Abstract
The new Roche Cobas Amplicor Mycobacterium tuberculosis assay, which is a semiautomated version of the manually performed Roche Amplicor M. tuberculosis test, was compared to culture and an IS6110-based in-house PCR protocol. A total of 1,681 specimens from 833 patients, including specimen types other than sputum, were tested in parallel by both the in-house PCR and the Cobas Amplicor M. tuberculosis assay. After we resolved discrepant PCR results, the sensitivity, specificity, and positive and negative predictive values for the Cobas Amplicor M. tuberculosis assay were 66.33, 99.71, 94.36, and 97.66%, respectively. The corresponding values for the in-house PCR were 91.08, 99.85, 97.87, and 99.37%, respectively. For culture- and smear-positive specimens, the sensitivity of the Cobas Amplicor M. tuberculosis test was 96.42% (in-house PCR, 100%). If only smear-negative sputum specimens were considered, the Cobas Amplicor M. tuberculosis assay exhibited a sensitivity of 45.45% (in-house PCR, 63.63%) relative to that of culture. With a modified protocol for DNA extraction (washing of samples plus ultrasonication), both PCR methods performed better with gastric aspirates than with sputum samples (sensitivity of the Cobas Amplicor M. tuberculosis assay with smear-negative gastric aspirates, 70.00%; sensitivity of in-house PCR, 90.00%). With dithiothreitol being used for liquefaction of specimens in this study, the Cobas Amplicor M. tuberculosis assay exhibited an inhibition rate of 9.16%. In our view, the new Cobas Amplicor M. tuberculosis test (i) is well suited for typing of smear-positive specimens, (ii) may also be applied to gastric aspirates and other types of specimens if DNA extraction methods are modified appropriately, and (iii) exhibits a sensitivity with smear-negative sputum specimens which makes it recommendable that a minimum of three samples from the same patient be tested.
Since the beginning of this decade, PCR and other amplification techniques have been introduced into the diagnosis of infections with Mycobacterium tuberculosis (1, 6, 9, 14–16, 22, 25, 26). Although no amplification system known today provides sufficient sensitivity to replace culture as a reliable screening tool, an increasing number of diagnostic laboratories have established amplification techniques as supplementory tests, because they provide good rates of positive results with better turnaround times than culture (days versus weeks) and they can specify positive smear results. The latter feature is essential for laboratories dealing with a high portion of infections with mycobacteria other than M. tuberculosis (MOTT) (9). Because establishing and maintaining in-house protocols for any amplification method requires highly specialized personnel and enormous logistic efforts to control the well-known contamination problem, there is an obvious need of commercially available, easy-to-use diagnostic kits which have the potential of automation. To date, a few diagnostic tests for detection of M. tuberculosis in clinical specimens after DNA or RNA amplification have been marketed. Of these, the Roche Diagnostics (Grenzach-Whylen, Germany) Cobas Amplicor M. tuberculosis test (CA) exhibits the highest degree of automation. The CA amplifies, hybridizes, and detects amplicons in one run without the need of any manual intervention and offers the possibility of processing PCR mixtures for different targets in parallel. The aim of this study was to evaluate this new test system by comparing it to culture and an in-house PCR protocol. In addition, we tested the CA with specimen types other than sputum and evaluated the diagnostic value of the PCR techniques investigated in this study in a case-oriented fashion.
MATERIALS AND METHODS
Clinical specimens.
All specimens submitted to our institution for diagnosis of tuberculosis (TB) from January to November 1996 were examined in parallel by conventional analysis (fluorochrome stain and culture; see below) and PCR (CA and in-house PCR; see below). The following restrictions applied: (i) if the actual work load exceeded the capacities of the PCR working group, PCR was performed on a subset of specimens which guaranteed that at least one specimen per patient was examined by both PCR methods, (ii) only one smear-positive specimen per smear-positive patient was tested in order to minimize the input of genomic DNA into the DNA extraction area, (iii) small-volume specimens were tested only by conventional methods, and (iv) stool and blood samples were not examined by either PCR system. Specimens inhibitory to CA were not included for calculation of sensitivity, specificity, and predictive values.
Specimen processing for conventional analysis.
Sputa were mechanically homogenized and liquified with 1 volume of 0.1% (wt/vol) dithiothreitol (DTT) in distilled water. The liquified material was split for further separate processing by conventional analysis and PCR (approximately 1 ml for each method). Subsequently, bacteria were sedimented in a refrigerated centrifuge at 4,500 × g for 10 min. A loopful of the resulting pellet was used to prepare a slide for acid-fast staining, and the remaining material was further processed by the Zephirol-trisodium phosphate method for decontamination (23). After neutralization, the suspension was centrifuged at 4,500 × g for 10 min and resuspended in 1 ml of distilled water. Urine and other liquid samples were centrifuged at 4,500 × g for 10 min, and the resulting pellets were treated as described above for sputum sediments after liquefaction. Swab samples were squeezed into sterile distilled water and centrifuged; the sediment was treated as described above. Cerebrospinal fluid was concentrated only by centrifugation; there was no further treatment. Biopsy samples were mechanically homogenized, and the homogenate was directly inoculated onto culture media. If the presence of a contaminating flora was expected (e.g., in biopsies from abscess walls), the homogenate was decontaminated and neutralized as described above. The numbers of each specimen type studied are given in the first row of Table 5.
TABLE 5.
