Abstract
Objective
Stimulator of interferon genes (STING) is a key regulator in initiating innate immune response from sensing cytosolic DNA. Recent studies have revealed that the cGAS-STING signaling pathway has a crucial role in tumor development and progression across cancer types. Herein, we conducted a meta-analysis to explore the relationship between the immunoexpression of STING and the survival outcome of patients in various solid tumors. Studies relevant to the subject were searched from PubMed, Embase, and Web of Science.
Results
Eleven studies including 2,345 patients were eligible for the analysis. STING expression in tumor cells was related to improved disease-free survival/recurrence-free survival (DFS/RFS) (HR = 0.656, 95% CI = 0.455–0.946, p = 0.024) but not with overall survival (OS) (HR = 0.779, 95% CI = 0.534–1.136, p = 0.194). STING expression in stromal cells, however, did not show significant correlation with DFS/RFS and OS (HR = 0.979, 95% CI = 0.565–1.697, p-value = 0.940 and HR = 1.295, 95% CI = 0.845–1.985, p = 0.235, respectively). In a subgroup analysis, STING expression in tumor cells was associated with better DFS (HR = 0.622, 95% CI = 0.428–0.903, p = 0.012). In tumor cells, favorable DFS/RFS were also related to studies from univariate analysis and the gastrointestinal system (HR = 0.667, 95% CI = 0.482–0.923, p = 0.015 and HR = 0.566, 95% CI = 0.330–0.971, p = 0.039).
Conclusions
STING expression in tumor cells is associated with favorable outcome in solid tumors.
Systematic review registration
https://www.crd.york.ac.uk/prospero/, registration number: CRD42023427027
Keywords: meta-analysis, prognosis, STING, carcinoma, immunohistochemistry
1. Introduction
The ability of cancer cells to evade the immune system has been regarded as a crucial feature of tumorigenesis and tumor progression in human malignancies (1, 2). The cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) signaling pathway, responsible for sensing cytosolic double-strand DNA (dsDNA) and initiating innate immune response, has been considered as a potential driver of immune-mediated initiation, growth, and metastasis in cancer (3, 4). Accumulation of cytosolic DNA induced by DNA damage activates cGAS, leading to the production of cGAMP. cGAMP in tumor cells and antigen-presenting cells (APCs) activates STING, which triggers a cascade to recruit kinases IKK and TBK1 and leads to phosphorylation of IRF3 (5–8). Phosphorylated IRF3 acts as a transcription factor and mediates expression of immune-stimulated genes (ISGs), type 1 interferons (IFNs), and senescence-associated secretory phenotype (SASP) (3, 9, 10).
Cytokines released from tumor cells and APCs could activate cytotoxic CD8+ T cells and natural killer (NK) cells to facilitate tumor clearance (8, 11, 12). Furthermore, the activation of STING and its downstream cascade increases autophagy, induces senescence and chronic inflammation, and regulates differentiation of myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs) (10, 13–15). Alteration of the tumor microenvironment by the signaling pathway could either suppress or promote tumor cells. As a modulator of anti-tumor immune response, activation of cGAS-STING signaling pathway by radiation induces interferon production in colorectal cancer (16, 17). In lung cancer, STING promotes activation of lymphocytes or promotes M2 macrophage to be re-educated as M1 macrophage (18, 19). STING also enhances CD8+ T-cell recruitment and/or mediates APC response in glioma, head and neck carcinoma, melanoma, pancreatic cancer, and prostate cancer (4). Apart from these anti-tumor effects, STING also has a pro-tumor role. Chronic STING activation leading to chronic SASP and chronic type I IFN signaling can cause immune suppression and promote metastasis (20, 21).
Tumor cells must evade or adapt to the cGAS-STING signaling pathway to proliferate and survive (22). This facilitates STING protein as a putative target for cancer therapy. Classic cancer therapies, such as radiation and chemotherapy, increase DNA damage to promote tumor clearance through the cGAS-STING signaling pathway (23). However, persistent stimulation could lead to resistance and diminishing effect (8). STING has been considered as an ideal adjuvant for immune checkpoint inhibitors due to its ability to improve T-cell response via type I IFN (20). Recent studies have shown that activation of STING increases expression of PD-1 pathway components in murine carcinomas, and the combination of STING agonist with CTLA-4 and PD-1 antibodies has a survival advantage in mouse tumor models (24, 25). STING agonists are also being designed for tumor vaccines and chimeric antigen receptor (CAR) T-cell therapies (26). When developing STING agonists, the tumor microenvironment should be vastly considered since prolonged usage of STING agonist could potentially lead to a pro-tumor effect (27).
