Abstract
The Hippo pathway is a master regulator of organ growth, stem cell renewal, and tumorigenesis, its activation is tightly controlled by various post-translational modifications, including ubiquitination. While several E3 ubiquitin ligases have been identified as regulators of Hippo pathway, the corresponding E2 ubiquitin-conjugating enzymes (E2s) remain unknown. Here, we performed a screen in Drosophila to identify E2s involved in regulating wing overgrowth caused by the overexpression of Crumbs (Crb) intracellular domain and identified Bruce as a critical regulator. Loss of Bruce downregulates Hippo target gene expression and suppresses Hippo signaling inactivation induced tissue growth. Unexpectedly, our genetic data indicate that Bruce acts upstream of Expanded (Ex) but in parallel with the canonical Hippo (Hpo) -Warts (Wts) cascade to regulate Yorkie (Yki), the downstream effector of Hippo pathway. Mechanistically, Bruce synergizes with E3 ligase POSH to regulate growth and ubiquitination-mediated Ex degradation. Moreover, we demonstrate that Bruce is required for Hippo-mediated malignant tumor progression. Altogether, our findings unveil Bruce as a crucial E2 enzyme that bridges the signal from the cell surface to regulate Hippo pathway activation in Drosophila.
Subject terms: Cell proliferation, Cancer models, Ubiquitylation
Introduction
Hippo signaling pathway was initially identified in Drosophila using genetic screens aiming to uncover genes whose disruption would cause distinct overgrowth phenotype [1–3]. Following studies using various animal models revealed evolutionarily conserved roles of Hippo pathway in regulating numerous biological activities under both physiological and pathological conditions, including cell death, cell proliferation, embryogenesis, stem cell renewal, immune surveillance, and not surprisingly, tumorigenesis [4, 5]. The core components of the Drosophila Hippo pathway consist of Expended (Ex), warts (wts), hippo (hpo), yorkie (yki), and scalloped (sd). Ex, Merlin, and Kibra form a complex that recruits the adapter protein Salvador, which in turn recruits the core kinase Hpo [reviewed in [4]]. Hpo phosphorylates Wts, which in turn phosphorylates and inactivates Yki, once Hippo signaling is inactivated, Yki localizes to nuclei and binds to Sd to initiate transcription of target genes such us expanded (ex), Drosophila inhibitor of apoptosis 1 (diap1), and four-jointed (fj). Apart from the classical kinase cascade-mediated phosphorylation, the most abundant type of post-translational modifications (PTMs) in regulating Hippo pathway is ubiquitination [6], a reversible event sequentially catalyzed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s). Numerous E3s have been identified as regulators of Hippo pathway in both Drosophila and mammal [6–11], however, it remains unknown which E2 is essential for the regulation of Hippo pathway.
Unlike many other conventional pathways, Hippo signaling is not activated by dedicated ligand/receptor complexes. Still, several transmembrane proteins are recognized as crucial upstream inputs that can initiate Hippo signal transduction, among which crumbs (crb), the apical-basal polarity protein, is one of the most intensively studied one in Drosophila [12–19]. Crb contains a large extracellular domain and a short intracellular domain, the function of Crb largely depends on the intracellular domain (crbintra). Crb promotes apical localization of Ex through a direct interaction, and overexpression of crbintra inactivates Hippo pathway and induces Yki-dependent overgrowth through ubiquitination-dependent degradation of Ex, at least in part, through E3s including Plenty of SH3s (POSH) and supernumerary limbs (slmb) [12–16, 20].
Here, we performed a genetic candidate screen in Drosophila aiming to identify E2 coding genes whose disruption could suppress wing phenotype caused by ectopically express crbintra during wing development (Fig. 1A). We identified BIR repeat containing ubiquitin-conjugating enzyme (Bruce) as a crucial regulator of Hippo signaling. Unexpectedly, we found the Bruce-Ex axis acts in parallel with Hpo-Wts cascade to regulate Yki activity and showed that Bruce synergizes with POSH to regulate Ex ubiquitination and degradation. We further demonstrated that Bruce is essential for Hippo pathway-dependent malignant tumor progression.
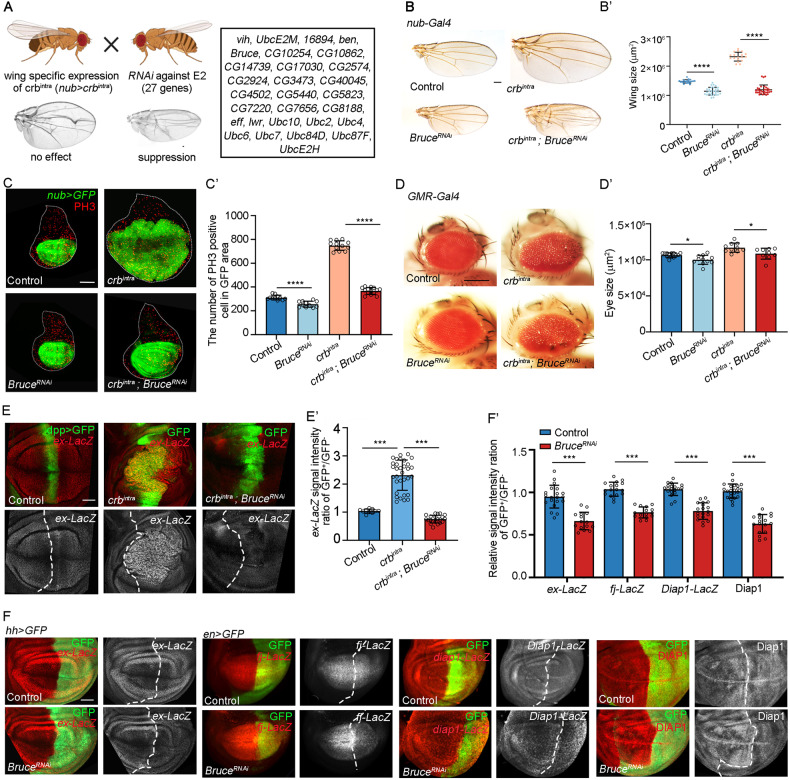
Fig. 1. Bruce is required for crbintra-induced overgrowth and Yki target gene expression.
A A scheme of RNAi screen strategy to identify E2s whose depletion could suppress crbintra-induced wing overgrowth, candidate E2 enzymes screened are listed. B Light micrographs of adult wings bearing indicated genotypes are shown. Quantification of wing size (B’), n = 19, 29, 18, 32 (from left to right). C GFP labeled wing discs bearing indicated genotypes. (C’) Quantification of PH3 positive cells of indicated genotypes, n = 12, 10, 12, 14 (from left to right). D Light micrographs of adult eyes bearing indicated genotypes are shown. Quantification of eye area (D’), n = 13, 11, 10, 10 (from left to right). E Wing pouch regions from larvae with indicated genotypes stained for expression of ex-lacZ. Quantification of relative ex-lacZ intensity (E’), n = 11, 32, 21 (from left to right). F Third instar wing discs expressing BruceRNAi by hh promoter or en promoter stained for expression of ex-lacZ, fj-lacZ, diap1-lacZ, and DIAP1. Quantification of relative staining intensity (F’), n = 20, 15, 17, 15, 19, 19, 23, 17 (from left to right). Mean ± SD, *p < 0.05, ***p < 0.001, ****p < 0.0001; one-way ANOVA tests (B’, C’, D’, and E’) or two-tailed Student’s t tests (F’). Scale bars: 200 μm for (B) and (D), 50 μm for (F); 20 μm for (E).
