Abstract
Nuclear receptors (NRs) are important transcriptional modulators in metazoans. Typical NRs possess a conserved DNA binding domain (DBD) and a ligand binding domain (LBD). Since we discovered a type of novel NRs each of them has two DBDs and single LBD (2DBD-NRs) more than decade ago, there has been very few studies about 2DBD-NRs. Recently, 2DBD-NRs have been only reported in Platyhelminths and Mollusca and are thought to be specific NRs to lophotrochozoan. In this study, we searched different databases and identified 2DBD-NRs in different animals from both protostomes and deuterostomes. Phylogenetic analysis shows that at least two ancient 2DBD-NR genes were present in the urbilaterian, a common ancestor of protostomes and deuterostomes. 2DBD-NRs underwent gene duplication and loss after the split of different animal phyla, most of them in a certain animal phylum are paralogues, rather than orthologues, like in other animal phyla. Amino acid sequence analysis shows that the conserved motifs in typical NRs are also present in 2DBD-NRs and they are gene specific. From our phylogenetic analysis of 2DBD-NRs and following the rule of Nomenclature System for the Nuclear Receptors, a nomenclature for 2DBD-NRs is proposed.
Introduction
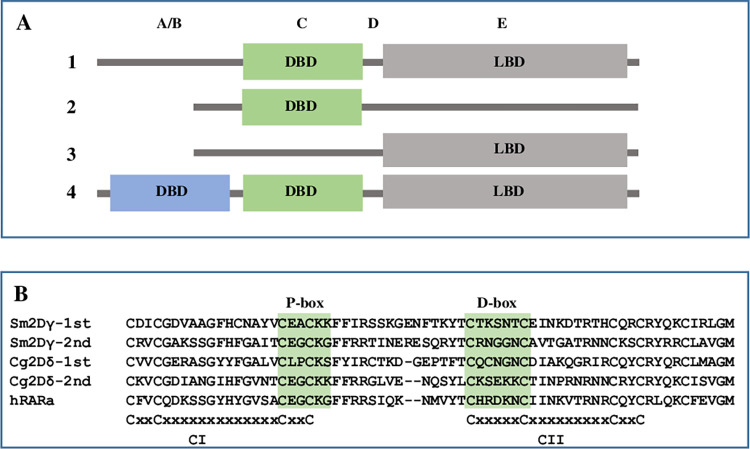
Nuclear receptors (NRs) are important transcriptional modulators in metazoans, members of the NR superfamily are characterized by a modular structure: typical NRs contain an N terminal A/B domain, a C domain (DNA binding domain, DBD), a D domain (hinge) and an E domain (ligand binding domain, LBD) (Fig 1A). NRs regulate transcription through binding to the promoter region of their target gene by the DBD and activation or repression of mRNA synthesis through co-regulators bound to the LBD [1–5]. Atypical NRs exist in some animals, NRs with DBD but no LBD are found in arthropods and nematodes [6–9], NRs missing DBD but contain a LBD are present in vertebrates [10, 11] (Fig 1A).
Fig 1. Modular structure of nuclear receptor.
A. Modular structure of nuclear receptors. 1. Typical NR with single DBD and a LBD, 2. Atypical NR with only a DBD but without LBD, 3. Atypical NR with only a LBD but without DBD, 4. Atypical NR with two DBDs and a LBD. B. DBD sequence alignment shows the two zinc fingers and the conserved P-box and D-Box. The first zinc finger (CI) with a conserved motif sequence of C-X2-CX13-C-X2-C; the second zinc finger (CII) with a conserved motif sequence of C-X5-C-X9-C-X2-C. C: cysteine residue, X followed by a number that indicates the number of amino acids between the Cs (Cys). Sm2Dγ-1st: the first DBD of Schistosoma mansoni 2DBD-NRγ (Sm2DBD-NRγ, GenBank: AAW88550), Sm2Dγ-2nd: the second DBD of Sm2DBD-NRγ, Cg2Dδ-1st: the first DBD of Crassostrea gigas 2DBD-NRδ (Cg2DBD-NRδ, GenBank: XP_011428801), Cg2Dδ-2nd: the second DBD of Cg2DBD-NRδ, hRARα: human RARα (GenBank: AAD05222.1).
In 2006, we reported our result of identification and isolation of partial cDNAs of three NRs from blood fluke Schistosoma mansoni, each of them possesses two tandem DBDs (2DBD-NRs) [12], they were then verified by the S. mansoni Genome Project [13] and the full length cDNAs were isolated [14]. This was the first time to demonstrate that NR possesses a novel modular structure: A/B-DBD-DBD-hinge-LBD organization [14] (Fig 1A). By an extensive search of whole genomic sequence (WGS) databases, we further demonstrated 2DBD-NRs were present in other animals including Platyhelminths Schmidtea mediterranea, Dugesia japonica and Mollusca Lottia gigantean. Phylogenetic analysis of DBD sequences showed that all of these 2DBD-NRs belonged to a monophyletic group and suggested that 2DBD-NRs originated from a common ancestor gene [14]. Recently, 2DBD-NRs were only identified and/or isolated in Platyhelminths [12, 14–21] and in Mollusca [22, 23]. Until now, there are fewer studies on 2DBD-NRs. Our study showed that Sm2DBD-NRα could form a homodimer but could not form a heterodimer with RXRs [14]. By searching structurally homologous sequences in the protein data bank (PDB), Alvite et al. showed that unsaturated fatty acids were preferred ligands by a Echinococcus granulosus 2DBD-NR (Eg2DBDa.1) [15]. Tharp et al. showed that S. mediterranea 2DBD-NR (nhr-1) was only detected in male and female accessory reproductive organs, and they suggested that S. mediterranea 2DBD-NR (nhr-1) was required for planarian reproductive maturation [18].
In this study, 2DBD-NRs are mined from different databases and are phylogenetically analyzed. From our phylogenetic analysis of 2DBD-NRs and following the rule of Nomenclature System for the Nuclear Receptors, a nomenclature for 2DBD-NRs is proposed.
Materials and methods
1. Data mining
2DBD-NRs were mined from The National Center for Biotechnology Information (NCBI) protein database (https://www.ncbi.nlm.nih.gov/), the Ensembl Genomes project Capitella_teleta database (https://metazoa.ensembl.org/Capitella_teleta/Tools/Blast) [24], Notospermus geniculatus database (https://marinegenomics.oist.jp/nge_v2/blast/search?project_id=52) and Phoronis australis database (https://marinegenomics.oist.jp/pau_v2/viewer/info?project_id=51) [25]. Amino acid sequences of both DBDs of Sm2DBD-NRα (AH013462) and Cg2DBD-NR (XP_019919868) were used as the query to pblast (with E-value threshold: 1e-1) against all available NCBI protein databases, and tblastn (with E-value threshold: 1e-1) against C. teleta, N. geniculatus and P. australis genome databases. Any sequence that contains a zinc finger structure of the DBD of NRs (Cys-X2-Cys-X13-Cys-X2-Cys or Cys-X5-Cys-X9-Cys-X2-Cys) was retained (Fig 1B). After careful check by eye, all amino acid sequences containing two DBDs, partial two DBDs or highly conserved sequence to 2DBD-NRs were retained.
2. Phylogenetic analysis
Phylogenetic trees of 2DBD-NRs were constructed from deduced amino acid sequences of both the first and the second DBDs. The amino acid sequences were aligned with ClustalW [26], phylogenetic analysis of the data set was carried out using Bayesian inference MrBAYES v3.1.1 [27] as in our previous study [21]. Only Bayesian inference was carried out in this study because our previous study demonstrated that Bayesian inference highly supported phylogenetic analysis of NRs more than other methods [21]. The trees were started randomly with a mix amino acid replacement model + gamma rates. Two sets of four simultaneous Markov chains were run for 5 million generations and the trees were sampled every 100 generations. The Bayesian posterior probabilities (BPPs) were calculated using a Markov chain Monte Carlo (MCMC) sampling approach implemented in MrBAYES v3.1.1 and the burn-in value was set at 12,500.
3. Amino acid sequence analysis
Amino acid sequences of every 2DBD-NRs including full length or partial sequences were aligned using ClustalW [26] and the conserved sequences were identified by the most common amino acid residue at each position. Sequence Logos were created online (https://weblogo.berkeley.edu/logo.cgi) [28].
Results and disscussion
1. Identification and phylogenetic analysis of 2DBD-NRs
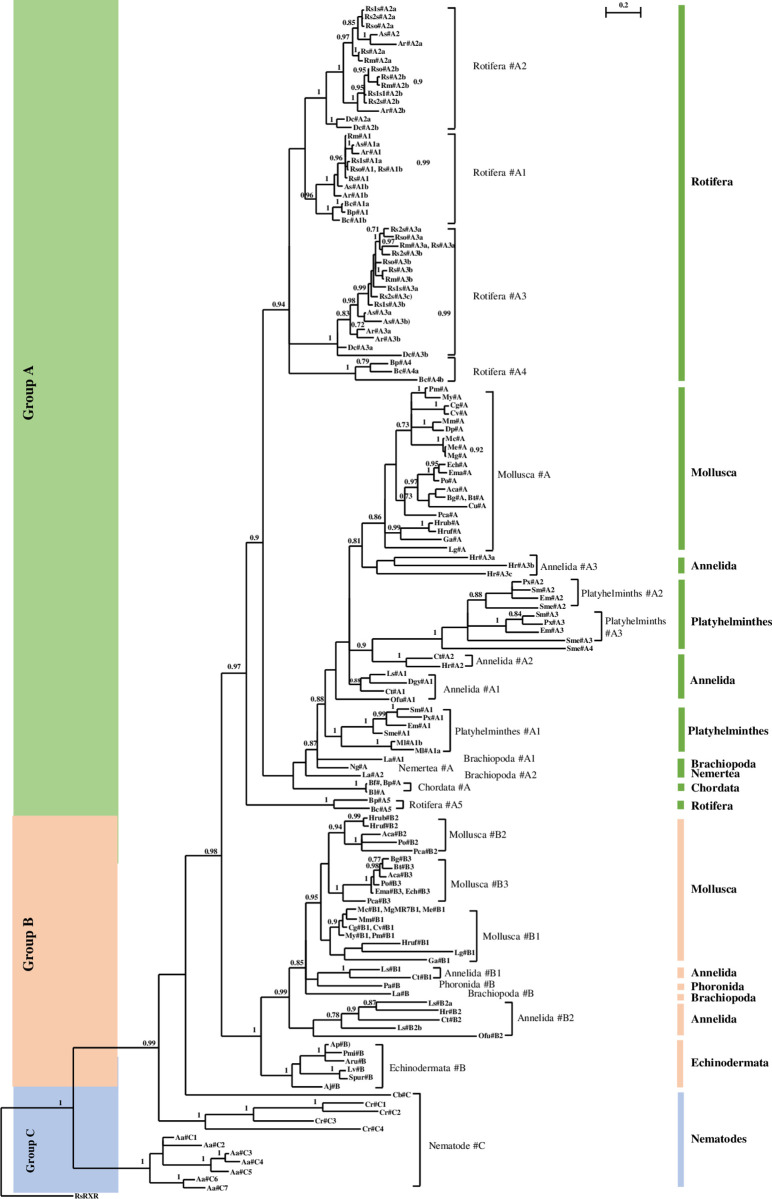
2DBD-NRs are identified in different animal species including those from protostome Spiralia (Rotifera, Brachiopoda, Mollusca, Annelida, Platyhelminths, Brachiopoda, Nemertea and Phoronida) and Ecdysozoa (Nematoda), and from deuterostome Unchordata (Echinodermata and Chordata (Cephalochordata). Previously, 2DBD-NRs are thought to be lophotrochozoan-specific [18], this study shows that 2DBD-NRs broadly exist in protostome and deuterostome species, this further indicates that 2DBD-NR gene was already present in the urbilaterian, a common ancient ancestor of protostomes and deuterostomes.