The association between STING and survival outcome has shown variability in previous studies. Some studies have suggested that the expression level of STING or STING-related genes is related to a favorable prognosis, while other studies indicate that STING expression in tumor or stromal cells is associated with the worst prognosis (28–33). A pan-cancer study utilizing data from The Cancer Genome Atlas (TCGA), however, has suggested that STING mRNA levels are not a prognostic factor for most tumor types (34). This may reflect the ambivalent nature of the cGAS-STING signaling pathway, where both anti-tumor and tumor-promoting mechanisms could be expected (1, 22). Nevertheless, a systematic review of immunoexpression of STING across solid tumors has not been addressed.
In this study, therefore, we conducted a meta-analysis to determine the prognostic significance of STING-positive cells in solid tumors. The aim of this study is to clarify the prognostic role of STING expression as a biomarker across multiple tumors and to verify which cell components (tumor cell or stromal cell) and survival outcome could be represented as a prognostic marker in human solid malignancies.
2. Materials and methods
2.1. Publication search strategy
Three electronic databases, PubMed, Web of Science, and Embase, were searched for relevant publications until 1 June 2023. Search terms were (“STING” or “STING1” or “Stimulator of interferon genes” or “TMEM173), (“prognosis” or “prognostic” or “outcome” or “survival”), and (“tumor” or “carcinoma” or “malignancy” or “malignant tumor”). Each source and collection period of tumor samples was carefully recorded to avoid studies with identical patient populations. Investigation was conducted by two pathologists (YK and GHK), and consensus was reached for any discrepancies for the cases. This review was reported under the Preferred Reporting Program for Systematic Reviews and Meta-Analysis (PRISMA). The protocol was registered on the International Prospective Register of Systematic Reviews (registration number: CRD42023427027).
2.2. Inclusion and exclusion criteria
Inclusion criteria for systematic review were as follows: (1) studies published in English; (2) original articles that report the correlation between immunohistochemical expression of STING and outcome; and (3) studies that offer hazard ratio (HR) and 95% confidence intervals (CIs) directly from the main article or supplementary material or studies that provide alternative values that could estimate HR and 95% CIs. The following studies were excluded from the systematic review: (1) studies that were from abstracts of conferences, reviews, and comments; (2) studies that were based on xenograft models or human hematopoietic or lymphoid malignancies; and (3) studies that had insufficient pathological or survival data that led to unavailability to extract HR and 95% Cis.
2.3. Data extraction and quality assessment
Two reviewers independently collected data from eligible articles according to the criteria. The Newcastle–Ottawa Scale (NOS) was used to evaluate the quality of each study. Studies with a score ≥6 out of 9 were defined as qualified for further analysis. The following parameters were extracted from each study: surname of first author, year of publication, primary organ, type of cancer, cell type (tumor cell or stromal cell) assessed for STING expression, cutoff method for STING-positivity, type of outcome, and number of analyzed patients. HR and 95% CIs for overall survival (OS), disease-free survival (DFS), and recurrence-free survival (RFS) were initially extracted from univariate Cox analysis when available. If HR and 95% CIs were not available, they were extracted from multivariate Cox analysis or estimated from Kaplan–Meier curves using Engauge Digitizer software (version 9.8, http://markummitchell.github.io/engauge-digitizer/) and methods provided by Tierney et al. (35).
2.4. Statistical analysis
The correlation between STING expression of tumor cell or stromal cell of solid tumor and prognosis of patients was measured via meta-analysis. Meta-analysis of OS and DFS/RFS was conducted with R programming (version 4.3.0) packages “meta” and “dmetar”. Cochran’s Q and I 2 were used to determine statistical heterogeneity. Random-effects model was used due to significant heterogeneity in most pooled analysis (I 2 > 50%). Subgroup analysis was performed to explore sources of heterogeneity. To assess publication bias, a graphical funnel plot and Egger’s test were evaluated. Sensitivity analysis was conducted to find out the effect of a single study to the pooled analysis. Statistical significance was set at p < 0.05.