Results
Bruce is required for crbintra-induced overgrowth and Yki target gene expression
We have previously identified the E3 ubiquitin ligase POSH as a regulator of Hippo pathway and showed that POSH is essential for overexpression of Crbintra-induced growth in Drosophila wing [15], however, the E2 ubiquitin-conjugating enzyme that modulates Hippo pathway has remained unknown. To uncover the corresponding E2s, we performed RNAi screen against Drosophila coding E2s that have homologs in human, aiming to find candidates whose depletion could suppress crbintra-induced wing overgrowth phenotype (Fig. 1A). We focused on one strong candidate named Bruce, a giant Baculovirus IAP repeat (BIR) domain containing E2 that plays essential roles in development, apoptosis, and autophagy [21–24]. Depletion of Bruce under the control of nubbin (nub) promoter efficiently reduces the size in both adult and developing wing and dramatically suppressed crbintra-induced tissue overgrowth and cell proliferation in both stages (Fig. 1B-C’). Although Bruce knockdown did not result in a noticeable phenotype in the eyes, it effectively mitigated the enlarged rough eye phenotype caused by overexpressing crbintra in the developing eye using GMR-Gal4 (Fig. 1D-D’). Notably, we also observed a comparable suppression phenotype using an independent Bruce RNAi strain (Fig. S1), indicating that Bruce is essential for crbintra overexpression induced overgrowth.
As Crbintra overexpression regulates organ size in imaginal discs through inhibiting Hippo signaling [12–14, 16, 20], next, we asked whether Bruce plays a role in regulating Hippo target gene expression. Ectopic expression of crbintra along the anterior-posterior boundary of wing disc under the control of dpp-Gal4 strongly upregulated ex transcription, as monitored by the ex-lacZ staining (Fig. 1E-E’), whereas coexpression of Bruce RNAi significantly inhibited crbintra-driven ex upregulation (Fig. 1E-E’). Furthermore, we found that under physiological conditions, depletion of Bruce specifically in the posterior region of wing epithelium robustly downregulated the endogenous expression of multiple Yki targets, including ex, fj, and diap1 (Fig. 1F-F’). Together, these data indicate that Bruce is required for Yki-dependent target gene expression.
Bruce genetically acts upstream of Ex but in parallel with Hpo-Wts cascade to regulate Yki activity
To further pinpoint where Bruce executes its function in Hippo signaling, we performed genetic epistasis analysis between Bruce and different Hippo pathway components. As previously demonstrated, inhibition of Hippo signaling by knocking down wts, hpo, kibra, ex or overexpression of yki under the nubbin (nub) promoter induced overgrowth phenotype in the developing wing or adult wing (Fig. 2A-B’). We found that depletion of Bruce significantly suppressed loss of kibra, hpo, or wts, and overexpression of yki-induced overgrowth phenotype (Fig. 2A-B’), as well as wts-RNAi induced ex-lacZ upregulation (Fig. 2C-C’), indicating that genetically Bruce might act in parallel or downstream of Yki. Similarly, the overgrowth and ex upregulation induced by depletion of cell polarity gene scribble (scrib), another upstream regulator of Hippo pathway, was also suppressed by knockdown of Bruce (Fig. 2C-C’). Although overexpression of full-length Bruce (BruceFL) completely suppresses reaper-induced apoptotic eye phenotype [23], it has no effect on the small eye phenotype caused by Hpo overexpression (Fig. S2A, B). Furthermore, overexpression of Bruce did not lead to a further increase in excessive proliferation caused by the ex or kibra mutants, suggesting that Bruce overexpression alone is not sufficient to activate Yki in eye epithelium (Fig. S2C-C’). Remarkably, we noticed an unexpected genetic interaction between Ex and Bruce, unlike the rest of the Hippo components, ex loss-induced wing overgrowth phenotype remained unaffected upon the depletion of Bruce (Fig. 2A-A’). Furthermore, we confirmed these intriguing findings in Drosophila eye-antennal epithelium using mosaic analysis with a repressible cell marker (MARCM) based clone induction system (Fig. 2D-D’). Given that Ex could inhibit Yki activation by tethering it in the cytosol [25, 26], these data suggest that Bruce positively regulates Yki activity by acting upstream of Ex but in parallel with the Kibra-Hpo-Wts cascade.
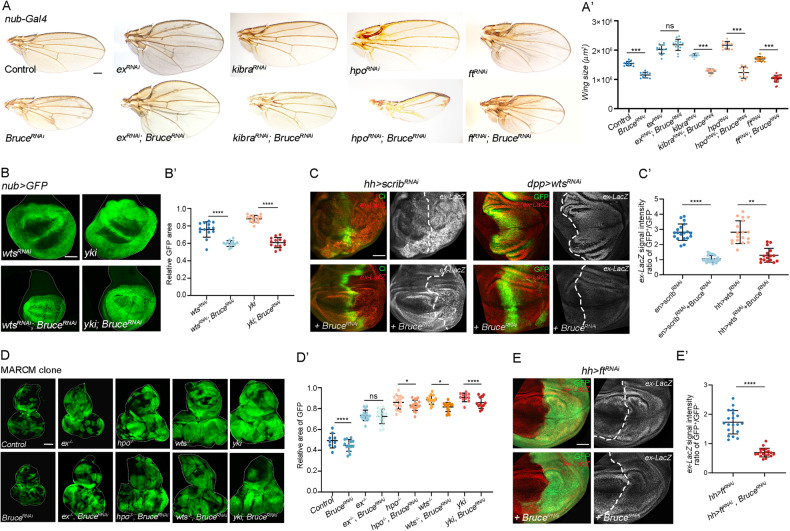
Fig. 2. Bruce acts upstream of Ex but in parallel with Hpo-Wts cascade.
