR with primers with the appropriate genetic specificities. PCR protocols for identifying the Leishmania genus (8, 30, 48) or individual complexes, including L. braziliensis (14, 20, 28, 33), L. mexicana (15, 33), and L. donovani (13, 23, 33, 43, 47, 51), have been described. However, none of the assays allow simultaneous detection of all three complexes. As an alternative approach to distinguish Leishmania complexes, additional steps, such as restriction enzyme analysis, single-strand conformation polymorphism (54), and DNA probe hybridization of the amplified products (10, 39, 45, 46, 53), have been used in conjunction with PCR. Last, RAPD analysis has been reported for characterization of New World Leishmania at the species level, but it is extremely sensitive to minor variations in reaction parameters and is therefore not well suited to the difficult conditions often prevailing in developing-country laboratories (38). In addition, RAPD analysis can be conducted only with purified DNA from cultured parasites and it has a sensitivity (1 to 10 ng) much lower than that of standard PCR. Here we describe a single-step assay designed to detect all the New World Leishmania species and simultaneously differentiate the three complexes without requiring three separate reactions or further manipulation of the amplified products.
We investigated several repetitive regions of Leishmania genome DNA sequences, including the SL RNA gene repeat (16), the kDNA minicircles (50), and the SSU rDNA (54), and the SL RNA gene repeat region best satisfied the criteria chosen for design of the primers. There are approximately 200 copies of the SL RNA gene repeat per haploid genome, and each copy contains both a conserved and a variable region (16, 22). The conserved region is transcribed and contains an invariant 39-bp SL sequence followed by a fairly well-conserved region of variable size (55 to 101 bp, depending on the species). The nontranscribed variable regions differ in both sequence and size (140 to 350 bp) among the complexes of Leishmania (Fig. 1). We developed a multiplex PCR assay targeting the SL RNA gene repeat that employs a single conserved 5′ primer common to all Leishmania species and three distinct 3′ primers, each of which is specific to a different complex. Thus, the identification of the Leishmania complexes is based on both the sizes of the products and the sequence specificities of the primers. This assay yields a range of sizes due to the variably sized nontranscribed region. For the L. donovani complex, products range in size from 351 to 397 bp; for the L. mexicana complex, products range in size from 218 to 240 bp; and for the L. braziliensis complex, products range in size from 146 to 149 bp.
The assay was specific to the Leishmania genus and correctly identified species representing broad taxonomic and geographic diversity. Strains tested included two species from the L. donovani complex, two species from the L. mexicana complex, and five species from the L. braziliensis complex (Table 1). The multiplex assay distinguished Leishmania from T. cruzi, which generated no product. This is an important feature, since mixed infections of Leishmania and T. cruzi are known to occur (12) and since serological tests are subject to antibody cross-reactivities to related antigens in the two parasites (41). C. fasciculata has been known to contaminate laboratory cultures of Leishmania and can be difficult to distinguish morphologically from Leishmania parasites; therefore, we tested C. fasciculata and found that it yielded no PCR product. Two other members of the order Kinetoplastida, T. brucei gambiense and T. brucei rhodesiense, also generated no product in the multiplex assay.
The sensitivity of the assay ranged from 1 fg (∼0.01 parasite) to 10 pg (∼100 parasites) of total DNA. Crude extracts of cultured parasites, prepared by simply boiling dilutions of the cultures, also served as excellent templates for amplification. In addition, the multiplex assay detected parasites directly from clinical specimens. Crude preparations of dermal scrapings from ulcerated leishmanial cutaneous lesions were analyzed and were shown to contain parasites from the L. braziliensis complex. When dermal scrapings from patients presenting with atypical cutaneous nodules were analyzed, the amplification results demonstrated that the patients were infected with parasites from the L. donovani complex. Boiled lesion aspirates from hamsters inoculated with Leishmania isolates from Bolivia were also amplified, confirming that the isolated parasites belonged to the L. mexicana complex. These results indicate that crude preparations of skin scrapings or aspirates from cutaneous leishmaniasis can serve as suitable specimens for the multiplex assay. Dermal scrapings are less invasive than lesion biopsies and have been shown to serve as effective samples for PCR diagnosis of cutaneous leishmaniasis (5). Lesion aspirates present another less invasive sampling strategy that can be used to evaluate the presence of parasites around an active or scarred lesion by PCR.