3. Results
3.1. Literature search and study characteristics
A total of 853 studies were found from PubMed, Embase, and Web of Science after removing duplicate records. The detailed process of study selection is depicted in Figure 1 . A total of 842 studies were excluded because they are not a complete article, they are irrelevant to the current subject, they lack information about STING expression and survival outcome, or they only have mRNA data about STING. In the end, 11 studies fulfilled the selection criteria and were included for the meta-analysis (30, 31, 33, 35–42). All eligible studies were retrospective and included STING expression of tumor cells ( Table 1 ). Four studies also included STING expression in stromal cells (33, 39, 41, 43). Among these studies, the study by Biesaga et al. examined STING expression of all stromal cells while three other studies examined its expression for immune cells only. Two studies included two independent cohorts, although each cohort included carcinoma from identical organs (30, 37).
Figure 1.
Flowchart of literature search and study selection.
Table 1.
Characteristics of studies eligible for examination of STING in tumor cells and stromal cells.
| Study | Year | Availability | Sources | Survival | Total N | Tumor cell positivity (%) |
Stromal cell positivity (%) |
Organ | Subtype | Stages | Cutoff | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Biesaga et al. [37] | 2022 | Cox | Univariate | OS/DFS | 77 | 71.4 | 71.4 | Head and neck | SQ | I–IV | NA | 7 |
| Chon et al. [30] | 2019 | Cox | Multivariate | OS/RFS | 225 | 50.2 | NA | Colorectum | AD | I–IV | Median | 8 |
| Kol et al. [31] | 2021 | Cox | Univariate | DFS | 251/255 | 50.4 | NA | Cervix | AD,SQ | I–IV | Median | 7 |
| Lohinai et al. [32] | 2021 | Cox | Univariate | OS | 421 | 55.6 | NA | Lung | AD,SQ | I–IV | H-score (>50) | 7 |
| Marletta et al. [25] | 2023 | Cox | Multivariate | RFS | 146 | 36.3 | NA | Kidney | RCC | I–IV | H-score (>5) | 8 |
| Parkes et al. [33] | 2021 | Cox | Univariate | RFS | 156 | 50.0 | 50.0 | Breast | IDC, ILC, mixed | Early stage | Median | 7 |
| Song et al. [34] | 2017 | Cox | Univariate | OS | 217 | 35.5 | NA | Stomach | Gastric cancer | I–IV | Modified H-score (>6) | 7 |
| Sun et al. [24] | 2021 | Cox | Univariate | OS | 76/210 | 53.1 | NA | Stomach | Gastric cancer | I–IV | Modified H-score (>6) | 8 |
| Wang et al. [27] | 2017 | Cox | Univariate | OS/DFS | 112 | 50.9 | 50.9 | Esophagus | SQ | I–IV | Median | 8 |
| Zhang et al. [35] | 2021 | Cox, KM | Uni-/multivariate | OS/DFS | 112 | 42.9 | 67.9 | Liver | HCC | NA | Median | 7 |
| Zhong et al. [36] | 2018 | KM | Univariate | OS | 87 | 9.2 | NA | Colorectum | AD | I–III | Any expression | 8 |
* NOS, Newcastle–Ottawa Scale; KM, Kaplan–Meier curve; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; SQ, squamous cell carcinoma; AD, adenocarcinoma; RCC, renal cell carcinoma; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; HCC, hepatocellular carcinoma.
3.2. STING expression in tumor cells and survival
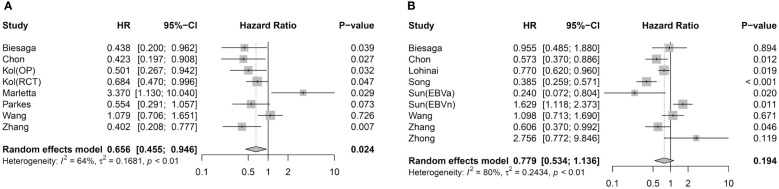
The meta-analysis between STING expression in tumor cells and the prognostic value of DFS/RFS involved seven articles with eight effect sizes (ESs) ( Figure 2A ). Pooled analysis demonstrated that STING-positive tumor cells are associated with better DFS/RFS in solid malignancies (HR = 0.656, 95% CI = 0.455–0.946, p = 0.024). Eight studies with nine ESs included the correlation between STING expression in tumor cells and OS ( Figure 2B ). Unlike DFS/RFS, OS was not significantly associated with STING expression in tumor cells (HR = 0.779, 95% CI = 0.534–1.136, p = 0.194).