Inhibition rates of CA with different specimen types
| Specimen typea | Total no. of specimens | No. of noninhibitory specimens | No. of inhibitory specimens | Inhibition rate (%) |
|---|---|---|---|---|
| Pus | 47 | 45 | 2 | 4.25 |
| Swab | 32 | 31 | 1 | 3.12 |
| Sputum or BALa | 821 | 733 | 88 | 10.71 |
| Gastric aspirate | 354 | 309 | 45 | 12.71 |
| Urine | 157 | 153 | 4 | 2.54 |
| Biopsy | 86 | 83 | 3 | 3.48 |
| Pleural fluid | 122 | 113 | 9 | 7.37 |
| CSFb | 29 | 29 | 0 | 0.00 |
| Other | 33 | 31 | 2 | 6.06 |
| Total | 1,681 | 1,527 | 154 | 9.16 |
BAL, bronchoalveolar lavage.
CSF, cerebrospinal fluid.
Smear examination.
Acid-fast stains of the homogenized specimens were prepared with auramine-rhodamine and Ziehl-Neelsen stain and examined according to standard procedures (23). Urine samples were examined by microscopy only if urogenital TB was suspected.
Culture and identification.
After decontamination as described above, 0.2 ml of the resulting suspension was inoculated into one BACTEC 12B bottle (Becton Dickinson, Heidelberg, Germany) supplemented according to the manufacturer’s instructions, 0.2 ml was inoculated onto one Stonebrink slant (Becton Dickinson), and 0.2 ml was inoculated into 2 ml of brain heart infusion broth as a contamination control. Since July 1996, MGIT fluorescent liquid medium (Becton Dickinson) has been used instead of the BACTEC system. All culture media were read twice weekly; all cultures suspected of growth were immediately examined by acid-fast staining, the p-nitro-acetylamino-hydroxy-propiophenon (NAP) test, and a PCR-restriction fragment length polymorphism technique suited for typing of mycobacteria (28). In addition, all isolates were identified to species level by standard techniques (23).
Specimen processing for in-house PCR.
After homogenization and centrifugation, sediments were washed twice with an equal volume of Tris-EDTA buffer (10 mM Tris–HCl [pH 8.0], 0.5 mM EDTA) at 16,000 × g for 5 min. The resulting pellet was resuspended in an equal volume of Tris-EDTA buffer and subjected to 5 min of ultrasonication in an ultrasonication water bath (80 W). The sample was then boiled for 10 min, chilled on ice, and centrifuged at 16,000 × g for 10 min, and 5 μl of the supernatant was analyzed by PCR in a 50-μl reaction mixture (see below).
DNAs from biopsy samples were extracted with a DNA tissue extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions except that the sample was sonicated in the protease digestion mixture as described above. The resulting eluate was heated to 100°C for 10 min and chilled on ice, and 5 μl was used as input for a 50-μl PCR mixture.
Specimen processing for CA.
A portion (0.1 ml) of the sample as obtained after liquefaction was washed and further processed by alkaline lysis as recommended by the manufacturer with the reagents provided in the sample preparation kit. For biopsy samples, DNAs were extracted as described above; 5 μl of each resulting DNA solution was added to 50 μl of a mixture of lysis and neutralization reagents (1:1 [vol/vol]). After being mixed, 50 μl was added to 50 μl of the master mix.
In-house PCR.
An IS6110-based PCR was performed with primers developed by Kolk et al. (18). The reaction mixture was modified to prevent amplicon carryover by adding uracil-N-glycosylase (UNG) and using dUTP instead of dTTP (21). In brief, after treatment of the specimens as described above, a PCR with primers INS1 (5′-CGTGAGGGCATCGAGGTGGC-3′) and INS2 (5′-GCGTAGGCGTCGGTGACAAA-3′) was performed. The reaction mixture (final volume, 50 μl) consisted of 0.067 M Tris–HCl (pH 8.8), 0.016 M ammonium sulfate, 0.01 M 2-mercaptoethanol, 0.02% (wt/vol) gelatin, 3 mM MgCl2, 1 U of Taq polymerase (Pharmacia, Freiburg, Germany) per 50 μl, 1 U of UNG (Boehringer Mannheim, Mannheim, Germany) per 50 μl, 200 μM dGTP, 200 μM dATP, 200 μM dCTP, 600 μM dUTP, and 0.5 μM each primer (final concentrations). The temperature profile consisted of an initial 20 min at 25°C and then an initial denaturation step at 95°C for 10 min, followed by 40 cycles at 94°C for 30 s (denaturation), 65°C for 30 s (annealing), and 72°C for 1 min (extension). After completion of the amplification reaction the temperature was set to 72°C until the reaction vessels were removed from the thermal cycler and immediately chilled on ice. Five microliters of the reaction mixture was further analyzed by agarose gel electrophoresis and Southern blotting according to standard protocols. A DNA probe labelled with digoxigenin (DIG) as described in the following paragraph was used for hybridization, and a commercially available kit (DIG DNA luminescent kit; Boehringer Mannheim) was used for detection. All recommendations of the manufacturer were strictly followed, except that a ready-for-use hybridization solution (QuickHyb; Stratagene, Heidelberg, Germany) was used. The complete PCR working area was organized according to standard recommendations, including separation of the entire workspace into three distinct areas, use and frequent change of gloves, and use of disposable filter pipette tips. Inhibition testing was carried out by adding 10,000 molecules of amplicons generated with INS1 and INS2 to the reaction mixture and performing the PCR in the same run as the unspiked reaction. Inhibition testing was performed retrospectively only on culture-positive samples where the in-house PCR failed to detect M. tuberculosis DNA.
Preparation of the probe for the in-house PCR.
Approximately 104 copies of amplification product obtained with the primers INS1 and INS2 were subjected to 40 amplification cycles with primers pt3 (5′-GAACGGCTGATGACCAAACT-3′) and pt6 (5′-ACGTAGGCGAACCCTGCCCA-3′). The reaction conditions were the same as described above for primers INS1 and INS2 except that the UNG was omitted and all of the dUTP was replaced by 150 μM dTTP and 50 μM DIG-dUTP. After completion of the PCR, the reaction mixtures were stored at 4°C; 5-μl samples were used as probes without any further purification in 5 ml of hybridization buffer.