In our experience, this assay detects parasites directly from clinical samples with high numbers of parasites but is less effective in amplifying Leishmania from samples with low numbers of parasites. That this assay is not as sensitive as kDNA-based PCR assays is not surprising, since there are 50-fold fewer copies of the SL RNA gene than of the kinetoplastid minicircle. We recommend the present protocol as a tool for rapid characterization of both cultured parasites and selected clinical specimens with high numbers of parasites. Such a characterization tool is necessary to complement diagnostic assays, since most of the latter do not furnish the taxonomic information about the parasite required to determine the appropriate therapeutic regimens and control measures.
Traditionally, diagnosis of leishmaniasis has relied on clinical and epidemiological criteria, since certain disease manifestations have been associated with particular species or complexes. However, there have been accounts of atypical infections which call into question these classifications. These reports include a new variant of cutaneous disease caused by L. chagasi in Honduras (44) and Nicaragua (4); mucosal leishmaniasis caused by L. amazonensis (3) and L. mexicana complex (55); and visceral disease caused by L. braziliensis (26), L. amazonensis (2), and a putative L. braziliensis-L. mexicana hybrid (24). Since many of these atypical infections are clinically identical to typical infections and occur in geographical areas where multiple Leishmania complexes are found, laboratory methods are all the more necessary for distinguishing between the complexes and defining foci of disease caused by the different complexes. In addition, Leishmania is increasingly recognized as an opportunistic pathogen during coinfection with human immunodeficiency virus, where visceral disease has been associated with parasite species commonly identified with cutaneous disease (19, 24). Thus, in the AIDS era, quick assays for the simultaneous detection and identification of all the Leishmania complexes are needed.
This multiplex PCR assay has been used to clarify two situations of atypical infections in Nicaragua. Several patients with cutaneous facial lesions were referred to the Centro Nacional de Diagnóstico y Referencia from an area in northwestern Nicaragua (Leon) where visceral leishmaniasis is endemic but where no cutaneous leishmaniasis had been reported. The lesions were papular and nonulcerated, similar to those described by Ponce et al. (44) that have been associated with L. chagasi, the agent of visceral leishmaniasis in the New World. Using the multiplex assay, investigators at the Centro Nacional de Diagnóstico y Referencia determined that the lesions were caused by L. chagasi, and approximately 100 patients with confirmed leishmanial lesions from this region have been examined to date. Another group of patients, who presented with lesions that were also papular and nonulcerated but that were smaller in size and located on the extremities as well as on the face, was examined. These patients were from an area in central Nicaragua (Apompua) with no history of visceral leishmaniasis. The multiplex PCR assay again demonstrated that the etiologic agent belonged to the L. donovani complex. To date, 56 patients have been diagnosed in Apompua with similar lesions of ACL. Epidemiological, parasitological, and entomological characteristics of these situations are under investigation.