Figure 2.
Association between STING expression in tumor cells and survival outcome. (A) Forest plot for DFS/RFS. (B) Forest plot for OS.
3.3. STING expression in stromal cells and survival
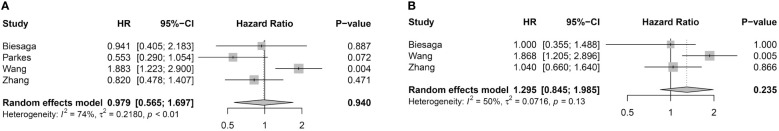
Four studies were available for pooled analysis of STING expression in stromal cells and DFS/RFS ( Figure 3A ). Analysis did not show significant correlation between STING-positive immune cells and DFS/RFS (HR = 0.979, 95% CI = 0.565–1.697, p-value = 0.940). Pooled analysis of OS also had no association with STING expression in stromal cells (HR = 1.295, 95% CI = 0.845–1.985, p = 0.235) ( Figure 3B ).
Figure 3.
Association between STING expression in stromal cells and survival outcome. (A) Forest plot for DFS/RFS. (B) Forest plot for OS.
3.4. Subgroup analysis
Subgroup analysis was conducted for expression of STING in tumor cells ( Table 2 and Supplementary Figure S1 ). High STING expression in tumor cells was associated with good DFS (HR = 0.622, 95% CI = 0.428–0.903, p = 0.012) but not with RFS (HR = 0.868, 95% CI = 0.260–1.057, p = 0.818). Pooled analysis of STING-positive tumor cells from univariate Cox analysis was associated with better DFS/RFS (HR = 0.667, 95% CI = 0.482–0.923, p = 0.015) but those from multivariate analysis were not associated (HR = 0.783, 95% CI = 0.211–2.906, p = 0.714). Inversely, STING-positive tumor cells were associated with better OS for multivariate studies (HR = 0.587, 95% CI = 0.424–0.814, p = 0.001), but not univariate studies (HR = 0.853, 95% CI = 0.517–1.408, p = 0.535). STING-positive tumor cells of the gastrointestinal system was associated with improved DFS/RFS (HR = 0.566, 95% CI = 0.330–0.971, p = 0.039), but not with OS (HR = 0.576, 95% CI = 0.185–1.796, p = 0.342). Correlation between OS and STING expression of tumor cells was not significant despite considering the difference in cutoff method (HR = 0.730, 95% CI = 0.481–1.109, p = 0.140 and HR = 0.642, 95% CI = 0.297–1.388, p = 0.260 for median and H-score, respectively).
Table 2.
Subgroup analysis with STING expression in tumor cells.
| Type of survival |
Subgroup | No. of studies | Hazard ratio | Lower limit of 95% CI | Upper limit of 95% CI | p-value |
|---|---|---|---|---|---|---|
| RFS/DFS | DFS | 5 | 0.622 | 0.428 | 0.903 | 0.012* |
| RFS/DFS | RFS | 3 | 0.868 | 0.260 | 1.057 | 0.818 |
| RFS/DFS | Univariate | 5 | 0.667 | 0.482 | 0.923 | 0.015* |
| RFS/DFS | Multivariate | 3 | 0.783 | 0.211 | 2.906 | RFS/DFS |
| RFS/DFS | GI system | 4 | 0.566 | 0.330 | 0.971 | 0.039* |
| OS | Univariate | 7 | 0.853 | 0.517 | 1.408 | 0.535 |
| OS | Multivariate | 2 | 0.587 | 0.424 | 0.814 | 0.001* |
| OS | GI system | 7 | 0.576 | 0.185 | 1.796 | 0.342 |
| OS | Median | 3 | 0.730 | 0.481 | 1.109 | 0.140 |
| OS | (Modified) H-score | 4 | 0.642 | 0.297 | 1.388 | 0.260 |
*p < 0.05.
