Abstract
Background
anti-Programmed Death-1 (anti-PD-1) immunotherapy has shown promising manifestation in improving the survival rate of patients with advanced melanoma, with its efficacy closely linked to Programmed cell death-Ligand 1 (PD-L1) expression. However, low clinical efficacy and drug resistance remain major challenges. Although the metabolic alterations from tricarboxylic acid (TCA) cycle to glycolysis is a hallmark in cancer cells, accumulating evidence demonstrating TCA cycle plays critical roles in both tumorigenesis and treatment.
Methods
The plasma levels of metabolites in patients with melanoma were measured by nuclear magnetic resonance (NMR) spectroscopy. The effect of pyruvate dehydrogenase subunit 1 (PDHA1) and oxoglutarate dehydrogenase (OGDH) on immunotherapy was performed by B16F10 tumor-bearing mice. Flow cytometry analyzed the immune microenvironment. RNA sequencing analyzed the global transcriptome alterations in CPI613-treated melanoma cells. The regulation of PD-L1 and glycolysis by PDHA1/OGDH-ATF3 signaling were confirmed by Quantitative real-time polymerase chain reaction (qRT-PCR), western blotting, dual-luciferase reporter gene, Chromatin immunoprecipitation (ChIP)-quantitative PCR and Seahorse assay. The relationship between PDHA1/OGDH-ATF3-glycolysis and the efficacy of melanoma anti-PD-1 immunotherapy was verified in the clinical database and single-cell RNA-seq (ScRNA-Seq).
Results
In our study, the results showed that significant alterations in metabolites associated with glycolysis and the TCA cycle in plasma of patients with melanoma through NMR technique, and then, PDHA1 and OGDH, key enzymes for regulation TCA cycle, were remarkable raised in melanoma and negatively related to anti-PD-1 efficacy through clinical database analysis as well as ScRNA-Seq. Inhibition of PDHA1 and OGDH by either shRNA or pharmacological inhibitor by CPI613 dramatically attenuated melanoma progression as well as improved the therapeutic efficacy of anti-PD-1 against melanoma. Most importantly, suppression of TCA cycle remarkably raises PD-L1 expression and glycolysis flux through AMPK-CREB-ATF3 signaling.
Conclusions
Taken together, our results demonstrated the role of TCA cycle in immune checkpoint blockade and provided a novel combination strategy for anti-PD-1 immunotherapy in melanoma treatment.
Keywords: Melanoma, Immunotherapy
WHAT IS ALREADY KNOWN ON THIS TOPIC
Accumulating evidence showed that mitochondrial metabolism (tricarboxylic acid (TCA) cycle/oxidative phosphorylation) is required for tumor progression. Tumor cells in various states, including but not limited to drug-resistant tumor cells, metastatic tumor cells, tumor stem cells, and tumor-initiating cells, exhibit varying degrees of reliance on the TCA cycle for their survival.
WHAT THIS STUDY ADDS
Inhibition of TCA cycle by knocking down of PDHA1 and oxoglutarate dehydrogenase or application of pharmacological inhibitor (CPI613) upregulate PD-L1 expression as well as glycolysis flux through AMPK-CREB-ATF3 axis, sensitizing anti-PD-1 immunotherapy in melanoma.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These findings revealed the novel mechanism of TCA cycle effect on glycolysis as well as the efficacy of anti-PD-1 immunotherapy, and provided a novel promising strategy to improve anti-PD-1 clinical efficacy in melanoma treatment.
Introduction
Metabolic reprogramming is a key feature of tumors, playing an essential role in not only tumor development but also tumor immunity.1 Glycolysis and tricarboxylic acid (TCA) cycle are the two main metabolic pathways underlying tumor cell metabolism; they are critical for providing energy to the cell. According to current studies, tumor cells tend to use glycolysis while mitochondrial function is dysregulated,2 however, studies also exhibited that TCA cycle/oxidative phosphorylation (OXPHOS) is significantly increased in some tumors, such as melanoma, breast cancer.3 4 Melanoma cells possess remarkable metabolic flexibility, contributing to their high malignancy and resistance to treatment.5 When subjected to metabolic stress or drug therapy, the metabolism of these cells shifts from glycolysis to the TCA cycle in order to sustain tumor progression and evade drug treatment.6 7 Moreover, melanoma cells lead to tumor microenvironment (TME) acidification via glycolysis, and the acidified TME in turn increases melanoma OXPHOS and inhibits glycolysis, ultimately inducing a suppressive immune microenvironment.8 Therefore, targeting the TCA cycle is a novel strategy for melanoma treatment.5 9 However, the details of the TCA cycle in melanoma have not been fully elucidated.
Malignant melanoma, derived from melanocytes, is a highly invasive and metastatic tumor that is notoriously difficult to treat.10 Recently, immunotherapy with the Anti-PD-1 antibody has been successful in cancer treatment; however, its response rate is low (less than 30%) and can lead to drug resistance.11 12
Two key enzymes (pyruvate dehydrogenase complex (PDHc) and oxoglutarate dehydrogenase complex (OGDHc)) orchestrate the carbon flow into the TCA cycle.13 The PDHc irreversibly catalyzes decarboxylating pyruvate to acetyl coenzyme A that could enter the TCA cycle, which provides an important intermediate metabolite for TCA cycle. The OGDHc is the rate-limiting enzyme in the TCA cycle, which irreversibly drives the TCA cycle.14 The structures and catalytic mechanisms of the two rate-limiting enzymes are very similar. Therefore, inhibitors that specifically target the two enzymes, such as CPI613, which acts as an analog of α-lipoic acid (LA), have been developed to inhibit the PDHc/OGDHc function by competitively binding to the complex and ultimately inhibiting the TCA cycle.15 In vitro and in vivo studies demonstrated that CPI613 is a promising candidate for non-small cell lung cancer and pancreatic cancer therapy.16 Several phase I–III clinical trials already demonstrated the safety and efficacy of CPI613 in pancreatic cancer and hematologic malignancies.14 17
Here, we found that the pyruvate dehydrogenase subunit 1 (PDHA1) and oxoglutarate dehydrogenase (OGDH) genes, subunits of two key enzymes associated with TCA, were significantly increased in patients with melanoma. Subsequent intervention with the two key enzymes or application CPI613 significantly inhibited the growth of melanoma cells, meanwhile, inhibition of the TCA cycle enhanced PD-L1 and glycolysis through ATF3 signaling, sensitizing the anti-PD-1 immunotherapy in melanoma.
Methods
Plasma collection and nuclear magnetic resonance spectroscopy analysis
After obtaining ethics approval for human study we collected plasma samples and analyzed them according to the previous manipulations.11
Cell culture
Mouse-derived B16F10 and YUMM1.7, human-derived Sk-Mel-5, A375, and Sk-Mel-28 cells were of American Type Culture Collection (ATCC) origin. 293T cells were purchased from Clontech (Mountain View, California, USA). B16F10 cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (BI, #01-100-1A, Israel) containing 10% fetal bovine serum (FBS) (Gibco, #10099141C, USA), YUMM1.7 were cultured in DEM/F-12 (1:1) containing 10% FBS and NEAA (100×) (Gibco), and human-derived melanoma cells were cultured in high glucose Dulbecco’s Modified Eagle’s Medium (BI) containing 10% FBS, and incubated in a constant temperature incubator containing 5% CO2 at 37°C.
Lentiviral packaging and infection
The plasmids (Sh-mock, Sh-PDHA1, Sh-OGDH, and Sh-ATF3) used in this study were purchased from GeneChem (GeneChem, China), and the plasmids (PSPAX2 and PMD2G) used for lentiviral packaging were stored in laboratory constructs.18 For lentiviral constructs, plasmids and transfection reagent TurboFect (Thermo Fisher Scientific, #R0532, USA) were added when the 293T cells reached 50–60% fusion, transfected overnight, and the cell supernatant (including virus) was collected at 48–72 hours. For lentiviral infection, virus solution and medium were added at a ratio of 1:1 when the cells reached 30–40%, and then the infection reagent polybrene (Yeasen, #40804ES76, China) was added, infected overnight, and then changed to normal medium. After changing to normal medium, cells were screened by adding 2 µg/mL puromycin (BasalMedia, #S250J0, China) after 24 hours of culture until the normal uninfected cells died.