The multiplex assay can identify mixtures of Leishmania complexes and generate the appropriate products for each combination of complexes (Fig. 4). Thus, one potential application of the multiplex assay is the detection of mixed infections caused by parasites of different complexes. Given the geographical overlap of multiple complexes and the atypical infections discussed above, such coinfection is indeed a possibility. New World Leishmania species of different complexes can be found in the same types of clinical specimen; for example, L. mexicana and L. chagasi are found in cutaneous nodules (37), L. braziliensis and L. amazonensis are found in lesions of cutaneous leishmaniasis (3, 19), and L. braziliensis (5, 21) and L. chagasi (6) are found in human peripheral blood mononuclear cells. Several cases of concurrent infections with Leishmania from two different complexes involving New World (49) and Old World species (25, 31, 40) have been reported. However, only by using PCR have two different complexes been isolated from the same clinical sample (25). Such mixed infections may well be underreported due to the requirement for large-scale cultivation of parasites for classical species identification techniques. It is known that certain complexes (e.g., L. mexicana) grow better in culture than others (e.g., L. braziliensis) (41) and that during in vitro cultivation certain species can outgrow other species from an initially mixed infection (1). Therefore, the ability to identify Leishmania complexes by PCR directly from clinical specimens, or at least from minute amounts of cultured material immediately after isolation, will be useful for detecting infections with more than one complex and determining the real incidence of these mixed infections.
With the SL RNA gene repeat as a target, a PCR assay was developed to differentiate all three complexes of New World Leishmania in one step. We have demonstrated the direct detection of Leishmania in clinical specimens; further testing of clinical samples is in progress in Nicaragua and Bolivia. To our knowledge, this is the first report of a single-step PCR amplification for sequence-specific differentiation of the three complexes of New World Leishmania spp. There are many potential uses for this assay, including clinical diagnosis, rapid identification of Leishmania complexes for epidemiological purposes, and analysis of Leishmania in animal reservoirs (e.g., dogs, rodents, and foxes) and in its sand fly vector (42). Since this multiplex PCR assay was designed for in-country application in areas where leishmaniasis is endemic, we have maximized the simplicity, rapidity, and informative content of the assay while retaining the appropriate sensitivity and specificity.
ACKNOWLEDGMENTS
We are grateful to Martín López, Susana Revollo, Carlos Ponce, and Rafael Bonfante-Garrido for Leishmania strains. Many thanks go to Adil Wakil for clinical material and for editorial comments and to George Newport and Cristián Orrego for editorial comments. We are grateful to Steven Embury for his support.
This work was supported in part by the American Society for Biochemistry and Molecular Biology, the John D. and Catherine T. MacArthur Foundation, grant TW00905 from the Fogarty International Center, the Fondation Damien, grant ERBICI8CT960028 from the European Union, and grant AI21975 from the National Institutes of Health.
REFERENCES
- 1.Armijos R X, Chico M E, Cruz M E, Guderian R H, Kreutzer R D, Berman J D, Rogers M D, Grogl M. Human cutaneous leishmaniasis in Ecuador: identification of parasites by enzyme electrophoresis. Am J Trop Med Hyg. 1990;42:424–428. doi: 10.4269/ajtmh.1990.42.424. [DOI] [PubMed] [Google Scholar]
- 2.Barral A, Badaro R, Barral-Netto M, Grimaldi G, Jr, Momen H, Carvalho E M. Isolation of Leishmania mexicana amazonensis from the bone marrow in a case of American visceral leishmaniasis. Am J Trop Med Hyg. 1986;35:732–734. doi: 10.4269/ajtmh.1986.35.732. [DOI] [PubMed] [Google Scholar]
- 3.Barral A, Pedral-Sampaio D, Grimaldi G J, Momen H, McMahon-Pratt D, Ribeiro de Jesus A, Almeida R, Badaro R, Barral-Netto M, Carvalho E M, Johnson W D., Jr Leishmaniasis in Bahia, Brazil: evidence that Leishmania amazonensis produces a wide spectrum of clinical disease. Am J Trop Med Hyg. 1991;44:536–546. doi: 10.4269/ajtmh.1991.44.536. [DOI] [PubMed] [Google Scholar]
- 4.Belli, A., D. García, X. Palacios, B. Rodriguez, F. Marín, and E. Harris. Unpublished results.
- 5.Belli A, Rodriguez B, Avilés H, Harris E. Simplified PCR detection of New World Leishmania from clinical specimens. Am J Trop Med Hyg. 1998;58:102–109. doi: 10.4269/ajtmh.1998.58.102. [DOI] [PubMed] [Google Scholar]
- 6.Belli, A., and E. Videa. Unpublished results.
