Abstract
Our previous studies have shown that Actinobacillus actinomycetemcomitans isolates of a given arbitrarily primed PCR (AP-PCR) genotype belong to the same serotype (of serotypes a through e). In the present study we investigated whether the AP-PCR genotypes of nonserotypeable A. actinomycetemcomitans isolates match those of the serotypeable isolates. The isolates were additionally characterized by restriction analysis of the apaH PCR amplification products. The material included 75 nonserotypeable and 18 serotypeable A. actinomycetemcomitans isolates from 34 epidemiologically unrelated subjects. The serotypeable isolates were obtained from subjects who also harbored nonserotypeable isolates. Eight AP-PCR genotypes were distinguished among the isolates; six genotypes matched those detected in our previous studies, whereas two genotypes were new. Intraindividually, the A. actinomycetemcomitans isolates produced identical AP-PCR banding patterns, regardless of whether they were serotypeable or nonserotypeable, in 22 of 23 subjects participating with multiple isolates. AP-PCR genotype 3, corresponding to serotype c, was by far the most common among the nonserotypeable isolates (62% of subjects). Results obtained with the apaH restriction analysis confirmed the results obtained with AP-PCR for 31 of the 34 subjects. The results suggest that nonserotypeable A. actinomycetemcomitans isolates originate from serotypeable isolates, especially from serotype c isolates, and the likelihood of the existence of additional serotypes is small.
Actinobacillus actinomycetemcomitans, a gram-negative capnophilic coccobacillus, is a major pathogen in the initiation and progression of periodontitis (9, 27, 28). A. actinomycetemcomitans has also been sporadically isolated in nonoral infections such as endocarditis, pericarditis, pneumonia, septicemia, and abscesses (5, 7, 12, 13, 24, 26, 27). There are five known serotypes of A. actinomycetemcomitans, designated a, b, c, d, and e (8, 18). The serotype-specific antibody binding of A. actinomycetemcomitans was previously reported to be mediated by the O-antigen carbohydrate chains of the lipopolysaccharide (15, 25). However, a recent report contradicts these findings by stating that serotype-specific epitopes are on the amorphous material on the cell surface (14). Thus, the exact nature of the serotype-specific antigens still remains unclear.
Of the five known A. actinomycetemcomitans serotypes, the most prevalent in the oral cavity are serotypes a, b, and c (16, 18). A. actinomycetemcomitans isolates that do not react with any of the five serotype-specific antisera have occasionally been detected; they comprise 3 to 9% of isolates (3, 16, 18). Only a few nonserotypeable A. actinomycetemcomitans isolates have been further characterized genotypically, and these studies suggest that nonserotypeable isolates are serotype antigen variants originating from isolates of known serotypes (3, 16, 19, 23).
Intraindividual colonization by the same A. actinomycetemcomitans serotype(s) can be stable over several years (18). In our studies we have found no indication that spontaneous A. actinomycetemcomitans serotype switching might occur in an individual in the course of time (reference 18 and unpublished data). Antigenic variation resulting in serotype switching has been reported in other species, such as Borrelia hermsii, the causative agent of relapsing fever (4).
Arbitrarily primed PCR (AP-PCR) has proved an applicable technique for the genotyping of A. actinomycetemcomitans isolates. Nine to 17 different AP-PCR genotypes have been reported among A. actinomycetemcomitans isolates, depending on both the primers used in the amplification and the number of isolates tested (2, 3, 6, 17, 19, 21). Our previous studies have shown that A. actinomycetemcomitans isolates of different serotypes also have different AP-PCR genotypes (3, 19). The few nonserotypeable A. actinomycetemcomitans isolates previously analyzed by AP-PCR had genotypes similar to those of the serotypeable isolates (3, 19). However, the AP-PCR genotype distribution of nonserotypeable isolates has not been studied. In another Actinobacillus species, A. pleuropneumoniae, genotyping with AP-PCR has proved a rapid and accurate method in serotype identification of serologically cross-reactive or nontypeable field isolates (10).