Cell viability assay
In 96-well plates, 3000–4000 tumor cells were grown, incubated overnight, and 450 nm absorbance value (OD450) was detected using a spectrophotometer (Beckman, USA) after 0–72 hours by adding CCK-8 reagent (Selleck, #B34302, USA) for 1.5 hours.
Cell cycle assay
Tumor cells to be tested were trypsin-digested and collected into 1.5 mL eppendorf (EP) tubes, resuspended into single cell, washed three times with ice phosphate buffered saline (PBS), and centrifuged at 800 rpm for 3 min; 200 µL PBS resuspended cells were added to 1 mL of 70% ice ethanol for fixation, fixed overnight at 4°C, and then transferred to −20°C for storage or 300 µL PI/RNase Staining Buffer Solution (BD, #550825, USA) was added directly for detection.
qRT-PCR
RNA was extracted from the cells by adding 1 mL of TriPure, chloroform, isopropanol, and 75% anhydrous ethanol. The RNA was then reverse transcribed to complementary DNA using the Reverse Transcription Kit (Yeasen, #11141ES60, China) and finally detected using fluorescent quantitative PCR (qPCR) using the SYRB Green qPCR Master Mix (Bimake, #B21703, USA). The PCR primers used are listed in online supplemental table S1.
jitc-2023-007146supp001.pdf (1.4MB, pdf)
Western blotting
The supernatant was collected using centrifugation (12,000 rpm, 10 min) at 4°C after the cells were fully lysed using cell lysate (Beyotime, #P0013D, China) with ultrasound. The supernatant was quantified by BCA (Beyotime, #P0010S), added to 5×loading buffer (Ncmbio, #WB2001, China), denatured, and gel electrophoresis was performed using a PVDF membrane (Millipore, #IPVH00010, Billerica, Massachusetts, USA) and incubated with primary antibody (anti-PDHA1, Abcam, #ab110330, 1:1,000; anti-OGDH, CST, #26865S, 1:1,000; anti-PD-L1, Abcam, #ab213524, 1:1,000; anti-α-tubulin, ProteinTech, #1124–1-AP, 1:5,000; anti-ATF3, Abcam, #ab216569, 1:1,000) overnight; the second antibody (anti-rabbit, Abclonal, #AS014, 1:8,000; anti-mouse, Abclonal, #AS003, 1:8,000) was incubated on the second day, and the membrane was washed and imaged using a gel imager (Bio-Rad, USA).
Animal experiments
The C57BL/6 mice used in the animal experiments were purchased from the Department of Zoology of Central South University. The animal experiments were approved by the Ethics Committee of Xiangya Hospital and ethical principles were observed. To construct tumor-bearing mice, 5×106/mL B16F10 or YUMM1.7 cells were first collected and 100 µL tumor was implanted in the right abdominal dorsum of each mouse; when the tumor grew to 50 mm3, IgG2a (Bio X Cell, USA; #BE0089), PD-1 monoclonal antibody (mAb) (Bio X Cell; #BE0146), CPI613 (MCE, #HY-15453, USA), and PD-1 mAb combined with CPI613 were administered to treat the tumor-bearing mice, and the tumor volume and body weight of the mice were measured every other day. When the tumor grew to 1000 mm3, the administration was stopped and the mice were sacrificed. The tumor tissues were collected for multicolor flow cytometry assay.
Multicolor flow cytometry assay
The collected tumor tissues were prepared as single cell suspensions, stained with Zombie Aqua Fixable Viability Kit (anti-BV510, BioLegend, #423102, USA), incubated for 15 min at room temperature, and then terminated and centrifuged. The fragment crystallizable (FC) was blocked with anti-CD16/32 (BioLegend, #101320) and stained with surface antibody after 15 min (in two tubes, the first tube was stained as follows: anti-PE-NK1.1 (BioLegend, #108708); anti-PC5.5-CD4 (BioLegend, #100434); anti PECY7-CD8 (BioLegend, #100722); anti-APC-CD3 (BioLegend, #100236); anti-APCCY7-CD45 (BioLegend, #103116); anti-BV711-interferon (IFN)-γ (BioLegend, #505836). Tube 2: anti-fluoresceine isothiocyanate (FITC)-major histocompatibility complex (MHC)-II (BioLegend, #107606); anti-PE-CD11B (BioLegend, #101208); anti-PC5.5-Gr1; anti-APC-F4/80 (BioLegend, #123116); anti-APCCY7-CD45), incubated at 4°C for 30 min, followed by fixed membrane breaking solution incubation for 30 min, and stained with intracellular antibodies (tube 1: anti-FITC-granzyme B (BioLegend, #372206); anti-BV711-IFN-γ, tube 2: anti-PECY7-CD206 (BioLegend, #372206)) incubated at 4°C for 30 min, followed by flow cytometry detection (BD FACS LSRFortessa, USA).
Co-culture experiment
Peripheral blood mononuclear cells (PBMC) from mouse spleens were extracted using lymphocyte isolate (DAKEWE, #DKW33-R0100/DKW33-R0400, China), co-cultured with B16F10 cells (cell ratio 15:1), and treated with 5 µg/mL PD-1 mAb and/or 120 µM CPI613 for 48 hours. Cell survival was detected using crystalline violet staining and CCK8.
Oxidative phosphorylation and glycolysis assay
First, 8,000–10,000 tumor cells were grown in a Seahorse 96-well assay plate. After overnight incubation, the cells were washed twice with pre-prepared assay solution, the pretreated probe plate was placed in the cell plate, OXPHOS (Agilent Technologies, #103015–100, USA) or glycolysis assay (Agilent Technologies, #103020–100) reagents were added sequentially, and the intracellular metabolic changes were detected in real time using the Agilent Seahorse XFe96 (Agilent Technologies).
RNA sequencing
The extracted cellular messenger RNA (mRNA) was submitted to Shanghai Majobio Bio-Pharm Technology for sequencing. Briefly, mRNA was sequenced using the Illumina NovaSeq 6000 sequencing platform, and the library was constructed using the Illumina TruSeq RNA Sample Prep Kit method. After sequencing, quality control, reference genome alignment, and transcript assembly were performed, followed by differential gene data and advanced analyses. The data were analyzed on the online platform of Majorbio Cloud Platform (www.majorbio.com).
ELISA
For the detection of AMP or ATP levels in melanoma cells, the lysate was added to a 96-well plate precoated with AMP or ATP capture antibodies, followed by antibody biotinylation, Horseradish Peroxidase (HRP) labeling, and Tetramethylbenzidine (TMB) color development (Enzyme-linked Biotechnology, China). The absorbance values were finally detected by a spectrophotometer (Beckman, USA). The intracellular AMP or ATP levels and AMP/ATP ratio were calculated according to the standard curve.
Luciferase reporter gene assay
Tumor cells (8×104) were grown in 24-well plates overnight, and the next day, PD-L1-Luc and SV-40Renilla Luc and transfection reagent were added under the condition of CPI613 treatment. The cells were lysed and collected using a lysis solution 24 hours after incubation, and the promoter firefly and Renilla luciferase activities were detected using a dual luciferase reporter system (Promega, #E1910, Wisconsin, USA).