- 7.Belli A A, Miles M A, Kelly J M. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology. 1994;109:435–442. doi: 10.1017/s0031182000080689. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharyya R, Das K, Sen S, Roy S, Majumder H K. Development of a genus specific primer set for detection of Leishmania parasites by polymerase chain reaction. FEMS Microbiol Lett. 1996;135:195–200. doi: 10.1111/j.1574-6968.1996.tb07989.x. [DOI] [PubMed] [Google Scholar]
- 9.Bonfante-Garrido R, Melendez E, Barroeta S, de Alejos M A, Momen H, Cupolillo E, McMahon-Pratt D, Grimaldi G., Jr Cutaneous leishmaniasis in western Venezuela caused by infection with Leishmania venezuelensis and L. braziliensis variants. Trans R Soc Trop Med Hyg. 1992;86:141–148. doi: 10.1016/0035-9203(92)90544-m. [DOI] [PubMed] [Google Scholar]
- 10.Bozza M, Fernandes O, Degrave W M, Lopes U G. Characterization of “Old World” Leishmania species using amplified minicircle variable regions as molecular probes. Trans R Soc Trop Med Hyg. 1995;89:333–334. doi: 10.1016/0035-9203(95)90569-3. [DOI] [PubMed] [Google Scholar]
- 11.Chevet E, Lemaitre G, Katinka M D. Low concentrations of tetramethylammonium chloride increase yield and specificity of PCR. Nucleic Acids Res. 1995;23:3343–3344. doi: 10.1093/nar/23.16.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiaramonte M G, Zwirner N W, Caropresi S L, Taranto N J, Malchiodi E L. Trypanosoma cruzi and Leishmania spp. human mixed infection. Am J Trop Med Hyg. 1996;54:271–273. doi: 10.4269/ajtmh.1996.54.271. [DOI] [PubMed] [Google Scholar]
- 13.Costa J-M, Durand R, Deniau M, Rivollet D, Izri M, Houin R, Vidaud M, Bretagne S. PCR enzyme-linked immunosorbant assay for diagnosis of leishmaniasis in human immunodeficiency virus-infected patients. J Clin Microbiol. 1996;34:1831–1833. doi: 10.1128/jcm.34.7.1831-1833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Brujin M H, Barker D C. Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop. 1992;52:45–58. doi: 10.1016/0001-706x(92)90006-j. [DOI] [PubMed] [Google Scholar]
- 15.Eresh S, McCallum S M, Barker D C. Identification and diagnosis of Leishmania mexicana complex isolates by polymerase chain reaction. Parasitology. 1994;109:423–433. doi: 10.1017/s0031182000080677. [DOI] [PubMed] [Google Scholar]
- 16.Fernandes O, Murthy V K, Kurath U, Degrave W M, Campbell D A. Mini-exon gene variation in human pathogenic Leishmania species. Mol Biochem Parasitol. 1994;66:261–271. doi: 10.1016/0166-6851(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 17.Grimaldi G, McMahon-Pratt D. Monoclonal antibodies for the identification of New World Leishmania species. Mem Inst Oswaldo Cruz Rio J. 1996;91:37–42. doi: 10.1590/s0074-02761996000100006. [DOI] [PubMed] [Google Scholar]
- 18.Grimaldi G, Jr, David J R, McMahon-Pratt D. Identification and distribution of New World Leishmania species characterized by serodeme analysis using monoclonal antibodies. Am J Trop Med Hyg. 1987;36:270–287. doi: 10.4269/ajtmh.1987.36.270. [DOI] [PubMed] [Google Scholar]
