Abstract
Background
Deletions or loss-of-function mutations in phosphatase and tensin homolog (PTEN) are common in glioblastoma (GBM) and have been associated with defective DNA damage repair. Here we investigated whether PTEN deficiency presents a vulnerability to a simultaneous induction of DNA damage and suppression of repair mechanisms by combining topoisomerase I (TOP1) and PARP inhibitors.
Methods
Patient-derived GBM cells and isogenic PTEN-null and PTEN-WT glioma cells were treated with LMP400 (Indotecan), a novel non-camptothecin TOP1 inhibitor alone and in combination with a PARP inhibitor, Olaparib or Niraparib. RNAseq analysis was performed to identify treatment-induced dysregulated pathways.
Results
We found that GBM cells lacking PTEN expression are highly sensitive to LMP400; however, rescue of the PTEN expression reduces sensitivity to the treatment. Combining LMP400 with Niraparib leads to synergistic cytotoxicity by inducing G2/M arrest, DNA damage, suppression of homologous recombination-related proteins, and activation of caspase 3/7 activity significantly more in PTEN-null cells compared to PTEN-WT cells. LMP400 and Niraparib are not affected by ABCB1 and ABCG2, the major ATP-Binding Cassette (ABC) drug efflux transporters expressed at the blood-brain barrier (BBB), thus suggesting BBB penetration which is a prerequisite for potential brain tumor treatment. Animal studies confirmed both an anti-glioma effect and sufficient BBB penetration to prolong survival of mice treated with the drug combination.
Conclusions
Our findings provide a proof of concept for the combined treatment with LMP400 and Niraparib in a subset of GBM patients with PTEN deficiency.
Key Points.
1. PTEN deficiency sensitizes glioblastoma cells to LMP400, a novel TOP1 inhibitor.
2. Combining LMP400 with PARP inhibitor Niraparib leads to synergistic cytotoxicity.
3. LMP400 and Niraparib are not affected by ABC transporters, unlike their current analogs.
Importance of the Study.
Perturbations in PTEN signaling have been detected in nearly half of glioblastoma patients and are typically associated with a worse prognosis. This is the first study to report that PTEN deficiency sensitizes glioblastoma cells to simultaneous inhibition of TOP1 and PARP leading to cell growth arrest and activation of cell death. These findings support the clinical investigation of a new targeted strategy that should have a broad impact on precision medicine in a subset of glioblastoma patients with PTEN deficiency.
Treatment of GBM has proven to be challenging due to tumor heterogeneity and the blood-brain barrier (BBB) that impedes exposure to therapeutic agents.1,2 Integrated strategies provided insights into genomic subtyping of the disease with the hope of identifying therapeutic targets for a precision medicine approach for GBM.3,4 Hence, it is crucial to investigate candidate drugs, which penetrate the BBB and develop a precision medicine approach tailored to target existing tumor-specific genetic alterations and provide selective sensitization to the treatment to improve outcomes for patients.
DNA topoisomerase I (TOP1) is an essential enzyme that relaxes DNA supercoiling generated by transcription and replication. It nicks and re-ligates DNA by forming transient DNA single-strand breaks referred to as “cleavage complexes” (TOP1ccs).5 Camptothecins such as topotecan and irinotecan (also known as CPT-11, a prodrug for its active metabolite SN-38) act by stabilizing TOP1ccs leading to lethal DNA double-strand breaks and replication stress.5 Despite potent anti-cancer activity, use of camptothecins is prevented due to dose-limiting toxicities, such as severe diarrhea, as well as chemical instability, and drug resistance.6–10 Drug screening by the National Cancer Institute (NCI) Developmental Therapeutics Program discovered the indenoisoquinolines, new non-camptothecin TOP1 inhibitors that overcome the limitations of camptothecins.7 LMP400 (Indotecan) exhibited potent activity, and compared to camptothecins is chemically stable, and relatively well-tolerated with much less diarrhea in patients with advanced solid tumors and lymphomas, and in patient-derived xenograft mouse models of triple-negative breast cancer.11–13
Poly(ADP-ribose) polymerase (PARP) enzymes function as DNA damage sensors that recognize DNA damage and form mono(ADP-ribose) or poly(ADP-ribose) (PAR) chains to facilitate DNA damage repair.14,15 Moreover, PARP can facilitate pro-survival responses to TOP1 inhibition, including reversal of TOP1ccs and DNA damage repair.16,17
PTEN deficiency is a common genetic alteration in gliomagenesis and is typically associated with a worse prognosis.4 PTEN deficiency can be caused by several mechanisms, including but not limited to loss-of-function mutations or deletions of PTEN that were detected in about 40% of GBM and represent a significant fraction of GBM patients.4 In addition, PTEN has been found to play a critical role in DNA damage repair and its loss compromises homologous recombination (HR) repair, thereby sensitizing tumor cells to ionizing radiation, DNA alkylating agents, and PARP inhibitors.18–21
In this study, we hypothesized that PTEN deficiency will present a vulnerability to combined inhibition of TOP1 and PARP to enhance the cytotoxic effects in GBM. Overall, our preclinical results and screening for potential mechanisms of drug resistance indicate that this novel therapy may be a promising approach in the treatment of GBM with PTEN deficiency.
Materials and Methods
Drugs and Antibodies
LMP400 was kindly provided by Dr. Yves Pommier. Olaparib (#S1060) was purchased from Selleckchem (Houston, TX). Niraparib was received from the Developmental Therapeutics Program at NCI (NSC 754355). The antibodies used in this study are described in Supplementary Materials and Methods document.
Cell Culture
The cell lines’ sources and culture conditions are described in Supplementary Materials and Methods. All cell lines were cultured at 37°C and 5% CO2. The cell lines were routinely checked and tested negative for mycoplasma contamination using MycoAlert Mycoplasma kit (#LT07-318, Lonza).
Generation of PTEN-KO and PTEN-WT Cells
Constructs for PTEN knockout (KO) or rescue were obtained from Genome Modification Core Laboratory, Leidos Biomedical Inc/ Frederick National Laboratory for Cancer Research (Frederick, MD), and described in Supplementary Materials and Methods.
Cell Viability Assay
Cells were seeded in 12-well plates at 3–4 × 104 cells/well in duplicates. The next day, cells were treated with increasing concentrations of LMP400, Olaparib or Niraparib, and drug combination for 72 hours before direct cell counting using a Beckman Coulter Vi-CELL TM XR cell viability analyzer or Celigo Image Cytometer, as described in Supplementary Materials and Methods.
Synergism Analysis
CompuSyn software (ComboSyn, Inc., Paramus, NJ) was used to calculate the combination index (CI) values from the % drug effect data, where CI = 1 indicates an additive effect, CI > 1 indicates antagonism, and CI < 1 indicates synergism.22
Colony Formation Assay
Cells were seeded at 200 or 400 cells/well in 6-well plates and treated with drugs for 72 hours followed by culturing in fresh drug-free media for 6 days before fixation. Colonies were fixed with 4% PFA in PBS, washed, and stained with 0.5% crystal violet before counting using ImageJ.
Cell Cycle Analysis
At least 1 × 106 cells per sample were fixed in 70% ethanol at −20°C. Prior to staining, the cells were washed with cold PBS and incubated with 500 μL FxCycle PI/RNase Staining Solution (#F10797, Thermo Fisher Scientific) for 30 minutes at room temperature in the dark. The data were acquired on a BD LSR Fortessa flow cytometer and analyzed using FlowJo software.