A Light micrographs of adult wings bearing indicated genotypes are shown. Quantification of wing size (A’), n = 12, 12, 14, 14, 16, 18, 11, 11, 15, 23 (from left to right). B GFP labeled wing pouch regions with indicated genotypes are shown. Quantification of relative GFP region size (B’), n = 14, 13, 15, 13 (from left to right). C Wing pouch regions from larvae with indicated genotypes stained for expression of ex-lacZ. Quantification of relative ex-lacZ intensity (C’), n = 19, 22, 18, 16 (from left to right). D Eye-antennal discs of ey-Flp-MARCM-induced GFP positive mosaics clones with indicated genotypes. Quantification of clone size was shown in (D’), n = 9, 13, 16, 16, 18, 18 15, 15, 11, 12 (from left to right). E Wing pouch regions from larvae with indicated genotypes stained for expression of ex-lacZ. Quantification of relative ex-lacZ intensity (E’), n = 15, 14 (from left to right). Mean ± SD, n.s. not significant; *p < 0.05, **p < 0.01, ****p < 0.0001; two-tailed Student’s t tests (E’) or ordinary one-way ANOVA tests (A’, B’, C’, and D’). Scale bars: 200 μm for (A), 100 μm for (B, D); 50 μm for (E).
Apart from Crb, fat (ft) represents another well-studied transmembrane regulator of the Hippo pathway, loss of ft resulted in Ex-dependent overgrowth and increased Yki-driven ex transcription (Fig. 2E-E’), but paradoxically, it also decreases Ex protein levels in the subapical cell cortex, indicating a post-transcriptional effect on Ex [27–30]. Consistent with the genetic epistatic evidence that places Ft upstream of Ex, we found that loss of ft induced ex-lacZ upregulation was significantly suppressed by co-depletion of Bruce (Fig. 2E-E’). Overexpression of Dachs ligand with SH3s (Dlish), an SH3 domain containing protein that physically interacts with Fat, could also cause overgrowth by destabilizing Ex [31]. Consistently, inhibition of Bruce significantly suppressed Dlish-induced wing overgrowth phenotype (Fig. S2D-D’).
Bruce is required for ubiquitination-mediated Ex degradation
Our epistasis experiments placed Bruce downstream of Crbintra and upstream of Ex to regulate Yki activation. Given that crb is known to function upstream in Hippo signaling, at least in part by regulating stability and localization of Ex [12–14, 16, 18, 20], we hypothesized that Bruce could modulate Ex stability. Firstly, we analyzed in vivo Ex protein level in the wing disc using immunostaining and did not observe distinct change upon Bruce depletion under the control of dpp-Gal4 (Fig. 3A-A’). However, we noticed that although inhibition of endogenous Bruce is not sufficient to increase Ex abundance and accumulation at the apical junctional region in vivo (Fig. S3), strong upregulation of exogenously expressed Ex (Ex-Myc) was detected in Drosophila S2 cells treated with Bruce dsRNA (Fig. 3B). In accordance with this, knockdown of Bruce significantly suppressed dpp > crbintra-induced Ex degradation (Fig. 3A-A’). Furthermore, it has been reported that slmb, an E3 ubiquitin ligase, interacts with Crb to regulate the degradation of Ex [20]. Given that reducing Bruce alone did not fully reverse the decrease in Ex protein induced by crbintra, we simultaneously downregulated the expression of both Bruce and slmb in dpp > crbintra. Remarkably, this combined downregulation led to a significant increase in Ex protein level (Fig. 3A-A’).
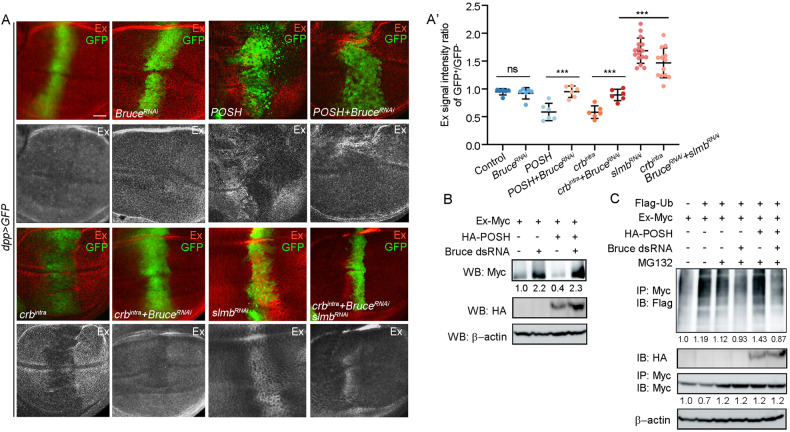
Fig. 3. Bure regulates POSH-dependent Ex ubiquitination and degradation.
A Fluorescent micrographs of wing pouch region with gene expression under the control of dpp promoter stained for Ex expression. Quantification of relative Ex signal intensity ratio of GFP+ region/GFP− region (A’), n = 6, 6, 6, 6, 6, 6, 16, 13 (from left to right). B S2 cells transfected with HA-tagged POSH and Myc-tagged Ex were treated with or without dsRNAs of Bruce, and the lysates were analyzed by Western blotting. C Bruce is required for POSH-induced Ex ubiquitination in S2 cells. Mean ± SD; n.s. not significant; ****p < 0.0001; ordinary one-way ANOVA (A’). Scale bars: 20 μm for (A).
As we have previously uncovered POSH as an Ex-binding E3 ligase that regulates Crbintra-induced Ex ubiquitination and degradation [15], we further explored the potential genetic interactions between POSH and Bruce. We found that POSH overexpression-induced Ex protein reduction phenotype was dramatically reversed by knocking down Bruce (Fig. 3A-A’). Furthermore, transfection of S2 cells with POSH-induced Ex ubiquitination and degradation were also suppressed by dsRNA-mediated Bruce knockdown (Fig. 3B, C), suggesting that ectopic POSH activity crucially depends on the E2 function of endogenous Bruce to regulate ubiquitination-mediated Ex degradation.
Bruce synergizes with POSH to induce overgrowth and tumorigenesis
Next, we further explored the in vivo roles of Bruce and POSH in different contexts. Ectopic expression of POSH induces overgrowth phenotype in the wing epithelium under the control of nub-Gal4, which was significantly suppressed by depletion of Bruce (Fig. 4A-A’). Conversely, we noticed that co-expression of BruceFL and POSH synergistically induces wing overgrowth (Fig. 4A-A’). POSH is known to cooperate with oncogenic Ras (RasV12) to induce tumor overgrowth and Hippo inactivation in the eye epithelium utilizing MARCM technique (Fig. 4B-B’) [32]. Similarly, we found that co-expression of BruceFL also synergistically enhances RasV12-induced clonal overgrowth (Fig. 4B-B’). Consistent with the notion that Bruce is required for POSH induced growth in the wing disc, depletion of Bruce significantly inhibited RasV12/POSH-induced tumor overgrowth in the eye epithelium (Fig. 4B-B’). Moreover, bioinformatic analysis of The Cancer Genome Atlas (TCGA) data revealed a high positive correlation across multiple human cancers between the mRNA level of BIRC6 and SH3RF1, the human orthologs of Bruce and POSH, respectively (Fig. S4). Together, these data imply that Bruce collaborates with POSH to drive overgrowth and tumorigenesis.