Our recent study on apaH gene polymorphism in A. actinomycetemcomitans clinical isolates indicated that all isolates, regardless of serotype, had apaH, a gene which in Escherichia coli confers ability to invade KB oral epithelial cells in vitro, but restriction analysis of the gene revealed serotype-specific differences among the isolates. Serotype c isolates and a subpopulation (genogroup 2) of serotype e isolates could be clearly differentiated from the other isolates on the basis of differences in the SphI and NheI restriction patterns of the apaH PCR amplification product (20).
The aim of the present study was to investigate, by AP-PCR and apaH restriction analysis, whether nonserotypeable A. actinomycetemcomitans isolates have genotypes matching those of serotypeable isolates. Finding additional, previously unknown genotypes could indicate the existence of new serotypes.
MATERIALS AND METHODS
Subjects and bacterial isolates.
The study material comprised 75 nonserotypeable oral A. actinomycetemcomitans isolates from 34 epidemiologically unrelated subjects (age range, 14 to 68 years). Six of the subjects also harbored serotypeable A. actinomycetemcomitans isolates, and these isolates (n = 18) were included in the study in order to compare the genotypes of intraindividual serotypeable and nonserotypeable isolates. The isolates were chosen from our collection of about 1,300 previously serotyped isolates at the Institute of Dentistry, University of Helsinki (references 3, 18, and 19 and unpublished data). Serotyping had been performed by an immunodiffusion technique with polyclonal serotype-specific rabbit antisera against serotypes a through e (18). Each of the 34 subjects contributed 1 to 12 (mean, 2.7) A. actinomycetemcomitans isolates, of which 1 to 7 (mean, 2.2) were serologically nontypeable. The isolates originated from samples of subgingival plaque and saliva and from samples from the tongue surface or oral mucosa.
In addition, three A. actinomycetemcomitans reference strains (ATCC 29523 for serotype a, ATCC 43718 for serotype b, and ATCC 33384 for serotype c) were included in the material as reference strains for AP-PCR.
A. actinomycetemcomitans isolates were grown on tryptic soy-serum-bacitracin-vancomycin (TSBV) agar plates (22) incubated in 5% CO2 in air at 37°C for 2 to 3 days. Subcultures starting from a single colony were preserved in 20% skim milk at −70°C until used.
AP-PCR genotyping.
Chromosomal DNA was extracted from A. actinomycetemcomitans isolates by a previously described method for ribotyping (19). Five microliters of a 1:300 dilution of extracted DNA was used as a template DNA in AP-PCR. AP-PCR was performed in a 50-μl reaction volume consisting of 0.2 mM deoxynucleoside triphosphates (Pharmacia Biotech, Piscataway, N.J.), 0.4 μM primer, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 4 mM MgCl2, and 2.5 U of AmpliTaq (Roche Molecular Systems, Inc., Branchburg, N.J.) overlaid with mineral oil. The random sequence oligonucleotide OPA-13 (5′-CAGCACCCAC-3′) (Operon Technologies, Inc., Alameda, Calif.) was used as a primer for all A. actinomycetemcomitans isolates, and two additional primers, OPA-03 (5′-AGTCAGCCAC-3′) and OPA-07 (5′-GAAACGGGTG-3′), were used for selected isolates indistinguishable with the OPA-13 primer. The temperature profile in a thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.) was 35 cycles of 94°C for 1 min, 32°C for 2 min, and 72°C for 2 min. The initial denaturation was carried out at 94°C for 5 min, and the final extension was carried out at 72°C for 5 min. Amplification products were analyzed electrophoretically in a 1% (wt/vol) agarose gel containing ethidium bromide (0.5 μg/ml) and were visualized under UV light.
PCR amplification of apaH and restriction analysis of the amplification product.