- 19.Grimaldi G, Jr, Tesh R B. Leishmaniasis of the New World: current concepts and implications for future research. Clin Microbiol Rev. 1993;6:230–250. doi: 10.1128/cmr.6.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guevara P, Alonso G, da Silveira J F, de Mello M, Scorza J V, Anez N, Ramirez J L. Identification of new world Leishmania using ribosomal gene spacer probes. Mol Biochem Parasitol. 1992;56:15–26. doi: 10.1016/0166-6851(92)90150-i. [DOI] [PubMed] [Google Scholar]
- 21.Guevara P, Rojas E, Gonzalez N, Scorza J V, Anez N, Valera M, Ramirez J L. Presence of Leishmania braziliensis in blood samples from cured patients or at different stages of immunotherapy. Clin Diagn Lab Immunol. 1994;1:385–389. doi: 10.1128/cdli.1.4.385-389.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan M Q, Das S, Adhya S. Mini-exon derived RNA gene of Leishmania donovani: structure, organization, and expression. J Biosci (Bangalore) 1992;17:55–66. [Google Scholar]
- 23.Hassan M Q, Ghosh A, Ghosh S S, Gupta M, Basu D, Mallik K K, Adhya S. Enzymatic amplification of mini-exon-derived RNA gene spacers of Leishmania donovani: primers and probes for DNA diagnosis. Parasitology. 1993;107:509–517. doi: 10.1017/s0031182000068086. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez D E, Rodriguez N, Wessolossky M, Convit J. Visceral leishmaniasis due to a Leishmania variant that shares kinetoplast DNA sequences with Leishmania braziliensis and Leishmania mexicana in a patient infected with human immunodeficiency virus: identification of the Leishmania species with use of the polymerase chain reaction. Clin Infect Dis. 1991;21:701–702. doi: 10.1093/clinids/21.3.701. [DOI] [PubMed] [Google Scholar]
- 25.Ibrahim M E, Smyth A J, Ali M H, Barker D C, Kharazmi A. The polymerase chain reaction can reveal the occurrence of naturally mixed infections with Leishmania parasites. Acta Trop. 1994;57:327–332. doi: 10.1016/0001-706x(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, W. D., Jr. Personal communication.
- 27.Kreutzer R D, Christensen H A. Characterization of Leishmania spp. by isozyme electrophoresis. Am J Trop Med Hyg. 1980;29:199–208. doi: 10.4269/ajtmh.1980.29.199. [DOI] [PubMed] [Google Scholar]
- 28.López M, Inga R, Cangalaya M, Echevarria J, Llanos-Cuentas A, Orrego C, Arévalo J. Diagnosis of Leishmania using the polymerase chain reaction: a simplified procedure for field work. Am J Trop Med Hyg. 1993;49:348–356. doi: 10.4269/ajtmh.1993.49.348. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, E. Personal communication.
- 30.Mathis A, Deplazes P. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J Clin Microbiol. 1995;33:1145–1149. doi: 10.1128/jcm.33.5.1145-1149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mebrahtu Y B, Lawyer P G, Hendricks L D, Muigai R, Oster C N, Perkins P V, Koech D K, Pamba H, Roberts C R. Concurrent infection with Leishmania donovani and Leishmania major in a Kenyan patient: clinical description and parasite characterization. Am J Trop Med Hyg. 1991;45:290–296. doi: 10.4269/ajtmh.1991.45.290. [DOI] [PubMed] [Google Scholar]
- 32.Melby P C, Kreutzer R D, McMahon-Pratt D, Gam A A, Neva F A. Cutaneous leishmaniasis: review of 59 cases seen at the National Institutes of Health. Clin Infect Dis. 1992;15:924–937. doi: 10.1093/clind/15.6.924. [DOI] [PubMed] [Google Scholar]