Apoptosis Assays
Apoptosis was analyzed by flow cytometry using the FITC Annexin V Apoptosis Detection Kit with PI (#640914, Biolegend). At 72 hours post-treatment with LMP400, Niraparib, or both, cells were initially washed with PBS and then twice with Cell Staining Buffer (Biolegend), resuspended in 100 μL Annexin V binding buffer, 5 μL of FITC Annexin V, and 10 μL of PI solution. After 15-minute incubation at room temperature in the dark, 400 μL of Annexin V Binding Buffer was added, and the samples were analyzed using BD LSR Fortessa flow cytometer and FlowJo software. Unstained and single-stained samples were used to calculate compensation.
The caspase 3/7 activity assay was performed using the Caspase-Glo 3/7 assay system (Promega, Madison, WI), per manufacturer’s instructions.
Western Blots
Protein concentration was determined using a DC (detergent compatible) protein assay (Bio-Rad, Hercules, CA) and western blot was performed according to standard protocol.
Immunofluorescent Staining
Treated cells were washed in cold PBS and fixed in 4% PFA for 20 min followed by permeabilization with 0.5% Triton X-100. After probing with antibodies, coverslips were mounted using Fluoroshield with DAPI (Sigma-Aldrich, St. Louis, MO). Fluorescent γ-H2AX foci were captured by confocal microscopy. For quantification, cells containing more than 5 foci were considered positive.
Single Cell Electrophoresis Assay (Comet Assay)
The comet assay (neutral) was performed using a comet assay kit (Abcam, Cambridge, UK), per manufacturer’s instructions.
RNA Sequencing and Gene Set Enrichment Analysis Analysis
Cells were plated into 100 mm dishes overnight and then treated with 10 nM LMP400, 1 μM Olaparib, or both for 48 hours. Cells were harvested and RNA was extracted using PureLink RNA Mini Kit (#12183018A, Thermo Fisher Scientific) following manufacturer’s instructions, including on-column DNase treatment to remove contaminating DNA (#12185010, PureLink DNase Set, Invitrogen). More information on RNAseq and gene set enrichment analysis can be found in Supplementary Materials and Methods.
Orthotopic Mouse Model
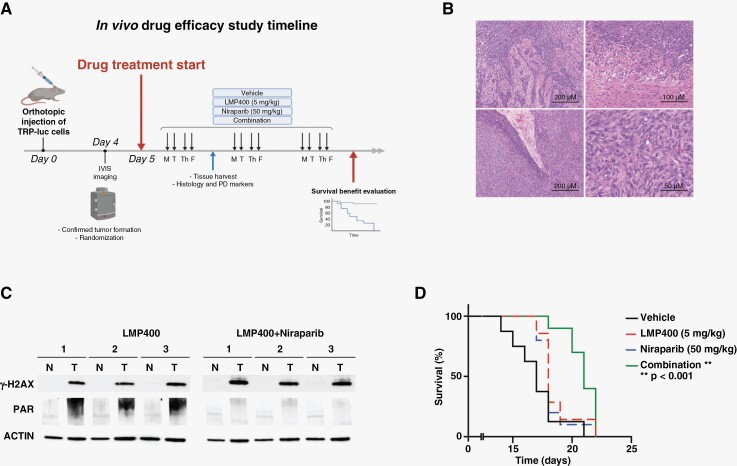
Eight-week-old female C57BL/6 mice were injected intracranially (2 mm anterior and 1 mm lateral to bregma; 3.5 mm deep from the dura) using stereotactic apparatus with TRP-luc cells (5000 cells/2μL) resuspended in serum-free DMEM medium with 1% methylcellulose. Bioluminescent imaging (BLI) was performed on day 4 after intracranial injection of TRP-luc cells on IVIS Lumina Series III using Living Image software (PerkinElmer, Waltham, MA). Based on BLI confirmation of tumor formation, the mice were included in the study and randomized to experimental groups. LMP400 was dissolved as described previously.23 Niraparib was dissolved according to the manufacturer’s instructions. The drugs were administered i.p. at 5 mg/kg for LMP400 and 50 mg/kg for Niraparib 4 days a week, repeated until the endpoint. All animal experiments were approved by the NCI Animal Care and Use Committee and in accordance with federal regulatory requirements and standards. All components of the intramural NIH ACU program are accredited by AAALAC International.
Histology and Pharmacodynamic (PD) Biomarkers Evaluation
At 6 days of treatment and 4 hours after last treatment dose, 4 mice per group were sacrificed to harvest whole brains containing non-tumor and tumor regions, which were fixed in 10% formalin, embedded in paraffin, sectioned at 5 μm, and mounted on slides. Hematoxylin and eosin (H&E) staining was performed per standard protocol. Pathology evaluation was conducted by 2 independent pathologists. The protein lysates extracted from brain tissues were examined for the expression of PD markers by Western blot.
The ATP-Binding Cassette (ABC) Transporters and Cell Survival Assays
Stably transfected HEK293 cells overexpressing ATP-binding cassette transporters were generated and verified as described previously.24 Briefly, cells were plated at a density of 5000 cells/well in opaque white 96-well plates and allowed to attach overnight. The next day, drugs were added, and plates were incubated for 3 days after which CellTiterGlo (Promega) was used according to the manufacturer’s instructions.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (version 9.3.1, San Diego, CA). The data is expressed as mean ±SEM from at least 2 independent experiments. A 2-tailed t-test was used to evaluate a difference between experimental groups. P < .05 was considered statistically significant. The Kaplan–Meier analysis and log-rank test were employed to analyze survival benefits in the in vivo study.
Data Availability
The RNA sequencing data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE225210.
Results
PTEN Deficiency Sensitizes GBM Cells to LMP400
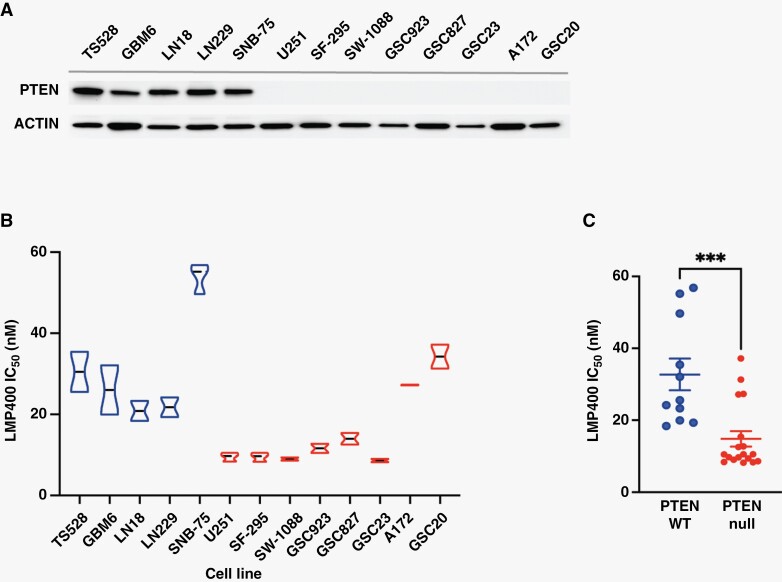
To investigate whether sensitivity to LMP400 is linked to PTEN expression, we first screened a panel of patient-derived GBM cells and glioma stem-like cells (GSCs) (Figure 1A). Western blot analysis revealed that 8 out of 11 cell lines were deficient for PTEN expression, while 5 cell lines expressed WT PTEN25–27 (Figure 1A). In cell viability assays, LMP400 IC50 in most PTEN-deficient cells ranged from 8 nM to 15 nM compared to PTEN-expressing cells which showed a higher IC50 range from 18 nM to 57 nM (Figure 1B). Overall, LMP400 IC50 was significantly lower in PTEN-deficient cells (P < .001) (Figure 1C and Supplementary Figure S1A), suggesting that deficiency in PTEN protein expression may correlate with increased sensitivity to LMP400.