Phylogenetic analysis of identified 2DBD-NRs using amino acid sequence of both the first and the second DBD was carried out by Bayesian inference. The result shows that all of the 2DBD-NRs from protostome Spiralia and deuterostomes are clustered together forming a Spiralia/deuterostomes group (Bayesian posterior probabilities (BPPs) = 0.98), while all Ecdysozoa Nematoda 2DBD-NRs are clustered outside of the Spiralia/deuterostomes group. This result suggests that protostome Spiralia and deuterostomes share a close common ancestor 2DBD-NR gene (Fig 2). In protostome spiralia/deuterostomes group, 2DBD-NRs are clustered in two groups: 2DBD-NRA and 2DBD-NRB with BBP = 0.97 and 1, respectively (Fig 2). Both 2DBD-NRA and 2DBD-NRB groups contain members from protostome and deuterostome species, this result suggests that two ancient 2DBD-NR genes (2DBD-NRA and 2DBD-NRB) were present in a common ancestor of protostomes and deuterostomes. Phylogenetic analysis further shows that most 2DBD-NR genes underwent duplications after the split of different animal phyla. Thus, 2DBD-NR in a certain animal phylum may be a paralogue, rather than an orthologue, of that in another animal phylum (Fig 2). For example, Rotifera 2DBD-NRs are clustered into five subgroups in the 2DBD-NRA group, and another four subgroups are clustered together without members from any other Phylum.
Fig 2. Bayesian phylogenetic analysis of 2DBD-NRs.
Bayesian phylogenetic tree is constructed with the amino acid sequence of both the first and second DBD. The BPP values are shown above each branch or after the name of the NR, branches under PPs of 0.5 are shown as polytomies. Aa: Aphelenchus avenae, Aca: Aplysia californica, Aj: Anneissia japonica, Ap: Acanthaster planci, Ar: Adineta ricciae, Aru: Asterias rubens, As: Adineta steineri, Bc: Brachionus calyciflorus, Bf: Branchiostoma floridae, Bg: Biomphalaria glabrata, Bl: Branchiostoma lanceolatum, Bt: Bulinus truncatus, Bp: Brachionus plicatilis, Cb: Caenorhabditis brenneri, Cg: Crassostrea gigas, Cr: Caenorhabditis remanei, Cu: Candidula unifasciata, Cv: Crassostrea virginica, Ct: Capitella teleta, Dc: Didymodactylos carnosus, Dgy: Dimorphilus gyrociliatus, Dp: Dreissena polymorpha, Ech: Elysia chlorotica, Em: Echinococcus multilocularis, Ema: Elysia marginata, Ga: Gigantopelta aegis. Hr: Helobdella robusta, Hrub: Haliotis rubra, Hruf: Haliotis rufescens, La: Lingula anatina, Lg: Lottia gigantea, Ls: Lamellibrachia satsuma, Lv: Lytechinus variegatus, Mco: Mytilus coruscus, Me: Mytilus edulis, Mg: Mytilus galloprovincialis, Ml: Macrostomum lignano, Mm: Mercenaria mercenaria, My: Mizuhopecten yessoensis, Ofu: Owenia fusiformis, Pa: Phoronis australis, Pca: Pomacea canaliculata, Pm: Pecten maximus, Pmi: Patiria miniata, Po: Plakobranchus ocellatus, Px: Protopolystoma xenopodis, Rm: Rotaria magnacalcarata, Rs: Rotaria socialis, Rso: Rotaria sordida, Rs1s: Rotaria sp. Silwood1, Rs2s: Rotaria sp. Silwood2, Sm: Schistosoma mansoni, Sme: Schmidtea mediterranea, Spur: Strongylocentrotus purpuratus. #: 2DBD-NR. The capital letter (A, B or C) after # (2DBD-NR) indicates 2DBD-NR group, Arabic numeral after capital letter indicates individual gene, and a lowercase letter at the end of the gene indicates variant. *: All member of Rotifers 2DBD-NRA3 and 2DBD-NRA5 groups, **: All member of Echinodermata 2DBD-NRB group. GenBank Accession number of analyzed NRs see Table 6.
2. 2DBD-NRs in different animals
Members from both 2DBD-NRA and 2DBD-NRB groups are identified in Mollusca, Annelida and Brachiopoda; while 2DBD-NRA is not found in Phoronida and Echinodermata, 2DBD-NRB is not identified in Platyhelminths, Nemertea, Rotifera and Chordata. Since most 2DBD-NRs are paralogues among different animal phylum, we present our findings below by animal phyla.
1) 2DBD-NRs in Mollusca
2DBD-NRs are identified in Mollusca species from Class Bivalvia and Class Gastropoda (Table 1). Phylogenetic analysis shows that Mollusca 2DBD-NRs are clustered in both 2DBD-NRA and 2DBD-NRB groups. 2DBD-NRA group contains only one member from each analyzed Mollusca species, it suggests that one 2DBD-NRA is present in Mollusca species from Class Bivalvia and Class Gastropoda. Mollusca 2DBD-NRB group contains three subgroups (2DBD-NRB1, 2DBD-NRB2 and 2DBD-NRB3), it suggests that three 2DBD-NRBs are present in analyzed Mollusca species (Fig 2). All of the three 2DBD-NRBs (2DBD-NRB1, 2DBD-NRB2 and 2DBD-NRB3) are found in species from Gastropoda, but only one 2DBD-NRB (2DBD-NRB1) is identified in species from Class Bivalvia. This result suggests that four 2DBD-NRs (2DBD-NRA, 2DBD-NRB1, 2DBD-NRB2 and2DBD-NRB3) are present in Gastropoda and two 2DBD-NRs (2DBD-NRA and 2DBD-NRB1) are present in Bivalvia. Since Mollusca 2DBD-NRB2 and 2DBD-NRB3 subgroups share a shallower node, it suggests that 2DBD-NRB2 and 2DBD-NRB3 were formed by recent gene duplication (Fig 2 and Table 1).
Table 1. 2DBD-NRs identified in Mollusca.
| Class | Species | 2DBD-NRA | 2DBD-NRB1 | 2DBD-NRB2 | 2DBD-NRB3 | Total |
|---|---|---|---|---|---|---|
| Bivalvia | Crassostrea virginica, C. gigas | 1 | 1 | 2 | ||
| Pecten maximus | 1 | 1 | 2 | |||
| Mytilus coruscus, M. edulis, M. galloprovincialis | 1 | 1 | 2 | |||
| Dreissena polymorpha | 1 | 1 | 2 | |||
| Mercenaria mercenaria | 1 | 1 | 2 | |||
| Mizuhopecten yessoensis | 1 | 1 | 2 | |||
| Gastropoda | Batillaria attramentaria | 1 | 1 | 2 | ||
| Lottia gigantea | 1 | 1 | 2 | |||
| Gigantopelta aegis | 1 | 1 | 2 | |||
| Haliotis rufescens | 1 | 1 | 1 | 3 | ||
| Haliotis rubra | 1 | 1 | 2 | |||
| Plakobranchus ocellatus | 1 | 1 | 1 | 3 | ||
| Aplysia californica | 1 | 1 | 1 | 3 | ||
| Pomacea canaliculata | 1 | 1 | 1 | 3 | ||
| Elysia chlorotica, E. marginata, | 1 | 1 | 2 | |||
| Biomphalaria glabrata | 1 | 1 | 2 | |||
| Bulinus truncatus | 1 | 1 | 2 | |||
| Candidula unifasciata | 1 | 1 |
Mining genome database of Mollusca Crassostrea gigas, Vogeler et al. [23] identified two C. gigas 2DBD-NRs (Cg2DBDγ and Cg2DBDδ). They showed that Cg2DBDγ contained the same P-box sequences, CEACKK, as S. mansoni 2DBD-NRs in the first DBD, but Cg2DBDδ contained a different P-box sequence, CLPCKS, in the first DBD, this P-box sequence was not found in any other nuclear receptor. Amino acid sequence alignment shows that 2DBD-NRBs from Annelida, Brachiopoda and Phoronida and Mollusca possess the same P-box sequence CLPCKS in their first DBD with few members demonstrating divergent sequences (S1 File).
2) 2DBD-NRs in Brachiopoda
Three members were identified in Brachiopoda Lingula anatine. MrBayes inference shows that two of them belong to 2DBD-NRA group (La2DBD-NRA1 and La2DBD-NRA2) and one is clustered in 2DBD-NRB group (La2DBD-NRB) (Fig 2).
3) 2DBD-NRs in Phoronida
One member is identified in Phoronida Phoronis australis that belongs to 2DBD-NRB group (Pa2DBD-NRB) (Fig 2).
4) 2DBD-NRs in Nemertea
One member is identified in Nemertea Notospermus geniculatus and it is clustered in 2DBD-NRA group (Ng2DBD-NRA) (Fig 2).
5) 2DBD-NRs in Annelida
2BDB-NRs are identified in Annelida species from Class Polychaeta and Class Clitellata, they are clustered in five subgroups. Three subgroups are clustered in 2DBD-NRA group and two subgroups are in the 2DBD-NRB group. This result suggests that at least five 2DBD-NRs exist in Annelida. 2DBD-NRA2 and 2DBD-NRB2 are identified in both Class Polychaeta and Clitellata, 2DBD-NRA1 and 2DBD-NRB1 are identified only in Class Polychaeta, while 2DBD-NRA3 is only identified in Class Clitellata (Fig 2, Table 2).
Table 2. 2DBD-NRs in Annelida.
| Class | Species | 2DBD-NRA1 | 2DBD-NRA2 | 2DBD-NRA3 | 2DBD-NRB1 | 2DBD-NRB2 | Total |
|---|---|---|---|---|---|---|---|
| Polychaeta | Owenia fusiformis | 1 | 1 | 2 | |||
| Capitella teleta | 1 | 1 | 1 | 1 | 4 | ||
| Lamellibrachia satsuma | 1 | 1 | 2 | 4 | |||
| Dimorphilus gyrociliatus | 1 | 1 | |||||
| Clitellata | Helobdella robusta | 1 | 3 | 1 | 5 |
6) 2DBD-NRs in Rotifera
NRs were found genome-wide in Rotifera in different species of Brachionus, no 2DBD-NR were identified from Rotifera in a previous study [29]. In this study, 2DBD-NRs were identified in ten species from two subclasses (Bdelloidea and Monogononta) of Class Eurotatoria (Table 3) in Rotifera. These 2DBD-NRs are clustered in five subgroups in 2DBD-NRA group. This result suggests that five 2DBD-NRAs are present in Rotifera Eurotatoria Class. 2DBD-NRA1 are identified in both subclasses, 2DBD-NRA2 and 2DBD-NRA3 are only identified in subclass Bdelloidea, while 2DBD-NRA4 and 2DBD-NRA5 are only identified in subclass Monogononta. The significant character of Rotifera 2DBD-NRs is that two or more divergent copies of every 2DBD-NR gene are present, this is consistent with the previous study of the genome of Bdelloidea [30, 31] (Fig 2 and Table 3).