PCR amplification of the apaH gene was performed as previously described (20) in a total volume of 50 μl consisting of 0.2 mM deoxynucleoside triphosphates (Perkin-Elmer Cetus), 10× Taq buffer, 1 μM each primer, 1.25 U of AmpliTaq (Perkin-Elmer Cetus), and 0.5 to 2 μl (approximately 50 ng) of the template DNA overlaid with mineral oil. Primers used in the amplification of a 771-bp internal fragment of the 825-bp coding sequence of the apaH gene were 5′-ATTTAATCGGCGACCTGCAC-3′ and 5′-TGTCTTCCCAACGTAGCATG-3′. The temperature profile in a thermocycler (Perkin-Elmer Cetus) was 35 cycles of 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min. The initial denaturation was carried out at 94°C for 3 min, and the final extension was carried out at 72°C for 5 min.
Amplification products (an 8-μl sample) were characterized by restriction analysis using the restriction endonucleases SphI and NheI according to the manufacturer’s instructions (Gibco BRL) as previously described (20). SphI digestion yields DNA fragments of 347 and 424 bp from the 771-bp amplification product of isolates of serotypes a, b, and d and genogroup 1 of serotype e. NheI digestion produces DNA fragments of 104 and 667 bp for isolates of the same groups. Serotype c isolates have an additional SphI site in their apaH, resulting in fragments of 129, 218, and 424 bp, whereas they have lost the unique NheI site. Serotype e isolates of genogroup 2 have apaH with neither an SphI nor an NheI site (20).
RESULTS
AP-PCR genotypes.
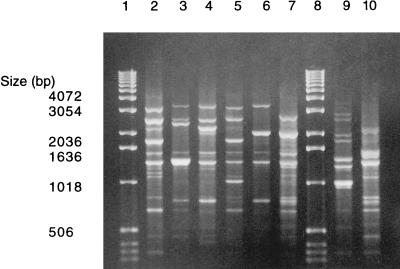
The OPA-13 primer distinguished eight AP-PCR genotypes among the 75 nonserotypeable A. actinomycetemcomitans isolates from 34 subjects (Fig. 1). Genotypes 1, 2, 3, 5, 11, and 16 were similar to those found in our previous studies (1–3) (Fig. 1, lanes 2 through 7), whereas genotypes 18 and 19 within two subjects were new (Fig. 1, lanes 9 and 10; Table 1).
FIG. 1.
Eight different AP-PCR genotypes, obtained with OPA-13, were found among the 75 nonserotypeable A. actinomycetemcomitans isolates from 34 subjects. Lanes 1 and 8, molecular size markers; lane 2, AP-PCR type 1; lane 3, AP-PCR type 2; lane 4, AP-PCR type 3; lane 5, AP-PCR type 5; lane 6, AP-PCR type 11; lane 7, AP-PCR type 16; lane 9, AP-PCR type 18; lane 10, AP-PCR type 19.
TABLE 1.
Nonserotypeable A. actinomycetemcomitans isolates of 34 subjects grouped into “serotypes” based on AP-PCR typing
| AP-PCR typea | “Serotype”b | No. (%) of subjects | No. of isolates |
|---|---|---|---|
| 1 | a | 6 (18) | 12 |
| 2 | b | 2 (6) | 3 |
| 3 | c | 21 (62) | 45 |
| 5 | d | 1 (3) | 2 |
| 16 | b | 1 (3) | 1 |
| 11 | Nonserotypeable | 2 (6) | 9 |
| 18 | Nonserotypeable | 1 (3) | 1 |
| 19 | Nonserotypeable | 1 (3) | 2 |
| Total | 34c | 75 |
AP-PCR type designations 1 through 16 are given according to the work of Asikainen et al. (2). AP-PCR types 18 and 19 are new.
“Serotype” designation based on AP-PCR type.
One subject harbored A. actinomycetemcomitans isolates of AP-PCR types 5 and 18.
Twenty-three of the 34 subjects participated with multiple A. actinomycetemcomitans isolates, and six of these harbored both nonserotypeable and serotypeable A. actinomycetemcomitans isolates (two subjects with serotype a, one with serotype b, and three with serotype c). Intraindividually, the A. actinomycetemcomitans isolates produced identical AP-PCR banding patterns regardless of whether they were serotypeable or nonserotypeable in 22 of the 23 subjects. A single subject had two AP-PCR genotypes within the nonserotypeable isolates.