3.5. Evaluation of publication bias and sensitivity analysis
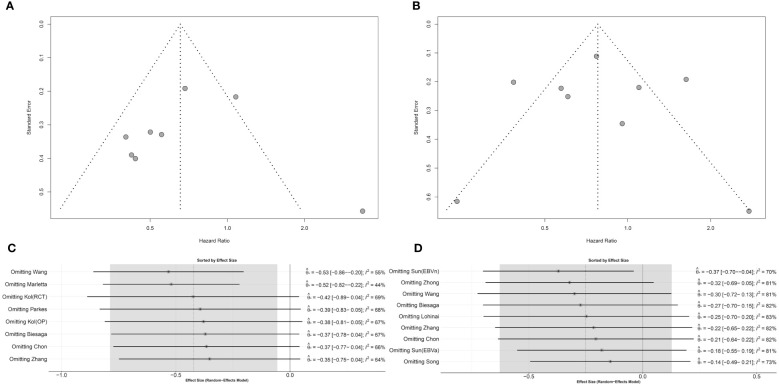
Publication bias in the eligible studies was evaluated by a graphical funnel plot and Egger’s test. Studies with DFS/RFS and tumor cells showed potential asymmetry in the funnel plot, but according to Egger’s test, a significant publication bias was not suspected (p = 0.828) ( Figure 4A ). Studies with OS and tumor cells showed symmetric distribution (p = 0.999 in Egger’s test) ( Figure 4B ). Omitting individual ES in DFS/RFS and OS has demonstrated that robustness of the pooled analysis could be limited ( Figures 4C, D ). Identical analyses were shown for the correlation between STING expression in stromal cells and DFS/RFS or OS ( Supplementary Figure S2 ).
Figure 4.
Publication bias and sensitivity analysis for STING expression in tumor cells. (A) Funnel plot for DFS/RFS. (B) Funnel plot for OS. (C) Sensitivity analysis for DFS/RFS. (D) Sensitivity analysis for OS.
4. Discussion
The cGAS-STING signaling pathway, triggered by sensoring dsDNA, activates kinases IKK and TBK1, leading to a cascade that transcripts pro-inflammatory cytokines including type I IFN, which recruits cytotoxic T cells and NK cells for tumor cell clearance (44). Consequently, expression of STING has been suggested as a therapeutic target as well as a prognostic biomarker in solid tumors. However, the evaluation of STING immunoexpression in various cell types, such as tumor cells, T cells, and macrophages, has shown varying degrees of association with survival outcome. This study, for the first time, explores the correlation between STING-positive tumor cells or stromal cells and prognosis in solid tumors through meta-analysis.
In a pooled analysis, STING expression of tumor cells was associated with improved DFS/RFS. Subgroup analysis revealed that excluding studies from multivariate analysis did not impact the overall association between STING expression and DFS/RFS. A subgroup analysis of the gastrointestinal system maintained the correlation between STING expression in tumor cells and DFS/RFS. However, a subgroup analysis with a smaller number of studies (equal to or fewer than three studies) was determined insignificant with DFS/RFS. In the pooled analysis for stromal STING, the correlation with survival was not significant. However, it should be emphasized that only a small number of studies were obtained compared with that of tumoral STING (four and three for DFS/RFS and OS, respectively).
STING expression in tumor cells was largely unassociated with OS in overall pooled analysis and in subgroup analysis. These results align with previous studies analyzing TCGA data, in which mRNA expression of STING was mostly unrelated to OS regardless of carcinoma type (34). Intriguingly, high STING mRNA expression was associated with worse OS in renal cell carcinomas, and a similar trend has been observed for immunohistochemical STING expression in renal cell carcinoma (31). These similarities suggest that STING mRNA in renal carcinoma strongly recapitulates STING expression in tumor cells, rather than that in stromal cells despite estimated tumor purity of renal cell carcinomas in TCGA only being intermediate (45).