Table 3. 2DBD-NRs in Rotifera.
| Class | Subclass | Species | 2DBD-NRA1 | 2DBD-NRA2 | 2DBD-NRA3 | 2DBD-NRA4 | 2DBD-NRA5 | Total |
|---|---|---|---|---|---|---|---|---|
| Eurotatoria | Bdelloidea | Rotaria magnacalcarata | 1 | 2 | 2 | 5 | ||
| Adineta steineri | 2 | 1 | 2 | 5 | ||||
| Adineta ricciae | 2 | 2 | 2 | 6 | ||||
| Rotaria sp. Silwood1 | 1 | 3 | 2 | 6 | ||||
| Rotaria sp. Silwood2 | 2 | 3 | 5 | |||||
| Rotaria sordida | 1 | 2 | 2 | 5 | ||||
| Rotaria socialis | 1 | 2 | 2 | 5 | ||||
| Didymodactylos carnosus | 2 | 2 | 4 | |||||
| Monogononta | Brachionus calyciflorus | 2 | 2 | 1 | 5 | |||
| Brachionus plicatilis | 1 | 1 | 1 | 3 |
7) 2DBD-NRs in Platyhelminths
Our previous study showed that two 2DBD-NRs were present in Rhabditophora Macrostomum lignano [16, 21] and four in Schmidtea mediterranea. Three members were present in parasitic species from Platyhelminths including Class Monogenea, Cestoda and Trematoda, respectively [12, 14, 15, 17–21]. Phylogenetic analysis of Platyhelminths 2DBD-NRs in this study shows that all Platyhelminths 2DBD-NRs are clustered in 2DBD-NRA group, which suggests that 2DBD-NRB gene is missing in Platyhelminths. All parasitic 2DBD-NRs are clustered in three subgroups: 2DBD-NRA1 (Schistosoma mansoni 2DBD-NRγ orthologues), 2DBD-NRA2 (S. mansoni 2DBD-NRα orthologues) and 2DBD-NRA3 (S. mansoni 2DBD-NRβ orthologues), each subgroup contains members from species of each Class of Monogenea, Cestoda and Trematoda. For free-living flatworms, both M. lignano 2DBD-NRs are clustered in Platyhelminths 2DBD-NRA1 subgroup. For the four S. mediterranea 2DBD-NRs, one is clustered in Platyhelminths 2DBD-NRA1 subgroup, one is in Platyhelminths 2DBD-NRA2 subgroup and one is clustered in Platyhelminths 2DBD-NRA3 subgroup. The fourth S. mediterranea 2DBD-NR (Sme2DBD-NRA4) is on the base of 2DBD-NRA2 and 2DBD-NRA3 subgroups (Fig 2). This result suggests that 2DBD-NR underwent another round of duplication in a common ancestor of S. mediterranea and parasitic Platyhelminths and then one 2DBD-NR was lost in a common ancestor of parasitic Platyhelminths. This result is consistent with our previous study [21]. In this study, 2DBD-NRs are identified in more parasitic Platyhelminths including the lung flukes Paragonimus westermani, P. skrjabini miyazakii and P. heterotremus, liver fluke Fasciola gigantica, giant intestinal fluke Fasciolopsis buski and tapeworm Sparganum proliferum. Sequence alignment shows that all of them are highly conserved with known Platyhelminths 2DBD-NRs.
8) 2DBD-NRs in Echinodermata
One member was identified in each analyzed species of Echinodermata including those from Class Asteroidea, Class Echinoidea and Class Crinoidea. MrBayes inference analysis shows that all Echinodermata 2DBD-NRs form a monophyletic subgroup in 2DBD-NRB group (Fig 2 and Table 4). Previously, 33 NRs were identified in the genome database of Strongylocentrotus purpuratus, but until this report no 2DBD-NR was reported [32].
Table 4. 2DBD-NRs in deuterostome.
| Phyla | Class | Species | 2DBD-NRA | 2DBD-NRB |
|---|---|---|---|---|
| Echinodermata | Asteroidea | Acanthaster planci | 1 | |
| Asterias rubens | 1 | |||
| Patiria miniata | 1 | |||
| Echinoidea | Lytechinus variegatus | 1 | ||
| Strongylocentrotus purpuratus | 1 | |||
| Crinoidea | Anneissia japonica | 1 | ||
| Chordata | Leptocardii | Branchiostoma belcheri | 1 | |
| Branchiostoma floridae | 1 | |||
| Branchiostoma lanceolatum | 1 |
9) 2DBD-NRs in Chordata
One member is identified in each species of analyzed Chordata Amphioxi including Branchiostoma belcheri, B. floridae and B. lanceolatum. MrBayes inference analysis shows that all of them form a monophyletic subgroup in 2DBD-NRA group (Fig 2 and Table 4). Though 33 NRs were identified in B. floridae previously, but until this report no 2DBD-NR was reported in Amphioxi [33].
10) 2DBD-NRs in Nematoda
2DBD-NRs are present in nematode species Aphelenchus avenae, Caenorhabditis brenneri and Caenorhabditis remanei. Four 2DBD-NRs are found in C. remanei, one in C. brenneri and seven in A. avenae. Phylogenetic analysis shows that all these 2DBD-NRs are clustered outside of Spiralia/deuterostomes 2DBD-NR group (Fig 2). Sequence alignment of DBDs shows that the P-box sequence is highly divergent in Nematoda 2DBD-NRs (S1 File). These results suggests that Nematode 2DBD-NRs underwent extensive divergence as found in other nematode NRs [34–37].
See Table 5 for a summary of the numbers of 2DBD-NRs identified in different animal species.
Table 5. Numbers of 2DBD-NRs identified in different animals.
| Animal | Class | 2DBD-NRA | 2DBD-NRB | Total |
|---|---|---|---|---|
| Rotifera | Eurotatoria | 5 | 5 | |
| Mollusca | Bivalvia | 1 | 1 | 2 |
| Gastropoda | 1 | 3 | 4 | |
| Annelida | Clitellata | 3 | 1 | 4 |
| Polychaeta | 2 | 3 | 5 | |
| Platyhelminthes | Rhabditophora | 4 | 4 | |
| Monogenea | 3 | 3 | ||
| Cestoda | 3 | 3 | ||
| Trematoda | 3 | 3 | ||
| Brachiopoda | Lingulata | 2 | 1 | 3 |
| Phoronida | 1 | 1 | ||
| Nemertea | Pilidiophora | 1 | 1 | |
| Echinodermata | Asteroidea | 1 | 1 | |
| Echinoidea | 1 | 1 | ||
| Crinoidea | 1 | 1 | ||
| Chordata | Leptocardii | 1 | 1 |
3. Amino acid sequence analysis of 2DBD-NRs
1) AB domain
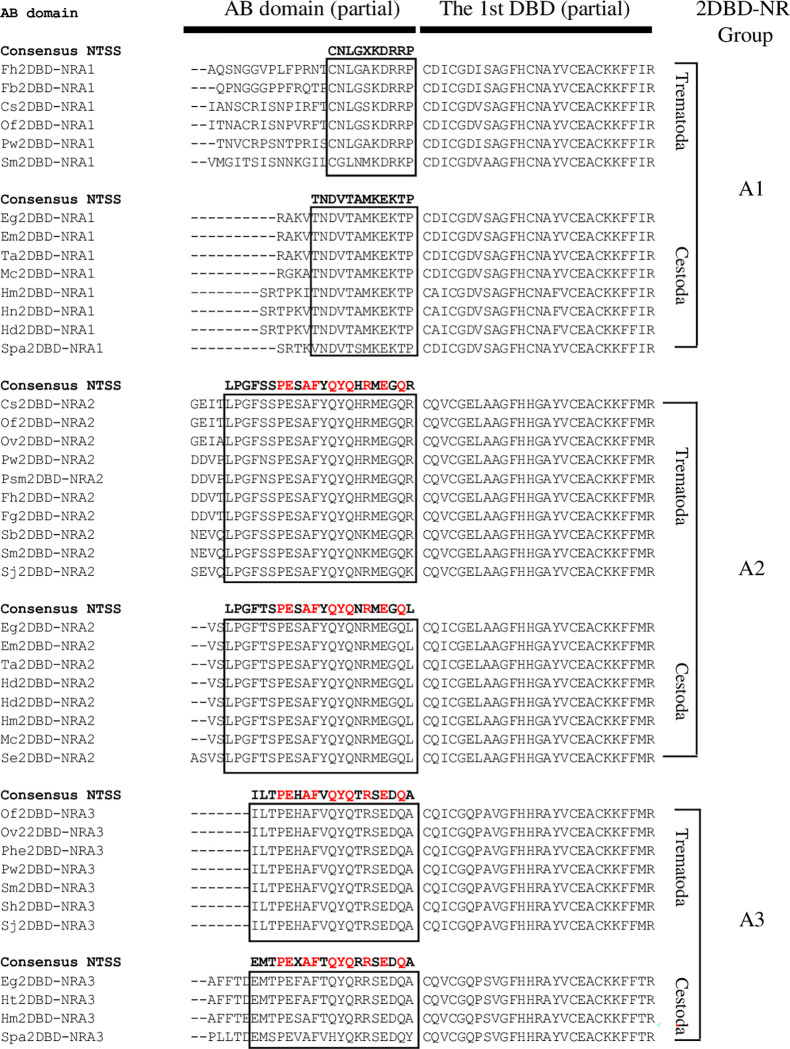
A highly conserved ’N-terminal signature sequence’ (NTSS) [38] is found at the 3’end of the A/B domain of parasitic Platyhelminths 2DBD-NRs. A NTSS of CNLGXKDRRP is present in Platyhelminth Trematode 2DBD-NRA1s, and a NTSS of TNDVTAMKEKTP is present in Cestoda 2DBD-NRA1s. A NTSS of (S/T)PEXAFXQYQXR(M/S)EGQX represents both Platyhelminths 2DBD-NRA2s and 2DBD-NRA3s (Fig 3). The fact that Platyhelminth 2DBD-NRA2 and 2DBD-NRA3 members share a conserved NTSS further supports our phylogenetic analysis.
Fig 3. Amino acid sequence alignment shows the conserved NTSS in parasitic Platyhelminths 2DBD-NRs.
Cs: Clonorchis sinensis, Eg: Echinococcus granulosus, Em: Echinococcus multilocularis, Fb: Fasciolopsis buski, Fh: Fasciola hepatica, Fg: Fasciola gigantica, Hd: Hymenolepis diminuta, Hm: Hymenolepis microstoma, Hn: Hymenolepis nana, Ht: Hydatigera taeniaeformis, Of: Opisthorchis felineus, Ov: Opisthorchis viverrini, Phe: Paragonimus heterotremus, Mc: Mesocestoides corti, Pw: Paragonimus westermani, Psm: Paragonimus skrjabini miyazakii, Se: Spirometra erinaceieuropaei, Sb: Schistosoma bovis, Sh: Schistosoma haematobium, Sj: Schistosoma japonicum, Spa: Sparganum proliferum, Sm: Schistosoma mansoni, Ta: Taenia asiatica. The red letters indicate the amino acid residues that are conserved in parasitic Platyhelminths 2DBD-NRA2 and 2DBD-NRA3, it suggests that parasitic Platyhelminths 2DBD-NRA2 and 2DBD-NRA3 underwent recent duplication.
2) DBDs
DBD is the most conserved region in NRs, this region contains two highly conserved C4 type zinc fingers with a module of C-X2-C-X13-C-X2-C for the first Zinc finger (CI) and C-X5-C-X9-C-X2-C for the second Zinc finger (CII), where C represents cysteine, X represents any variable and the next number indicates the number of amino acids between the cysteines [39]. Amino acid alignment of all identified 2DBD-NRs in this study shows that both DBDs of most of the 2DBD-NRs contain the conserved zinc finger modules as above with very little exception (Fig 1B and S2 File). Recently, we reported a novel zinc finger CHC2 motif (C-X6-C-X9-H-X2-C) in DBD of parasitic Platyhelminth NRs [21], this motif is not identified in any 2DBD-NRs.