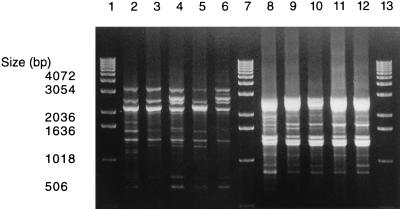
The most common AP-PCR genotype among the present nonserotypeable A. actinomycetemcomitans isolates was genotype 3 (corresponding to serotype c) (3), which was detected in 21 subjects. AP-PCR genotype 3, obtained with OPA-13, consisted of four groups of banding patterns exhibiting only slight variation (Fig. 2). The OPA-03 primer generated only minor additional differences in the banding patterns (Fig. 2), and the OPA-07 primer did not distinguish the isolates any further.
FIG. 2.
The slightly different AP-PCR banding patterns of the nonserotypeable A. actinomycetemcomitans isolates representing AP-PCR type 3 obtained with OPA-13 (lanes 2 through 6) and OPA-03 (lanes 8 through 12). Lanes 1, 7 and 13, molecular size markers; lanes 2 and 8, reference strain A. actinomycetemcomitans ATCC 33384 (serotype c); lanes 3 through 6 and 9 through 12, clinical isolates from four subjects, shown in the same order.
When the nonserotypeable A. actinomycetemcomitans isolates were grouped into AP-PCR genotypes corresponding to different serotypes, the distribution was as follows: “serotype a” in 6 subjects (18%), “serotype b” in 3 subjects (9%), “serotype c” in 21 subjects (62%), and “serotype d” in 1 subject (3%) (Table 1). Twelve isolates from four subjects (12%) remained nonserotypeable with AP-PCR.
apaH restriction analysis.
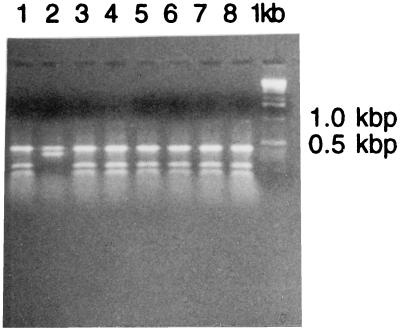
Based on restriction analysis of the apaH amplification product, 24 of the 34 subjects harbored A. actinomycetemcomitans isolates with a serotype c-like restriction pattern (20). All other subjects had isolates with the “common” restriction pattern representing serotypes a, b, and d and genogroup 1 of serotype e (Fig. 3).
FIG. 3.
SphI restriction patterns of apaH PCR amplification products of nonserotypeable A. actinomycetemcomitans isolates. Lanes 1 and 3 through 8 represent the SphI restriction pattern obtained from isolates of AP-PCR type 3. The same restriction pattern was also found among isolates of AP-PCR types 11 and 16. Lane 2 represents the SphI restriction pattern obtained from an isolate of AP-PCR type 19; the same pattern is also obtained from isolates of AP-PCR types 1, 2, 5, and 18. Right lane, molecular size markers.
The results obtained by apaH restriction analysis coincided with AP-PCR results for 31 of the 34 subjects. For two subjects AP-PCR genotyping classified the isolates as AP-PCR group 11, corresponding to a nonserotypeable group, and for one subject it classified the isolate as AP-PCR type 16, corresponding to serotype b (1), whereas according to apaH restriction analysis, these three isolates represented serotype c.