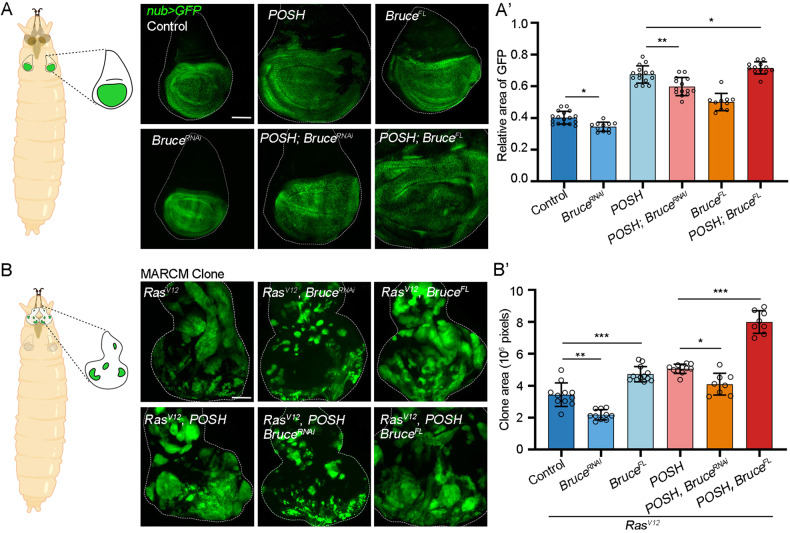
Fig. 4. Bure synergizes with POSH to induce overgrowth and tumorigenesis.
A Cartoon elucidation of wing discs from a Drosophila larva (left). GFP labeled wing pouch regions with indicated genotypes are shown (right). Quantification of relative GFP region size (A’), n = 17, 11, 15, 14, 10, 12 (from left to right). B Cartoon elucidation of eye-antennal discs from a Drosophila larva (left). Eye-antennal discs of ey-Flp-MARCM-induced GFP positive mosaic clones with indicated genotypes. Quantification of relative clone size (B’), n = 11, 10, 12, 11, 8, 8 (from left to right). Mean ± SD; *p < 0.05, **p < 0.01, ****p < 0.0001; ordinary one-way ANOVA (A’ and B). Scale bars: 100 μm for (A) and (B).
Bruce is essential for Hippo signaling-mediated malignant tumor progression
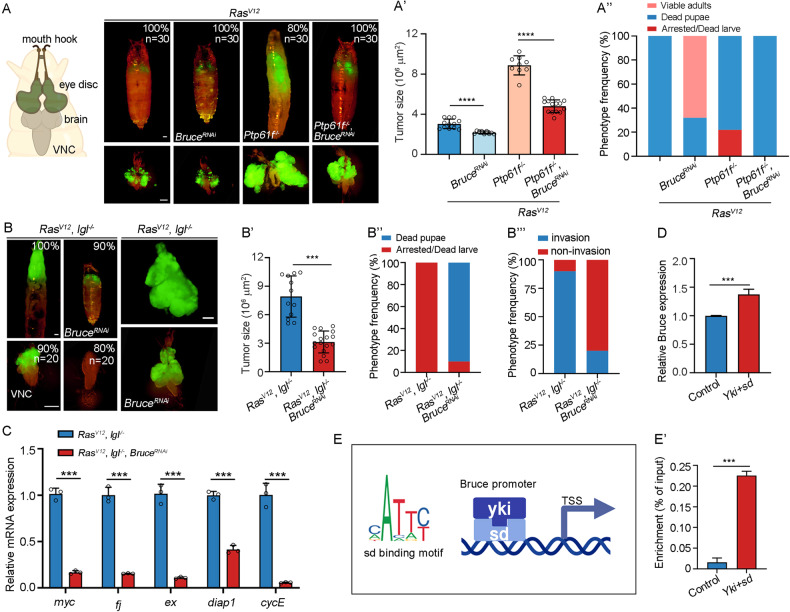
To further dissect the physiological function of Bruce during tumorigenesis, we depleted Bruce in several Drosophila malignant tumor models. Recently, we uncovered the tyrosine phosphatase Ptp61F as a crucial regulator of three-dimensional organ size under both physiological and pathological conditions and demonstrated that loss of Ptp61F synergizes with RasV12 to induce Hippo-dependent tumorigenesis through Ex [33]. Clonal expression of RasV12 alone in the eye-antennal disc induces hyperproliferation and forms benign tumors [32, 33], whereas simultaneously removal of Ptp61F results in malignant tumor formation (Fig. 5A-A’) [33], and 80% of the animals were unable to pupate or died as giant larvae (Fig. 5A”). Remarkably, we found that depletion of Bruce drastically inhibited the tumor overgrowth and related phenotypes (Fig. 5A-A”). Apart from Ptp61F, disruption of cell polarity gene lethal (2) giant larvae (l(2)gl) could also collaborate with RasV12 to induce aggressive malignant tumors [34–36], with invasive migration into the ventral nerve cord (VNC) of the central nervous system (Fig. 5B). Consistently, inhibiting Bruce activity strongly impeded RasV12/lgl−/− triggered tumor overgrowth (Fig. 5B-B”), invasion (Fig. 5B”’), and transcriptional upregulation of multiple Yki target genes (Fig. 5C). Together, these data suggest that Bruce plays a significant role in the malignant tumor progression.
Fig. 5. Bruce is required for Hippo pathway mediated malignant tumor progression.
A Cartoon elucidation of a third-instar larvae cephalic complex (left). Dorsal views of ey-Flp-MARCM-induced GFP positive tumor bearing pupae or larvae (upper) and the corresponding cephalic complexes (bottom), % indicate representative phenotype penetrance. Quantification of tumor size (A’, n = 12, 9, 9, 14) and larvae phenotype (A”, n = 30, 30, 30, 30). B Dorsal views of ey-Flp-MARCM-induced GFP positive tumor bearing pupae or larvae (upper) and the corresponding ventral nerve cord (NVC) (bottom), % indicate representative phenotype penetrance. Quantification of tumor size (B’, n = 12, 17), larvae phenotype (B”, n = 20, 20), and invasion percentage (B”’, n = 20, 20). C qRT-PCR analysis of Yki target gene expression of tumors dissected from RasV12/lgl−/− and RasV12/lgl−/− + BruceRNAi. D Quantification of relative Bruce mRNA level of S2 cells transfected with or without HA-Sd + Yki-Myc, n = 3. E Scheme of the Bruce promoter region and transcription start site (TSS), with the potential Sd binding motif (CATTT/C). (E’) S2 cells transfected with HA-Sd and Yki-Myc were used for Yki and Sd enrichment quantification on target region (−1326 to −1249) that contains Sd binding site, n = 3. Mean ± SD; *p < 0.05, ***p < 0.001, ****p < 0.0001; ordinary one-way ANOVA (A’, B’ and F’), two-tailed Student’s t tests (C’, D’ and E’). Scale bars: 500 μm for (A, C) (upper); 200 μm for (A, C) (bottom).