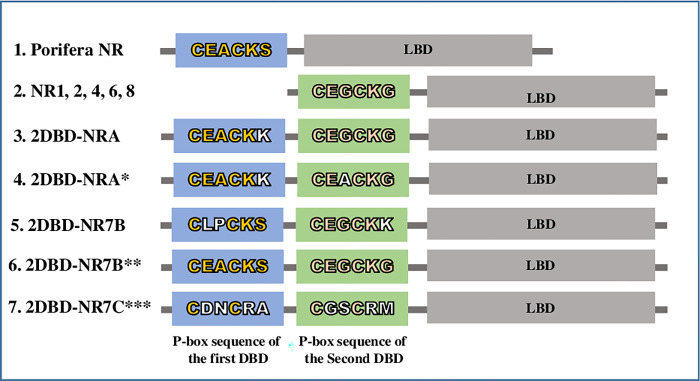
In 1989, [40] identified two conserved motifs in DBD of NRs. One motif was defined as a proximal box (P-box) that follows the third Cysteine of zinc finger I (CI) including five amino acids. The other motif was a distal box (D-box) which was located between the fifth and sixth Cysteine in zinc finger II (CII) of DBD (Fig 1B). The P-box is critical for identifying the primary nucleotide sequence of the half-sites and the D-box is important for protein dimerization. Previously, we demonstrated that 2DBD-NRs possess a P-box sequence of EACKK in the first DBD, which is similar but different from the P-box of ERRs (EACKA). The P-box sequence of the second DBD (EGCKG) followed by the amino acid sequence FFRR (EGCKGFFRR) is identical to that of most members in NR subfamily 1 (NR1) [12, 14]. Recently, Vogeler et al. [23] showed that a different P-box sequence (LPCKS) was present in the first DBD of a 2DBD-NR in Mollusca C. gigas [23]. In this study, various different P-box sequences are found in 2DBD-NRs. The P-box sequence in the first DBD and the second DBD of 2DBD-NRs (P-P module) was found to be different among 2DBD-NRAs, 2DBD-NRBs and 2DBD-NRCs (Fig 4 and S1 and S2 Files). The P-P module of 2DBD-NRA is EACKK-EGCKG (Fig 4(3) and 4(4)) with a few divergence sequences in the second DBD (S1 and S2 Files). Two types of P-P modules are found in 2DBD-NRB, one is LPCKS-EGCKK (Fig 4(5), and it is present in 2DBD-NRBs of Mollusca, Annelida, Brachiopoda and Phoronida, with a divergent sequence in the P-box of the first DBD in Annelida and Brachiopoda 2DBD-NRB (S1 and S2 Files). The P-box sequence of LPCKS was first identified in Cg2DBDδ [23]. The P-P module EACKS-EGCKG is only found in Echinodermata 2DBD-NRBs (Fig 4(6)). Interestingly, the P-box sequence of the first DBD of Echinodermata 2DBD-NRBs (EACKS) is identical to that of basal metazoans Porifera NRs (SdRXR and RsNR1) (Fig 4(1)) and the P-box sequence of the second DBD is identical to that of most members of 2DBD-NRA, and some members from NR subfamily 1, 2, 4, 6 and 8 (Fig 4(2)). The P-P module of 2DBD-NRCs is highly variable, not only from 2DBD-NRA and 2DBD-NRB, but also variable from the members in 2DBD-NRC group (Fig 4(7), S1 and S2 Files). The different P-P module among 2DBD-NRs suggests that the mechanism of DNA binding may be different in 2DBD-NRAs, 2DBD-NRBs and 2DBD-NRCs and it suggests that 2DBD-NRAs, 2DBD-NRBs or 2DBD-NRCs recognize and regulate different kinds of target genes.
Fig 4. P-P module of 2DBD-NRs (the P-box sequence in the first DBD and the second DBD of 2DBD-NRs).
1. Shows the P-box sequence of a Porifera NRs (Suberites domuncula RXR, SdRXR, GenBank: CAD57002.1) that is the same as the first DBD of Echinodermata 2DBD-NRB. 2. Shows the P-box sequence of NRs in subfamily 1, 2 4, 6 and 8 that is the same as the P-box of the second DBD of 2DBD-BRA. 3. P-P module of 2DBD-NRA (CEACKK-CEGCKG) which is found in most of 2DBD-NRA except members of Rotifers NR7A3 and NR7A5 groups. 4. P-P module of 2DBD-NRA (CEACKK-CEACKG) which is only found in the members of Rotifers NR7A3 and NR7A5 groups. 5. P-P module of 2DBD-NRB (CLPCKS-CEGCKK) which is found in most of 2DBD-NRB except members of Echinodermata 2DBD-NRB. 6. P-P module of 2DBD-NRB (CEACKS-CEGCKG) which is only found in Echinodermata 2DBD-NRB. 7. P-P module of the member of 2DBD-NRC group is highly variable. An example (Cr2DBD-NRC1) is shown in the picture. *: P-P module sequences only found in the members of Rotifers 2DBD-NRA3 and 2DBD-NRA5. **: P-P module sequences only found in the members of Echinodermata 2DBD-NRB. ***: P-P module of the member of 2DBD-NRC group is highly variable, here is only an example (Cr2DBD-NRC1). Letters in yellow color in the P-box sequence indicate the conserved amino acids and the letters in white color in the P-box sequence indicate the divergent amino acids.
The D-box of NR is located between the fifth and sixth Cysteine in zinc finger II (CII) of DBD with five amino acid residue between the two Cysteines (C-X5-C) [40]. Amino acid sequence alignment of 2DBD-NRs shows that most of 2DBD-NRs have a conserved D box with C-X5-C in both DBDs (S2 File).
3) Amino acid sequence between the first and second DBDs
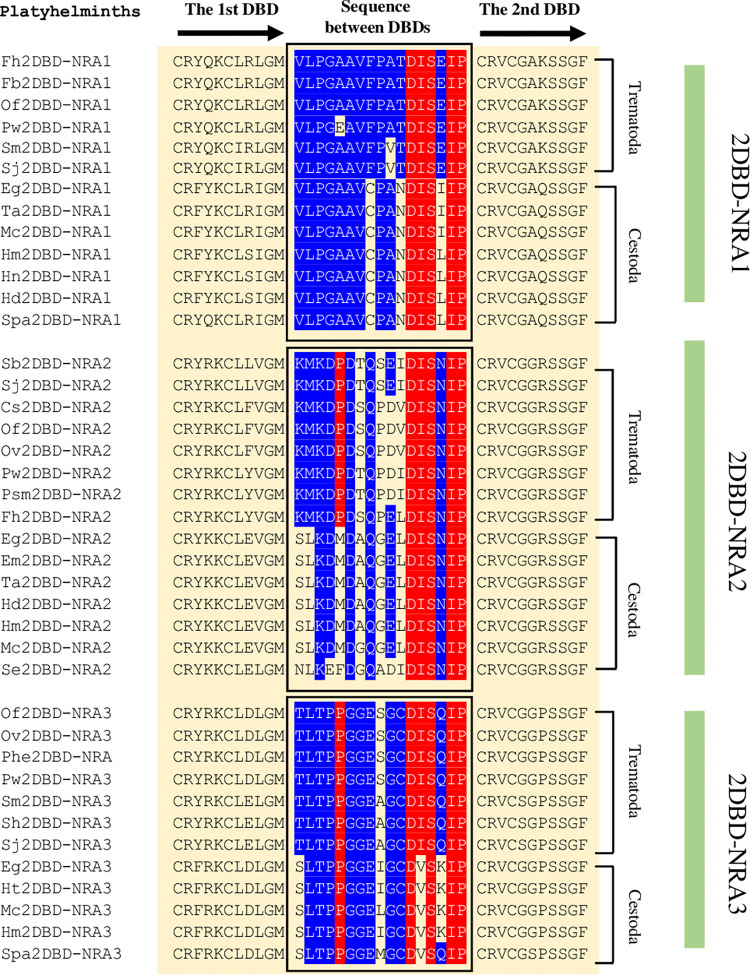
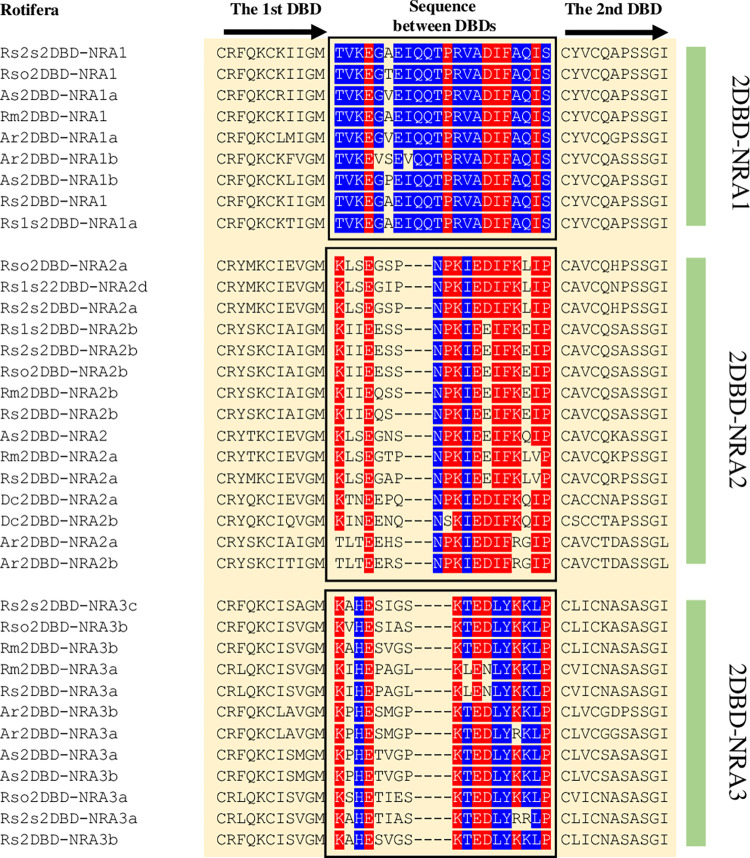
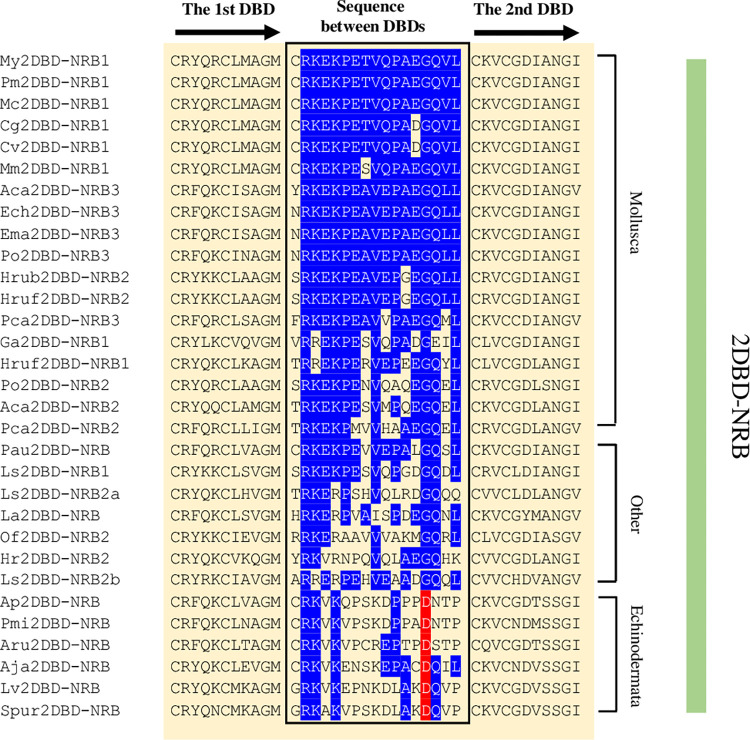
Amino acid sequence alignment shows that most members of the 2DBD-NRA and 2DBD-NRB possess 17–22 amino acids between two DBDs. Highly conserved sequences in this region were found in parasitic Platyhelminths 2DBD-NRAs (Fig 5), Rotifera 2DBD-NRAs (Fig 6) and in 2DBD-NRBs (Fig 7), respectively.
Fig 5. Amino acid sequence between the first and second DBDs in Platyhelminth 2DBD-NRs.
Cs: Clonorchis sinensis, Eg: Echinococcus granulosus, Em: Echinococcus multilocularis, Fb: Fasciolopsis buski, Fh: Fasciola hepatica, Hd: Hymenolepis diminuta, Hm: Hymenolepis microstoma, Hn: Hymenolepis nana, Ht: Hydatigera taeniaeformis, Mc: Mesocestoides corti, Of: Opisthorchis felineus, Ov: Opisthorchis viverrini, Pw: Paragonimus westermani, Psm: Paragonimus skrjabini miyazakii, Sj: Schistosoma japonicum, Se: Spirometra erinaceieuropaei, Sh: Schistosoma haematobium, Sm: Schistosoma mansoni, Spa: Sparganum proliferum, Ta: Taenia asiatica. Blue highlighted letters indicate the conserved amino acid residues in each subgroup and red highlighted letters indicate the conserved amino acid residues among subgroups.
Fig 6. Amino acid sequence between the first and second DBDs in Rotifera 2DBD-NRs.
Ar: Adineta ricciae, As: Adineta steineri, Rotaria magnacalcarata, Dc: Didymodactylos carnosus, Rm: Rotaria magnacalcarata, Rs: Rotaria socialis, Rs1s: Rotaria sp. Silwood1, Rs2s: Rotaria sp. Silwood2, Rso: Rotaria sordida, Rs1s: Rotaria sp. Silwood1. Blue highlighted letters indicate the conserved amino acid residues in each subgroup and red highlighted letters indicate the conserved amino acid residues among subgroups.