ChIP assay
Tumor cells were grown in 10 cm dishes overnight and treated with CPI613 for 24 hours. Then, the cells were collected and assayed using the ChIP kit (CST, #9003, USA). Briefly, 4×106 cells were treated with 37% formaldehyde to cross-link proteins to DNA, followed by nucleus preparation and chromatin digestion; 10 µg of digested cross-linked chromatin was immunoprecipitated overnight (anti-ATF3, Abcam, #ab207434, 1:50; 1–5 µL Rabbit IgG, CST, #2729S), then chromatin was eluted from the precipitate and uncross-linked; the DNA was then purified and real-time qPCR was performed to detect the enrichment of DNA.
Single-cell sequencing
Single-cell sequencing was performed by Wuhan Huada Sequencing Company. Simply, fresh tissues were dissociated into single-cell suspensions and tested for cell viability. After passing the test, samples were labeled and cells were captured using BD Rhapsody platform microplates, and mRNA was captured by magnetic beads after cell lysis. After reverse transcription, magnetic beads were denatured to separate different library templates and the library was constructed. After library quality control was completed, DNBSEQ sequencing was performed. Finally, sequencing data were analyzed according to the previous manipulations.19
Statistics
Unpaired Student’s t-test was used to compare the differences between the two groups, and one-way or two-way analysis of variance tests were used to compare the differences between multiple groups. GraphPad software (V.6.01) was used to analyze the results; statistical differences were considered when the p value was <0.05, and the asterisk represents the degree of difference (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
Results
PDHA1 and OGDH is highly expressed in melanoma and negatively correlated with anti-PD-1 immunotherapy responsiveness
To study metabolism alteration in melanoma, we examined metabolites in the plasma of 20 patients with melanoma and 20 normal subjects through nuclear magnetic resonance spectroscopy, and found significant alterations of metabolites involved in TCA cycle and glycolysis such as pyruvate, fumarate, 2-oxoglutarate, and glucose (figure 1A). Among those metabolites, pyruvate serves as the link between glycolysis and the TCA cycle, which converted to acetyl coenzyme A via pyruvate dehydrogenase (PDH) before entering the TCA cycle, and it is essential for sustaining TCA activity.14 In addition, fumarate and 2-oxoglutarate are crucial intermediate metabolites of the TCA cycle, indicated TCA cycle may have essential function in melanoma. The glycolysis in melanoma had been intensively studied, whereas the role of TCA cycle is not fully elucidated. Given PDHc and OGDHc orchestrate the main carbon flow into the TCA cycle, we analyzed the expression of PDHA1 and OGDH in the Gene Expression Omnibus
Figure 1.
Combined intervention of OGDH and PDHA1 inhibits melanoma cell proliferation. (A) Nuclear magnetic resonance assay for altered metabolites in patients with melanoma (n=20) versus normal participants (n=20). (B–C) Data from the GEO database (GSE 3189, nevus (n=18), melanoma (n=45)) were used to analyze the PDHA1 (B) and OGDH (C) expression in patients with melanoma. (D) The correlation of patient with PDHA1 expression and melanoma survival were analyzed in the GEPIA website. (E–F) CCK8 was used to detect the proliferation of A375 (E) and Sk-Mel-5 (F) cells after PDHA1 and OGDH gene knockdown (n=6 samples per group). (G–H) CCK8 was used to detect the proliferation of A375 (G) and Sk-Mel-5 (H) cells after CPI613 treatment for 0–72 hours (n=6 samples per group). (I–J) Flow cytometry was used to detect the cell cycle of A375 (I) and Sk-Mel-5 (J) cells after PDHA1 and OGDH gene knockdown (n=3 samples per group). (K–L) Flow cytometry was used to detect the cell cycle of A375 (K) and Sk-Mel-5 (L) cells after CPI613 treatment for 24 hours (n=3 samples per group). Multiple samples were compared for differences using mean±SD, and the differences were marked according to the statistical markers in the methods. GEO, gene expression omnibus; OGDH, oxoglutarate dehydrogenase; PDHA1, pyruvate dehydrogenase subunit 1.
(GEO) public database and Gene Expression Profiling Interactive Analysis (GEPIA),20 and found that the expression of PDHA1 and OGDH were significantly raised in melanoma (figure 1B,C). Moreover, patients with melanoma with elevated PDHA1 expression exhibited poor prognosis (figure 1D), implying PDHA1 and OGDH might have oncogenic features in melanoma.
To investigate the role of the TCA cycle in melanoma, we knocked down PDHA1/OGDH expression in melanoma cells, as shown in online supplemental figure 1, knock down of either PDHA1 or OGDH significantly inhibited the proliferation of melanoma cells (online supplemental figures S1A-F) and induced cell cycle arrest in the G0/G1 phase (online supplemental figures S1G-J). Furthermore, inhibition of both PDHA1 and OGDH expression or their pharmacological inhibitor (CPI613) significantly reduced melanoma cells proliferation (figure 1E–H) and arrested cell cycle in G0/G1 phase (figure 1I–L).
Next, we assessed the expression of PDHA1 and OGDH in patients receiving anti-PD-1 immunotherapy, and the results showed that in two GEO data sets, PDHA1 expression was significantly lower in complete response groups than in progressive disease groups (online supplemental figures S2A,B), and 67–89% of patients with partial or complete remission had a lower PDHA1 (8/9) and OGDH (6/9) expression after anti-PD-1 treatment, compared with pretreated stage (online supplemental figures S2C,D), indicating that PDHA1 and OGDH were associated with immunotherapy response. Furthermore, we analyzed the relationship between PDHA1 and the immune score in GSE91061 by the ESTIMATE method. As shown in online supplemental figure 2E, PDHA1 expression was negatively correlated with stromal, immune, and ESTIMATE scores, while exhibiting a strong positive correlation with tumor purity, suggesting that PDHA1 is linked to a suppressed immunological microenvironment (online supplemental figure S2E). Additionally, to investigate the association between PDHA1 and OGDH expression and the immune state of patients with melanoma undergoing anti-PD-1 therapy, we performed the ImmuCellAI-human method,21 and found that PDHA1 expression showed an inverse correlation with the abundance of CD4+ T cells, while OGDH expression was associated with lower state of immune cell infiltration as well as decreased number of CD8 cytotoxic T cells (online supplemental figures S2F,G), indicating PDHA1 and OGDH are associated with immunosuppressive microenvironment. Interestingly, we also found that PDHA1 and OGDH expression was decreased in the tumor tissues of YUMM1.7 tumor-bearing mice (sensitive to anti-PD-1 treatment) compared with B16F10 tumor-bearing mice (relatively resistant to anti-PD-1 therapy) in a single-cell sequencing data (online supplemental figure S2H).
Inhibition of TCA cycle enhances anti-PD-1 immunotherapy efficacy in melanoma cells
To study involvement of PDHA1 and OGDH in melanoma immune responsiveness, we established an in vitro co-culture system of melanoma cells and PBMCs to simulate the in vivo microenvironment, as shown in online supplemental figure 3A, although pharmacological inhibitor (CPI613) could reduce melanoma cells, we did not observe any difference in tumor cell death between the pharmacological inhibitor (CPI613) treatment and the CPI613 plus anti-PD-1 combination treatment without PBMC (online supplemental figure S3A), however, CPI613 significantly enhanced the efficacy of anti-PD-1 mAb on melanoma cells in present of PBMC (online supplemental figures S3B,C).