As multiple upstream components of Hippo pathway (e.g., Mer, Ex, Kibra, Ptp61F) can be transcriptionally activated by Yki/Sd to form a negative feedback loop to fine tune the pathway [4, 33], we examined whether Bruce could be a downstream target of Yki. Indeed, transfection of S2 cells with Yki and Sd significantly upregulated Bruce transcription level (Fig. 5D). Furthermore, our cleavage under targets and tagmentation (CUT&Tag) analysis in the S2 cells revealed that precipitated DNA was appreciably enriched in the promoter region of Bruce that contains putative Sd binding motif (Fig. 5E-E’). Collectively, these results demonstrate that Bruce is a direct transcriptional target of Yki/Sd.
A putative conserved role of BIRC6 in Hippo-dependent human cancer
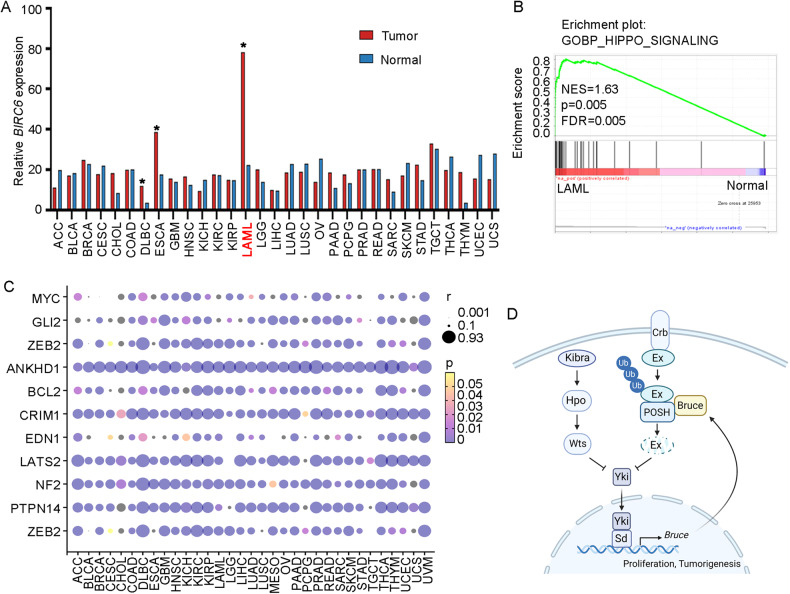
As majority of Hippo pathway components are highly functional conserved between Drosophila and humans, we further explored whether Bruce-mediated Hippo signaling regulation is conserved during human tumor progression. We used GEPIA2 (Gene Expression Profiling Interactive Analysis, version 2, http://gepia2.cancer-pku.cn) [37] to analyze mRNA expression level of BIRC6 (human Bruce homolog) in various human cancers, and found that BIRC6 mRNA levels are strongly increased in several cancer type, with highest expression in acute myeloid leukemia (LAML) patients (Fig. 6A). Consistent with genetic correlation between Bruce and Hippo pathway in Drosophila, our gene set enrichment analysis (GSEA) results revealed a significant enrichment of genes associated with Hippo pathway in LAML patients (Fig. 6B). Moreover, we also observed strong positive correlations between the mRNA of BIRC6 and multiple Hippo pathway target genes across human cancers (Fig. 6C). Collectively, these data indicate that BIRC6 might play a conserved role in regulating Hippo pathway in human cancers.
Fig. 6. A putative conserved role of BIRC6 in Hippo-dependent human cancer.
A Comparison of Bruce mRNA expression in different tumor samples (red) and paired normal tissues (blue) using the GEPIA2 (http://gepia2.cancer-pku.cn). B GSEA enrichment of Hippo pathway-related genes in LAML patients. C Heat map of the correlation coefficients between Bruce and multiple Hippo pathway target genes in TCGA patients, which are displayed in different colors and size. The size scale indicates the degree of correlation. The color of the points is based on the p value. D A model for Bruce-mediated Hippo signaling regulation.
Discussion
Ubiquitination is an essential post-translational modification process that determines protein stability or subcellular localization, dysregulation of which affects various cellular activities, including cell cycle, cell death, and tumorigenesis [38]. Despite the identification of numerous E3 ligases as regulators of key Hippo pathway components [7–11, 15, 20, 39], the responsible E2 that regulates ubiquitin-mediated Hippo signaling activation remains unknown. Here, we have conducted an RNAi screen against E2 coding genes in Drosophila and found that Bruce positively regulates the Hippo pathway by genetically acting upstream of Ex and in parallel with Hpo-Wts cascade (Fig. 6D). First, we demonstrate that depletion of Bruce downregulated endogenous Yki target gene expression and suppressed Crbintra-induced tissue growth. Second, our genetic epistasis data indicate that Bruce acts upstream of Ex but in parallel with the conical Kibra-Hpo-Wts cascade. Third, we show that Bruce is required for POSH overexpression induced Ex ubiquitination and degradation and reveal a collaborative role between POSH and Bruce in growth control. We noticed that depletion of Bruce alone did not significant impact the level of endogenous Ex protein, suggesting that Bruce may play a pivotal role in stress or pathological conditions. It is probable that Bruce actively engages in cellular stress response mechanisms to maintain or restore the normal levels of Ex protein. Consequently, additional studies should be carried out to explore the interaction mechanism between Bruce and Ex, as well as their specific functions in regulating Ex protein degradation. Apart from being a master regulator of organ growth, the Hippo signaling pathway has been implicated in various cancer development in mice and humans. As in mammals, Hippo pathway-dependent tumors can be genetically induced in various Drosophila tissues, including eye epithelium, gut, and ovary [40, 41]. For instance, overexpression of RasV12 in lgl mutant clones (RasV12/lgl−/−) in eye-antennal discs resulted in tumor-like growth with invasive migration into adjacent regions [32, 35, 36]. We show that inhibition of Bruce dramatically impeded RasV12/lgl−/− triggered tumor overgrowth, invasion, as well as activation of Hippo pathway target genes. Similarly, RasV12/Ptp61F−/−-induced malignant tumor progression was also suppressed by knocking down Bruce, suggesting that Bruce is a putative oncogene. Remarkably, a recent study demonstrated that the Bruce’s homolog BIRC6 is selectively required for the survival of a subset of epithelial tumors with a high degree of aneuploidy, and BIRC6 inhibition reduced cell fitness by activating the integrated stress response (ISR) [42]. Consistent with BIRC6’s role as a putative oncogene [43, 44], we observe strong enrichment of Hippo signature genes in LAML patients and reveal intensively strong correlations between BRIC6 mRNA and Hippo pathway target genes across numerous cancer types. Collectively, given the conservation of Hippo pathway and overwhelming evidence that Drosophila is an excellent model to study human cancer biology [4, 40, 41, 45], our findings here indicate that similar mechanisms might exist between the E2 enzyme BIRC6 and Hippo signaling in mammals during both normal development and cancer progression.