Fig 7. Amino acid sequence between the first and second DBDs in 2DBD-NRBs.
Aca: Aplysia californica, Aj: Anneissia japonica, Ap: Acanthaster planci, Aru: Asterias rubens, Cg: Crassostrea gigas, Cv: Crassostrea virginica, Ech: Elysia chlorotica, Ema: Elysia marginata, Hr: Helobdella robusta, Hrub: Haliotis rubra, Hruf: Haliotis rufescens, La: Lingula anatina, Ls: Lamellibrachia satsuma, Lv: Lytechinus variegatus, Mco: Mytilus coruscus, Mm: Mercenaria mercenaria, My: Mizuhopecten yessoensis, Ofu: Owenia fusiformis, Pa: Phoronis australis, Pca: Pomacea canaliculata, Pm: Pecten maximus, Pmi: Patiria miniata, Po: Plakobranchus ocellatus, Spur: Strongylocentrotus purpuratus. Blue highlighted letters indicate the conserved amino acid residues in members of 2DBD-NRB group and red highlighted letters indicate the conserved amino acid residues in Echinodermata 2DBD-NRBs.
4) The C-terminal Extension (CTE)
The C-terminal Extension (CTE) of DBD is important for DNA sequence recognition and binding. In 1992, two boxes in CTE of human NGFI-B (hNR4A1) were identified [41]. One box, termed T-Box, consisted of 12 amino acids that determined binding to tandem repeats of the half-site. The adjacent C-terminal seven amino acids, termed A-box, was required for recognition of the DNA binding element [41]. In 1998, a subunit, termed Grip Box (G-box)in the CTE of human RevErbA-α was identified [42]. The G-box formed a significant minor groove in the DNA binding surface [42]. They further showed that the G-box sequence was conserved in different orphan receptors despite the length of the pre-Grip sequences was different. The consensus G-box sequence they identified as RXGRZP (where X is a F, R, or G and Z usually contains a hydrophobic side chain). In this study, amino sequence alignment showed that all 2DBD-NRs contain a conserved G-box with the consensus sequence of RXGRQ(P/S) in 2DBD-NRAs, KXGR(P/H) in 2DBD-NRBs and RDRRGP in Nematoda A. avenae 2DBD-NRCs (Fig 8). The different conserved G-box sequence among 2DBD-NRA, 2DBD-NRB and 2DBD-NRC may represent the different DNA binding ability of these NRs.
Fig 8. Amino acid sequence alignment of the C-terminal Extension (CTE) of 2DBD-NRs.
Amino acid sequence alignment shows conserved G-box sequence in 2DBD-NRs. The deep green highlighted letters indicate 2DBD-NR gene specific amino acid residues in CTE, the blue highlighted letters indicate conserved amino acid residues in the G-box of 2DBD-NRs and other NRs, the red highlighted letters indicate conserved amino acid residues amino acid (H) after the G-box. They are ancient signal amino acids among 2DBD-NRAs.
Amino acid sequence alignment shows that the fifth amino acid (H) after the G-box are conserved in most 2DBD-NRAs including members from Nemertea, Annelida, Brachiopoda, Mollusca and Chordata, members from Rotifer 2DBD-NRA1 and 2DBD-NRA2 subgroup and members from Platyhelminthes 2DBD-NRA1 subgroup. However, the members from Rotifer 2DBD-NRA3 and 2DBD-NRA4 subgroups and the members from Platyhelminth 2DBD-NRA2 and 2DBD-NRA3 subgroups possess a different amino acid at this position. This suggests that the fifth amino acid residue (H) after the G-box are ancient signal amino acids among 2DBD-NRAs. The G-box sequence is also conserved in human NGFI-B that is localized in the 5’ end of the A-box of NGFI-B (Fig 8), the pre-Grip sequences represents the T-box of NGFI-B which contains 12 residues [41]. The pre-Grip sequences in 2DBD-NR only contains five amino acids in 2DBD-NRA and 2DBD-NRBs, and eight amino acids in 2DBD-NRC, this data suggests that no conserved T-box is present in 2DBD-NRs.
5) LBD
LBD of NRs contain 12 helices and has two conserved regions, one region is known as “signature”, it contains 34 amino acid residues between the C terminus of H3 and the middle of H5. Within this region, a motif containing 20 amino acid residues was defined as an LBD specific signature (Ti) for the NR superfamily, it’s consensus sequence is ((F,W,Y)(A,S,I) (K,R,E,G)xxxx(F,L)xx(L,V,I)xxx(D,S)(Q,K)xx(L,V)(L,I,F)) [43]. In the C-terminus of LBD, there is an inducible transcription activation function TAF-2 (AF2) [44, 45]. The amino sequence of AF-2 region was conserved among many nuclear hormone receptors, the common consensus AF2-AD core structure is ФФxEФФ, where Ф denotes a hydrophobic residue [46–48]. Amino acid sequence alignment of 2DBD-NRs shows that Ti is conserved in 2DBD-NR (Fig 9 and S3 File). AF2-AD core sequence is identified in most of 2DBD-NRs, but it is missing in the nematode Aphelenchus avenae 2DBD-NRs. Sequence alignment shows that AF2-AD core sequence is conserved among 2DBD-NRs with ФФx(E,Q,R)ФФ in 2DBD-NRAs, ФФx(E,K)Фh (where h denotes a Hydrophilic residue) in 2DBD-NRBs and xxФФФФ in Caenorhabditis brenneri 2DBD-NRCs (Fig 9 and S4 File).
Fig 9. Conserved amino acid sequence of LBD specific signature (Ti) and activation function TAF-2 (AF2) in 2DBD-NRs.
Sequence logo shows that both Ti and AF2 are conserved in 2DBD-NRs, and the pattern is similar to that of NR subfamily 1 (NR1). For the sequences used for generation of the logo see S3 and S4 Files. Blue letter indicates a hydrophilic residue (RKDENQ), green letter indicates a neutral residue (SGHTAP) and red letter indicates a Hydrophobic residue (YVMCLFIW).
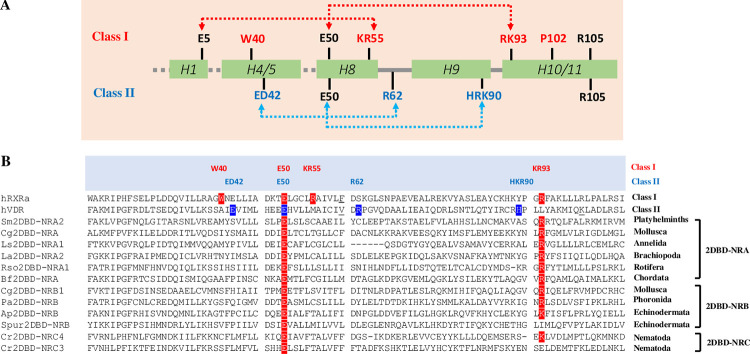
The LBD of NRs are required for NR homodimerization and/or heterodimerization ([43, 49]). In 2004, Brelivet et. al. proposed two functional classes of NRs (class I and class II) based on the conservation of amino acids in the region of LBD [50], they showed that LBD of NRs in class I could form homodimers, while LBD of members in Class II could form heterodimers with RXR. The main features of class I NRs was that their LBD possess conserved amino acids E5, E50, KR55 and RK93, these conserved amino acids formed two salt bridges, the first salt bridge was formed by amino acids E5 and KR55, and the second bridge was formed by E50 and KR93. While class II NRs possess conserved amino acids ED42, E50, R62 and HRK90, the first salt bridge was formed by amino acid ED42 and R62, and the second bridge was formed by E50 and HKR90. They further showed that RK93 was strictly conserved in class I NRs, R62 was strictly conserved in class II NRs, and E50 was conserved in both classes of NRs (Fig 10A). Our previous study showed that LBD of Sm2DBD-NRα (Sm2DBD-NRA2) could form homodimers but not heterodimers with RXR ([14]),a similar result was reported for Echinococcus granulosus 2DBD-NRα1 (Eg2DBDα1, Eg2DBD-NRA2) [51]. Paper [51] was published while the present paper was under review. The fact that Sm2DBD-NRα or Eg2DBDα1 could form homodimers but not heterodimers with RXRs suggested that 2DBD-NRs belongs to NR class I. Analysis of the amino acid sequence of all available 2DBD-NRs showed that all 2DBD-NRs possessed a conserved amino acid E50 and most of them possessed a strictly conserved amino acid of class I KR93. No conserved amino acids for class II NR were identified in 2DBD-NRs (Fig 10B and S5 File). This result suggested that 2DBD-NRs belong to class I NRs. Although 2DBD-NRs could form homodimers, the conserved amino acid KR55 was missing in 2DBD-NRs. KR55 was known to form the first salt bridge with E5. The absence of KR55 in 2DBD-NRs suggests that the first class I salt bridge is absent from 2DBD-NRs. The first salt bridge of class I NRs was missing in Branchiostoma lanceolatum NR7 (of note, NR7 was recently used to describe another group of NRs other than 2DBD-NRs, see next section) [52]. Experiments showed that 2DBD-NRs could form homodimers in a DNAindependent way [14, 51], which suggests that other factors may be involved in regulation of homodimerization of 2DBD-NRs. The formation of homodimers of 2DBD-NRs may result in four DBDs binding to a specific DNA element, the short region between two DBDs (17–22 amino acids in this region) may constraint the flexibility of their DBDs and then limit them to bind DNA in a certain region of the target gene. It is not clear whether 2DBD-NRs possessed the first salt bridge and lost it, or other class I NRs gained this salt bridge after their common ancestor split from the ancient 2DBD-NR gene.
Fig 10. Identification of amino acids in LBD of 2DBD-NRs that are conserved in class I NRs.
A. Secondary structure diagram showing NR class-specific features [50]. The red letters indicate conserved amino acids in class I NRs, the blue letters indicate conserved amino acids in class II NRs, and the black letters indicate no class-specific amino acids. Arrows indicate the salt bridges in the 3D structures. B. Sequence alignment of LBD of 2DBD-NRs shows the conserved amino acids according to class I and class II NRs described in [50]. The red or red highlighted letters indicate conserved amino acids found in class I NRs, the blue or blue highlighted letters indicate conserved amino acids found in class II NRs. Blocks indicate the variable inserts are deleted according to [50].
4. A proposed nomenclature for 2DBD-NRs
A nomenclature for the NR superfamily was proposed, typical NRs (with both DBD and LBD) include 6 subfamilies (I-VI), and the NRs that are missing either DBD or LBD are placed in subfamily 0, irrespective of their evolutionary origin [53]. Yet the 2DBD-NRs are not included in this classification system. According to our phylogenetic analysis of 2DBD-NRs, we propose a nomenclature for 2DBD-NRs following the rule of Nomenclature System for Nuclear Receptors [53]:
1) NR subfamilies are designated by Arabic numerals (NR7, 2DBD-NR)
Previously, we suggested placing 2DBD-NRs in a new NR subfamily (NR subfamily 7, NR7) [12, 14] and this was accepted by some other studies [16, 54, 55]. However, NR7 was recently used to describe NRs [52] previously known as NR8 members [55] and amphioxus NR1Hs [33, 56]. To avoid confusion of 2DBD-NRs with other NRs named as NR7, we propose to designate2DBD-NRs as a subfamily instead of using NR7.
2) Groups are designated by capital letters (2DBD-NRA, 2DBD-NRB and 2DBD-NRC)
Phylogenetic analysis shows that 2DBD-NRs contain three groups: 2DBD-NRA, 2DBD-NRB and 2DBD-NRC (Fig 2). Both 2DBD-NRA and 2DBD-NRB groups contain members from protostome Spiralia and deuterostomes, while 2DBD-NRC only contains members from Nematoda.
3) Individual genes are designated by Arabic numerals
For example, three 2DBD-NRs are identified in Mollusca Aplysia californica. One of them clustered in 2DBD-NRA group and is defined as A. californica 2DBD-NRA gene (Ac2DBD-NRA); two of A. californica 2DBD-NRs are clustered in 2DBD-NRB group, one is in Mollusca 2DBD-NRB2 subgroup 2, and it is defined as A. californica 2DBD-NRB2 (Ac2DBD-NRB2). The other one is clustered in Mollusca mono-phylogenetic 3 subgroup and it is defined as A. californica 2DBD-NRB3 (Ac2DBD-NRB3) (Fig 2).