Currently, there is no clear explanation for the discrepancies shown between DFS/RFS and OS in tumor cells. STING activity has been identified as a source of suppressor of spontaneous outbreak from disseminated cancer cells in lung adenocarcinoma (46). Expression of STING enhances tumor cell clearance by T cells and NK cells, thereby inhibiting metastasis and tumor relapse. Considering that a relapse of carcinoma would directly affect DFS and RFS but not OS per se, it could be hypothesized that the mechanism of immune recognition and evasion associated with STING during metastasis contributes to the difference in results observed between the survival outcomes.
STING is a pivotal regulator of cancer immunity and initiates innate immune response (47). As an upstream signaling molecule, various methods for the evaluation of active STING could be considered. In this study, immunohistochemical expression of STING was considered as a putative marker for STING activation. A more direct method would be examining phosphorylated STING, which consequently activates TBK1 and recruits IRF3, but to the best of our knowledge, a study that includes immunohistochemical staining of phospho-STING in tumors has yet been published (48). Another way to evaluate STING activation would include assessment of downstream effector molecules. STING-related gene signatures were associated with prognosis in breast, prostate, and colorectal cancers (28, 29, 32). Therefore, developing alternative protein markers alongside STING could provide a more comprehensive understanding of STING activation and its functional implications even in cases when upstream signaling pathway is compromised by genetic alterations.
There are several limitations in this study. The primary tumor sites analyzed for DFS/RFS and OS are not identical and, therefore, may not have an identical impact compared with when all organs are matched. Subgroup analysis for gastrointestinal systems was identical to the overall pooled analysis for both survival outcomes. Nevertheless, STING expression in tumor cells could have an anti-tumor and a pro-tumor effect according to specific tumor context, and therefore, trying to analyze STING expression of all solid malignancy in a uniform manner might undermine the distinct role of the cGAS-STING signaling pathway for each tumor. Another limitation involves the sensitivity analysis revealing limitations in robustness. However, there was no significant publication bias despite the relatively small number of studies available for each pooled analysis of tumor cells. Lastly, our analysis of STING was restricted to cytoplasmic and membranous expression, with only one study describing subcellular location other than cytoplasm and membrane as a region associated with prognosis (39).
In conclusion, this meta-analysis demonstrated that STING expression in tumor cells is associated with improved DFS/RFS but not OS, while in small available studies, STING expression in stromal cells has no association with survival outcome when evaluating malignancies from various organs. Further studies should investigate whether the prognostic value of STING expression in tumor cells can serve as a viable target for predicting therapeutic response and guiding personalized treatment strategies in solid tumors.
Data availability statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
Author contributions
YK and GHK: protocol, data collection, and funding acquisition; YK: conceptualization and writing the first manuscript; GHK: reviewing the manuscript and supervision; HJ: quality assessment; N-YC and LJ: figure legends. All authors contributed to the article and approved the submitted version.
Funding Statement
This study was supported by a grant from the Research Fund of Seoul St. Mary’s Hospital, the Catholic University of Korea (ZC22TISI0753) and from the National Research Foundation of Korea (NRF) funded by the Korean Government (MSIT) (Nos. 2021R1A2C1003542 and RS-2023-00238446).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1244962/full#supplementary-material
References
- 1. Ng KW, Marshall EA, Bell JC, Lam WL. cGAS-STING and cancer: dichotomous roles in tumor immunity and development. Trends Immunol (2018) 39(1):44–54. doi: 10.1016/j.it.2017.07.013 [DOI] [PubMed] [Google Scholar]
- 2. He L, Xiao X, Yang X, Zhang Z, Wu L, Liu Z. STING signaling in tumorigenesis and cancer therapy: A friend or foe? Cancer Lett (2017) 402:203–12. doi: 10.1016/j.canlet.2017.05.026 [DOI] [PubMed] [Google Scholar]