Figure 1.
PTEN-deficient glioblastoma cells are highly sensitive to LMP400. (A) PTEN protein expression in a panel of patient-derived GBM cells and GSC cells by Western blot. (B) The LMP400 IC50 in each cell line was examined at 72 hours post-treatment. (C) Comparison of LMP400 IC50 values between PTEN-expressing and PTEN-deficient cells.
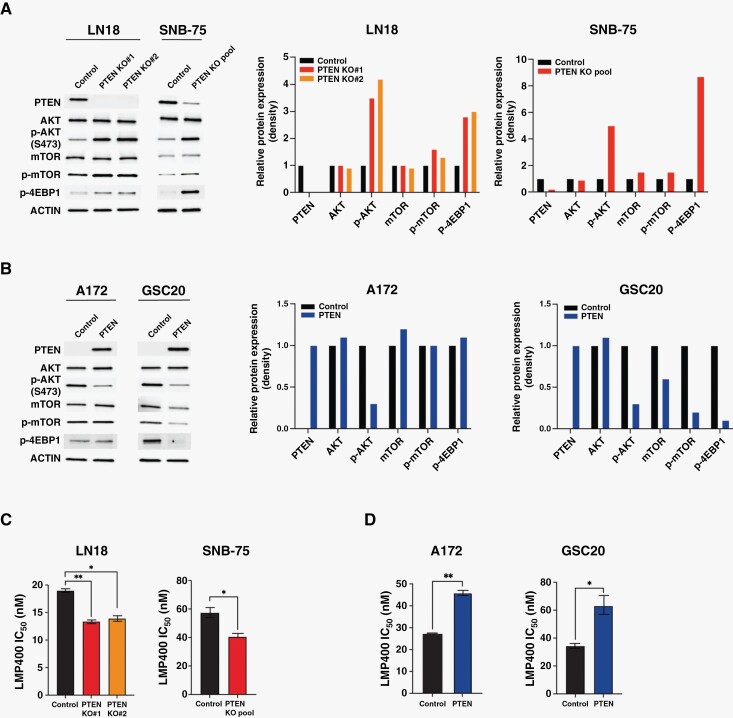
To further test whether sensitivity to LMP400 is dependent on PTEN expression, PTEN was knocked out in LN18 and SNB-75 cells. PTEN targeting and subsequent loss of protein expression were confirmed by DNA sequencing (Supplementary Figure S1B and C) and Western blot, respectively (Figure 2A). An increased expression of the downstream targets p-AKT, p-mTOR, and, p-4EBP1 was demonstrated in LN18 PTEN-KO single clones as well as in the SNB-75 PTEN-KO pool, consistent with loss-of-function effect of PTEN KO (Figure 2A). In cell viability assays, PTEN-KO cells exhibited a significantly lower IC50 to LMP400 compared to PTEN-expressing control cells (P < .05 in all) (Figure 2C), suggesting that PTEN deficiency sensitizes cells to LMP400 treatment.
Figure 2.
PTEN-KO sensitizes GBM cells to LMP400, while rescuing PTEN expression confers drug resistance. (A) Protein expression of PI3K/AKT/mTOR pathway-related genes in PTEN-KO cells. Protein expression density was measured by ImageJ and normalized to ACTIN and compared to the Control. (B) Protein expression of PI3K/AKT/mTOR pathway signals in A172 and GSC20 cells following transduction with lentivirus to re-express PTEN. (C) IC50 of LMP400 measured in LN18 and SNB-75 cells after PTEN KO. (D) IC50 of LMP400 measured in A172 and GSC20 cells after PTEN rescue.
Next, we examined whether PTEN rescue in the cells lacking PTEN expression can reverse the enhanced sensitivity to LMP400. The PTEN expression and downstream signals were demonstrated by Western blot (Figure 2B). Intriguingly, although the LMP400 IC50 in A172 and GSC20 was comparable to that of some PTEN-expressing cells (Figure 1B), the sensitivity to LMP400 was markedly reduced in both cells after PTEN rescue, leading to a higher IC50 (P < .05 in both) (Figure 2D) and supporting PTEN involvement in determining response to LMP400.
Additionally, to test whether treatment response could be reversed after PTEN-KO/rescue genetic manipulation, we used a pharmacologic approach as a method to functionally rescue PTEN. Addition of GDC0084, a dual PI3K/mTOR inhibitor, interfered with sensitivity to LMP400 after PTEN KO/rescue in LN18 single clones. Additionally, there was a decrease in the percentage of G2/M phase population when treated with LMP400 plus GDC0084 compared to LMP400 alone (P = .098 in PTEN KO#1 and P = .028 in PTEN KO#2) (Supplementary Figure S2). Of note, GDC0084 was used at a 70 nM dose that is sufficient to inhibit PI3K/mTOR, but insufficient to induce drug-related cytotoxicity.28,29 Collectively, these findings suggest that PTEN deficiency renders an increased sensitivity to LMP400.
LMP400 Synergizes With Olaparib and Niraparib to Induce Cytotoxicity in GBM Cells
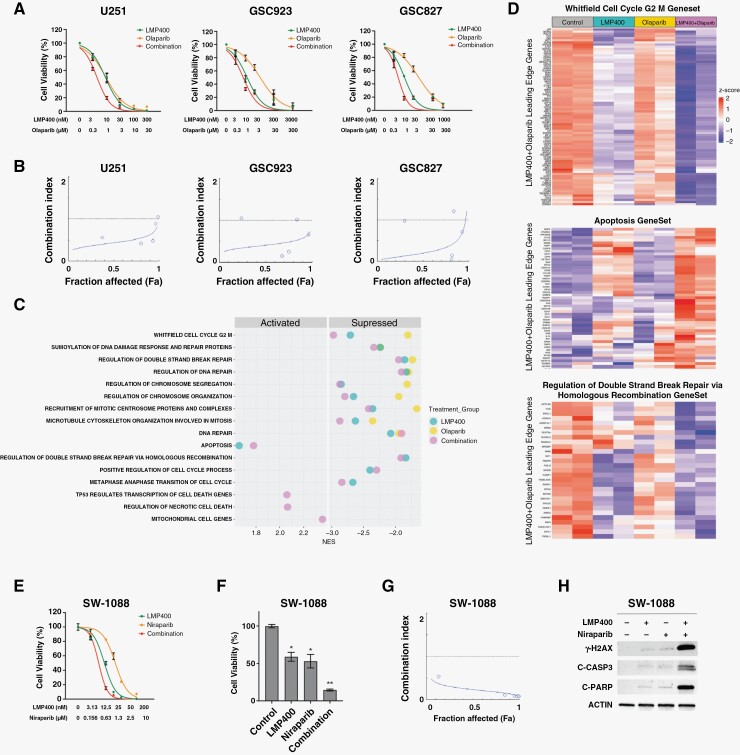
PARP is known to be involved in repair of TOP1-mediated DNA damage,30 providing a rationale for combining TOP1 and PARP inhibitors to enhance antitumor effects. As Olaparib, a PARP inhibitor, has been extensively tested in the clinical trials of brain tumors, we evaluated the effect of combining LMP400 with Olaparib in GBM cells in vitro. The combination of LMP400 and Olaparib led to a more profound suppression of cell growth in U251, GSC923, and GSC827 cells compared to either single agent (Figure 3A). To quantitatively analyze the effect of this drug combination, we determined the CI values from the % drug effect data. CI values were <1 in most of the concentrations tested in all 3 cell lines (Figure 3B and Supplementary Figure S3). Importantly, a CI value of <0.3 indicating strong synergism31 was observed in GSC923 and GSC827 cell lines at LMP400 concentrations ranging from 10 nM to 30 nM and Olaparib concentrations ranging from 1 μM to 3 μM (Supplementary Figure S3). Thus, the dose combination of 10 nM LMP400 and 1 μM Olaparib was used in the following experiments. Overall, these findings suggest that LMP400 and Olaparib lead to synergistic inhibition of cell growth in GBM cells.