PCR by CA.
After adding 50 μl of sample to 50 μl of the master mix, PCR amplification tubes were closed and the amplification ring was transferred into the CA amplification system. One positive and one negative control per vessel ring (12 vessels) provided with the kit were included in each run. The CA automates the amplification and detection procedure for PCR. It should be noted that the CA uses a UNG carryover prevention system (21) and a coamplified internal control (50 targets per reaction). For discrimination between positive and negative results, the results were calculated by the CA software with the cutoff set to an optical density at 650 nm of 0.35.
Handling of discrepant results and definition of an adapted gold standard.
Specimens with discrepant results by any of the PCR techniques were retested by the same system; however, we used the primary results for calculating sensitivity, specificity, and positive and negative predictive values (PPV and NPV, respectively). False-negative specimens by in-house PCR were tested for inhibition as outlined above, and the isolated strains were directly subjected to in-house PCR in serial 10-fold dilutions to detect IS6110-negative strains. A discrepant positive PCR result was considered a true positive if one or more of the following criteria were met: (i) the sample originated from a patient from whom other samples were culture positive, (ii) the sample originated from a patient under successful therapy for TB (iii) the specimen was positive by both PCR methods, and (iv) the patient’s clinical history, chest roentgenograms, and actual clinical presentation were sufficiently indicative of TB for an empirical antituberculous therapy. Additionally, all culture-negative, smear-positive specimens from patients with culture-proven TB were regarded true DNA positives. A positive culture result was defined as a false positive if the specimen was negative by both PCR methods and there was no clinical or further laboratory evidence of TB for the corresponding patient. All patients’ medical records were reviewed as far as possible.
Assessment of the number of specimens necessary for diagnosis of smear-negative TB patients by CA.
For this analysis, we included from smear-negative TB patients only specimens from the individual site of infection drawn before initiation of therapy. Specimens were ordered by their time of receipt and sequentially numbered, and the number of sequential specimens that had to be tested until CA gave a positive result was determined.
RESULTS
Culture and acid-fast stain.
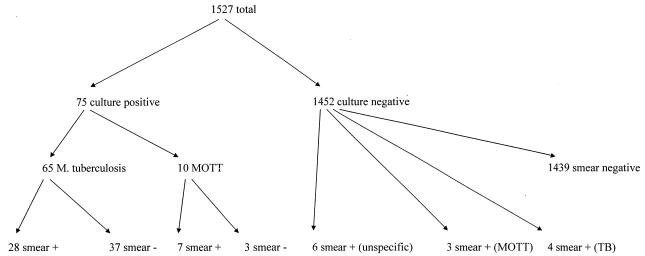
Of 1,681 specimens tested, 154 (9.16%) were inhibitory for CA and thus were not further regarded for calculation of sensitivity, specificity, and predictive values. From the remaining 1,527 specimens from 833 patients, 65 samples from 27 patients were culture positive for M. tuberculosis and 10 samples from 4 patients grew MOTT (these samples were considered culture negative with regard to growth of M. tuberculosis). Thirty-five of the culture-positive samples were also smear positive (53.8%, of which 28 samples were culture positive for M. tuberculosis and 7 samples exhibited MOTT). Thirteen samples were positive by acid-fast stain but negative by culture; of these, four were from patients under antituberculous therapy, three were from patients under therapy for MOTT infection, and six were considered unspecific positive smear results, because they were negative by both PCR systems (Fig. 1). For culture-positive specimens, the average turnaround time was 14.9 days. From the above data, levels of prevalence of 4.25% for M. tuberculosis and 0.65% for MOTT were calculated. These figures are representative for all of the specimens examined by conventional methods in our laboratory (approximately 4,500 to 5,000 samples per year).
FIG. 1.
Results of conventional testing of noninhibitory specimens.
Comparison of CA and in-house PCR with culture.
Of 65 culture-positive samples, 50 were positive by CA (sensitivity, 76.92%; NPV, 98.97%) and 59 were positive by in-house PCR (sensitivity, 90.76%; NPV, 99.58%). Fifteen culture-positive samples were missed by CA, and six culture-positive samples were missed by in-house PCR. Twenty-one culture-negative samples were positive by CA (specificity, 98.56%; PPV, 70.42%), and 35 culture-negative samples were positive by in-house PCR (specificity, 97.60%; PPV, 62.72%). Of 32 specimens with a positive smear result, CA detected 27 of 28 culture-positive samples (sensitivity, 96.42%; NPV, 96.42%) and in-house PCR detected all of them (sensitivity, 100%; NPV, 100%). If only smear-negative samples are considered, the figures are as follows: 23 of 37 culture-positive samples were correctly identified as positive by CA (sensitivity, 62.16%; NPV, 99.03%) and 31 were correctly identified as positive by in-house PCR (sensitivity, 83.78%; NPV, 99.58%). Fourteen culture-positive samples were missed by CA, and six were missed by in-house PCR. There were 20 discordantly positive results by CA (specificity, 98.62%; PPV, 53.48%) and 32 dis cordantly positive results by in-house PCR (specificity, 97.80%; PPV, 46.20%) (Table 1).
TABLE 1.
Comparison of CA and in-house PCR with culture
| Specimens | Amplification system | No. of culture-positive specimensa that were PCR:
|
No. of culture-negative specimensb that were PCR:
|
Sensitivity (%) | Specificity (%) | NPV (%) | PPV (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||||
| All (n = 1,527) | CA | 50 | 15 | 21 | 1,441 | 76.92 | 98.56 | 98.97 | 70.42 |
| In house | 59 | 6 | 35 | 1,427 | 90.76 | 97.60 | 99.58 | 62.72 | |
| Smear positive | CA | 27 | 1 | 1 | 3 | 96.42 | 75.00 | 75.00 | 96.42 |
| (n = 32)c | In house | 28 | 0 | 3 | 1 | 100.00 | 25.00 | 100.00 | 90.32 |
| Smear negatived | CA | 23 | 14 | 20 | 1,438 | 62.16 | 98.56 | 99.03 | 53.48 |
| (n = 1,495) | In house | 31 | 6 | 32 | 1,426 | 83.78 | 97.60 | 99.58 | 46.20 |
Includes only specimens growing M. tuberculosis.