Materials and methods
Key resource table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| mouse monoclonal anti-β-gal | Developmental Studies Hybridoma Bank | Cat# 40-1a; RRID: AB_2314509 |
| rat monoclonal anti-Ci | Developmental Studies Hybridoma Bank | Cat# 2A1; RRID: AB_2109711 |
| rabbit polyclonal anti-Phospho-Histone H3 | Cell Signaling Technology | Cat# 9701; RRID: AB_331535 |
| rabbit polyclonal anti- Cleaved Drosophila Dcp-1 | Cell Signaling Technology | Cat# 9578; RRID: AB_2721060 |
| HA-Tag (C29F4) Rabbit | Cell Signaling Technology | Cat# 3724; RRID: AB_1549585 |
| Myc-tag mouse monoclonal | Cell Signaling Technology | Cat# 2276; RRID: AB_331783 |
| mouse monoclonal anti-β-actin | Cell Signaling Technology | Cat# 4968, RRID: AB_2313904 |
| guinea-pig monoclonal anti-Ex | Gift from Richard Fehon (University of Chicago) | Maitra et al. [46] |
| Ubiquitin Antibody Rabbit | Cell Signaling Technology | Cat# 3933, RRID: AB_2180538 |
| Goat anti-Mouse IgG (H + L) Alexa FluorTM Plus 555 | Invitrogen | Cat# A32727; RRID: AB_2633276 |
| Goat anti-Rabbit IgG (H + L) Alexa FluorTM Plus 555 | Invitrogen | Cat# A32732; RRID: AB_2633281 |
| Chemicals, peptides, and recombinant proteins | ||
| Trizol | Thermo Fisher Scientific | Cat# 15596026 |
| Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H-1800 |
| Critical commercial assays | ||
| ESF 921 Insect Cell Culture Medium | Expression System | Cat# 96-001-01 |
| Fetal bovine Serum | CellMax | Cat# SA112 |
| Penicillin-Streptomycin (100X) | Thermo Fisher Scientific | Cat# 15070063 |
| Antibiotic-Antimycotic | Thermo Fisher Scientific | Cat# 15240096 |
| Effectene Transfection Reagent | QIAGEN | Cat# 301427 |
| Protease inhibitor cocktail | Sigma | Cat# P8340 |
| BCA Protein Quantification Kit | Vazyme | Cat# E112-01 |
| PVDF membrane | Merck Millipore | Cat# IPVH00010 |
| Hyperactive Universal CUT&Tag Assay Kit | Vazyme | Cat# TD903-00 |
| Taq Pro Universal SYBR qPCR Master Mix | Vazyme | Cat# Q712-02 |
| MEGAscriptTM RNAi Kit | Invitrogen | Cat# AM1626 |
| Pierce™ Protein A/G Magnetic Beads | Thermo Fisher Scientific | Cat# 88802 |
| FastPure Plasmid Mini Kit | Vazyme | Cat# DC201-01 |
| HiScript II 1st Strand cDNA Synthesis Kit | Vazyme | Cat# R211-01 |
| Oligonucleotides | ||
| qPCR for Bruce F | GGGTGACTTTCTCGCTGACA | N/A |
| qPCR for Bruce R | CCCAGCGAGCTGCTGATTTA | N/A |
| qPCR for CUT&Tag Bruce F | TTTAAGCGGAATTATCGCG | N/A |
| qPCR for CUT&Tag Bruce R | CCAGTTATTCAACCGTTAC | N/A |
| DsRNA for Bruce F | TAATACGACTCACTATAGGGAGAGCATGAAGACCTGTGTGGAT | N/A |
| Oligonucleotides | ||
| DsRNA for Bruce R | CTCCCTATAGTGAGTCGTATTATACAGAACTACTAAAGCTGAGG | N/A |
| qPCR for rp49 F | CCACCAGTCGGATCGATATGC | N/A |
| qPCR for rp49 R | CTCTTGAGAACGCAGGCGACC | N/A |
| qPCR for ex F | GGATCTACCGTAAGCCACCG | N/A |
| qPCR for ex R | TCATCATGACAGCGGGATCG | N/A |
| qPCR for diap1 F | AAATGCTTTTTCTGCGGCGT | N/A |
| qPCR for diap1 R | CTCATCTCCAGCGTCGAGTC | N/A |
| qPCR for fj F | AGGGATGCGGAAGAATGCAA | N/A |
| qPCR for fj R | GGCTGAAAGGGCTGTGGTAT | N/A |
| qPCR for myc F | AGCCAGAGATCCGCAACATC | N/A |
| qPCR for myc R | CGCGCTGTAGAGATTCGTAGAG | N/A |
| qPCR for cycE F | GAATGGAGAGCGTACTCCCG | N/A |
| qPCR for cycE R | TCCAAGAGAATGGCACGCAT | N/A |
| Experimental models: Cell lines | ||
| Drosophila S2 cells | Gift from Jose’ C. Pastor-Pareja (Tsinghua University) | FBtc9000001 |
| Experimental models: Organisms/strains | ||
| The Drosophila strains used in this study are listed in Supplementary Table 1 | N/A | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.0 | GraphPad Software | RRID: SCR_002798 |
| Fiji/ImageJ software | https://fiji.sc | Schindelin et al. [47] |
| Adobe Photoshop | https://www.adobe.com/cn/products/photoshop. | RRID: SCR_014199 |
Fly stocks and genetics
Drosophila stocks and crosses were raised on standard cornmeal-yeast-agar medium at 25 °C. The standard cornmeal-yeast food including 9 g agar, 24.5 g dry yeast, 50 g corn flour, 7.25 g white sugar, 4.4 ml propionic acid, 30 g brown sugar, 1.25 g nipagin per liter and 12.5 ml ethanol. All Drosophila lines used in this study were listed and described in Supplementary Table 1, and detailed genotypes for each figure are described in Supplementary data.
Fluorescently labeled mitotic clones were produced in the eye-antennal discs using the mosaic analysis with a repressible marker (MARCM) system with the following strains: y, w, eyFLP1; Act5C > y + > Gal4, UAS-GFP; FRT82B, Tub-Gal80 (82B tester); yw, ey-Flp; act > y + > GAL4, UAS-GFP; Tub-GAL80, FRT 80B (80B tester); yw, ey-Flp; tub-Gal80, FRT 40 A; act > y + > GAL4 UAS-GFP (40A tester).
Drosophila cell culture
Drosophila S2 cells were cultured at 25 °C in an atmosphere and in Cell Culture Medium supplemented with 10% fetal bovine serum and 1% Antibiotic-Antimycotic.