Figure 3.
LMP400 synergizes with Olaparib and Niraparib to induce cytotoxic effects in GBM. (A) The dose–response curves for LMP400, Olaparib, or both in U251, GSC923, and GSC827 cells treated with increasing drug concentrations for 72 hours prior to cell viability detection. (B) Synergy plots for U251, GSC923, and GSC827 cells generated by CompuSyn software. (C) Gene set enrichment analysis was performed with data from GSC827 cells treated with 10 nM LMP400 or 1 μM Olaparib or both drugs combination for 48 hours. The summary plot demonstrates representative pathways altered upon LMP400/Olaparib combined treatment. (D) Representative heatmaps showing altered expression of genes related to cell cycle G2/M, apoptosis, and regulation of double-strand break repair via homologous recombination pathways. Leading edge genes of the combination group for each gene set are exhibited. (E) Dose–response curves for LMP400, Niraparib or both at 72 hours post-treatment in SW-1088 cells. (F) Cell viability was measured at 72 hours after treatment with 10 nM LMP400 or 1 μM Niraparib or both. (G) Synergism plot for LMP400/Niraparib combination assessed by CompuSyn software. (H) Western blot demonstrating expression of DNA damage and apoptosis markers in SW-1088 cells treated with 10 nM LMP400 or 1 μM Niraparib or both for 72 hours.
To investigate signaling pathways affected by LMP400 and Olaparib as a single agent or combined treatment, gene set enrichment analysis (GSEA) was performed. From this analysis, we identified that multiple pathways related to cell cycle regulation were significantly suppressed in the LMP400-treated group with a greater enrichment of suppression following its combination with Olaparib (Figure 3C-D and Supplementary Figure S4). Moreover, pathways linked to DNA repair such as regulation of double-strand breaks repair via HR and regulation of DNA repair were also considerably suppressed in the single agents and combination groups. Importantly, activation of cell death pathways such as apoptosis, TP53-regulated transcription of cell death genes, and regulation of necrotic cell death was observed in the combination group. Representative heatmaps and enrichment plots of the dysregulated pathways are included in Figure 3D and Supplementary Figure S4. Collectively, these results suggest that synergistic cytotoxicity induced by the LMP400/Olaparib combination could be accounted for enhanced suppression of cell cycle progression and attenuated DNA damage repair, ultimately leading to cell death.
Recent studies reported several advantages of Niraparib over Olaparib, including higher potency in trapping PARP on DNA and reducing cell viability in vitro and in vivo.32–35 In addition, Niraparib crosses the BBB, showing good delivery and sustainability in the brain.32,33 Therefore, using more cell lines from our GBM cell panel, we randomly selected and tested SW-1088 cells to investigate whether LMP400 synergizes with Niraparib. Both single agents inhibited cell viability (LMP400 IC50 8.5–10 nM and Niraparib IC50 1.3–1.7 μM) (Figure 3E). However, combined treatment using each drug IC50 (LMP400 10 nM and Niraparib 1 μM) led to a greater suppression of cell viability (LMP400: 53%–65%, P = .023, Niraparib: 44%–62%, P = .037, Combination: 13%–16%, P < .001) (Figure 3F). Synergism analysis revealed that CI values at all concentrations were <1, most of which ranged from 0.08 to 0.11 (Figure 3G and Supplementary Figure S3), suggesting a strong synergistic effect. In addition, combined treatment led to strong expression of γ-H2AX, indicating DNA damage (Figure 3H). Further exploring whether this treatment can induce cell death, we found that LMP400 or Niraparib as single agents induced a modest cleavage of caspase 3 and PARP, while their combination triggered a substantial activation of caspase 3 and propagation of an apoptotic signal leading to cleavage of PARP (Figure 3H). These results suggest that LMP400/Niraparib combination can synergistically induce suppression of cell viability and activation of cell death pathways in GBM cells.
Synergistic LMP400/Niraparib Combination Leads to Growth Arrest Selectively in PTEN-Deficient GBM Cells
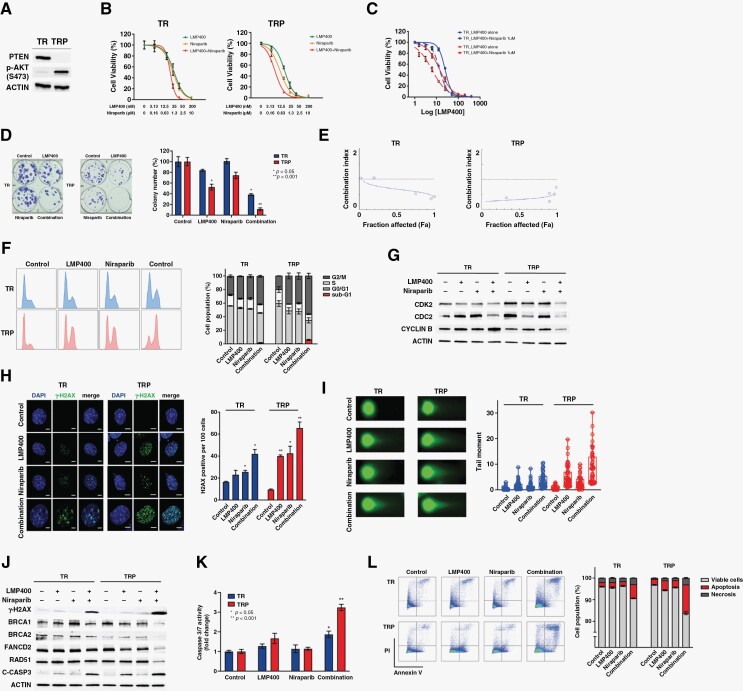
To investigate the role of PTEN in determining response to the LMP400/Niraparib combination, the isogenic cell lines with (TRP) and without (TR) Pten deletion derived from a genetically engineered murine model (GEM) of GBM were used.36 PTEN protein expression in TR and its absence in TRP cells were confirmed by Western blot (Figure 4A). The LMP400 IC50 in PTEN-WT TR cells was in 25.8–32.1 nM range compared to 12.4–15.1 nM in PTEN-null TRP cells. The Niraparib IC50 in TR cells was in 1–1.5 μM range compared to 0.3–0.5 μM in TRP cells. When both drugs were combined, the IC50 for LMP400 in TRP cells dropped to 3.8–5.6 nM compared to 17–20 nM in TR cells, and for Niraparib—to 0.2–0.3 μM in TRP versus 0.9–1 μM in TR cells (Figure 4B).
Figure 4.