Includes 10 culture-positive specimens growing MOTT.
Includes specimens from TB patients only.
Includes six unspecific positive smears and 10 smear-positive specimens from patients infected with MOTT.
Resolving discordant results.
Fifteen of 21 discordantly positive CA samples originated from patients with culture-proven TB, and 2 discordantly positive results could be confirmed by in-house PCR. The remaining four discrepantly positive CA results were obtained from patients 3, 58, 88, and 254. Patient 3 was a 4-month-old infant admitted to the hospital with an acute severe respiratory infection. Standard microbiological examination of pharyngeal swabs and nasal aspirates revealed an infection with respiratory syncythial virus. In consideration of this result, the age of the patient, and the baby’s clinical presentation, a clinically relevant infection with M. tuberculosis was considered highly improbable and the positive CA result from one gastric aspirate was considered a false positive. Patient 58 was a 31-year-old woman with notoriously frequent episodes of spontaneous staphylococcal abscesses. During the course of the study she was admitted to the hospital for a cesarean section; immediately thereafter she developed a paraspinal abscess at the fourth cervical vertebra causing progressive tetraplegia. Two independently drawn abscess aspirates revealed Staphylococcus aureus, and consequently, the positive CA result from one of the abscess aspirates was therefore considered a false positive. From patient 88 only one pus swab obtained upon lung surgery was submitted; bronchial carcinoma was suspected. The positive CA result was considered false positive, because there were no further data available supporting a history of TB for this patient. From patient 254 only one sputum sample was available; the clinical diagnosis was atypical pneumonia. This patient had a high titer of antibody against Coxiella burnetii by complement fixation (1:80), thus making rickettsial pneumonia probable. Consequently, we considered the positive CA result false. Taken together, four positive CA results could not be confirmed by either laboratory or clinical data and were considered false positives according to the criteria set forward in Materials and Methods (Table 2).
TABLE 2.
Adjustment of discrepant results to the adapted gold standard
| Test | No. of true-positive specimens for which:
|
No. of specimens for which positive PCR was not confirmableb | ||
|---|---|---|---|---|
| Positive PCR was confirmed by other culture-positive samples from same patient | Positive PCR was confirmed by other PCR | Negative PCR was confirmed by both PCRsa | ||
| CA | 15 | 2 | 4 | |
| In-house PCR | 31 | 2 | 2 | |
| Acid-fast staining | 1 | |||
Smear-negative, culture-negative specimen from a patient with culture-proven TB.
False-positive PCR.
For in-house PCR, there were 35 discordantly positive results. Thirty-one of these originated from patients with culture-proven TB, two were confirmed by CA, and two were obtained from patients 57 and 253. Patient 57 was a 56-year-old woman who was hospitalized with diffuse abdominal pain. Standard imaging techniques revealed multiple retroperitoneal abscesses. Several specimens from these abscesses obtained upon surgery were examined; histological examination suggested actinomycosis. However, no pathogen could be cultivated despite intense efforts with a broad spectrum of culture media and culturing conditions. The patient recovered during a prolonged stay at the hospital under broad antibiotic therapy with β-lactams and aminoglycosides. The final success of conventional antibiotic therapy led us to consider the positive in-house PCR result for one of the abscess specimens a false positive. Patient 253 was a 4-year-old boy with frequent episodes of pulmonary infections with common pathogens due to known multiple atelectases; there had never been any clinical suspicion of TB. According to the criteria outlined in Materials and Methods, the positive PCR result for one tracheal aspirate was considered false positive. Additionally, one smear-positive, culture-negative specimen from a patient with a previous series of smear-positive specimens growing M. tuberculosis was considered a true positive. None of the culture-positive and PCR-negative samples could be considered a false-positive culture result.
Comparison of CA and in-house PCR with the extended gold standard.
After resolution of discrepantly positive results, 101 samples had to be considered true DNA positives. Of these, the 67 recognized by CA gave a sensitivity of 66.33% and an NPV of 97.66% and the 92 samples recognized by in-house PCR gave a sensitivity of 91.08% and an NPV of 99.37%. From the remaining four unconfirmed positive CA results, a specificity of 99.71% and a PPV of 94.36% could be calculated. The corresponding figures for the in-house PCR were 99.85% for specificity and 97.87% for the PPV. For smear-positive samples, the sensitivity of CA was 87.5% and the sensitivity of the in-house PCR was 96.87%. Because there were no DNA-negative, smear-positive samples by definition, the specificities of both PCR techniques could not be calculated. If only smear-negative samples were considered, CA recognized 39 of 69 DNA-positive samples (sensitivity, 56.52%; NPV, 97.93%) and in-house PCR recognized 61 of these (sensitivity, 88.40%; NPV, 99.44%). The unconfirmed CA and in-house PCR results (four and two samples, respectively) mentioned above gave specificities of 99.71 and 99.85% and PPV of 90.69 and 96.82%, respectively (Table 3).
TABLE 3.