4) A lowercase letter is added at the end of the gene to designate variants
In Rotifera, 2DBD-NR genes underwent rounds of gene duplications and gave birth to different gene variations, thus, a lowercase letter is added at the end of the gene to designate variants. For example, two variations of Brachionus calyciflorus 2DBD-NRs are clustered in Rotifera 2DBD-NRA1 subgroup, thus, they are defined as B. calyciflorus 2DBD-NRA1a (Bc2DBD-NRA1a) and B. calyciflorus 2DBD-NRA1b (2DBD-NRA1b), respectively. All identified 2DBD-NRs, their name in this nomenclature and their GenBank Accession number are listed in Table 6.
Table 6. A proposed nomenclature for 2DBD-NRs.
| Group | Animal | Class | Species | NR/Gene | Trivial Names | Accession Number |
|---|---|---|---|---|---|---|
| 7A | Rotifera | Eurotatoria | Rotaria magnacalcarata (Rm) | Rm2DBD-NRA1 | CAF5052617 | |
| Rm2DBD-NRA2a | CAF1635244 | |||||
| Rm2DBD-NRA2b | CAF2064338 | |||||
| Rm2DBD-NRA3a | CAF1325921 | |||||
| Rm2DBD-NRA3b | CAF2083731 | |||||
| Adineta steineri (As) | As2DBD-NRA1a | CAF1578566 | ||||
| As2DBD-NRA1b | CAF1211849 | |||||
| As2DBD-NRA2 | CAF0905228 | |||||
| As2DBD-NRA3a | CAF3771316 | |||||
| As2DBD-NRA3b | CAF1331931 | |||||
| A. ricciae (Ar) | Ar2DBD-NRA1a | CAF1025086 | ||||
| Ar2DBD-NRA1b | CAF1622127 | |||||
| Ar2DBD-NRA2a | CAF0828206 | |||||
| Ar2DBD-NRA2b | CAF0844309 | |||||
| Ar2DBD-NRA3a | CAF1274457 | |||||
| Ar2DBD-NRA3b | CAF1396991 | |||||
| Rotaria sp. Silwood1 (Rs1s) | Rs1s2DBD-NRA1a | CAF3409410 | ||||
| Rs1s2DBD-NRA2d | CAF0869039 | |||||
| Rs1s2DBD-NRA2a | CAF0869039 | |||||
| Rs1s2DBD-NRA2b | CAF3389986 | |||||
| Rs1s2DBD-NRA3a | CAF3637304 | |||||
| Rs1s2DBD-NRA3b | CAF4677071 | |||||
| Rotaria sp. Silwood2 | Rs2s2DBD-NRA1 | CAF2401455 | ||||
| Rs2s2DBD-NRA2a | CAF275803 | |||||
| (Rs2s) | Rs2s2DBD-NRA3a | CAF2547262 | ||||
| Rs2s2DBD-NRA3b | CAF4430748 | |||||
| Rs2s2DBD-NRA3c | CAF2976618 | |||||
| Rs2s2DBD-NRA2b | CAF2373862 | |||||
| R. sordida (Rso) | Rso 2DBD-NRA1 | CAF0748647 | ||||
| Rso2DBD-NRA2a | CAF0805499 | |||||
| Rso2DBD-NRA2b | CAF1284752 | |||||
| Rso2DBD-NRA3a | CAF1379682 | |||||
| Rso2DBD-NRA3b | CAF1365057 | |||||
| R. socialis (Rs) | Rs 2DBD-NRA1 | CAF4632909 | ||||
| Rs2DBD-NRA2a | CAF3303110 | |||||
| Rs2DBD-NRA2b | CAF4354886 | |||||
| Rs2DBD-NRA3a | CAF3522866 | |||||
| Rs2DBD-NRA3b | CAF3629551 | |||||
| Didymodactylos carnosus (Dc) | Dc2DBD-NRA2a | CAF0923356 | ||||
| Dc2DBD-NRA2b | CAF0904316 | |||||
| Dc2DBD-NRA3a | CAF1101057 | |||||
| Dc2DBD-NRA3b | CAF1014202 | |||||
| Brachionus calyciflorus | Bc2DBD-NRA1a | CAF0776353 | ||||
| Bc2DBD-NRA1b | CAF0776371 | |||||
| Bc2DBD-NRA4a | CAF0820254 | |||||
| Bc2DBD-NRA4b | CAF1066507 | |||||
| Bc2DBD-NRA5 | CAF0986382 | |||||
| B. plicatilis | Bp2DBD-NRA1 | RNA38473 | ||||
| Bp2DBD-NRA4 | RNA21328 | |||||
| Bp2DBD-NRA5 | RNA34403 | |||||
| 7A | Mollusca | Bivalvia | Pecten maximus | Pm2DBD-NRA | XP_033734716 | |
| Mizuhopecten yessoensis | My2DBD-NRA | XP_021367015 | ||||
| Crassostrea gigas | Cg2DBD-NRA | Cg2DBDγ | XP_019919868 | |||
| Crassostrea virginica | Cv2DBD-NRA | XP_022331995 | ||||
| Mercenaria mercenaria | Mm2DBD-NRA | XP_045168098 | ||||
| Dreissena polymorpha | Dp2DBD-NRA | KAH3855004 | ||||
| Mytilus coruscus | Mco2DBD-NRA | CAC5379089 | ||||
| Mytilus edulis | Me2DBD-NRA | CAG2205142 | ||||
| Mytilus galloprovincialis | Mg2DBD-NRA | VDI76762 | ||||
| Gastropoda | Elysia chlorotica | Ech2DBD-NRA | RUS87101 | |||
| Elysia marginata | Ema2DBD-NRA | GFR71482 | ||||
| Plakobranchus ocellatus | Po2DBD-NRA | GFN82368 | ||||
| Aplysia californica | Aca2DBD-NRA | XP_005103360 | ||||
| Biomphalaria glabrata | Bg2DBD-NRA | XP_013065455 | ||||
| Bulinus truncatus | BT2DBD-NRA | KAH9507289 | ||||
| Candidula unifasciata | Cu2DBD-NRA | CAG5125116 | ||||
| Pomacea canaliculata | Pca2DBD-NRA | XP_025103483 | ||||
| Haliotis rubra | Hrub2DBD-NRA | XP_046571180 | ||||
| Haliotis rufescens | Hruf2DBD-NRA | XP_046364309 | ||||
| Gigantopelta aegis | Ga2DBD-NRA | XP_041351218 | ||||
| Lottia gigantea | Lg2DBD-NRA | Lg2DBD-NRγ | XP_009064814 | |||
| Annelida | Clitellata | Helobdella robusta | Hr2DBD-NRA3a | XP_009022576 | ||
| Hr2DBD-NRA3b | XM_009022049 | |||||
| Hr2DBD-NRA3c | XP_009022817 | |||||
| Hr2DBD-NRA2 | XM_009018880 | |||||
| Polychaeta | Capitella teleta | Ct2DBD-NRA2 | ELU14753 | |||
| Ct2DBD-NRA1 | ELU03757 | |||||
| Lamellibrachia satsuma | Ls2DBD-NRA | KAI0243018 | ||||
| Dimorphilus gyrociliatus | Dgy2DBD-NRA1 | CAD5121447 | ||||
| Owenia fusiformis | Ofu2DBD-NRA1 | CAC9635804 | ||||
| 7A | Platyhelminthes | Rhabditophora | Macrostomum lignano | Ml2DBD-NRA1 | PAA89302 | |
| Ml2DBD-NRA2 | PAA53885 | |||||
| Schmidtea mediterranea | Sme2DBD-NRA1 | nhr-1 | ||||
| Sme2DBD-NRA2 | nhr-2 | |||||
| Sme2DBD-NRA3a | nhr-3 | |||||
| Sme2DBD-NRA3b | nhr-6 | |||||
| Monogenea | Gyrodactylus salaris | Gs2DBD-NRA1 | ||||
| Gs2DBD-NRA2 | ||||||
| Gs2DBD-NRA3 | ||||||
| Protopolystoma xenopodis | Px2DBD-NRA1 | |||||
| Px2DBD-NRA2 | VEL20704, partial DBD | |||||
| Px2DBD-NRA3 | VEL22304, partial DBD | |||||
| Cestoda | Dibothriocephalus latus | Dl2DBD-NRA1 | VDK35285, 2nd DBD) | |||
| Dl2DBD-NRA2 | ||||||
| Dl2DBD-NRA3 | ||||||
| Echinococcus Canadensis | Ec2DBD-NRA1 | |||||
| Ec2DBD-NRA2 | ||||||
| Ec2DBD-NRA3 | ||||||
| Echinococcus granulosus | Eg2DBD-NRA1 | Eg2DBDg | KAH9284012 | |||
| Eg2DBD-NRA2 | Eg2DBDα | AZM65758 | ||||
| Eg2DBD-NRA3 | Eg2DBDb | KAH9280969.1 | ||||
| Echinococcus multilocularis | Em2DBD-NRA1 | CDI98537 | ||||
| Em2DBD-NRA2 | CDS36659 | |||||
| Em2DBD-NRA3 | CDS37339 | |||||
| Echinostoma caproni | Eca2DBD-NRA1 | VDP70475.1, Partial DBD | ||||
| Eca2DBD-NRA2 | VDP81705 | |||||
| Eca2DBD-NRA3 | ||||||
| Hydatigera taeniaeformis | Ht2DBD-NRA1 | VDM16170, Partial | ||||
| Ht2DBD-NRA2 | VDM26556, Partial | |||||
| Ht2DBD-NRA3 | VDM33972 | |||||
| Hymenolepis diminuta | Hd2DBD-NRA1 | VDL20127 | ||||
| Hd2DBD-NRA2 | VDL59771 | |||||
| Hd2DBD-NRA3 | VUZ44963 | |||||
| Hymenolepis microstoma | Hm2DBD-NRA1 | CDS27504 | ||||
| Hm2DBD-NRA2 | CUU99503 | |||||
| Hm2DBD-NRA3 | CDS31978 | |||||
| Hymenolepis nana | Hn2DBD-NRA1 | VDO05171 | ||||
| (Rodentolepis nana) | Hn2DBD-NRA2 | VDO08914 | ||||
| Hn2DBD-NRA3 | VDN97648 | |||||
| Mesocestoides corti | Mc2DBD-NRA1 | VDD74272 | ||||
| Mc2DBD-NRA2 | VDD74096 | |||||
| Mc2DBD-NRA3 | VDD77213 | |||||
| Schistocephalus solidus | Ss2DBD-NRA1 | VDL94711, 2nd DBD | ||||
| Ss2DBD-NRA2 | VDL97469, 2nd DBD | |||||
| Ss2DBD-NRA3 | VDL91072, partial DBD | |||||
| Sparganum proliferum | Spa2DBD-NRA1 | VZI04907 | ||||
| Spa2DBD-NRA2 | VZI51357, VZI24038, VZI24041 | |||||
| Spa2DBD-NRA3 | VZI32835 | |||||
| Spirometra erinaceieuropae i | Se2DBD-NRA1 | VDM16170 | ||||
| Se2DBD-NRA2 | VZI28417 | |||||
| Se2DBD-NRA3 | VZI27737 | |||||
| 7A | Platyhelminthes | Cestoda | Taenia asiatica | Ta2DBD-NRA1 | VDK37619 | |
| Ta2DBD-NRA2 | VUZ40124 | |||||
| Ta2DBD-NRA3 | VDK20940 | |||||
| Taenia multiceps | Tm2DBD-NRA1 | |||||
| Tm2DBD-NRA2 | ||||||
| Tm2DBD-NRA3 | ||||||
| Taenia saginata | Ts2DBD-NRA1 | |||||
| Ts2DBD-NRA2 | ||||||
| Ts2DBD-NRA3 | ||||||
| Taenia solium | Tso2DBD-NRA1 | |||||
| Tso2DBD-NRA2 | ||||||
| Tso2DBD-NRA3 | ||||||
| Trematoda | Clonorchis sinensis | Cs2DBD-NRA1 | KAG5448022 | |||
| Cs2DBD-NRA2 | GAA27896 | |||||
| Cs2DBD-NRA3 | GAA48469 | |||||
| Fasciolopsis buski | Fb2DBD-NRA1 | KAA0185392 | ||||
| Fb2DBD-NRA2 | KAA0191852, partial | |||||
| Fb2DBD-NRA3 | KAA0198729, partial | |||||
| Fasciola gigantica | Fg2DBD-NRA1 | TPP59537, partial | ||||
| Fg2DBD-NRA2 | TPP61208 | |||||