Synergistic LMP400/Niraparib combination leads to cell growth arrest and apoptosis in PTEN-deficient cells through enhanced DNA damage and attenuated DNA repair. (A) Western blot demonstrating the PTEN expression in TR, but not TRP cells. Increased p-AKT (S473) was observed upon PTEN deletion in TRP cells. (B) The dose–response curves of TR and TRP cells treated with increasing concentrations of LMP400, Niraparib, or both for 72 hours. (C) The dose–response curves of TR and TRP cells treated with increasing concentrations of LMP400 with or without addition of 1 μM Niraparib. (D) Colony formation assay in TR and TRP cells treated with 15 nM LMP400 or 0.5 μM Niraparib or both for 72 hours before releasing to drug-free media. (E) Synergism plots in TR and TRP cells generated by Compusyn software. (F) Cell cycle analysis performed by flow cytometry in TR and TRP cells after treatment with 50 nM LMP400 or 2 μM Niraparib or both for 48 hours. (G) Western blot of cell cycle-related proteins in TR and TRP cells treated with 30 nM LMP400 or 1 μM Niraparib or both for 72 hours. (H) Immunocytochemical staining of γ-H2AX in TR and TRP cells treated with 50 nM LMP400 or 2 μM Niraparib or both for 48 hours. (I) Single-cell electrophoresis (comet) assay performed in TR and TRP cells treated with 30 nM LMP400 or 1 μM Niraparib or both for 72 hours. Tail moment was measured in at least 50 cells per each experimental group. (J) Western blot analysis of DNA damage and repair proteins in TR and TRP cells treated with 30 nM LMP400 or 1 μM Niraparib or both for 72 hours. (K) Caspase 3/7 activity assay in TR and TRP cells treated with 30 nM LMP400 or 1 μM Niraparib or both for 72 hours. The data is expressed as mean ±SEM normalized to the average value of the control group. (L) Flow cytometry analysis of TR and TRP cells by double staining with Annexin V and PI after treatment with 30 nM LMP400 or 1 μM Niraparib or both for 72 hours. Apoptosis fraction includes both early (Annexin V+, PI−) and late apoptosis (Annexin V+, PI+). Necrosis was measured as Annexin V−, PI+.
In addition, since IC50 for Niraparib in TR cells was close to 1 μM, we compared sensitivity to LMP400 with and without adding 1 μM Niraparib in both cell lines. When Niraparib was added, the LMP400 IC50 in TRP cells decreased to 5.4–6.9 nM compared to 13.3–14.5 nM in TR cells (Figure 4C). Overall, these results indicate that PTEN-null TRP cells are more sensitive to LMP400 alone and LMP400/Niraparib combination than PTEN-WT TR cells.
Next, to examine the long-term effect on cell survival, we performed colony formation assays. In TRP cells, LMP400 reduced the number of surviving colonies to approximately 50% compared to control (P = .007), while combined treatment dramatically decreased the number of viable colonies to <10% (P < .001) (Figure 4D). In contrast, a high and moderate number of surviving colonies was observed in TR cells treated with either single agent (81%–100%, P > .05) or both drugs (35%–41%, P < .01), respectively. These results suggest that combining LMP400 with Niraparib in PTEN-null cells, but not PTEN-WT cells, inhibits long-term cell survival as the colonies in this treatment group were not able to recover and re-populate after release into drug-free media.
Synergism analysis revealed that the CI values <1 in TR cells were detected at the highest concentrations of LMP400 and Niraparib, while the CI values at the lowest concentrations of both were more than 1, suggesting a moderate synergy only at the highest concentrations. In contrast, the CI values in TRP cells were <1 at all concentrations, indicating a consistent synergistic effect. Moreover, the smallest CI values (CI = 0.27–0.39) in TRP observed at the lowest concentrations of drugs suggested a stronger synergy at lower concentrations of each drug (Figure 4E and Supplementary Figure S3). Together, these data indicate that the LMP400/Niraparib combination synergistically inhibits both short-term and long-term growth of PTEN-deficient GBM cells.
LMP400/Niraparib Combination Targets Specifically PTEN-Deficient GBM Cells Through Enhanced DNA Damage and Impaired DNA Repair
To further explore the effect of LMP400 and Niraparib on the cell cycle when PTEN is expressed or deficient, TR and TRP cells were analyzed by flow cytometry. TR cells demonstrated a modest increase in the G2/M phase population after single agent treatment (P = .033 in LMP400 and P = .105 in Niraparib) and a moderate increase after combined treatment (P = .001) compared to control (Figure 4F). In TRP cells, single agents’ treatment led to a significant increase in proportion of G2/M population (P = .007 in LMP400 and P = .013 in Niraparib). Notably, the LMP400/Niraparib combination led to a marked G2/M arrest in TRP cells (P < .001). Additionally, there was a significant decrease in S phase population after drug combination compared to control (P = .025) in TRP only (Figure 4F). Indeed, Western blot analysis of cell cycle-related proteins showed decreased expression of G2/M phase-related proteins such as CDC2 and Cyclin B as well as CDK2 in TRP cells, and only a modest decrease of CDC2 in TR cells after combined treatment (Figure 4G). These results suggest that the LMP400/Niraparib combination affects cell cycle progression mainly by arresting at the G2/M phase specifically in PTEN-deficient GBM cells.
Since LMP400 and Niraparib are known to induce DNA damage, we checked the expression of γ-H2AX. LMP400 induced a significant number of γ-H2AX positive cells in TRP cells (P = .001) (Figure 4H). In addition, Niraparib induced γ-H2AX expression in both cells with a greater extent in TRP cells (P < .05 in both) Notably, LMP400/Niraparib combination led to a dramatic increase of γ-H2AX positive cells, especially in TRP cells (38–46 positive per 100 cells, P = .024 in TR vs. 60-71 positive, P = .009 in TRP) (Figure 4H).
Next, we measured DNA damage at a single cell level and found that single-agent treatment led to a moderate increase in the tail moment in both TR and TRP cells (P < .05 in all), while combined treatment triggered a much greater elevation selectively in TRP cells (Figure 4I), indicating a more profound and likely unresolved DNA damage after treatment with the LMP400/Niraparib combination.
Because we observed increased DNA damage and cell cycle arrest upon treatment, we checked the expression of DNA repair-related proteins. As expected, the expression of γ-H2AX was moderately increased in TRP cells after either LMP400 or Niraparib (Figure 4J). Combined treatment induced a markedly higher expression of γ-H2AX, which was obviously more intense in TRP than in TR cells. Interestingly, LMP400 alone was able to decrease the expression of Fanconi anemia group D2 protein (FANCD2), which was considerably more suppressed specifically in TRP, but not in TR cells treated with both drugs. Importantly, protein expression of the critical HR-related proteins, such as BRCA1 and BRCA2, was significantly diminished after combined treatment only in TRP cells. Consistently, the expression of RAD51 was also decreased specifically in TRP cells (Figure 4J), suggesting an impairment of HR repair process in PTEN-deficient cells upon combined treatment.
Finally, we investigated the drug's effect on apoptosis. Cleaved caspase 3 was detected in LMP400-treated TRP cells as well as combination-treated TR and TRP cells (Figure 4J). Additionally, caspase 3/7 activity was significantly elevated in both cells upon drug combination with a much greater increase in TRP cells (P < .001) (Figure 4K). Moreover, measurements of apoptosis by flow cytometry demonstrated that combined treatment significantly increased the percentage of cells undergoing apoptosis as well as necrosis in both cells with a higher percentage in TRP than TR cells (P < .001 in all) (Figure 4L). Overall, these data suggest that hypersensitivity of PTEN-deficient cells to the LMP400/Niraparib combination is due to compromised HR repair leading to cell growth arrest, accumulation of DNA damage, and activation of apoptosis.