Comparison of CA and in-house PCR to the adapted gold standard
| Specimens | Amplification system | No. of adapted gold standard-positive specimensa that were PCR:
|
No. of adapted gold standard-negative specimensb that were PCR:
|
Sensitivity (%) | Specificity (%) | NPV (%) | PPV (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||||
| All (n = 1,527) | CA | 67 | 34 | 4 | 1,422 | 66.33 | 99.71 | 97.66 | 94.36 |
| In house | 92 | 9 | 2 | 1,424 | 91.08 | 99.85 | 99.37 | 97.87 | |
| Smear positive | CA | 28 | 4 | 0 | 0 | 87.50 | 99.37 | ||
| (n = 32)c | In house | 31 | 1 | 0 | 0 | 96.87 | 99.37 | ||
| Smear negative | CA | 39 | 30 | 4 | 1,422 | 56.52 | 99.71 | 97.93 | 90.69 |
| (n = 1,495)d | In house | 61 | 8 | 2 | 1,424 | 88.40 | 99.85 | 99.44 | 96.82 |
Specimens from patients with TB (for a definition, see Materials and Methods).
Specimens from patients without any evidence of TB.
Includes only specimens from TB patients.
Includes six unspecific positive smears and 10 smear-positive specimens from patients infected with MOTT.
Dependence of sensitivity on different types of specimens.
One aim of this study was to evaluate the sensitivity of CA when it was applied to specimen types other than sputum. The sensitivities of CA and in-house PCR compared to that of culture are shown in Table 4 and demonstrate that CA performed even better with gastric aspirates (sensitivity 70.00% with smear-negative samples) than with respiratory specimens (sensitivity, 45.45% with smear-negative samples). This effect was also observed with in-house PCR (90.00 versus 63.63%, respectively). For other specimen types the number of culture-positive samples was too low for a valid analysis; however, the data indicate that CA may also be suited for use on urine specimens (sensitivity, 100%).
TABLE 4.
Sensitivity of CA and in-house PCR compared to culture with different types of specimens
| Specimen type | No. of all samples (sensitivity [%]) that were:
|
No. of smear-negative samples (sensitivity [%]) that were:
|
||||
|---|---|---|---|---|---|---|
| Culture positive | CA positive | In-house PCR positive | Culture positive | CA positive | In-house PCR positive | |
| Sputum or BALa | 23 | 17 (73.91) | 19 (82.60) | 11 | 5 (45.45) | 7 (63.63) |
| Gastric aspirate | 34 | 27 (79.41) | 32 (94.11) | 20 | 14 (70.00) | 18 (90.00) |
| Urine | 4 | 4 (100.00) | 4 (100.00) | 4 | 4 (100.00) | 4 (100.00) |
| Other | 4 | 2 (50.00) | 4 (100.00) | 2 | 0 (0) | 2 (100.00) |
BAL, bronchoalveolar lavage.
Correlation between specimen type and inhibition of PCR.
A total of 154 specimens were inhibitory to the CA system (9.16%). Among these 154 samples were 9 culture-positive samples which would thus have been missed by CA; however, because multiple samples were examined from the same patient, no positive patient was missed because of PCR inhibition. When inhibition rates were calculated for individual specimen types (Table 5), sputum and gastric aspirates exhibited the highest inhibitory potential (10.71 and 12.71%, respectively). The inhibition rates for all other specimen types ranged between 2.54 and 7.37%. Among the six results falsely negative by in-house PCR, there were no inhibitory samples; however, it should be noted here that inhibition testing for the in-house PCR detects only complete inhibition; strains isolated from these samples exhibited a strong positive signal when they were tested directly by in-house PCR, indicating that none of these strains was IS6110 negative.
Patient-oriented analysis of the diagnostic value of in-house PCR and CA.
Among the 833 patients enrolled in this study, 46 exhibited positive results by at least one of the investigated methods. Four patients had already been diagnosed with TB at entry into this study by conventional methods and therefore cannot be considered in this paragraph. The roles of the two PCR systems for the diagnosis of the remaining 42 patients are summarized in Table 6.
TABLE 6.
Role of PCR for the primary diagnosis of TB and specification of positive smear results
| Test(s) performed for primary diagnosis | No. of specimens | Comment |
|---|---|---|
| Microscopy plus CA or in-house PCR | 4 | Smear-positive MOTT infections |
| 4 | Unspecific smear results | |
| 17 | Smear-positive TB cases | |
| CA or in-house PCR | 7 | Smear-negative TB cases (including 2 culture-negative cases) |
| In-house PCR only | 4 | Smear-negative TB cases (including 3 missed by CA) |
| In-house PCR | 2 | False-positive PCR results |
| CA | 4 | False-positive PCR results |
| Total | 42 | Cases with a positive result by any of the investigated methods |
For four smear-positive patients with MOTT infections, an infection with M. tuberculosis could be excluded by either PCR system; one of these four patients was not human immunodeficiency virus positive and presented with clinical signs typical of pulmonary TB. The negative PCR results gave the decisive clue for correct anti-MOTT therapy. For four additional patients with unspecific positive smear results, TB could also be ruled out by either PCR technique. This decision was essential for one of these patients, for whom an already initiated unnecessary antituberculous therapy could be withdrawn. All of the 17 smear-positive TB cases included in this study could be confirmed by either PCR system. Of the 11 smear-negative TB patients, 7 (including 2 culture-negative patients) were diagnosed by either PCR system before positive culture results were available, and 4 patients were diagnosed first by in-house PCR (including 3 patients whose TB was missed by CA). No case was missed by in-house PCR. CA and in-house PCR gave false-positive results for four and two patients, respectively. Because CA results were not given to our clinical colleagues, we cannot estimate retrospectively if unnecessary therapy would have been initiated. The two unconfirmed positive in-house PCR results did not induce initiation of antituberculous therapy after the corresponding cases had been thoroughly discussed with the clinical colleagues.
Number of specimens necessary for diagnosis of smear-negative TB patients by CA.