To further validate the role of PDHA1 and OGDH in the melanoma immunotherapy in vivo, we constructed B16F10 tumor-bearing C57BL/6 mouse model and treated them with an inhibitor of PDHA1 and OGDH (CPI613) in combination with PD-1 mAb (figure 2A, left panel). As shown in figure 2A, the combination treatment significantly improved the therapeutic efficacy of anti-PD-1 (figure 2A, right panel), and prolonged the survival of mice (figure 2B). Similarly, immunotherapy efficacy had been also enhanced in YUMM1.7 melanoma-bearing mice (online supplemental figure S4A). The body weight of the mice was not significantly changed during the treatment period (online supplemental figures S4B,C). To assess the effect of the combination treatment on the immune microenvironment, we analyzed tumor-infiltrating leukocytes in tumorous tissues. As shown in figure 2C–J, the combination therapy significantly reduced the proportion of (Gr1+CD11B+) myeloid-derived suppressor cells and (F4/80+CD11B+) macrophages in the TME (figure 2C,D), whereas, the infiltration of (MHC-II+F4/80+CD11B+) M1 macrophages had been elevated (figure 2E). In addition, the combination treatment markedly induced infiltration of functional CD4+ and CD8+ T cells (figure 2F–J). While, the infiltration of natural killer (NK)+ T cells and Regulatory T (Treg) cells had not been altered, indicating the combination therapy’s efficacy was not primarily influenced by NK+ T cells or Treg cells (online supplemental figures S4D,E). Consistent with the above results, knock down both PDHA1 and OGDH significantly altered the immune microenvironment, which benefited to that improve the therapeutic efficacy of anti-PD-1 (figure 2K and online supplemental figure S4F-KS4F-K), and the combination treatment had no significant effect on mouse body weight (online supplemental figure S4L). In conclusion, our findings demonstrated that targeting TCA cycle by PDHA1 and OGDH remarkably improved the efficacy of anti-PD-1 therapy in melanoma, indicating the TCA cycle inhibitor (CPI613) is a potential therapy strategy.
Figure 2.
Inhibition of PDHA1 and OGDH enhanced the efficacy of melanoma anti-PD-1 immunotherapy. (A) Schematic diagram of CPI613 combined with PD-1 mAb for melanoma treatment (left panel). Tumor growth curves for B16F10 tumor-bearing mice receiving the designated treatments (right panel). (B) Survival curves of B16F10 tumor-bearing C57BL/6 mice receiving the designated treatments. (C–J) The percentage of Gr1+CD11B+ cells (C), F4/80+CD11B+ cells (D) F4/80+CD11B+MHC-II+ cells (E) CD8+/CD4+ T cells (F) IFN-γ+CD4+ T cells (G) granzyme B+CD4+ T cells (H) IFN-γ+CD8+ T cell (I) and granzyme B+CD8+ T cell (J) in tumors determined by FACS. (K) Tumor growth curves for B16F10 tumor-bearing mice receiving the designated treatments. Multiple samples (n=5 mice per group) were compared for differences using mean±SD, and the differences were marked according to the statistical markers in the methods. PD-1, programmed death 1; mAb, monoclonal antibody; IFN, interferon; MHC, major histocompatibility complex; FACS, fluorescence activating cell sorter; OGDH, oxoglutarate dehydrogenase; PDHA1, pyruvate dehydrogenase subunit 1.
Inhibition of PDHA1 and OGDH expression raises PD-L1 expression in melanoma cells
PD-L1 expression in tumorous cells is a key factor for the efficacy of anti-PD-1 immunotherapy,22 therefore, we examined the effects of PDHA1 and OGDH on PD-L1 expression through simultaneous knocking down or treatment with pharmacological inhibitor (CPI613). As expected, suppression of PDHA1 and OGDH gene or pharmacological inhibition resulted in upregulation of PD-L1 mRNA (figure 3A,B) as well as protein expression (figure 3C–F) in melanoma cells.
Figure 3.
Combined intervention of OGDH and PDHA1 upregulates PD-L1 expression in melanoma cells. (A) The mRNA levels of PD-L1 in A375 (left panel) and Sk-Mel-5 (right panel) cells were measured using qRT-PCR after PDHA1 and OGDH gene knockdown (n=3 samples per group). (B) The mRNA levels of PD-L1 in A375 (left panel) and SK-MEL-5 (right panel) cells were detected using qRT-PCR after CPI613 (300 µm) treatment for 48 hours (n=3 samples per group). (C–D) The protein levels of PD-L1, OGDH, and PDHA1 in A375 (C) and Sk-Mel-5 (D) cells were detected using western blotting after PDHA1 and OGDH gene knockdown. (E–F) The protein levels of PD-L1, OGDH, and PDHA1 in A375 (E) and SK-MEL-5 (F) cells were detected using western blotting after CPI613 (200–300 µm) treatment for 24 hours. Multiple samples were compared for differences using mean±SD, and the differences were marked according to the statistical markers in the methods. PD-L1, programmed cell death-ligand 1; mRNA, messenger RNA; qRT-PCR, quantitative real-time polymerase chain reaction; OGDH, oxoglutarate dehydrogenase; PDHA1, pyruvate dehydrogenase subunit 1.
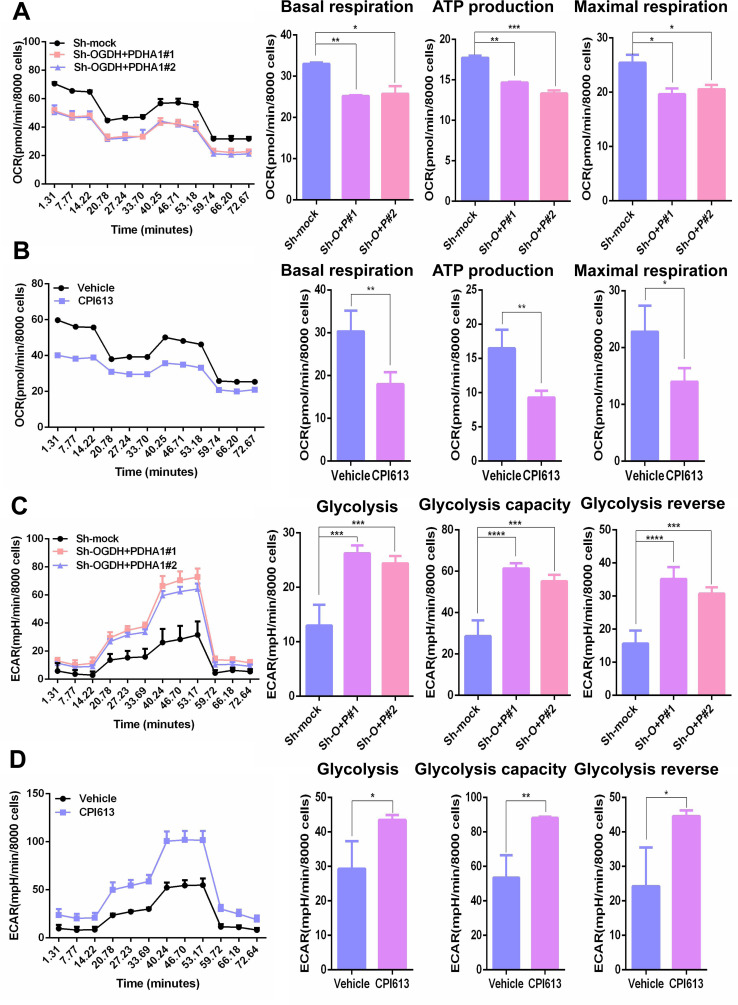
Given PDH and OGDH are crucial enzymes for the initiation and efficient execution of the TCA cycle, we investigated oxygen consumption rate alteration in melanoma cells following PDHA1 and OGDH inhibition. As shown in figure 4A,B, knock down of PDHA1 plus OGDH expression or pharmacological inhibition significantly reduced basal OXPHOS, ATP production, and maximal respiratory capacity (figure 4A,B). It was well known that TCA cycle inhibition induces the upregulation of glycolysis flux,23 and increasing glycolysis flux has been documented to raise PD-L1 expression,24 25 therefore, we also tested glycolysis in melanoma cells following PDHA1 and OGDH suppression. The results exhibited that glycolysis levels, glycolytic capacity, and glycolytic reverse were notably elevated in melanoma cells after suppression of PDHA1 and OGDH (figure 4C,D), which suggested that inhibition of TCA may sensitize anti-PD-1 immunotherapy through regulation of PD-L1.
Figure 4.