LMP400/Niraparib Combination Prolongs Survival in a PTEN-Deficient Mouse Model of GBM
To evaluate the efficacy of the LMP400/Niraparib combination in vivo, a TRP mouse model was used. This syngeneic orthotopic mouse model of GBM has been developed to harbor perturbations crucial to human GBM, including PTEN deletion.36 As shown in Figure 5A, tumor formation was confirmed on day 4 after intracranial injection of TRP-luc cells into immunocompetent mice. After randomization based on the tumor BLI signal and body weight, the mice started receiving either single agent or combined treatment. Tumor histology and PD markers were evaluated and 7–10 mice per group were followed until the endpoint to assess treatment-related survival benefit.
Figure 5.
LMP400/Niraparib combination prolongs survival in a GBM mouse model with PTEN deficiency. (A) An in vivo study scheme showing a timeline for the evaluation of the LMP400 and Niraparib efficacy in a GBM mouse model. (B) H&E staining of the tumor tissues harvested from the TRP mice confirmed histopathological features of glioblastoma, such as infiltrative tumor edge (upper left, 10×, scale bar 200 μm and upper right, 20×, scale bar 100 μm) and pseudopalisading necrosis (lower left, 10×, scale bar 200 μm). High cellularity, nuclear atypia, pleomorphism, and numerous mitotic figures were also observed (lower right, 40×, scale bar 50 μm). (C) Western blot of PD markers in mouse brain tumor tissues and contralateral non-tumor counterparts. (D) Kaplan–Meier survival curves of the mice treated with vehicle or LMP400 (5 mg/kg via i.p.) or Niraparib (50 mg/kg via i.p.) or combination.
Histologically, we confirmed the formation of glioblastoma in all experimental groups. In H&E-stained tumor slides, high cellularity, hyperchromasia, marked nuclear atypia, and pleomorphism along with numerous mitotic figures were indicative of malignant features. In addition, the infiltrative tumor edge and pseudopalisading necrosis resembling human GBM were observed (Figure 5B).
Since γ-H2AX and PAR are known PD markers for LMP400 and Niraparib, respectively,12,37 we examined their expression in mouse brain tissues. LMP400 induced the expression of γ-H2AX in tumor tissues as compared to contralateral non-tumor counterpart, reflecting induction of DNA damage in tumors. Consistently, PAR was markedly upregulated in these samples upon LMP400-induced DNA damage. When Niraparib was added to LMP400, γ-H2AX remained high compared to the non-tumor tissues and appeared even more elevated compared to the LMP400 single agent treatment. Moreover, addition of Niraparib significantly suppressed PAR, indicating inhibition of PARylation (Figure 5C and Supplementary Figure S5). These results demonstrate that LMP400 and Niraparib could penetrate BBB to induce DNA damage and block PARP-mediated repair in brain tumor tissues in vivo.
Finally, upon confirming the formation of GBM in the mouse model used and BBB penetration of the drugs, we examined whether this combined treatment could impact survival of PTEN-deficient mice. Median survival in LMP400- or Niraparib-treated groups was equally 18 days (n = 7 and n = 10, respectively), which was not significantly different from the one of the vehicle-treated group (17 days, n = 8), demonstrating lack of survival benefit by single agent treatment (Figure 5D). However, there was a statistically significant prolonged survival in mice receiving combined treatment (21 days, n = 10, P < .001) (Figure 5D). Collectively, these data suggest that LMP400 and Niraparib can penetrate the BBB and lead to an anti-glioma effect in vivo, resulting in a survival benefit in PTEN-deficient GBM.
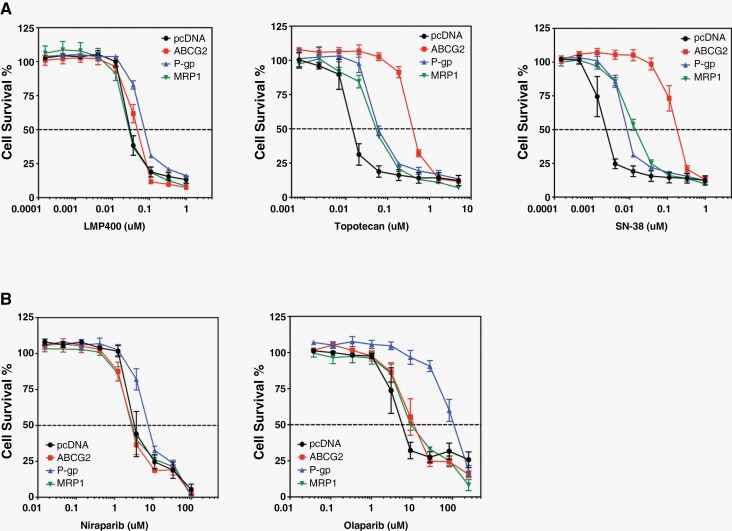
LMP400 and Niraparib Are Not Substrates for ABC Transporters
To determine whether LMP400 and Niraparib are substrates for the multidrug ABC transporters, 2 of which are known to be part of the BBB,2 we used stably transfected HEK293 cells overexpressing P-glycoprotein (P-gp, ABCB1), multidrug resistance-associated protein 1 (MRP1, ABCC1), and breast cancer resistance protein (ABCG2). Upon treatment with LMP400 or Niraparib, there was no change in survival of these cells overexpressing ABC transporters, except for P-gp showing a minimal increase (Figure 6A-B). In contrast, survival of cells treated with Topotecan and SN-38, other TOP1 inhibitors, was increased by all 3 transporters (Figure 6A), whereas treatment with Olaparib was mainly affected by P-gp (Figure 6B). Collectively, these findings suggest that LMP400 and Niraparib are likely not affected by ABC transporters.
Figure 6.
LMP400 and Niraparib offer improved bioavailability than current analogs. (A) Comparison of survival of HEK293 cells stably expressing common ATP-Binding Cassette (ABC) transporters upon treatment with LMP400, Topotecan, or SN-38. (B) Comparison of survival of HEK293 cells stably expressing common ABC transporters upon treatment with Niraparib or Olaparib.
Discussion
In the present study, we investigated the effects of LMP400 alone and in combination with PARP inhibitors on GBM in vitro and in vivo. In a panel of human GBM cell lines, we found that most of them are sensitive to LMP400, showing IC50 values in the low nanomolar range. Genetic and pharmacologic manipulations of GBM cells confirmed that PTEN-deficient cells are particularly sensitive to LMP400 when compared to PTEN-expressing cells. The combination of LMP400 with Niraparib, a newer PARP inhibitor with good BBB penetration, leads to synergistic cytotoxicity by inducing G2/M arrest, DNA damage, suppression of HR-related proteins, and activation of caspase 3/7 in PTEN-deficient GBM. These results have been confirmed in a syngeneic mouse model of GBM in which the combination significantly enhances survival of mice with implanted GBM. These findings support the development of clinical trials to test this combination in GBM patients and correlate the treatment response with PTEN status, to ultimately establish it as a therapy for GBM with PTEN deficiency, an aggressive and deadly disease.