Of the 11 smear-negative TB patients enrolled in this study, 6 could be identified by CA by testing two specimens; for an additional 2 patients, testing of 7 and 8 specimens was necessary. The conditions of three remaining patients were missed by CA (for more details, see Table 7). If the level of bacterial shedding (as assessed from the rate of true-positive specimens) is correlated with the number of CA testings necessary for a diagnosis, it can be demonstrated that all patients with 100% true-positive specimens except one (patient 354) could be diagnosed by testing two specimens with CA. For patients with lower bacterial counts (as concluded from the low rate of true-positive specimens), the number of necessary CA testings was seven or more; however, there were too few patients for an extact estimation.
TABLE 7.
Assessment of the minimum number of specimens necessary for CA for smear-negative TB patients
| Patient | No. of specimensa | No. of true-positive specimensb | Rate of true-positive specimens (%) | No. of CA-positive specimens | Sequential no. of first CA-positive specimenc | Localization of infection |
|---|---|---|---|---|---|---|
| 55 | 4 | 4 | 100 | 4 | 1 | Urogenital organs |
| 543 | 5 | 5 | 100 | 2 | 1 | Lungs |
| 266 | 5 | 5 | 100 | 2 | 1 | Lungs |
| 176 | 1 | 1 | 100 | 1 | 1 | Lungs |
| 222 | 1 | 1 | 100 | 1 | 1 | Lymph node |
| 505 | 3 | 3 | 100 | 2 | 2 | Lungs |
| 287 | 10 | 1 | 10 | 1 | 7 | Lungs |
| 356 | 9 | 5 | 55.6 | 1 | 8 | Lungs |
| 354 | 3 | 3 | 100 | 0 | Lungs | |
| 536 | 6 | 1 | 16.7 | 0 | Mediastinal lymph node | |
| 587 | 3 | 1 | 33.3 | 0 | Pulmonary lymph node |
Includes only specimens drawn from the site of infection before initiation of therapy. For details, see Materials and Methods.
See also the definition of the adapted gold standard in Materials and Methods.
As determined after ordering the specimens of each patient by the time of receipt.
DISCUSSION
The available data about the CA test show an almost 100% correlation of this test with the manually performed test (17), so it is justified in our view to compare our data with research evaluating the manually performed Amplicor MTB test, which has been extensively studied during the last few years (2–4, 7, 8, 10, 11–13, 16, 20, 24, 27, 29). Although a wide range of sensitivity has been reported for the manual Amplicor MTB test, most publications reporting investigations of more than 500 specimens and providing separate data for smear-negative samples demonstrate a sensitivity below 66% (D’Amato et al. [10], 51.2%; Cartuyvels et al. [8], 46%; Carpentier et al. [7], 76%; Moore and Curry [20], 66%; Bennedsen et al. [3], 60.9%; Bergmann and Woods [4], 40%; Wobeser et al. [29], 53%). Our data show a similar sensitivity for the CA test, indicating that this test works with a diagnostic efficiency comparable to that of the manually performed Amplicor MTB test. Bodmer et al. (5) have reported an overall sensitivity of 92.6% for the new CA relative to that of culture; however, 95.6% of all culture-positive samples were also smear positive. Although the sensitivity for the smear-negative samples is not explicitly given in the report of Bodmer et al. (5), the mean optical density for these samples was 0.01 (cutoff; 0.35), strongly suggesting that most of the culture-positive and smear-negative samples were negative by CA. The figures for our in-house PCR are in agreement with those of previously published reports comparing IS6110-based PCRs with culture in a large-scale format (6, 19, 24). In summary, these data indicate a lack of sensitivity for the Roche CA in comparison to the in-house PCR. The reasons for this might be as follows. (i) CA uses a single-copy gene as a target, whereas IS6110-based PCRs use a multiple-copy gene, which increases sensitivity. Of course, IS6110-based PCRs run a certain risk of missing M. tuberculosis strains lacking this insertion element. (ii) A larger sample volume was used for in-house PCR than for CA (1.0 versus 0.1 ml); however, the final volumes of DNA extract introduced into the amplification reaction were the reverse (5 μl for in-house PCR versus 50 μl for CA). As dilution effects varied with the sediment volume obtained by the extraction protocol for the in-house PCR, the impact of the larger sample volume on the in-house PCR cannot definitely be estimated. (iii) Competition could have suppressed amplification of mycobacterial DNA if it was present at concentrations far below the concentration of the internal control. (iv) The CA uses an enzyme-linked immunosorbent assay-based detection system, which is less sensitive than Southern blotting. In summary, the issues discussed above make it reasonable that the sensitivity of the CA was found to be lower than that of the in-house PCR, mainly for reasons related to the basic concept of the test.
The relatively high rate of inhibition of CA may be caused by the use of DTT for liquefaction of samples, as this method is not approved by the manufacturer; however, this is only speculative because we have not directly compared the results of use of DTT with the results of use of the approved liquefier (N-acetyl-cysteine–NaOH). Data indicating that use of benzalkonium chloride for decontamination decreases the sensitivity of the manually performed Roche Amplicor MTB test have been published (8, 19); however, for the present study we separated liquefaction and decontamination into two steps and performed PCR directly after liquefaction so that there was no benzalkonium chloride present in the samples analyzed by PCR. Interestingly, Cartuyvels et al. (8), who reported 46% sensitivity for the manually performed Amplicor assay with smear-negative specimens, also used benzalkonium chloride for decontamination of samples before culture but not for PCR; consequently the low sensitivity of PCR could also be a result of a relatively high sensitivity of culture resulting from a better yield of viable tubercle bacilli by the Zephirol-trisodium phosphate procedure (23).
We want to point out here that the specificities of both PCR methods never raised concern for clinical practice. Although the PPV of both methods were not 100%, no unnecessary therapy was initiated in response to PCR. Additionally, 8 of 26 smear-positive patients could be rapidly identified as having non-TB conditions and inappropriate treatment could be prevented. When we considered the cases where PCR made an essential contribution to rapid confirmation of positive smear results (17 of 28 confirmed TB cases) or rapid identification of smear-negative TB cases (in-house PCR, 11 of 28 confirmed TB cases; CA, 8 of 28 confirmed TB cases), the diagnostic benefit of either method by far outweighed the problems arising from unconfirmable positive PCR results.