Intervention in the tricarboxylic acid cycle inhibited oxidative phosphorylation and upregulated glycolysis in melanoma cells. (A) Seahorse was performed to analyze oxidative phosphorylation levels in Sk-Mel-5 cells (left panel). Basal oxidative phosphorylation levels, ATP production capacity and maximum respiration rate in Sk-Mel-5 cells after Sh-OGDH+PDHA1 (Sh-O+P, right panel). (B) Seahorse was performed to analyze oxidative phosphorylation levels (left panel) in Sk-Mel-5 cells. Basal oxidative phosphorylation levels, ATP production capacity and maximum respiration rate in Sk-Mel-5 cells after CPI613 (300 µm) treatment for 24 hours (right panel). (C) Seahorse was performed to analyze glycolysis levels in Sk-Mel-5 cells (left panel). The basal glycolysis level, the maximum capacity of glycolysis and glycolysis reverse in Sk-Mel-5 cells after Sh-OGDH+PDHA1 (Sh-O+P, right panel). (D) Seahorse was performed to analyze glycolysis levels in Sk-Mel-5 cells (left panel). The basal glycolysis level, the maximum capacity of glycolysis and glycolysis reverse in Sk-Mel-5 cells after CPI613 (300 µm) treatment for 24 hours (right panel). Multiple samples (n=3 samples per group) were compared for differences using mean±SD, and the differences were marked according to the statistical markers in the methods. OCR, oxygen consumption rate; ECAR, extracelluar acidification rate; OGDH, oxoglutarate dehydrogenase; PDHA1, pyruvate dehydrogenase subunit 1.
Inhibition of TCA elevates glycolysis via ATF3-HKDC1 signaling
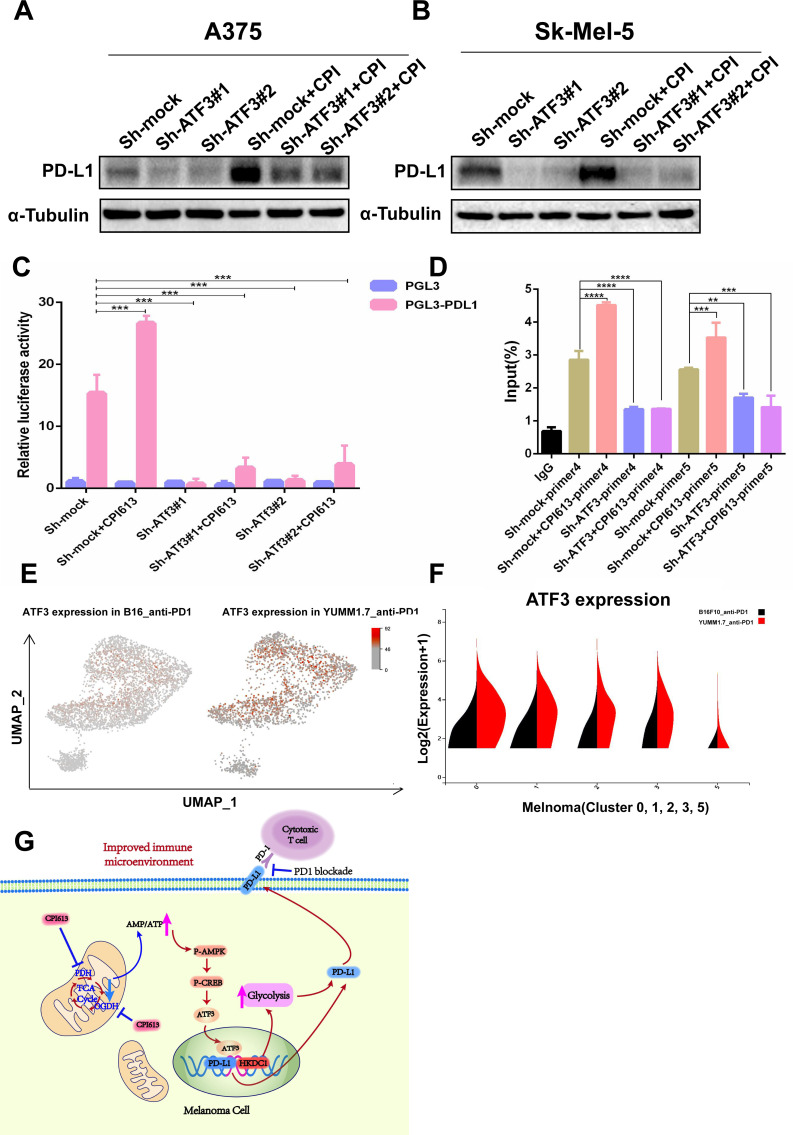
To further investigate the precise molecular mechanism underlying TCA inhibition-induced PD-L1 and glycolysis, RNA sequencing was conducted to analyze the transcriptomic alterations in B16F10 melanoma cells after CPI613 treatment. Principal component analysis and heat map clustering revealed pronounced distinctions between the CPI613-treated and control groups (online supplemental figures S5A,B). KEGG and Reactome annotation analyses showed that the major differential expression genes were clustered to signal transduction and metabolism (online supplemental figures S5C,D). As expected, GSEA analysis showed that the TCA cycle and OXPHOS pathway were dramatically altered in CPI613-treated cells, which was consistent with the previous cytological results (online supplemental figure S5E). Likewise, glycolysis-related genes were prominently elevated in the CPI613-treated group, including the classical transcription factors HIF and MYCL, which regulate glycolysis, and the transcription factor ATF3 (online supplemental figure S5F). ATF3 is a member of the ATF/CREB family of transcription factors,26 however its role in glycolysis remains unclear. AMPK is a metabolic energy sensor based on AMP/ATP ratio, directly perceiving intracellular ATP levels and decreased ATP production resulting from mitochondrial inhibition,27 28 meanwhile the transcriptional factor family of CREB/ATF exerts critical roles in AMPK triggering signaling pathway.29 30 Therefore, we speculated that inhibition of TCA cycle upregulates glycolysis and PD-L1 by activating the AMPK-CREB-ATF3 signaling. As expected, we verified that inhibition of PDHA1 and OGDH by either shRNA or CPI613 increased intracellular AMP/ATP ratio (figure 5A). Knock down of OGDH plus PDHA1 expression or administration of CPI613 significantly activated AMPK-CREB signaling pathway in melanoma cells (figure 5B). These findings indicated that inhibition of PDHA1 and OGDH increases glycolysis and PD-L1 through AMPK-CREB-ATF3 signaling. Meanwhile, suppression of PDHA1 and OGDH remarkably increased ATF3 expression at the mRNA and protein levels (figure 5C–E). Notably, inhibition of ATF3 expression significantly reduced glycolytic capacity and expression of key genes involved in glycolysis (figure 5F,G, online supplemental figures S6A-D), whereas ectopic ATF3 elevated those glycolytic genes expression (online supplemental figures S6E-H). Among those genes, HKDC1, encoding HK kinase, was the most significantly altered glycolytic gene after CPI613 treatment (online supplemental figure S5F). Therefore, we proposed that ATF3 might regulate melanoma glycolysis through HKDC1. As expected, our finding showed that ATF3 directly recognized the promoter region of HKDC1, while knockdown of ATF3 inhibited HKDC1 promoter activity (figure 5H).
Figure 5.