The analysis of additional GBM cell lines included in NCI-60 and NOB datasets demonstrated that LMP400 is more active in GBM cells with low PTEN expression (Supplementary Figure S6), supporting our observation that PTEN status may predict response to LMP400. Further investigation of the PTEN role in treatment response using isogenic cells with and without PTEN KO or rescue revealed that PTEN deficiency confers increased sensitivity to LMP400. These findings are in line with the previous studies reporting that sensitivity to camptothecin and topotecan was much higher in PTEN-null cells compared to PTEN-WT cells.19,38 Interestingly, PTEN rescue in SF-295 cell line produced less resistance to LMP400 compared to the other cell lines tested, such as A172, GSC20, GSC23, and SW-1088 (Figure 2 and Supplementary Figure S7). Thus, it is possible that other genetic factors, such as TP53 status, could also contribute to altering response to TOP1 inhibitors, as suggested by Wang et al.39 Therefore, in order to eliminate potential effects of various genetic backgrounds in patient-derived GBM cells, we used isogenic mouse cell lines derived from a GEM model of GBM that have identical genetic backgrounds, except for the Pten deletion, to investigate the role of PTEN in treatment response.
TOP1 inhibitors trap TOP1ccs that can readily be converted into DSBs as demonstrated by rapid phosphorylation of γ-H2AX, which is a marker for DSBs.40 Consistently, we found γ-H2AX expression upon LMP400 treatment in vitro and ex vivo. The DNA damage triggered by TOP1 inhibitors can be repaired through PARP and HR with RAD51.30,41 Niraparib is a newer FDA-approved PARP inhibitor for maintenance treatment of advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer, following the first PARP inhibitor—Olaparib—that is more potent and able to penetrate the BBB.32,35 Here, we found that the antitumor effects of LMP400 were significantly augmented when administered together with Niraparib specifically in GBM cells with PTEN deficiency. Combined treatment induced a significant short-term and long-term inhibition of cell growth, G2/M cell cycle arrest, suppression of DNA repair, and activation of cell death pathways, including apoptosis. PTEN-deficient cells displayed enhanced cell growth arrest, DNA damage, and suppressed HR repair capability as compared to isogenic PTEN-WT cells. These findings align with the previous studies reporting that PTEN deficiency compromises DNA damage repair through defective HR.19,42 On top of the PTEN deficiency that created vulnerability to DNA damaging agents through compromised HR repair, addition of a PARP inhibitor, Niraparib, led to a more profound inability to repair DNA damage induced by a TOP1 inhibitor, LMP400, leading to its accumulation and, ultimately, cell death. Overall, it is plausible that attenuated HR repair due to PTEN deficiency is responsible for selective sensitivity to the LMP400/Niraparib combination in GBM. Noteworthy, the LMP400/Niraparib combination was shown to be synergistic at lower doses of each drug, which would potentially reduce drug toxicity.
Since the superior response to LMP400 and Niraparib combination was observed in cell line models with PTEN deficiency, our goal was to verify the advantage of the drug combination in vivo using a PTEN-deficient mouse model. Single agent therapy did not produce a survival benefit, whereas the drug combination resulted in significantly prolonged survival. Importantly, both drugs dose/schedule used in the in vivo testing are consistent or even less than clinically reported doses, supporting the translational potential of this drug combination (NCT05076513, NCT05297864).12 Importantly, compared to the xenograft model, the use of TRP syngeneic model allowed for assessment of the drug efficacy in the adequate tumor microenvironment under the immunocompetent setting. However, the limitation of this mouse model stems from the extremely low penetrance of the TR model,36,43 which would have been a perfect comparison to be used. While recognizing the limitation of the preclinical models, the in vitro and in vivo results offer preclinical evidence of correlation between the PTEN status and treatment response and warrant further evaluation of the predictive value of a biomarker in the setting of a prospective clinical trial.
One of the challenging obstacles in delivery of cancer chemotherapy to treat brain tumors remains BBB penetration. ABC transporters have been implicated in multidrug resistance and a low bioavailability of drugs via efflux.44,45 Previous studies showed that camptothecin and irinotecan are affected by ABCG2.46 ABCG2 has been implicated in topotecan resistance in human ovarian cell lines and in lung cancer resistance to irinotecan.47,48 Here, we found that LMP400 and Niraparib are not affected by ABC transporters, suggesting an increased possibility of brain penetration and an advantage of higher bioavailability over current analogs. Importantly, evaluation of PD markers in the tumor tissues also indicated BBB penetration.
In conclusion, this study uncovers a vulnerability in GBM that is predicated on the deficiency of PTEN, which is typically associated with a worse prognosis. In tumors with PTEN deficiency, the combination of LMP400 and Niraparib had synergistic cytotoxic effects in vitro and in vivo. The pharmacology of both drugs is not impacted by ABC transporters which favors their use in the treatment of brain tumors. These preclinical findings provide a rationale for further clinical investigation of combining indenoisoquinolines and PARP inhibitors as a synergistic therapeutic strategy for a subset of GBM patients with PTEN deficiency.
Supplementary Material
Acknowledgments
We thank Dr. Yves Pommier and the Developmental Therapeutics Program at NIH/NCI for providing LMP400 and Niraparib for in vitro and in vivo studies. Also, we would like to thank Ryan E. Bash (The University of Alabama at Birmingham, School of Medicine, Birmingham, AL, USA) for providing technical support for the in vivo experiment. In addition, we thank CCR Collaborative Bioinformatics Resource at NIH/NCI for the support of bioinformatics analysis.
Contributor Information
Olga Kim, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Madison Butler, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Zach Sergi, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Robert W Robey, Laboratory of Cell Biology, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Meili Zhang, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Raj Chari, Genome Modification Core laboratory, Leidos Biomedical Inc/ Frederick National Laboratory for Cancer Research, Frederick, Maryland, USA.
Ying Pang, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Guangyang Yu, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Wei Zhang, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Hua Song, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Dionne Davis, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Robert G Hawley, Genetics Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Xinyu Wen, Genetics Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Herui Wang, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Martha Quezado, Laboratory of Pathology.
Bao Tran, Sequencing Facility, Leidos Biomedical Inc/ Frederick National Laboratory for Cancer Research, Frederick, Maryland, USA.
Mythili Merchant, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Alice Ranjan, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Frank B Furnari, University of California at San Diego, School of Medicine, La Jolla, California, USA.
Javed Khan, Genetics Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Mark R Gilbert, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Christopher Ryan Miller, The University of Alabama at Birmingham, School of Medicine, Birmingham, Alabama, USA.
Michael M Gottesman, Laboratory of Cell Biology, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Yves Pommier, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA; Developmental Therapeutics Branch and Laboratory of Molecular Pharmacology, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Jing Wu, Neuro-Oncology Branch, Center for Cancer Research, NCI, NIH, Bethesda, Maryland, USA.
Funding
This research was supported by the Intramural Research Program of the National Institutes of Health (1ZIABC011841 and 1ZIABC011840) and Lasker Clinical Research Scholar Program.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Authorship statement
Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft: O.K. Formal analysis, validation, investigation, methodology: M.B., R.R. Bioinformatic analysis: Z.S., R.G.H., X.W., J.K., B.T. In vivo experiments: O.K., M.Z., W.Z., H.S., D.D., H.W. Contribution of critical reagents/analytic tools, validation: R.C., C.R.M., F.B.F., M.M.G., Yv.P. Pathological evaluation and review: M.Q. Manuscript review and editing: O.K., M.B., R.R., M.M., A.R., Z.S., Y.P., G.Y., R.G.H., C.R.M., F.B.F., J.K., M.R.G., M.M.G., Yv.P., J.W. Conceptualization, resources, data curation, supervision, funding acquisition, manuscript review and editing: J.W.