One aim of this study was also to evaluate the performance of the CA with nonrespiratory specimens. Although there have been publications reporting a lower sensitivity of the manually performed Amplicor MTB for gastric aspirates than for respiratory specimens (11), in this study the sensitivity of CA with gastric aspirates compared to that of culture was even higher than its sensitivity with respiratory specimens. This finding may be explained by (i) a lower viscosity of gastric aspirates than sputa, increasing the yield of bacterial cells by centrifugation; (ii) high sample volumes (above 20 ml); and (iii) a high portion of bacterial cells killed by gastric acid that thus decreases the sensitivity of culture. Although we used different DNA extraction procedures for respiratory specimens and gastric aspirates (alkaline lysis versus washing plus ultrasonication), this does not seem to have played a major role in the better sensitivity of CA with gastric aspirates, because our in-house PCR also exhibited better sensitivity with gastric aspirates. However, for in-house PCR there were no differences in DNA extraction procedures for respiratory specimens and gastric aspirates.
If the three cases completely missed by CA are considered, it can be stated that all three samples (from three patients) probably contained low bacterial counts, as could be concluded from the fact that only the liquid media became positive after a prolonged period of incubation (>4 weeks). In addition, two of the three specimens were biopsies, so there might not have been an even splitting of the samples. Six of the 8 smear-negative TB patients identified by CA were recognized by testing one or two specimens; from this we conclude (also with regard to the overall sensitivity of CA) that testing three samples per patient with CA is a minimum requirement for smear-negative patients. Of course, this will identify only patients with considerable bacterial counts; in cases of lower numbers of bacteria, excessive CA testing may be required. For these cases, we cannot give any concrete recommendation based on our data, however.
Although DNA extraction still absorbs the major portion of the manpower involved, CA offers significant advantages with regard to the amount of hands-on time required after DNA extraction, compared to that of the manually performed Amplicor MTB test or our nonautomated in-house protocol. The possibility of running the system overnight made it possible to provide results at least by the morning after specimen receipt. However, as the sensitivity with smear-negative specimens is not satisfactory in our view, we recommend the use of this test only in addition to conventional methods (i) for identification of members of the M. tuberculosis complex in smear-positive specimens and (ii) with smear-negative specimens, only if a minimum of three samples from the same patient can be tested.
ACKNOWLEDGMENTS
We thank Annette Bauer, Monika Amrhein, and Martina Schwer for excellent technical assistance. We thank Roche Diagnostics for supplying the CA reagents and the CA system.
REFERENCES
- 1.Abe C, Hirano K, Wada M, Kazumi Y, Takahashi M, Fukasawa Y, Yoshimura T, Miyagi C, Goto S. Detection of Mycobacterium tuberculosis in clinical specimens by polymerase chain reaction and Gen-Probe Amplified Mycobacterium tuberculosis Direct Test. J Clin Microbiol. 1993;31:3270–3274. doi: 10.1128/jcm.31.12.3270-3274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beavis K G, Lichty M B, Jungkind D L, Giger O. Evaluation of Amplicor PCR for direct detection of Mycobacterium tuberculosis from sputum specimens. J Clin Microbiol. 1995;33:2582–2586. doi: 10.1128/jcm.33.10.2582-2586.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennedsen J, Thomsen V O, Pfyffer G E, Funke G, Feldman K, Beneke A, Jenkins P A, Hegginbothom M, Fahr A, Hengstler M, Cleator G, Klapper P, Wilkins E G L. Utility of PCR in diagnosing pulmonary tuberculosis. J Clin Microbiol. 1996;34:1407–1411. doi: 10.1128/jcm.34.6.1407-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann J S, Woods G L. Clinical evaluation of the Roche AMPLICOR PCR Mycobacterium tuberculosis test for detection of M. tuberculosis in respiratory specimens. J Clin Microbiol. 1996;34:1083–1085. doi: 10.1128/jcm.34.5.1083-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodmer T, Gurtner A, Scholkmann M, Matter L. Evaluation of the COBAS AMPLICOR MTB system. J Clin Microbiol. 1997;35:1604–1605. doi: 10.1128/jcm.35.6.1604-1605.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brisson-Noel A, Aznar C, Chureau C, Nguyen S, Pierre C, Bartoli M, Bonete R, Pialoux G, Gicquel B, Garrigue G. Diagnosis by DNA amplification in clinical practice evaluation. Lancet. 1991;338:364–366. doi: 10.1016/0140-6736(91)90492-8. [DOI] [PubMed] [Google Scholar]