| Fg2DBD-NRA3 | TPP62972, TPP57451 | |||||
| Fasciola hepatica | Fh2DBD-NRA1 | THD25668 | ||||
| Fh2DBD-NRA2 | THD25920 | |||||
| Fh2DBD-NRA3 | HD26972 | |||||
| Opisthorchis felineus | Of2DBD-NRA1 | TGZ59947 | ||||
| Of2DBD-NRA2 | TGZ60667 | |||||
| Of2DBD-NRA3 | TGZ71895 | |||||
| Opisthorchis viverrini | Ov2DBD-NRA1 | OON22325, 2nd DBD | ||||
| Ov2DBD-NRA2 | XP_009167226 | |||||
| Ov2DBD-NRA3 | XP_009175932 | |||||
| Paragonimus heterotremus | Phe2DBD-NRA1 | KAF5404614 | ||||
| Phe2DBD-NRA2 | KAF5404035 | |||||
| Phe2DBD-NRA3 | KAF5405364 | |||||
| Paragonimus skrjabini miyazakii | Psm2DBD-NRA1 | KAF7262107, KAF7262108 | ||||
| Psm 2DBD-NRA2 | KAF7261790 | |||||
| Psm 2DBD-NRA3 | KAF7232516 | |||||
| Paragonimus westermani | Pw2DBD-NRA1 | KAF8569986 | ||||
| Pw2DBD-NRA2 | KAA3679217 | |||||
| Pw2DBD-NRA3 | KAF8566917 | |||||
| Schistosoma bovis | Sb2DBD-NRA1 | RTG85541, partial DBD | ||||
| Sb2DBD-NRA2 | RTG87273 | |||||
| Sb2DBD-NRA3 | RTG84925, partial DBD | |||||
| Schistosoma curassoni | Sc2DBD-NRA1 | VDO64665 | ||||
| Sc2DBD-NRA2 | VDP76164, VDP65226 | |||||
| Sc2DBD-NRA3 | VDP48464 | |||||
| Schistosoma haematobium | Sh2DBD-NRA1 | KAH9594422 | ||||
| Sh2DBD-NRA2 | XP_035585440, XP_012799787 | |||||
| Sh2DBD-NRA3 | KAH9592582 | |||||
| Schistosoma japonicum | Sj2DBD-NRA1 | TNN12920 | ||||
| Sj2DBD-NRA2 | KAH8855859 | |||||
| Sj2DBD-NRA3 | TNN09743 | |||||
| Schistosoma margrebowiei | Sma2DBD-NRA1 | VDO62366 | ||||
| Sma2DBD-NRA2 | VDO64249, VDP52470 | |||||
| Sma2DBD-NRA3 | VDP26109 | |||||
| Schistosoma rodhaini | Sr2DBD-NRA1 | |||||
| Sr2DBD-NRA2 | ||||||
| Sr2DBD-NRA3 | ||||||
| Schistosoma mattheei | Smt2DBD-NRA1 | VDP64780, partial DBD | ||||
| Smt2DBD-NRA2 | VDP67915, VDP82804 | |||||
| Smt2DBD-NRA3 | VDP61516, VDP04037 | |||||
| Schistosoma mansoni | Sm2DBD-NRA1 | Sm2DBDγ | AY698061 | |||
| Sm2DBD-NRA2 | Sm2DBDα | AH013462 | ||||
| Sm2DBD-NRA3 | Sm2DBDβ | AY688251 | ||||
| Trichobilharzia regenti | Tr2DBD-NRA1 | |||||
| Tr2DBD-NRA2 | VDQ08843, partial | |||||
| Tr2DBD-NRA3 | VDP99859, VDQ10779 | |||||
| Brachiopoda | Lingulata | Lingula anatina | La2DBD-NRA1 | XP_013400324 | ||
| La2DBD-NRA2 | XP_013400322 | |||||
| Nemertea | Pilidiophora | Notospermus geniculatus | Ng2DBD-NRA | |||
| Chordata | Leptocardii | Branchiostoma belcheri | Bb2DBD-NRA | XM_019778353 | ||
| Branchiostoma floridae | Bf2DBD-NRA | XM_035818392 | ||||
| Branchiostoma lanceolatum | Bl2DBD-NRA | CAH1264236 | ||||
| 7B | Mollusca | Bivalvia | Crassostrea gigas | Cg2DBD-NRB1 | Cg2DBD-NRδ | XP_011428801 |
| Crassostrea virginica | Cv2DBD-NRB1 | XP_022329720 | ||||
| Pecten maximus | Pm2DBD-NRB1 | XP_033734800 | ||||
| Mytilus coruscus | Mco2DBD-NRB1 | CAC5405050 | ||||
| Mytilus edulis | Me2DBD-NRB1 | CAG2230085 | ||||
| Mytilus galloprovincialis | Mg2DBD-NRB1 | VDI78403 | ||||
| Mercenaria mercenaria | Mm2DBD-NRB1 | XP_045175060 | ||||
| Mizuhopecten yessoensis | My2DBD-NRB1 | XP_021378725 | ||||
| Gastropoda | Aplysia californica | Aca2DBD-NRB3 | XP_005112947 | |||
| Biomphalaria glabrata | Bg2DBD-NRB3 | XP_013069617 | ||||
| Bulinus truncatus | Btd2DBD-NRB3 | KAH9507198 | ||||
| Haliotis rufescens | Hruf2DBD-NRB1 | XP_046369823 | ||||
| Lottia gigantea | Lg2DBD-NRB1 | Lg2DBD-NRα/β | XP_009049651 | |||
| Gigantopelta aegis | Ga2DBD-NRB1 | XP_041351328 | ||||
| Aplysia californica | Aca2DBD-NRB2 | XP_005095909 | ||||
| Haliotis rubra | Hrub2DBD-NRB2 | XP_046577572 | ||||
| Haliotis rufescens | Hruf2DBD-NRB2 | XP_046379417 | ||||
| Pomacea canaliculata | Pca2DBD-NRB2 | PVD26031 | ||||
| Plakobranchus ocellatus | Po2DBD-NRB2 | GFO27501 | ||||
| Elysia marginata | Ema2DBD-NRB3 | GFS22057 | ||||
| Elysia chlorotica | Ech2DBD-NRB3 | RUS84791 | ||||
| Plakobranchus ocellatus | Pod2DBD-NRB3 | GFO07362 | ||||
| Pomacea canaliculata | Pca2DBD-NRB3 | PVD26170 | ||||
| Annelida | Clitellata | Helobdella robusta | Hr2DBD-NRB2 | XP_009031176 | ||
| Polychaeta | Lamellibrachia satsuma | Ls2DBD-NRB1 | KAI0214066 | |||
| Ls2DBD-NRB2a | KAI0207292 | |||||
| Ls2DBD-NRB2b | KAI0207833 | |||||
| Capitella teleta | Ct2DBD-NRB1 | ELT90493, 2nd DBD | ||||
| Ct2DBD-NRB2 | ||||||
| Owenia fusiformis | Ofu2DBD-NRB2 | CAH1791723 | ||||
| Brachiopoda | Lingulata | Lingula anatina | La2DBD-NRB | XM_024075933 | ||
| Phoronida | Phoronis australis | Pa2DBD-NRB | ||||
| Echinodermata | Asteroidea | Acanthaster planci | Ap2DBD-NRB | XM_022235619 | ||
| Asterias rubens | Aru2DBD-NRB | XM_033772834 | ||||
| Patiria miniata | Pmi2DBD-NRB | XM_038217445 | ||||
| Echinoidea | Lytechinus variegatus | Lv2DBD-NRB | XM_041613378 | |||
| Strongylocentrotus purpuratus | Spur2DBD-NRB | XM_030998347 | ||||
| Crinoidea | Anneissia japonica | Aja2DBD-NRB | XM_033246620 | |||
| 7C | Nematoda | Chromadorea | Aphelenchus avenae | Aa2DBD-NRC1 | KAH7714729 | |
| Aa2DBD-NRC2 | KAH7714119 | |||||
| Aa2DBD-NRC3 | KAH7695147 | |||||
| Aa2DBD-NRC4 | KAH7714341 | |||||
| Aa2DBD-NRC5 | KAH7721403 | |||||
| Aa2DBD-NRC6 | KAH7705661 | |||||
| Aa2DBD-NRC7 | KAH7711540 | |||||
| Caenorhabditis brenneri | Cb2DBD-NRC | EGT43828 | ||||
| Caenorhabditis remanei | Cr2DBD-NRC1 | KAF1767687 | ||||
| Cr2DBD-NRC2 | XM_003107303 | |||||
| Cr2DBD-NRC3 | KAF1767743 | |||||
| Cr2DBD-NRC4 | KAF1767675 |
Conclusion
In this study, 2DBD-NRs were identified in both protostomes and deuterostomes. Phylogenetic analysis shows that 2DBD-NRs consist of three groups, two groups are present in both protostomes and deuterostomes and the members of the third group are only found in Nematoda. Members of 2DBD-NRA and 2DBD-NRB are identified in both protostomes and deuterostomes, this result suggests that at least two 2DBD-NR genes were present in a common ancestor of the protostomes and deuterostomes. Phylogenetic analysis shows that 2DBD-NRs underwent gene duplication after the split of the different animal phyla. Thus, most of 2DBD-NR genes in a certain animal phylum are paralogues, rather than orthologues like in other animal phyla. 2DBD-NR gene losses occurred in different animal phyla, for example, 2DBD-NRA was missing in Phoronida and Echinodermata, and 2DBD-NRB was not identified in Nemertea, Rotifera, Platyhelminthes and Chordata. Sequence analysis shows that 2DBD-NRs possess highly conserved regions similar to that of typical NRs. The different P-P module (amino acid sequence of P-box in the first DBD and the second DBD) in different 2DBD-NR groups may affect their DBD binding abilities. Since very few studies have been carried out about 2DBD-NRs, little is known about their function. This study demonstrates that 2DBD-NR genes are widely distributed in both protostomes and deuterostomes, their role in regulation of the animal development awaits be revealed.
Supporting information
(PDF)
(PDF)
(PDF)
(NCBI accession number of 2DBD-NRs see Table 6, accession number of other NR is in the bracket after each NR name).
(PDF)
LBD sequence alignment of all 2DBD-NRs to identify the conserved amino acids in class I or class II NRs according to [50] (Signature of the oligomeric behavior of nuclear receptors at the sequence and structural level. Blocks indicate the variable inserts are deleted according to [50]).
(PDF)
Abbreviations
- 2DBD-NR
nuclear receptor with two DBDs
- DBD
DNA binding domain
- LBD
ligand binding domain
- NTSS
N-terminal signature sequence
- P-P module
the P-box sequence in the first DBD and the second DBD of 2DBD-NRs
Data Availability
Accession numbers are in WormBase ParaSite. The data base is hosted by Wellcome Sanger Institute (https://parasite.wormbase.org/index.html).