References
- 1. Aldape K, Brindle KM, Chesler L, et al. Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16(8):509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wijaya J, Fukuda Y, Schuetz JD.. Obstacles to brain tumor therapy: Key abc transporters. Int J Mol Sci . 2017;18(12):2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verhaak RGW, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brennan CW, Verhaak RGW, McKenna A, et al. ; TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pommier Y, Sun Y, Huang SN, Nitiss JL.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol. 2016;17(11):703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brangi M, Litman T, Ciotti M, et al. Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999;59(23):5938–5946. [PubMed] [Google Scholar]
- 7. Thomas A, Pommier Y.. Targeting topoisomerase I in the era of precision medicine. Clin Cancer Res. 2019;25(22):6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ardizzoni A. Camptothecin analogues in the treatment of non-small cell lung cancer. Lung Cancer. 1995;12(suppl 1):S177–S185. [DOI] [PubMed] [Google Scholar]
- 9. Pommier Y. Drugging topoisomerases: Lessons and challenges. ACS Chem Biol. 2013;8(1):82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slichenmyer WJ, Rowinsky EK, Donehower RC, Kaufmann SH.. The current status of camptothecin analogs as antitumor agents. J Natl Cancer Inst. 1993;85(4):271–291. [DOI] [PubMed] [Google Scholar]
- 11. Antony S, Agama K, Miao ZH, et al. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67(21):10397–10405. [DOI] [PubMed] [Google Scholar]
- 12. Kummar S, Chen A, Gutierrez M, et al. Clinical and pharmacologic evaluation of two dosing schedules of indotecan (LMP400), a novel indenoisoquinoline, in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2016;78(1):73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coussy F, El-Botty R, Chateau-Joubert S, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med. 2020;12(531):eaax2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pommier Y, O’Connor MJ, de Bono J.. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8(362):362ps317. [DOI] [PubMed] [Google Scholar]
- 15. Huang D, Kraus WL.. The expanding universe of PARP1-mediated molecular and therapeutic mechanisms. Mol Cell. 2022;82(12):2315–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murai J, Zhang Y, Morris J, et al. Rationale for PARP inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther. 2014;349(3):408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun Y, Chen J, Huang SN, et al. PARylation prevents the proteasomal degradation of topoisomerase I DNA-protein crosslinks and induces their deubiquitylation. Nat Commun. 2021;12(1):5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mendes-Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1(6-7):315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McEllin B, Camacho CV, Mukherjee B, et al. PTEN loss compromises homologous recombination repair in astrocytes: Implications for glioblastoma therapy with temozolomide or Poly(ADP-Ribose) polymerase inhibitors. Cancer Res. 2010;70(13):5457–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bassi C, Ho J, Srikumar T, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341(6144):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma J, Benitez JA, Li J, et al. Inhibition of nuclear PTEN tyrosine phosphorylation enhances glioma radiation sensitivity through attenuated DNA repair. Cancer Cell. 2019;35(3):504–518.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chou TC. Drug combination studies and their synergy quantification using the chou-talalay method. Cancer Res. 2010;70(2):440–446. [DOI] [PubMed] [Google Scholar]
- 23. Marzi L, Szabova L, Gordon M, et al. The indenoisoquinoline TOP1 inhibitors selectively target homologous recombination deficient- and Schlafen 11-positive cancer cells and synergize with olaparib. Clin Cancer Res. 2019;25(20):6206–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robey RW, Lin B, Qiu J, Chan LL, Bates SE.. Rapid detection of ABC transporter interaction: Potential utility in pharmacology. J Pharmacol Toxicol Methods. 2011;63(3):217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furnari FB, Lin H, Huang HS, Cavenee WK.. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci U S A. 1997;94(23):12479–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patil V, Pal J, Somasundaram K.. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget. 2015;6(41):43452–43471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ikediobi ON, Davies H, Bignell G, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5(11):2606–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao HF, Wang J, Shao W, et al. Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: Current preclinical and clinical development. Mol Cancer. 2017;16(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heffron TP, Ndubaku CO, Salphati L, et al. Discovery of clinical development candidate GDC-0084, a Brain Penetrant Inhibitor of PI3K and mTOR. ACS Med Chem Lett. 2016;7(4):351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yves P, Juana MB, Ashutosh Rao V. , et al. Repair of topoisomerase i‐mediated DNA damage. Prog Nucleic Acid Res Mol Biol. 2006;81:179–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. [DOI] [PubMed] [Google Scholar]
- 32. Kaiming Sun KM, Zebin W, Grace P, et al. A comparative pharmacokinetic study of PARP inhibitors demonstrates favorable properties for niraparib efficacy in preclinical tumor models. Oncotarget. 2018;9(98):37080–37096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maria J.Sambade AEDVS, Marni B. McClure, Allison MD, Charlene S, Kaiming S, Jing W, Keith M, and Carey KA.. Efficacy and pharmacodynamics of niraparib in BRCAmutant and wild-type intracranial triple-negative breast cancer murine models. Neuro Oncol Adv. 2019;1(1):vdz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murai J, Huang SYN, Das BB, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chornenkyy Y, Agnihotri S, Yu M, et al. Poly-ADP-ribose polymerase as a therapeutic target in pediatric diffuse intrinsic pontine glioma and pediatric high-grade astrocytoma. Mol Cancer Ther. 2015;14(11):2560–2568. [DOI] [PubMed] [Google Scholar]
- 36. Song YR, Zhang Q, Kutlu B, et al. Evolutionary etiology of high-grade astrocytomas. Proc Natl Acad Sci USA. 2013;110(44):17933–17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Do K, Chen AP.. Molecular pathways: Targeting PARP in cancer treatment. Clin Cancer Res. 2013;19(5):977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benitez JA, Finlay D, Castanza A, et al. PTEN deficiency leads to proteasome addiction: A novel vulnerability in glioblastoma. Neuro Oncol. 2021;23(7):1072–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y, Zhu S, Cloughesy TF, Liau LM, Mischel PS.. p53 disruption profoundly alters the response of human glioblastoma cells to DNA topoisomerase I inhibition. Oncogene. 2004;23(6):1283–1290. [DOI] [PubMed] [Google Scholar]
- 40. Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM.. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. [DOI] [PubMed] [Google Scholar]
- 41. Pommier Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat Rev Cancer. 2006;6(10):789–802. [DOI] [PubMed] [Google Scholar]
- 42. Vuono EA, Mukherjee A, Vierra DA, et al. The PTEN phosphatase functions cooperatively with the Fanconi anemia proteins in DNA crosslink repair. Sci Rep. 2016;6:36439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vitucci M, Karpinich NO, Bash RE, et al. Cooperativity between MAPK and PI3K signaling activation is required for glioblastoma pathogenesis. Neuro Oncol. 2013;15(10):1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen ZL, Shi TL, Zhang L, et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016;370(1):153–164. [DOI] [PubMed] [Google Scholar]
- 45. Gottesman MM, Fojo T, Bates SE.. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat Rev Cancer. 2002;2(1):48–58. [DOI] [PubMed] [Google Scholar]
- 46. Yang CJ, Horton JK, Cowan KH, E S.. Cross-resistance to camptothecin analogues in a mitoxantrone-resistant human breast carcinoma cell line is not due to DNA topoisomerase I alterations. Cancer Res. 1995;55(18):4004–4009. [PubMed] [Google Scholar]
- 47. Kawabata S, Oka M, Soda H, et al. Expression and functional analyses of breast cancer resistance protein in lung cancer. Clin Cancer Res. 2003;9(8):3052–3057. [PubMed] [Google Scholar]
- 48. Maliepaard M, van Gastelen MA, de Jong LA, et al. Overexpression of the BCRP/MXR/ABCP gene in a topotecan-selected ovarian tumor cell line. Cancer Res. 1999;59(18):4559–4563. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data generated in this study are publicly available in Gene Expression Omnibus (GEO) at GSE225210.