- 33.Meredith S E, Zijlstra E E, Schoone G J, Kroon C C, van Eys G J, Schaeffer K U, el-Hassan A M, Lawyer P G. Development and application of the polymerase chain reaction for the detection and identification of Leishmania parasites in clinical material. Arch Inst Pasteur Tunis. 1993;70:419–431. [PubMed] [Google Scholar]
- 34.Mytelka D S, Chamberlin M J. Analysis and suppression of DNA polymerase pauses associated with a trinucleotide consensus. Nucleic Acids Res. 1996;24:2774–2781. doi: 10.1093/nar/24.14.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navin T R, Arana B A, Arana F E, de Merida A M, Castillo A L, Pozuelos J L. Placebo-controlled clinical trial of meglumine antimoniate (Glucantime) vs. localized controlled heat in the treatment of cutaneous leishmaniasis in Guatemala. Am J Trop Med Hyg. 1990;42:43–50. doi: 10.4269/ajtmh.1990.42.43. [DOI] [PubMed] [Google Scholar]
- 36.Neva F, Sacks D. Leishmaniasis. In: Warren K S, Mahmoud A A F, editors. Tropical and geographical medicine. New York, N.Y: McGraw-Hill Information Services Co.; 1990. pp. 296–309. [Google Scholar]
- 37.Neva F A, Ponce C, Ponce E, Kreutzer R, Modabber F, Olliaro P. Non-ulcerative cutaneous leishmaniasis in Honduras fails to respond to topical paromomycin. Trans R Soc Trop Med Hyg. 1997;91:473–475. doi: 10.1016/s0035-9203(97)90290-x. [DOI] [PubMed] [Google Scholar]
- 38.Noyes H A, Belli A A, Maingon R. Appraisal of various random amplified polymorphic DNA-polymerase chain reaction primers for Leishmania identification. Am J Trop Med Hyg. 1996;55:98–105. doi: 10.4269/ajtmh.1996.55.98. [DOI] [PubMed] [Google Scholar]
- 39.Nuzum E, White F, Thakur C, Dietze R, Wages J, Grogl M, Berman J. Diagnosis of symptomatic visceral leishmaniasis by use of the polymerase chain reaction on patient blood. J Infect Dis. 1995;171:751–754. doi: 10.1093/infdis/171.3.751. [DOI] [PubMed] [Google Scholar]
- 40.Oliveira Neto M P, Marzochi M C A, Grimaldi G J, Pacheco R S, Toledo L M, Momen H. Concurrent human infection with Leishmania donovani and Leishmania braziliensis braziliensis. Ann Trop Med Parasitol. 1986;80:587–592. doi: 10.1080/00034983.1986.11812072. [DOI] [PubMed] [Google Scholar]
- 41.Pearson R D, de Querez Sousa A. Leishmania species: visceral (kala-azar), cutaneous, and mucosal leishmaniasis. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill Livingstone; 1995. pp. 2428–2442. [Google Scholar]
- 42.Pérez J E, Ogusuku E, Inga R, López M, Monje J, Paz L, Nieto E, Arévalo J. Natural Leishmania infection of Lutzomyia spp. in Peru. Trans R Soc Trop Med Hyg. 1994;88:161–164. doi: 10.1016/0035-9203(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 43.Piarroux R, Azaiez R, Lossi A M, Reynier P, Muscatelli F, Gambarelli F, Fontes M, Dumon H, Quilici M. Isolation and characterization of a repetitive DNA sequence from Leishmania infantum: development of a visceral leishmaniasis polymerase chain reaction. Am J Trop Med Hyg. 1993;49:364–369. doi: 10.4269/ajtmh.1993.49.364. [DOI] [PubMed] [Google Scholar]
- 44.Ponce C, Ponce E, Morrison A, Cruz A, Kreutzer R, McMahon-Pratt D, Neva F. Leishmania donovani chagasi: new clinical variant of cutaneous leishmaniasis in Honduras. Lancet. 1991;337:67–70. doi: 10.1016/0140-6736(91)90734-7. [DOI] [PubMed] [Google Scholar]