Cell transfection
Plasmid transfection and double-stranded RNA (dsRNA) were conducted using Effectene transfection reagent according to the manufacturer’s instructions. Plasmids used for Immunoprecipitation (IP) were: pUAST-attB-HA-POSH; pUAST-attB-Ex-Flag; pUAST-Ubi-myc. Plasmids used for Cut&Tag assay were: pUAST-HA-Sd and pUAST-Yki-myc. dsRNA of Bruce was prepared with MEGAscriptTM RNAi Kit according to the manufacturer’s instructions, and primers listed in Oligonucleotides.
IP assay and western blot
Cells were crushed with NP40 buffer (50 mM Tris (pH7.4), 150 mM NaCl, 1% NP-40, sodium pyrophosphate, β-glycerophosphate, sodium orthovanadate, sodium fluoride, EDTA and leupeptin). Cell lysate and pre-washed magnetic beads incubate with rotation for 20 min at room temperature. Separate the beads from the lysate using a magnetic separation rack, transfer the pre-cleared lysate to a clean tube. Then, add primary antibody to 200 μl cell lysate, and incubate overnight at 4 °C. After that, pre-washed magnetic bead was added to the lysate and antibody solution, Finally, the bound protein can be finally separated through the magnetic beads. Proteins were quantitated by using the BCA according to the manufacture’s instruction, followed by separation with SDS-PAGE and transferred to PVDF membranes. The membranes were blocked for 60 min with 5% bovine serum albumin in TBST and blotted with the following primary antibodies for 12–16 h at 4 °C: β-actin, HA, Flag and Myc. After washing with TBST three times, membranes were incubated with HRP-conjugated secondary antibodies for visualization.
Cut & Tag assay
Cut & Tag assay was performed with Hyperactive Universal CUT&Tag Assay Kit according to the manufacturer’s instructions. Briefly, S2 cells are bound by magnetic beads (Concanavalin A-coated Magnetic Beads Pro, ConA Beads Pro) and membrane permeabilization is performed using the nonionic detergent digitonin. Through the mediation of the primary antibody (Rabbit IgG or HA-tag or Mouse myc-tag), corresponding secondary antibody and Protein A/G against the target protein, transposons fused with Protein A/G can precisely target DNA sequences near the target protein. And enrichment of DNA sequences was detected using qPCR and primers listed in Oligonucleotides. Data were normalized and analyzed using fold enrichment analysis.
RNA isolation and qRT-PCR
Total RNA that extracted from S2 cell line was using Trizol. For mRNA expression analysis, the first-strand cDNA was generated using HiScript II 1st Strand cDNA Synthesis Kit. Quantitative Real-time PCR (q-PCR) was performed on triplicate samples in a reaction mix of Taq Pro Universal SYBR qPCR Master Mix with Jena Qtower384G Real-Time PCR System
Immunofluorescence, immunohistochemistry, and imaging
Third-instar larvae imaginal disc were dissected in cold PBS and fixed with 4% paraformaldehyde for 15 min at room temperature, then washed with PBS containing 0.1% Triton X-100 solution (PBST) for 5 min, three times. Samples were blocked in PBS with 5% normal donkey serum for 1 h and then incubated with primary antibodies at 4 °C overnight. After that, samples were incubated with secondary antibodies for 2 h at room temperature, then mounted with DAPI-containing medium. The images were performed with Zeiss Axio Observer with ApoTome.2. Images were processed with Fiji/Image J software (https://fiji.sc), Photoshop CS8 (Abode) for image merging and resizing.
Gene set enrichment analysis (GSEA)
GSEA analysis was performed with GSEA software (http://www.broadinstitute.org/gsea/index.jsp), using differentiation gene sets in LAML (Acute Myeloid Leukemia) and normal samples from TCGA (https://portal.gdc.cancer.gov/). GSEA results were considered significant when the FDR q value was less than 0.25 and p value was less than 0.05.
Quantification and statistical analysis
For clone-size quantification of wild type or mutant, the acquired pictures using fluorescence microscope were analyzed using ImageJ. Clone size was measured as “total GFP clone area/disc area (%)” by analyzing all the clones in at least 8 randomly selected eye discs (~10 discs on average) of each genotype. For antibody staining quantification within GFP or wild-type cells, a minimum of 10 wing discs was analyzed in ImageJ for the mean signal intensity of GFP cells versus neighboring wild-type cells. For quantification of adult eye phenotype, the eye area of female flies was measured manually using ImageJ.
Survival analysis was performed to investigate the associations between the censored outcomes and the expression measures of BIRC6 using GEPAI (http://gepia.cancer-pku.cn/). Pearson correlation analysis was employed to determine the correlation between the expression of Hippo targets and BIRC6 from TCGA. Statistical analyses were performed with GraphPad Prism 9.0. software. Unless indicated, statistical significance was calculated by unpaired, two-tailed Student’s t tests, ordinary ANOVA test (Tukey’s multiple comparisons test) for three or more groups and Mann–Whitney U tests. The number of animals used per experiment can be found in the figure legends. Data are mean ± s.d. ns (not significant), *p ≤ 0.05, **p ≤ 0.01, ***p < 0.001, ****p < 0.0001. n ≥ 3 independent experiments.
Supplementary information
Acknowledgements
We thank Richard Fehon, Jose C. Pastor-Pareja, Xing Wang, Elisabeth Knust, Tian Xu, Lei Xue, Duojia Pan, Hyung Don Ryoo, Zhiheng Xu, Bruce A Hay, Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, and DSHB for providing fly stocks and reagents. We thank Wenhan Liu for fly stock maintenance. This work was supported by startup funds from Westlake University and the grant from the National Natural Science Foundation of China (Grant 32170824) to XM, Westlake Laboratory of Life Sciences and Biomedicine (Grant 10128A092001), and “Team for Growth Control and Size Innovative Research” (Grant 201804016).
Author contributions
XM conceived and supervised the project and wrote the manuscript; XM and SS analyzed the data; SS performed experiments.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Dr Kim McCall
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-023-06130-2.