OGDH and PDHA1 increased glycolysis through AMPK-CREB-ATF3 signaling. (A) The ratio of AMP/ATP was detected using ELISA after Sh-OGDH+PDHA1(left panel) or CPI613 (300 µm, right panel) treatment for 24 hours (n=3 samples per group). (B) The protein levels of P-AMPK and P-CREB in Sk-Mel-5 cells were detected using western blotting after Sh-OGDH+PDHA1(left panel) or CPI613 (200–300 µm, right panel) treatment for 24 hours (B). (C) The messenger RNA levels of ATF3 in Sk-Mel-5 cells were detected using RT-PCR after Sh-OGDH+PDHA1(left panel) or CPI613 (300 µm, right panel) treatment for 24 hours (n=3 samples per group). (D) The protein levels of ATF3 in A375 (left panel) and Sk-Mel-5 (right panel) cells were detected using western blotting after Sh-OGDH+PDHA1. (E) Protein levels of ATF3 in A375 (left panel) and SK-MEL-5 (right panel) cells were detected using western blotting after CPI613 (300 µm) treatment for 24 hours. (F–G) Seahorse was performed to analyze glycolysis levels in Sk-Mel-5 cells (F). The basal glycolysis level and the maximum capacity of glycolysis in Sk-Mel-5 cells after Sh-ATF3 (G) (n=3 samples per group). (H) ChIP assay was performed to detect ATF3 binding to the HKDC1 promoter after ATF3 knocking down (n=3 samples per group). Multiple samples were compared for differences using mean±SD, and the differences were marked according to the statistical markers in the methods. OGDH, oxoglutarate dehydrogenase; PDHA1, pyruvate dehydrogenase subunit 1; ECAR, extracelluar acidification rate; RT-PCR, reverse transcription polymerase chain reaction; ChIP, chromatin immunoprecipitation.
Suppression of TCA cycle increases PD-L1 expression through ATF3
Consistent with our previous results,31 we confirmed the regulatory role of ATF3 on PD-L1 expression, and pharmacological inhibitors of PDHA1 and OGDH did not completely rescue the PD-L1 expression in ATF3 knocking down cells (figure 6A,B). Meanwhile, knock down of ATF3 expression significantly inhibited CPI613-induced PD-L1 promoter activity and the binding of ATF3 to the PD-L1 promoter (figure 6C,D), indicating inhibition of TCA cycle-induced PD-L1 expression depended on ATF3. Subsequently, we analyzed the correlation of ATF3 and glycolysis with anti-PD-1 treatment responsiveness in melanoma single-cell sequencing data and found that ATF3 and key glycolytic enzymes (HK/PFKM/PFKP) were expressed at higher levels in the tumor tissues of mice sensitive to anti-PD-1 treatment compared with the anti-PD-1 insensitive group (online supplemental figures 6E,F and S7).
Figure 6.
CPI613 regulates PD-L1 expression through ATF3. (A–B) The protein levels of PD-L1 in A375 (A) and Sk-Mel-5 (B) cells was detected using western blotting. (C) The effect of CPI613 and ATF3 on PD-L1 promoter activity was detected using a luciferase reporter gene. (D) ChIP assay was performed to detect the effect of CPI613 on the extent of ATF3 binding to the PD-L1 promoter. (E) Single-cell sequencing analyzed the expression of ATF3 in melanoma insensitive to PD-1 immunotherapy (left) versus sensitive melanoma (right). (F) Single-cell sequencing analyzed the expression of ATF3 in five melanoma clusters from single cell sequencing results (violin chart). (G) Diagram of molecular mechanism of the article. Multiple samples (n=3 samples per group) were compared for differences using mean±SD, and the differences were marked according to the statistical markers in the methods. PD-L1, programmed cell death-ligand 1; PD-1, programmed death 1; ChIP, chromatin immunoprecipitation.
Discussion
Metabolic reprogramming is one of the main features in tumor cells. Although, many years ago, Warburg et al made a significant discovery that tumor cells have a preference for using anaerobic glycolysis instead of OXPHOS for their energy supply, accumulating evidence showed that mitochondrial metabolism (TCA cycle/OXPHOS) is required for tumor progression.32 33 Tumor cells in various states, including but not limited to drug-resistant tumor cells, metastatic tumor cells, tumor stem cells, and tumor-initiating cells, exhibit varying degrees of reliance on the TCA cycle for their survival.34 This phenomenon was also found in melanoma cells; metastatic melanoma cells had been to shown to downregulate the glycolytic metabolites, whereas TCA circulating metabolites were significantly elevated compared with primary melanoma cells. Further mechanistic study found that this is due to inhibition of isoform of the short glycerol-3-phosphate dehydrogenase, which leads to reduced glycolysis and raise TCA-circulating metabolites, respectively, ultimately facilitating metastasis of melanoma cells.35
Actually, TCA cycle-related enzymes are crucial targets by oncogenes during carcinogenesis. Glucose-PDH-mediated TCA cycle/mitochondrial metabolism is required for Ras-driven non-small cell lung cancer formation.36 SIRT3 activates the activity of PDHA1 and pyruvate dehydrogenase phosphatase subunit 1 by regulating their deacetylation, thereby elevating the TCA cycle, and eventually enhancing lung cancer cell growth.37 Colorectal cancer cells with PIK3CA mutations catalyzes large amounts of glutamine to replenish the TCA cycle, which maintains tumor growth via glutamate pyruvate transaminase 2.38 In melanoma cells, PGC1-α promotes drug resistance and metastasis by converting the metabolism of melanoma from glycolysis to TCA cycle.39–41
Most importantly, the alteration of TCA cycle is also associated with tumor immune microenvironment. In the aggressively activated B-cell subtype of diffuse large B-cell lymphoma, glutamate enhances STAT3-induced PD-L1 expression in a TCA cycle-dependent manner.42 In gliomas with wild-type IDH1, PD-L1 was highly expressed, whereas IDH1 mutant gliomas inhibited PD-L1 expression via 2-hydroxyglutaric acid.43 It was also reported that α-ketoglutarate enhances PD-L1 expression in tumor cells via IFN-γ-STAT1-TET1-IRF1 signaling in hepatocellular carcinoma, sensitizing the therapeutic efficacy of anti-PD-1/PD-L1.44 Similarly, α-ketoglutarate enhances IFN-γ-STAT-PD-L1 signaling via the demethylase TET and enhances melanoma anti-PD-1 immunotherapy efficacy.19 The studies mentioned above indicate that the expression of PD-L1, which is induced by enzymes or metabolites related to TCA, could potentially enhance the responsiveness of tumors to anti-PD-1/PD-L1 therapy. On the other hand, it has been found that fumarate produced by tumors can inhibit the antitumor function of CD8+ T cells within the TME.45 The TCA cycle is a complex system, and the impact of changes in its critical enzymes or metabolites on tumor progression is intricate. Therefore, further research is necessary to fully understand the molecular mechanisms involved.
To explore the molecular mechanisms underlying the role of the TCA cycle in melanoma, we inhibited the TCA cycle by inhibiting PDHA1 and OGDH subunits of the PDH and OGDH enzyme complexes. The main sources of carbon flow in the TCA cycle are glycolytic-derived pyruvate and glutamine-derived α-ketoglutarate, both of which, PDH and OGDH, play key roles in the TCA cycle/OXPHOS, and they control the majority of the carbon flow into the TCA cycle.13 PDH supplies the TCA cycle by irreversibly catalyzing the conversion of pyruvate to acetyl coenzyme A, while OGDH facilitates the TCA cycle by catalyzing the conversion of a-ketoglutarate to succinyl coenzyme A. Given that this catalytic process is irreversible,14 therefore, inhibition of the TCA cycle/OXPHOS by inhibiting PDH and OGDH is the optimal choice for tumor therapy.
Our results confirmed that the knock down of PDHA1 and OGDH significantly inhibited OXPHOS levels and ATP production in tumor cells (figure 4). As mentioned above, although it was well known that melanoma cells enhance tumor metastasis through the suppression of glycolysis, which results in an upregulation of TCA-circulating metabolites implying a potential compensatory relationship between glycolysis and the TCA cycle,35 the details of molecular mechanisms responsible for the transition from glycolysis to the TCA cycle have been inadequately documented. This study investigated the mechanism behind the switch from the TCA cycle to glycolysis in melanoma and revealed that inhibiting the TCA cycle activates the AMPK-CREB-ATF3 signaling pathway, leading to a rise in both glycolysis and PD-L1 expression. We also investigated how the PD-L1 expression triggered by inhibiting the TCA cycle might enhance the efficacy of immunotherapy.