- 45.Qiao Z, Miles M A, Wilson S M. Detection of parasites of the Leishmania donovani-complex by a polymerase chain reaction-solution hybridization enzyme-linked immunoassay (PCR-SHELA) Parasitology. 1995;110:269–275. doi: 10.1017/s0031182000080859. [DOI] [PubMed] [Google Scholar]
- 46.Ramos A, Maslov D A, Fernandes O, Campbell D A, Simpson L. Detection and identification of human pathogenic Leishmania and Trypanosoma species by hybridization of PCR-amplified mini-exon repeats. Exp Parasitol. 1996;82:242–250. doi: 10.1006/expr.1996.0031. [DOI] [PubMed] [Google Scholar]
- 47.Ravel S, Cuny G, Reynes J, Veas F. A highly sensitive and rapid procedure for direct PCR detection of Leishmania infantum within human peripheral blood mononuclear cells. Acta Trop. 1995;59:187–196. doi: 10.1016/0001-706x(95)00079-t. [DOI] [PubMed] [Google Scholar]
- 48.Rodgers M R, Popper S J, Wirth D F. Amplification of kinetoplast DNA as a tool in the detection and diagnosis of Leishmania. Exp Parasitol. 1990;71:267–275. doi: 10.1016/0014-4894(90)90031-7. [DOI] [PubMed] [Google Scholar]
- 49.Silveira F T, Lainson R, Shaw J J, Ribeiro R S. Leishmaniose cutanea na Amazonia: registro do primeiro caso humano de infecção mista, determinado por duas especies distintas de leishmanias: Leishmania brasiliensis e Leishmania mexicana amazonensis. Rev Inst Med Trop Sao Paulo. 1984;26:272–275. doi: 10.1590/s0036-46651984000500008. [DOI] [PubMed] [Google Scholar]
- 50.Simpson L. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Annu Rev Microbiol. 1987;41:363–382. doi: 10.1146/annurev.mi.41.100187.002051. [DOI] [PubMed] [Google Scholar]
- 51.Smyth A J, Ghosh A, Hassan M Q, de Brujin M H L, Adhya S, Mallik K K, Barker D C. Rapid and sensitive detection of Leishmania kinetoplast DNA from spleen and blood samples of kala-azar patients. Parasitology. 1992;105:183–192. doi: 10.1017/s0031182000074096. [DOI] [PubMed] [Google Scholar]
- 52.Spithill T W, Grumont R J. Identification of species, strains and clones of Leishmania by characterization of kinetoplast DNA minicircles. Mol Biochem Parasitol. 1984;12:217–236. doi: 10.1016/0166-6851(84)90137-3. [DOI] [PubMed] [Google Scholar]
- 53.Uliana S R B, Nelson K, Beverley S M, Camargo E P, Floeter-Winter L M. Discrimination amongst Leishmania by polymerase chain reaction and hybridization with small subunit ribosomal DNA derived oligonucleotides. J Eukaryot Microbiol. 1994;41:324–330. doi: 10.1111/j.1550-7408.1994.tb06085.x. [DOI] [PubMed] [Google Scholar]
- 54.van Eys G J J M, Schoone G J, Kroon N C M, Ebeling S B. Sequence analysis of small subunit ribosomal RNA genes and its use for detection and identification of Leishmania parasites. Mol Biochem Parasitol. 1992;51:133–142. doi: 10.1016/0166-6851(92)90208-2. [DOI] [PubMed] [Google Scholar]
- 55.Walton B C, Intermill R D, Hajduk M E. Differences in biological characteristics of three Leishmania isolates from patients with espundia. Am J Trop Med Hyg. 1977;26:850–855. doi: 10.4269/ajtmh.1977.26.850. [DOI] [PubMed] [Google Scholar]
- 56.Wilson S M. DNA-based methods in the detection of Leishmania parasites: field applications and practicalities. Ann Trop Med Parasitol. 1995;89:95–100. [PubMed] [Google Scholar]
- 57.World Health Organization. The leishmaniases. WHO Tech Rep Ser. 1990;753:154. [Google Scholar]