Funding Statement
The author(s) received no specific funding for this work. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chambon P., The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol Endocrinol, 2005. 19(6): p. 1418–28. doi: 10.1210/me.2005-0125 [DOI] [PubMed] [Google Scholar]
- 2.Evans R.M., The nuclear receptor superfamily: a rosetta stone for physiology. Mol Endocrinol, 2005. 19(6): p. 1429–38. doi: 10.1210/me.2005-0046 [DOI] [PubMed] [Google Scholar]
- 3.Gronemeyer H., Gustafsson J.A., and Laudet V., Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov, 2004. 3(11): p. 950–64. doi: 10.1038/nrd1551 [DOI] [PubMed] [Google Scholar]
- 4.Gronemeyer H. and Laudet V., Transcription factors 3: nuclear receptors. Protein Profile, 1995. 2(11): p. 1173–308. [PubMed] [Google Scholar]
- 5.Laudet V. and Gronemeyer H., The nuclear receptor factsbook. 2002: Gulf Professional Publishing. [Google Scholar]
- 6.Nauber U., et al., Abdominal segmentation of the Drosophila embryo requires a hormone receptor-like protein encoded by the gap gene knirps. Nature, 1988. 336(6198): p. 489–92. doi: 10.1038/336489a0 [DOI] [PubMed] [Google Scholar]
- 7.Oro A.E., et al., The Drosophila gene knirps-related is a member of the steroid-receptor gene superfamily. Nature, 1988. 336(6198): p. 493–6. doi: 10.1038/336493a0 [DOI] [PubMed] [Google Scholar]
- 8.Rothe M., Nauber U., and Jackle H., Three hormone receptor-like Drosophila genes encode an identical DNA-binding finger. EMBO J, 1989. 8(10): p. 3087–94. doi: 10.1002/j.1460-2075.1989.tb08460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sengupta P., Colbert H.A., and Bargmann C.I., The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell, 1994. 79(6): p. 971–80. doi: 10.1016/0092-8674(94)90028-0 [DOI] [PubMed] [Google Scholar]
- 10.Seol W., Choi H.S., and Moore D.D., An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science, 1996. 272(5266): p. 1336–9. doi: 10.1126/science.272.5266.1336 [DOI] [PubMed] [Google Scholar]
- 11.Zanaria E., et al., An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature, 1994. 372(6507): p. 635–41. doi: 10.1038/372635a0 [DOI] [PubMed] [Google Scholar]
- 12.Wu W., et al., Schistosoma mansoni (Platyhelminthes, Trematoda) nuclear receptors: sixteen new members and a novel subfamily. Gene, 2006. 366(2): p. 303–15. doi: 10.1016/j.gene.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 13.Berriman M., et al., The genome of the blood fluke Schistosoma mansoni. Nature, 2009. 460(7253): p. 352–8. doi: 10.1038/nature08160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu W., et al., Evolution of a novel subfamily of nuclear receptors with members that each contain two DNA binding domains. BMC Evol Biol, 2007. 7: p. 27. doi: 10.1186/1471-2148-7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvite G., et al., Bioinformatic analysis of a novel Echinococcus granulosus nuclear receptor with two DNA binding domains. PLoS One, 2019. 14(11): p. e0224703. doi: 10.1371/journal.pone.0224703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Y., et al., Genome-Wide Characterization of the Nuclear Receptor Gene Family in Macrostomum lignano Imply Its Evolutionary Diversification. Front. Mar. Sci, 2021. 8. [Google Scholar]
- 17.Forster S., et al., Molecular characterisation of a serum-responsive, DAF-12-like nuclear hormone receptor of the fox-tapeworm Echinococcus multilocularis. J Cell Biochem, 2011. 112(6): p. 1630–42. doi: 10.1002/jcb.23073 [DOI] [PubMed] [Google Scholar]
- 18.Tharp M.E., Collins J.J. 3rd, and Newmark P.A., A lophotrochozoan-specific nuclear hormone receptor is required for reproductive system development in the planarian. Dev Biol, 2014. 396(1): p. 150–7. doi: 10.1016/j.ydbio.2014.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu W. and LoVerde P.T., Nuclear hormone receptors in parasitic helminths. Mol Cell Endocrinol, 2011. 334(1–2): p. 56–66. doi: 10.1016/j.mce.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu W. and LoVerde P.T., Nuclear hormone receptors in parasitic Platyhelminths. Mol Biochem Parasitol, 2019. 233: p. 111218. doi: 10.1016/j.molbiopara.2019.111218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu W. and LoVerde P.T., Identification and evolution of nuclear receptors in Platyhelminths. PLoS One, 2021. 16(8): p. e0250750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur S., et al., The nuclear receptors of Biomphalaria glabrata and Lottia gigantea: implications for developing new model organisms. PLoS One, 2015. 10(4): p. e0121259. doi: 10.1371/journal.pone.0121259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogeler S., et al., The nuclear receptor gene family in the Pacific oyster, Crassostrea gigas, contains a novel subfamily group. BMC Genomics, 2014. 15: p. 369. doi: 10.1186/1471-2164-15-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simakov O., et al., Insights into bilaterian evolution from three spiralian genomes. Nature, 2013. 493(7433): p. 526–31. doi: 10.1038/nature11696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo Y.J., et al., Nemertean and phoronid genomes reveal lophotrochozoan evolution and the origin of bilaterian heads. Nat Ecol Evol, 2018. 2(1): p. 141–151. doi: 10.1038/s41559-017-0389-y [DOI] [PubMed] [Google Scholar]
- 26.Larkin M.A., et al., Clustal W and Clustal X version 2.0. Bioinformatics, 2007. 23(21): p. 2947–8. doi: 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 27.Huelsenbeck J.P. and Ronquist F., MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 2001. 17(8): p. 754–5. doi: 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 28.Crooks G.E., et al., WebLogo: a sequence logo generator. Genome Res, 2004. 14(6): p. 1188–90. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D.H., et al., Genome-wide identification of nuclear receptor (NR) genes and the evolutionary significance of the NR1O subfamily in the monogonont rotifer Brachionus spp. Gen Comp Endocrinol, 2017. 252: p. 219–225. doi: 10.1016/j.ygcen.2017.06.030 [DOI] [PubMed] [Google Scholar]
- 30.Mark Welch D. and Meselson M., Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science, 2000. 288(5469): p. 1211–5. doi: 10.1126/science.288.5469.1211 [DOI] [PubMed] [Google Scholar]
- 31.Mark Welch D.B., et al., Divergent gene copies in the asexual class Bdelloidea (Rotifera) separated before the bdelloid radiation or within bdelloid families. Proc Natl Acad Sci U S A, 2004. 101(6): p. 1622–5. doi: 10.1073/pnas.2136686100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard-Ashby M., et al., Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol, 2006. 300(1): p. 90–107. doi: 10.1016/j.ydbio.2006.08.033 [DOI] [PubMed] [Google Scholar]
- 33.Schubert M., et al., Nuclear hormone receptor signaling in amphioxus. Dev Genes Evol, 2008. 218(11–12): p. 651–65. doi: 10.1007/s00427-008-0251-y [DOI] [PubMed] [Google Scholar]
- 34.Sluder A.E. and Maina C.V., Nuclear receptors in nematodes: themes and variations. Trends Genet, 2001. 17(4): p. 206–13. doi: 10.1016/s0168-9525(01)02242-9 [DOI] [PubMed] [Google Scholar]
- 35.Sluder A.E., et al., The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res, 1999. 9(2): p. 103–20. [PubMed] [Google Scholar]
- 36.Taubert S., Ward J.D., and Yamamoto K.R., Nuclear hormone receptors in nematodes: evolution and function. Mol Cell Endocrinol, 2011. 334(1–2): p. 49–55. doi: 10.1016/j.mce.2010.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Gilst M., Gissendanner C.R., and Sluder A.E., Diversity and function of orphan nuclear receptors in nematodes. Crit Rev Eukaryot Gene Expr, 2002. 12(1): p. 65–88. doi: 10.1615/critreveukaryotgeneexpr.v12.i1.40 [DOI] [PubMed] [Google Scholar]
- 38.Wu W., Niles E.G., and LoVerde P.T., Thyroid hormone receptor orthologues from invertebrate species with emphasis on Schistosoma mansoni. BMC Evol Biol, 2007. 7: p. 150. doi: 10.1186/1471-2148-7-150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson-Rechavi M., Escriva Garcia H., and Laudet V., The nuclear receptor superfamily. J Cell Sci, 2003. 116(Pt 4): p. 585–6. doi: 10.1242/jcs.00247 [DOI] [PubMed] [Google Scholar]
- 40.Umesono K. and Evans R.M., Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell, 1989. 57(7): p. 1139–46. doi: 10.1016/0092-8674(89)90051-2 [DOI] [PubMed] [Google Scholar]
- 41.Wilson T.E., et al., Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science, 1992. 256(5053): p. 107–10. doi: 10.1126/science.1314418 [DOI] [PubMed] [Google Scholar]
- 42.Zhao Q., et al., Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell, 1998. 1(6): p. 849–61. doi: 10.1016/s1097-2765(00)80084-2 [DOI] [PubMed] [Google Scholar]
- 43.Wurtz J.M., et al., A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol, 1996. 3(1): p. 87–94. doi: 10.1038/nsb0196-87 [DOI] [PubMed] [Google Scholar]
- 44.Kumar V., et al., Functional domains of the human estrogen receptor. Cell, 1987. 51(6): p. 941–51. doi: 10.1016/0092-8674(87)90581-2 [DOI] [PubMed] [Google Scholar]
- 45.Webster N.J., et al., The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell, 1988. 54(2): p. 199–207. doi: 10.1016/0092-8674(88)90552-1 [DOI] [PubMed] [Google Scholar]
- 46.Danielian P.S., et al., Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J, 1992. 11(3): p. 1025–33. doi: 10.1002/j.1460-2075.1992.tb05141.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy L., et al., Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev, 1999. 13(24): p. 3209–16. doi: 10.1101/gad.13.24.3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perissi V., et al., Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev, 1999. 13(24): p. 3198–208. doi: 10.1101/gad.13.24.3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Germain P., et al., Overview of nomenclature of nuclear receptors. Pharmacol Rev, 2006. 58(4): p. 685–704. doi: 10.1124/pr.58.4.2 [DOI] [PubMed] [Google Scholar]
- 50.Brelivet Y., et al., Signature of the oligomeric behaviour of nuclear receptors at the sequence and structural level. EMBO Rep, 2004. 5(4): p. 423–9. doi: 10.1038/sj.embor.7400119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanco V., Mozzo B., and Alvite G., Dimerization, host-parasite communication and expression studies of an Echinococcus granulosus 2DBD nuclear receptor. Parasitol Res, 2023. doi: 10.1007/s00436-023-07905-4 [DOI] [PubMed] [Google Scholar]
- 52.Beinsteiner B., et al., A novel nuclear receptor subfamily enlightens the origin of heterodimerization. BMC Biol, 2022. 20(1): p. 217. doi: 10.1186/s12915-022-01413-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nuclear Receptors Nomenclature C., A unified nomenclature system for the nuclear receptor superfamily. Cell, 1999. 97(2): p. 161–3. doi: 10.1016/s0092-8674(00)80726-6 [DOI] [PubMed] [Google Scholar]
- 54.Esteves A. and Alvite G., Perspective Chapter: Parasitic Platyhelminthes Nuclear Receptors as Molecular Crossroads, in Parasitic Helminths and Zoonoses, Morales-Montor G., Del Río-Araiza V., and Hernandéz-Bello R., Editors. 2022, IntechOpen: Croatia. [Google Scholar]
- 55.Huang W., et al., Evolution of a novel nuclear receptor subfamily with emphasis on the member from the Pacific oyster Crassostrea gigas. Gene, 2015. 567(2): p. 164–72. doi: 10.1016/j.gene.2015.04.082 [DOI] [PubMed] [Google Scholar]
- 56.Lecroisey C., Laudet V., and Schubert M., The cephalochordate amphioxus: a key to reveal the secrets of nuclear receptor evolution. Brief Funct Genomics, 2012. 11(2): p. 156–66. doi: 10.1093/bfgp/els008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(NCBI accession number of 2DBD-NRs see Table 6, accession number of other NR is in the bracket after each NR name).
(PDF)
LBD sequence alignment of all 2DBD-NRs to identify the conserved amino acids in class I or class II NRs according to [50] (Signature of the oligomeric behavior of nuclear receptors at the sequence and structural level. Blocks indicate the variable inserts are deleted according to [50]).
(PDF)
Data Availability Statement
Accession numbers are in WormBase ParaSite. The data base is hosted by Wellcome Sanger Institute (https://parasite.wormbase.org/index.html).