- 7.Carpentier E, Drouillard B, Dailloux M, Moinard D, Vallee E, Duthilh B, Maugein J, Bergogne-Berezin E, Carbonnelle B. Diagnosis of tuberculosis by Amplicor Mycobacterium tuberculosis test: a multicenter study. J Clin Microbiol. 1995;33:3106–3110. doi: 10.1128/jcm.33.12.3106-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartuyvels R, de Ridder C, Jonckheere S, Verbist L, van Eldere J. Prospective clinical evaluation of Amplicor Mycobacterium tuberculosis PCR test as a screening method in a low-prevalence population. J Clin Microbiol. 1996;34:2001–2003. doi: 10.1128/jcm.34.8.2001-2003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarridge J E, III, Shawar R M, Shinnick T M, Plikaytis B B. Large-scale use of polymerase chain reaction for detection of Mycobacterium tuberculosis in a routine mycobacteriology laboratory. J Clin Microbiol. 1993;31:2049–2056. doi: 10.1128/jcm.31.8.2049-2056.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Amato R P, Wallman A A, Hochstein L H, Colannino P M, Scardamaglia M, Ardila E, Ghouri M, Kim K, Patel R C, Miller A. Rapid diagnosis of pulmonary tuberculosis by using Roche AMPLICOR Mycobacterium tuberculosis PCR test. J Clin Microbiol. 1995;33:1832–1834. doi: 10.1128/jcm.33.7.1832-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devallois A, Legrand E, Rastogi N. Evaluation of Amplicor MTB test as adjunct to smears and culture for direct detection of Mycobacterium tuberculosis in the French Caribbean. J Clin Microbiol. 1996;34:1065–1068. doi: 10.1128/jcm.34.5.1065-1068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dilworth J P, Goyal M, Young D B, Shaw R J. Comparison of polymerase chain reaction for IS 6110 and Amplicor in the diagnosis of tuberculosis. Thorax. 1996;51:320–322. doi: 10.1136/thx.51.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doucet-Populaire F, Lalande V, Carpentier B, Bourgoin A, Dialloux M, Bollet C, Vachee A, Moinard D, Texier-Maugein J, Carbonnelle B, Grosset J. A blind study of the polymerase chain reaction for the detection of Mycobacterium tuberculosis DNA. Azay Mycobacteria Study Group. Tubercle Lung Dis. 1996;77:358–362. doi: 10.1016/s0962-8479(96)90102-1. [DOI] [PubMed] [Google Scholar]
- 14.Eisenach K D, Sifford M D, Cave M D, Bates J H, Crawford J T. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Respir Dis. 1991;144:1160–1163. doi: 10.1164/ajrccm/144.5.1160. [DOI] [PubMed] [Google Scholar]
- 15.Hermans P M W, Schuitema A R J, Van Soolingen D, Verstynen C P H J, Bik E M, Thole J E R, Kolk A H J, van Embden J D A. Specific detection of Mycobacterium tuberculosis complex strains by polymerase chain reaction. J Clin Microbiol. 1990;28:1204–1213. doi: 10.1128/jcm.28.6.1204-1213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang T-S, Liu Y-C, Lin H-H, Huang W-K, Cheng D-L. Comparison of the Roche AMPLICOR MYCOBACTERIUM assay and Digene SHARP Signal System with in-house PCR and culture for detection of Mycobacterium tuberculosis in respiratory specimens. J Clin Microbiol. 1996;34:3092–3096. doi: 10.1128/jcm.34.12.3092-3096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jungkind D, DiRenzo S, Beavis K G, Silverman N S. Evaluation of automated COBAS AMPLICOR PCR system for detection of several infectious agents and its impact on laboratory management. J Clin Microbiol. 1996;34:2778–2783. doi: 10.1128/jcm.34.11.2778-2783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolk A H J, Schuitema A R J, Kuijper S, Hermans P W M, van Embden J D A, Hartskeerl R A. Detection of Mycobacterium tuberculosis in clinical samples by using polymerase chain reaction and a nonradioactive detection system. J Clin Microbiol. 1992;30:2567–2575. doi: 10.1128/jcm.30.10.2567-2575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lalande V, Doucet-Populaire F, Chataigne R, Ghnassia J C, Petit J C. Comparison of two decontamination methods for direct detection by PCR of Mycobacterium tuberculosis from smear-positive specimens. Poster abstract at the Cinquièmes Journées de Mycobactériologie de Langue Francaise, Nice, France, 6 to 7 November 1996. 1996. [Google Scholar]
- 20.Moore D F, Curry J I. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by Amplicor PCR. 1995. J Clin Microbiol. 1995;33:2686–2691. doi: 10.1128/jcm.33.10.2686-2691.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persing D H, Cimino G D. Amplification product inactivation methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 105–121. [Google Scholar]
- 22.Pfyffer G E, Kissling P, Wirth R, Weber R. Direct detection of Mycobacterium tuberculosis complex in respiratory specimens by a target-amplified test system. J Clin Microbiol. 1994;32:918–923. doi: 10.1128/jcm.32.4.918-923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts G D, Koneman E W, Kim Y K. Mycobacterium. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 304–339. [Google Scholar]
- 24.Schirm J, Oostendorp L A B, Mulder J G. Comparison of Amplicor, in-house PCR, and conventional culture for detection of Mycobacterium tuberculosis in clinical samples. J Clin Microbiol. 1995;33:3221–3224. doi: 10.1128/jcm.33.12.3221-3224.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah J S, Liu J, Buxton D, Hendricks A, Robinson L, Radcliffe G, King W, Lane D, Olive D M, Klinger J D. Q-Beta replicase-amplified assay for detection of Mycobacterium tuberculosis directly from clinical specimens. J Clin Microbiol. 1995;33:1435–1441. doi: 10.1128/jcm.33.6.1435-1441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sjöbring U, Mecklenburg M, Andersen A B, Miorner H. Polymerase chain reaction for detection of Mycobacterium tuberculosis. J Clin Microbiol. 1990;28:2200–2204. doi: 10.1128/jcm.28.10.2200-2204.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stauffer F, Mutschlechner R, Hasenberger P, Stadlbauer S, Schinko H. Detection of Mycobacterium tuberculosis complex in clinical specimens by a commercial polymerase chain reaction kit. Eur J Clin Microbiol Infect Dis. 1995;14:1046–1051. doi: 10.1007/BF01590937. [DOI] [PubMed] [Google Scholar]
- 28.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wobeser W L, Krajden M, Conly J, Simpson H, Yim B, D’Costa M, Fuksa M, Hian-Cheong C, Patterson M, Phillips A, Bannatyne R, Haddad A, Brunton J L, Krajden S. Evaluation of Roche Amplicor PCR assay for Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:134–139. doi: 10.1128/jcm.34.1.134-139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]