CPI613 is an analog of α-LA, and only four enzyme complexes in the cell (PDH, OGDH, the branched chain oxidative acid dehydrogenase complex, and the glycine cleavage system) must be covalently bound to α-LA coenzymes to exert enzymatic activity.15 In addition to PDH and OGDH, the other two enzymes are only present in the mitochondrial matrix and feed the carbon produced from amino acids into the TCA cycle. Given PDH and OGDH are crucial enzymes regulating the carbon flux of the TCA cycle, CPI613 has been primarily employed as a specific inhibitor of two enzymes.4 13 CPI613 has been found to effectively inhibit the growth of several types of tumors. Specifically, in non-small cell lung cancer and pancreatic cancer, CPI613 has shown selective cytotoxicity towards tumor cells by disrupting mitochondrial metabolism.15 Notably, its mode of action has led to fewer systemic side effects, making it a promising candidate for targeted therapy.4
Here, we elucidate the molecular mechanisms by which CPI613 inhibits melanoma and enhances the efficacy of anti-PD-1 immunotherapy in melanoma. Our results showed that CPI613 significantly inhibited TCA cycling/OXPHOS in melanoma cells and revealed that CPI613 not only significantly inhibited melanoma growth but also upregulated melanoma cell glycolysis levels and PD-L1 expression through AMPK-CREB-ATF3 signaling and increased melanoma immune responsiveness to PD-1 mAb treatment. Our study demonstrated that CPI613 can significantly enhance the efficacy of melanoma immunotherapy as a TCA cycle inhibitor and has good clinical application.
Actually, CPI613 has been evaluated in several phase I–III clinical trials as a combination therapy for tumor treatment, with a focus on its efficacy, tolerability, and safety.14 According to the findings of a phase I clinical trial (NCT01835041), the combination of CPI613 and modified FOLFIRINOX (mFFX) reached a 61% objective response rate in patients with metastatic pancreatic cancer, indicating that the treatment was well-tolerated and deemed safe for participants.46 Furthermore, an ongoing phase III clinical trial registered as NCT03504423 is aimed at assessing the safety and effectiveness of CPI613 in combination with modified mFFX for managing patients with metastatic pancreatic cancer.17 In another phase I clinical trial (NCT01832857), the study focused on the inhibitory effects of CPI613 on the TCA cycle, as well as evaluated the pharmacokinetics and safety of CPI613 in patients with hematological malignancies that had relapsed or were refractory.14 These clinical trials indicated the great potential of CPI613 for clinical applications in melanoma immunotherapy.
There are certain limitations to consider in this study. Specifically, the enzymatic activity of PDH/OGDH in the TCA cycle is dependent on the acylation of lipids mediated by LA.47–49 In this study, we have elucidated that CPI613, an analog of LA, impairs the TCA cycle by inhibiting the activity of PDH/OGDH, however, we did not elucidate how CPI613 affects melanoma immunotherapy by inhibiting the lipid acylation of PDH/OGDH, and that is an interesting point for future study. In addition, although it has been well documented that the TCA cycle can affect AMPK-CREB signaling by regulating the AMP/ATP ratio,27–30 the specific molecular mechanism by which the TCA cycle regulates AMP/ATP remains to be further elucidated. Finally, further study is needed to investigate the effects of inhibiting the TCA cycle on metabolites and their potential impact on the immune microenvironment.
Conclusions
Taken together, our finding revealed the novel mechanism of TCA cycle effect on glycolysis as well as the efficacy of anti-PD-1 immunotherapy, inhibition of PDHA1 and OGDH or application of pharmacological inhibitor (CPI613) upregulate PD-L1 expression through AMPK-CREB-ATF3 axis, sensitizing anti-PD-1 immunotherapy (figure 6G), which provided a promising strategy to improve anti-PD-1 clinical efficacy.
Footnotes
XC and CP contributed equally.
Contributors: NL, MY and JW carried out in vitro and in vivo animal experiments. NL and JC analyzed the sequencing data and performed statistical analysis. NL and QT obtained and/or analyzed human data. XC and CP supervised the study. NL and CP conceptualized the study and wrote the manuscript. CP is responsible for the overall content of the paper as a guarantor. All authors read and approved the final manuscript.
Funding: This work was supported by National Natural Science, Grant No. 82073458, 82203024, 8213000715, 82221002, 82073018, and by the science and technology innovation Program of Hunan Province (2021RC4013), the Program of Introducing Talents of Discipline to Universities (111 Project, No. B20017).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Consent obtained from parent(s)/guardian(s).
Ethics approval
This study involves human participants and was approved by Ethics Committee of Xiangya Hospital (Central South University, China); Approval ID number: 201803363. Participants gave informed consent to participate in the study before taking part.
References
- 1. Sainero-Alcolado L, Liaño-Pons J, Ruiz-Pérez MV, et al. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ 2022;29:1304–17. 10.1038/s41418-022-01022-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016;2:e1600200. 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eniafe J, Jiang S. The functional roles of TCA cycle metabolites in cancer. Oncogene 2021;40:3351–63. 10.1038/s41388-020-01639-8 [DOI] [PubMed] [Google Scholar]
- 4. Bellio C, DiGloria C, Spriggs DR, et al. The metabolic inhibitor CPI-613 negates treatment enrichment of ovarian cancer stem cells. Cancers (Basel) 2019;11:1678. 10.3390/cancers11111678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avagliano A, Fiume G, Pelagalli A, et al. Metabolic plasticity of melanoma cells and their crosstalk with tumor microenvironment. Front Oncol 2020;10:722. 10.3389/fonc.2020.00722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruocco MR, Avagliano A, Granato G, et al. Metabolic flexibility in melanoma: a potential therapeutic target. Semin Cancer Biol 2019;59:187–207. 10.1016/j.semcancer.2019.07.016 [DOI] [PubMed] [Google Scholar]
- 7. Ratnikov BI, Scott DA, Osterman AL, et al. Metabolic rewiring in melanoma. Oncogene 2017;36:147–57. 10.1038/onc.2016.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peppicelli S, Toti A, Giannoni E, et al. Metformin is also effective on lactic acidosis-exposed melanoma cells switched to oxidative phosphorylation. Cell Cycle 2016;15:1908–18. 10.1080/15384101.2016.1191706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang C, Radi RH, Arbiser JL. Mitochondrial metabolism in melanoma. Cells 2021;10:3197. 10.3390/cells10113197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu N, Wang KS, Qi M, et al. Vitexin compound 1, a novel extraction from a Chinese herb, suppresses melanoma cell growth through DNA damage by increasing ROS levels. J Exp Clin Cancer Res 2018;37:269. 10.1186/s13046-018-0897-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu N, Zhang J, Yin M, et al. Inhibition of xCT suppresses the efficacy of anti-PD-1/L1 melanoma treatment through exosomal PD-L1-induced macrophage M2 polarization. Mol Ther 2021;29:2321–34. 10.1016/j.ymthe.2021.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lines JL, Sempere LF, Broughton T, et al. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer Immunotherapy. Cancer Immunol Res 2014;2:510–7. 10.1158/2326-6066.CIR-14-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stuart SD, Schauble A, Gupta S, et al. A strategically designed small molecule attacks alpha-Ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metab 2014;2:4. 10.1186/2049-3002-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pardee TS, Lee K, Luddy J, et al. A phase I study of the first-in-class antimitochondrial metabolism agent, CPI-613, in patients with advanced hematologic malignancies. Clin Cancer Res 2014;20:5255–64. 10.1158/1078-0432.CCR-14-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zachar Z, Marecek J, Maturo C, et al. Non-redox-active Lipoate Derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J Mol Med (Berl) 2011;89:1137–48. 10.1007/s00109-011-0785-8 [DOI] [PubMed] [Google Scholar]