DISCUSSION
In the present study we used AP-PCR and apaH restriction analysis to genotypically characterize nonserotypeable oral A. actinomycetemcomitans isolates. Our aim was to establish whether nonserotypeable isolates represent the same genotypes as serotypeable isolates, thus indicating that they have a common origin, or whether they represent previously unknown genotypes, possibly indicating the existence of new serotypes. This was accomplished by comparing the genotypes of the nonserotypeable isolates with those of the serotypeable isolates. The material included 75 nonserotypeable and 18 serotypeable A. actinomycetemcomitans isolates from our collection of about 1,300 serotyped A. actinomycetemcomitans isolates. Twenty-three of the 34 subjects included in the study participated with multiple A. actinomycetemcomitans isolates, and six of them harbored both nonserotypeable and serotypeable isolates.
Six of the eight AP-PCR genotypes detected among the present nonserotypeable A. actinomycetemcomitans isolates were similar to those found in our previous studies on serotypeable isolates (1, 2). Intraindividually, the A. actinomycetemcomitans isolates showed clonality within 22 of the 23 subjects with multiple isolates, whereas one subject had two AP-PCR genotypes within the nonserotypeable isolates. Six of the 23 subjects harbored both serotypeable and nonserotypeable A. actinomycetemcomitans isolates, and in each of these subjects clonality was apparent, suggesting a common origin for these isolates. Previously, clonality had been discovered by AP-PCR among the nonserotypeable and serotypeable A. actinomycetemcomitans isolates from two subjects (3). Clonality among intraindividual serotypeable and nonserotypeable isolates has also been detected for other pathogenic bacteria: identity between outer membrane protein profiles of encapsulated type b and nonserotypeable Haemophilus influenzae isolates has been demonstrated (11), suggesting that at least some nonserotypeable isolates might derive from the type b isolates.
The most common AP-PCR genotype among the nonserotypeable isolates was genotype 3 (corresponding to serotype c) (3), detected in 21 subjects. “Serotype” distribution (based on the AP-PCR genotyping results) among the nonserotypeable A. actinomycetemcomitans isolates was as follows: “serotype a” in 18% of the subjects, “serotype b” in 9%, “serotype c” in 62%, and “serotype d” in 3%. Twelve isolates from four subjects (12%) remained nonserotypeable with AP-PCR. This result differs from the actual serotype distribution among the A. actinomycetemcomitans isolates from 528 subjects in our culture collection: among these subjects serotypes a, b, and c are about equally represented (25, 31, and 27%, respectively), whereas serotypes d and e are rare (4 and 6%, respectively) (unpublished data). The AP-PCR genotype corresponding to serotype c is thus far more common in nonserotypeable isolates than could be expected on the basis of the actual serotype distribution of our A. actinomycetemcomitans isolates (62% versus 27%). The results obtained by restriction analysis of apaH amplification products confirmed the finding that “serotype c” predominates among nonserotypeable isolates. According to apaH restriction analysis, 71%, not 62%, of the subjects had isolates with a restriction pattern similar to that of serotype c isolates. The discrepancy between AP-PCR genotyping and apaH restriction analysis results was caused by three subjects whose isolates were classified as AP-PCR genotypes 11 (“nonserotypeable”) and 16 (“serotype b”), whereas according to apaH restriction analysis, these isolates represented serotype c. AP-PCR genotypes 11 and 16 are rare among A. actinomycetemcomitans isolates (3 and 1% of the subjects, respectively) (2). Thus, an explanation for the detected discrepancy might be that genotypes 11 and 16 are differently distributed among serotypes compared to the more commonly detected AP-PCR genotypes. Also, two of the three subjects with mismatching results were non-Caucasians, whereas our previous AP-PCR genotype distribution studies have been performed on Finnish subjects.
Our results suggest that nonserotypeable isolates represent a population of isolates that originally were serotypeable but later lost the ability to react with serotype-specific antisera. The predominance of genotypes corresponding to serotype c further suggests that serotype c isolates lose their ability to react with serotype-specific antisera more easily than isolates of the other serotypes. The results of Poulsen et al. (16) also suggest that nonserotypeable isolates originate from isolates of known serotypes. They characterized eight nonserotypeable A. actinomycetemcomitans isolates by several techniques, including multilocus enzyme electrophoresis, and found that these isolates were distributed in different evolutionary lines of the population and were genetically closely related to other isolates of the respective clusters. However, two of the eight genotypes found in the present study among the nonserotypeable A. actinomycetemcomitans isolates were not known from our previous studies (1–3), so the existence of a new serotype(s) still cannot be conclusively ruled out.