References
- 1.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–63. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 2.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–46. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 3.Pan D. The unfolding of the Hippo signaling pathway. Dev Biol. 2022;487:1–9. doi: 10.1016/j.ydbio.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Y, Pan D. The Hippo signaling pathway in development and disease. Dev Cell. 2019;50:264–82. doi: 10.1016/j.devcel.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu M, Hu Y, Lan T, Guan KL, Luo T, Luo M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target Ther. 2022;7:376. doi: 10.1038/s41392-022-01191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan F, Qian M, He Q, Zhu H, Yang B. The posttranslational modifications of Hippo-YAP pathway in cancer. Biochim Biophys Acta Gen Subj. 2020;1864:129397. doi: 10.1016/j.bbagen.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, Jho EH. Regulation of the Hippo signaling pathway by ubiquitin modification. BMB Rep. 2018;51:143–50. doi: 10.5483/BMBRep.2018.51.3.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim W, Cho YS, Wang XH, Park O, Ma XY, Kim H, et al. Hippo signaling is intrinsically regulated during cell cycle progression by APC/C-Cdh1. Proc Natl Acad Sci USA. 2019;116:9423–32. doi: 10.1073/pnas.1821370116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan B, Liu J, Shi A, Cao J, Yu Y, Zhu Y, et al. HERC3 promotes YAP/TAZ stability and tumorigenesis independently of its ubiquitin ligase activity. EMBO J. 2023;42:e111549. doi: 10.15252/embj.2022111549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho YS, Li S, Wang X, Zhu J, Zhuo S, Han Y, et al. CDK7 regulates organ size and tumor growth by safeguarding the Hippo pathway effector Yki/Yap/Taz in the nucleus. Genes Dev. 2020;34:53–71. doi: 10.1101/gad.333146.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma B, Chen Y, Chen L, Cheng H, Mu C, Li J, et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat Cell Biol. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- 12.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010;107:15810–5. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–90. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA. 2010;107:10532–7. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma XJ, Guo XW, Richardson HE, Xu T, Xue L. POSH regulates Hippo signaling through ubiquitin-mediated expanded degradation. Proc Natl Acad Sci USA. 2018;115:2150–5. doi: 10.1073/pnas.1715165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro P, Holder M, Frith D, Snijders AP, Tapon N. Crumbs promotes expanded recognition and degradation by the SCFSlimb/beta-TrCP ubiquitin ligase. Proc Natl Acad Sci USA. 2014;111:6862. doi: 10.1073/pnas.1315508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahey-Lozano N, La Marca JE, Portela M, Richardson HE. Drosophila models of cell polarity and cell competition in tumourigenesis. Adv Exp Med Biol. 2019;1167:37–64. doi: 10.1007/978-3-030-23629-8_3. [DOI] [PubMed] [Google Scholar]
- 18.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–81. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 19.Wu HG, Zhu NN, Liu JY, Ma J, Jiao RJ. Shaggy regulates tissue growth through Hippo pathway in Drosophila. Sci China Life Sci. 2022;65:2131–44. doi: 10.1007/s11427-022-2156-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang HT, Li CQ, Chen HQ, Wei CX, Dai F, Wu HG, et al. SCFSlmb E3 ligase-mediated degradation of Expanded is inhibited by the Hippo pathway in Drosophila. Cell Res. 2015;25:93–109. doi: 10.1038/cr.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, et al. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not hid-dependent cell death. Curr Biol. 2002;12:1164–8. doi: 10.1016/S0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- 22.Hou YCC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–39. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domingues C, Ryoo HD. Drosophila BRUCE inhibits apoptosis through non-lysine ubiquitination of the IAP-antagonist REAPER. Cell Death Differ. 2012;19:470–7. doi: 10.1038/cdd.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjorkoy G, Johansen T, et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–31. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su T, Ludwig MZ, Xu J, Fehon RG. Kibra and Merlin activate the Hippo pathway spatially distinct from and independent of expanded. Dev Cell. 2017;40:478–90.e3. doi: 10.1016/j.devcel.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, et al. The FERM-domain protein expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–20. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–10. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 28.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–50. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 29.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–9. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao CY, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Zhang Y, Blair SS. Fat-regulated adaptor protein Dlish binds the growth suppressor expanded and controls its stability and ubiquitination. Proc Natl Acad Sci USA. 2019;116:1319–24. doi: 10.1073/pnas.1811891116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma X, Lu JY, Moraru A, Teleman AA, Fang J, Qiu Y, et al. A novel regulator of ER Ca(2+) drives Hippo-mediated tumorigenesis. Oncogene. 2020;39:1378–87. doi: 10.1038/s41388-019-1076-z. [DOI] [PubMed] [Google Scholar]
- 33.Liu P, Guo Y, Xu W, Song S, Li X, Wang X, et al. Ptp61F integrates Hippo, TOR, and actomyosin pathways to control three-dimensional organ size. Cell Rep. 2022;41:111640. doi: 10.1016/j.celrep.2022.111640. [DOI] [PubMed] [Google Scholar]
- 34.Ma X, Lu JY, Dong Y, Li D, Malagon JN, Xu T. PP6 disruption synergizes with oncogenic Ras to promote JNK-dependent tumor growth and invasion. Cell Rep. 2017;19:2657–64. doi: 10.1016/j.celrep.2017.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra-Gorur K, Li D, Ma X, Yarman Y, Xue L, Xu T. Spz/Toll-6 signal guides organotropic metastasis in Drosophila. Dis Model Mech. 2019;12:dmm039727. doi: 10.1242/dmm.039727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo X, Sun Y, Azad T, Janse van Rensburg HJ, Luo J, Yang S, et al. Rox8 promotes microRNA-dependent yki messenger RNA decay. Proc Natl Acad Sci USA. 2020;117:30520–30. doi: 10.1073/pnas.2013449117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–60. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senft D, Qi J, Ronai ZA. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat Rev Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fulford AD, Holder MV, Frith D, Snijders AP, Tapon N, Ribeiro PS. Casein kinase 1 family proteins promote Slimb-dependent Expanded degradation. Elife. 2019;8:e46592. doi: 10.7554/eLife.46592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enomoto M, Siow C, Igaki T. Drosophila as a cancer model. Adv Exp Med Biol. 2018;1076:173–94. doi: 10.1007/978-981-13-0529-0_10. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Saavedra P, Perrimon N. Cancer cachexia: lessons from Drosophila. Dis Model Mech. 2022;15:dmm049298. doi: 10.1242/dmm.049298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cervia LD, Shibue T, Borah AA, Gaeta B, He L, Leung L, et al. A ubiquitination cascade regulating the integrated stress response and survival in carcinomas. Cancer Discov. 2023;13:766–95. doi: 10.1158/2159-8290.CD-22-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li YP, Tan YA, Wen LJ, Xing ZH, Wang CX, Zhang LH, et al. Overexpression of BIRC6 driven by EGF-JNK-HECTD1 signaling is a potential therapeutic target for triple-negative breast cancer. Mol Ther Nucl Acids. 2021;26:798–812. doi: 10.1016/j.omtn.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu TT, Weng SQ, Tang WQ, Xue RY, Chen S, Cai GX, et al. Overexpression of BIRC6 is a predictor of prognosis for colorectal cancer. PLoS ONE. 2015;10:e0125281. doi: 10.1371/journal.pone.0125281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonoshita M, Cagan RL. Modeling human cancers in Drosophila. Curr Top Dev Biol. 2017;121:287–309. doi: 10.1016/bs.ctdb.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 46.Maitra S, Kulikauskas RM, Gavilan H, Fehon RG. The tumor suppressors merlin and expanded function cooperatively to modulate receptor endocytosis and signaling. Curr Biol. 2006;16:702–9. doi: 10.1016/j.cub.2006.02.063. [DOI] [PubMed] [Google Scholar]
- 47.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.