- 16. Egawa Y, Saigo C, Kito Y, et al. Therapeutic potential of CPI-613 for targeting tumorous mitochondrial energy metabolism and inhibiting Autophagy in clear cell sarcoma. PLoS One 2018;13:e0198940. 10.1371/journal.pone.0198940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Philip PA, Buyse ME, Alistar AT, et al. A phase III open-label trial to evaluate efficacy and safety of CPI-613 plus modified FOLFIRINOX (mFFX) versus FOLFIRINOX (FFX) in patients with metastatic adenocarcinoma of the pancreas. Future Oncol 2019;15:3189–96. 10.2217/fon-2019-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X, Lin J, Kanekura T, et al. A small interfering Cd147-targeting RNA inhibited the proliferation, invasiveness, and metastatic activity of malignant Melanoma. Cancer Res 2006;66:11323–30. 10.1158/0008-5472.CAN-06-1536 [DOI] [PubMed] [Google Scholar]
- 19. Liu N, Zhang J, Yan M, et al. Supplementation with alpha-Ketoglutarate improved the efficacy of anti-Pd1 melanoma treatment through epigenetic modulation of PD-L1. Cell Death Dis 2023;14:170. 10.1038/s41419-023-05692-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li C, Tang Z, Zhang W, et al. Gepia2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res 2021;49:W242–6. 10.1093/nar/gkab418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miao Y-R, Zhang Q, Lei Q, et al. Immucellai: A unique method for comprehensive T-cell Subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh) 2020;7:1902880. 10.1002/advs.201902880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia-Diaz A, Shin DS, Moreno BH, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep 2017;19:1189–201. 10.1016/j.celrep.2017.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Molina JR, Sun Y, Protopopova M, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med 2018;24:1036–46. 10.1038/s41591-018-0052-4 [DOI] [PubMed] [Google Scholar]
- 24. Chen D-P, Ning W-R, Jiang Z-Z, et al. Glycolytic activation of peritumoral monocytes fosters immune privilege via the Pfkfb3-PD-L1 axis in human hepatocellular carcinoma. J Hepatol 2019;71:333–43. 10.1016/j.jhep.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 25. Xia Q, Jia J, Hu C, et al. Tumor-associated Macrophages promote PD-L1 expression in tumor cells by regulating Pkm2 nuclear translocation in pancreatic ductal adenocarcinoma. Oncogene 2022;41:865–77. 10.1038/s41388-021-02133-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fan F, Jin S, Amundson SA, et al. Atf3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of Atf3 protein suppresses cells growth. Oncogene 2002;21:7488–96. 10.1038/sj.onc.1205896 [DOI] [PubMed] [Google Scholar]
- 27. Hardie DG. AMP-activated protein kinase: maintaining energy homeostasis at the cellular and whole-body levels. Annu Rev Nutr 2014;34:31–55. 10.1146/annurev-nutr-071812-161148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adachi Y, De Sousa-Coelho AL, Harata I, et al. L-alanine activates hepatic AMP-activated protein kinase and modulates systemic glucose metabolism. Mol Metab 2018;17:61–70. 10.1016/j.molmet.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Merlen G, Gentric G, Celton-Morizur S, et al. AMPKα1 controls hepatocyte proliferation independently of energy balance by regulating cyclin A2 expression. J Hepatol 2014;60:152–9. 10.1016/j.jhep.2013.08.025 [DOI] [PubMed] [Google Scholar]
- 30. Salminen A, Kaarniranta K, Kauppinen A. Age-related changes in AMPK activation: role for AMPK phosphatases and inhibitory phosphorylation by upstream signaling pathways. Ageing Res Rev 2016;28:15–26. 10.1016/j.arr.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 31. Liu H, Kuang X, Zhang Y, et al. Adora1 inhibition promotes tumor immune evasion by regulating the Atf3-PD-L1 axis. Cancer Cell 2020;37:324–39. 10.1016/j.ccell.2020.02.006 [DOI] [PubMed] [Google Scholar]
- 32. Tan AS, Baty JW, Dong L-F, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 2015;21:81–94. 10.1016/j.cmet.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 33. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016;23:27–47. 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corbet C, Feron O. Cancer cell metabolism and mitochondria: nutrient plasticity for TCA cycle fueling. Biochim Biophys Acta Rev Cancer 2017;1868:7–15. 10.1016/j.bbcan.2017.01.002 [DOI] [PubMed] [Google Scholar]
- 35. Gill JG, Leef SN, Ramesh V, et al. A short Isoform of Spermatogenic enzyme GAPDHS functions as a metabolic switch and limits metastasis in melanoma. Cancer Res 2022;82:1251–66. 10.1158/0008-5472.CAN-21-2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davidson SM, Papagiannakopoulos T, Olenchock BA, et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metabolism 2016;23:517–28. 10.1016/j.cmet.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan J, Shan C, Kang H-B, et al. Tyr Phosphorylation of Pdp1 Toggles recruitment between Acat1 and Sirt3 to regulate the pyruvate dehydrogenase complex. Molecular Cell 2014;53:534–48. 10.1016/j.molcel.2013.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hao Y, Samuels Y, Li Q, et al. Oncogenic Pik3Ca mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun 2016;7:11971. 10.1038/ncomms11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gopal YNV, Rizos H, Chen G, et al. Inhibition of Mtorc1/2 overcomes resistance to MAPK pathway inhibitors mediated by Pgc1Alpha and oxidative phosphorylation in melanoma. Cancer Res 2014;74:7037–47. 10.1158/0008-5472.CAN-14-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang G, Frederick DT, Wu L, et al. Targeting mitochondrial Biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest 2016;126:1834–56. 10.1172/JCI82661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vazquez F, Lim J-H, Chim H, et al. Pgc1Α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 2013;23:287–301. 10.1016/j.ccr.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xia X, Zhou W, Guo C, et al. Glutaminolysis mediated by Malt1 protease activity facilitates PD-L1 expression on ABC-DLBCL cells and contributes to their immune evasion. Front Oncol 2018;8:632. 10.3389/fonc.2018.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mu L, Long Y, Yang C, et al. The Idh1 mutation-induced oncometabolite, 2-Hydroxyglutarate, may affect DNA methylation and expression of PD-L1 in gliomas. Front Mol Neurosci 2018;11:82. 10.3389/fnmol.2018.00082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lv H, Lv G, Chen C, et al. NAD+ metabolism maintains inducible PD-L1 expression to drive tumor immune evasion. Cell Metab 2021;33:110–27. 10.1016/j.cmet.2020.10.021 [DOI] [PubMed] [Google Scholar]
- 45. Cheng J, Yan J, Liu Y, et al. Cancer-cell-derived fumarate suppresses the anti-tumor capacity of Cd8+ T cells in the tumor microenvironment. Cell Metab 2023;35:961–78. 10.1016/j.cmet.2023.04.017 [DOI] [PubMed] [Google Scholar]
- 46. Alistar A, Morris BB, Desnoyer R, et al. Safety and tolerability of the first-in-class agent CPI-613 in combination with modified FOLFIRINOX in patients with metastatic pancreatic cancer: a single-centre, open-label, dose-escalation, phase 1 trial. Lancet Oncol 2017;18:770–8. 10.1016/S1470-2045(17)30314-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Paredes F, Sheldon K, Lassègue B, et al. Poldip2 is an oxygen-sensitive protein that controls PDH and alphaKGDH lipoylation and activation to support metabolic adaptation in hypoxia and cancer. Proc Natl Acad Sci U S A 2018;115:1789–94. 10.1073/pnas.1720693115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burr SP, Costa ASH, Grice GL, et al. Mitochondrial protein Lipoylation and the 2-Oxoglutarate dehydrogenase complex controls Hif1Α stability in aerobic conditions. Cell Metab 2016;24:740–52. 10.1016/j.cmet.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375:1254–61. 10.1126/science.abf0529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007146supp001.pdf (1.4MB, pdf)
Data Availability Statement
Data are available upon reasonable request.