In conclusion, the results suggest that nonserotypeable A. actinomycetemcomitans isolates originate from serotypeable isolates, especially from serotype c isolates, and that the possibility of finding additional serotypes is small.
ACKNOWLEDGMENTS
This study was supported by grants from the Academy of Finland (10131015, awarded to S. Asikainen, and 36226, awarded to M. Saarela), the Finnish Dental Society (awarded to S. Paju), and the National Institutes of Health (RO1DE09760, awarded to P. Fives-Taylor).
REFERENCES
- 1.Asikainen, S., C. Chen, M. Saarela, L. Saxén, and J. Slots. 1997. Clonal specificity of Actinobacillus actinomycetemcomitans in destructive periodontal disease. Clin. Infect. Dis. 25(Suppl. 2):S227–S229. [DOI] [PubMed]
- 2.Asikainen S, Chen C, Slots J. Likelihood of transmitting Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in families with periodontitis. Oral Microbiol Immunol. 1996;11:387–394. doi: 10.1111/j.1399-302x.1996.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 3.Asikainen S, Chen C, Slots J. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol Immunol. 1995;10:65–68. doi: 10.1111/j.1399-302x.1995.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 4.Barbour A G, Burman N, Carter C J, Kitten T, Bergstrom S. Variable antigen genes of the relapsing fever agent Borrelia hermsii are activated by promoter addition. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 5.Braconier J H, Söderström C. Indirect immunofluorescence as a diagnostic tool in a prosthetic heart valve endocarditis due to Actinobacillus actinomycetemcomitans and Staphylococcus aureus. Scand J Infect Dis. 1990;22:739–741. doi: 10.3109/00365549009027130. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., and J. Slots. 1995. Arbitrarily primed polymerase chain reaction analysis of periodontal pathogens: discriminative primers and genetic diversity. Clin. Infect. Dis. 20(Suppl. 2):S301–S303. [DOI] [PubMed]
- 7.Chen Y-C, Chang S-C, Luh K-T, Hsieh W-C. Actinobacillus actinomycetemcomitans endocarditis: a report of four cases and review of the literature. Q J Med. 1991;81:871–878. [PubMed] [Google Scholar]
- 8.Gmür R, McNabb H, van Steenbergen T J M, Baehni P, Mombelli A, van Winkelhoff A J, Guggenheim B. Seroclassification of hitherto nontypeable Actinobacillus actinomycetemcomitans strains: evidence for a new serotype e. Oral Microbiol Immunol. 1993;8:116–120. doi: 10.1111/j.1399-302x.1993.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 9.Haffajee A D, Socransky S S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 10.Hennessy K J, Iandolo J J, Fenwick B W. Serotype identification of Actinobacillus pleuropneumoniae by arbitrarily primed polymerase chain reaction. J Clin Microbiol. 1993;31:1155–1159. doi: 10.1128/jcm.31.5.1155-1159.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoiseth S K, Gilsdorf J R. The relationship between type b and nontypeable Haemophilus influenzae isolated from the same patient. J Infect Dis. 1988;158:643–645. doi: 10.1093/infdis/158.3.643. [DOI] [PubMed] [Google Scholar]
- 12.Horowitz E A, Pugsley M P, Turbes P G, Clark R B. Pericarditis caused by Actinobacillus actinomycetemcomitans. J Infect Dis. 1987;155:152–153. doi: 10.1093/infdis/155.1.152. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan A H, Weber D J, Odone E Z, Perfect J R. Infection due to Actinobacillus actinomycetemcomitans: 15 cases and review. Rev Infect Dis. 1989;11:46–63. doi: 10.1093/clinids/11.1.46. [DOI] [PubMed] [Google Scholar]
- 14.McArthur W P, Stroup S, McClellan S, Leung K-P. Differentiation of the serotype b and species-specific antigens of Actinobacillus actinomycetemcomitans recognized by monoclonal antibodies. Oral Microbiol Immunol. 1996;11:209–219. doi: 10.1111/j.1399-302x.1996.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 15.Page R C, Sims T J, Engel L D, Moncla B J, Bainbridge B, Stray J, Darveau R P. The immunodominant outer membrane antigen of Actinobacillus actinomycetemcomitans is located in the serotype-specific high-molecular-mass carbohydrate moiety of lipopolysaccharide. Infect Immun. 1991;59:3451–3462. doi: 10.1128/iai.59.10.3451-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poulsen K, Theilade E, Lally E T, Demuth D R, Kilian M. Population structure of Actinobacillus actinomycetemcomitans: a framework for studies of disease-associated properties. Microbiology. 1994;140:2049–2060. doi: 10.1099/13500872-140-8-2049. [DOI] [PubMed] [Google Scholar]
- 17.Preus H R, Haraszthy V I, Zambon J J, Genco R J. Differentiation of strains of Actinobacillus actinomycetemcomitans by arbitrarily primed polymerase chain reaction. J Clin Microbiol. 1993;31:2773–2776. doi: 10.1128/jcm.31.10.2773-2776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saarela M, Asikainen S, Alaluusua S, Pyhälä L, Lai C-H, Jousimies-Somer H. Frequency and stability of mono- and poly-infection by Actinobacillus actinomycetemcomitans serotypes a, b, c, d or e. Oral Microbiol Immunol. 1992;7:277–279. doi: 10.1111/j.1399-302x.1992.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 19.Saarela M, Asikainen S, Chen C, Alaluusua S, Slots J. Comparison of arbitrarily primed polymerase chain reaction and ribotyping for subtyping Actinobacillus actinomycetemcomitans. Anaerobe. 1995;1:97–102. doi: 10.1006/anae.1995.1004. [DOI] [PubMed] [Google Scholar]
- 20.Saarela, M., S. Asikainen, S. Alaluusua, and P. Fives-Taylor. apaH polymorphism in clinical Actinobacillus actinomycetemcomitans isolates. Anaerobe, in press. [DOI] [PubMed]
- 21.Slots J, Liu Y B, DiRienzo J M, Chen C. Evaluating two methods for fingerprinting genomes of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1993;8:337–343. doi: 10.1111/j.1399-302x.1993.tb00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slots J. Selective medium for isolation of Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1982;15:606–609. doi: 10.1128/jcm.15.4.606-609.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Steenbergen T J M, Bosch-Tijhof C-J, van Winkelhoff A J, Gmür R, de Graaff J. Comparison of six typing methods for Actinobacillus actinomycetemcomitans. J Clin Microbiol. 1994;32:2769–2774. doi: 10.1128/jcm.32.11.2769-2774.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhaaren H, Claeys G, Verschraegen G, de Niel C, Leroy J, Clement D. Endocarditis from a dental focus. Importance of oral hygiene in valvar heart disease. Int J Cardiol. 1989;23:343–347. doi: 10.1016/0167-5273(89)90194-0. [DOI] [PubMed] [Google Scholar]
- 25.Wilson M E, Schifferle R E. Evidence that the serotype b antigenic determinant of Actinobacillus actinomycetemcomitans Y4 resides in the polysaccharide moiety of lipopolysaccharide. Infect Immun. 1991;59:1544–1551. doi: 10.1128/iai.59.4.1544-1551.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan A, Yang P-C, Lee L-N, Chang D-B, Kuo S-H, Luh K-T. Actinobacillus actinomycetemcomitans pneumonia with chest wall involvement and rib destruction. Chest. 1992;101:1450–1452. doi: 10.1378/chest.101.5.1450. [DOI] [PubMed] [Google Scholar]
- 27.Zambon J J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 28.Zambon J J, Slots J, Genco R J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]