- 3. Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat Rev Mol Cell Biol (2020) 21(9):501–21. doi: 10.1038/s41580-020-0244-x [DOI] [PubMed] [Google Scholar]
- 4. Zhu Y, An X, Zhang X, Qiao Y, Zheng T, Li X. STING: a master regulator in the cancer-immunity cycle. Mol Cancer (2019) 18(1):152. doi: 10.1186/s12943-019-1087-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galluzzi L, Vanpouille-Box C, Bakhoum SF, Demaria S. SnapShot: CGAS-STING signaling. Cell (2018) 173(1):276–276 e1. doi: 10.1016/j.cell.2018.03.015 [DOI] [PubMed] [Google Scholar]
- 6. Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate immune signaling and regulation in cancer immunotherapy. Cell Res (2017) 27(1):96–108. doi: 10.1038/cr.2016.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balka KR, Louis C, Saunders TL, Smith AM, Calleja DJ, D’Silva DB, et al. TBK1 and IKKepsilon act redundantly to mediate STING-induced NF-kappaB responses in myeloid cells. Cell Rep (2020) 31(1):107492. doi: 10.1016/j.celrep.2020.03.056 [DOI] [PubMed] [Google Scholar]
- 8. Gan Y, Li X, Han S, Liang Q, Ma X, Rong P, et al. The cGAS/STING pathway: A novel target for cancer therapy. Front Immunol (2021) 12:795401. doi: 10.3389/fimmu.2021.795401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loo TM, Miyata K, Tanaka Y, Takahashi A. Cellular senescence and senescence-associated secretory phenotype via the cGAS-STING signaling pathway in cancer. Cancer Sci (2020) 111(2):304–11. doi: 10.1111/cas.14266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Decout A, Katz JD, Venkatraman S, Ablasser A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol (2021) 21(9):548–69. doi: 10.1038/s41577-021-00524-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sivick KE, Desbien AL, Glickman LH, Reiner GL, Corrales L, Surh NH, et al. Magnitude of therapeutic STING activation determines CD8(+) T cell-mediated anti-tumor immunity. Cell Rep (2018) 25(11):3074–3085 e5. doi: 10.1016/j.celrep.2018.11.047 [DOI] [PubMed] [Google Scholar]
- 12. Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, Raulet DH. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity (2018) 49(4):754. doi: 10.1016/j.immuni.2018.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu D, Wu H, Wang C, Li Y, Tian H, Siraj S, et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ (2019) 26(9):1735–49. doi: 10.1038/s41418-018-0251-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng H, Xu Q, Lu X, Yuan H, Li T, Zhang Y, et al. Activation of STING by cGAMP Regulates MDSCs to Suppress Tumor Metastasis via Reversing Epithelial-Mesenchymal Transition. Front Oncol (2020) 10:896. doi: 10.3389/fonc.2020.00896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q, Bergholz JS, Ding L, Lin Z, Kabraji SK, Hughes ME, et al. STING agonism reprograms tumor-associated macrophages and overcomes resistance to PARP inhibition in BRCA1-deficient models of breast cancer. Nat Commun (2022) 13(1):3022. doi: 10.1038/s41467-022-30568-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen J, Markelc B, Kaeppler J, Ogundipe VML, Cao Y, McKenna WG, et al. STING-dependent interferon-lambda1 induction in HT29 cells, a human colorectal cancer cell line, after gamma-radiation. Int J Radiat Oncol Biol Phys (2018) 101(1):97–106. doi: 10.1016/j.ijrobp.2018.01.091 [DOI] [PubMed] [Google Scholar]
- 17. Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity (2014) 41(5):843–52. doi: 10.1016/j.immuni.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Invest (2019) 129(3):1211–28. doi: 10.1172/JCI123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Downey CM, Aghaei M, Schwendener RA, Jirik FR. DMXAA causes tumor site-specific vascular disruption in murine non-small cell lung cancer, and like the endogenous non-canonical cyclic dinucleotide STING agonist, 2’3’-cGAMP, induces M2 macrophage repolarization. PloS One (2014) 9(6):e99988. doi: 10.1371/journal.pone.0099988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang M, Chen P, Wang L, Li W, Chen B, Liu Y, et al. cGAS-STING, an important pathway in cancer immunotherapy. J Hematol Oncol (2020) 13(1):81. doi: 10.1186/s13045-020-00916-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature (2018) 553(7689):467–72. doi: 10.1038/nature25432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Du JM, Qian MJ, Yuan T, Chen RH, He QJ, Yang B, et al. cGAS and cancer therapy: a double-edged sword. Acta Pharmacol Sin (2022) 43(9):2202–11. doi: 10.1038/s41401-021-00839-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yum S, Li M, Chen ZJ. Old dogs, new trick: classic cancer therapies activate cGAS. Cell Res (2020) 30(8):639–48. doi: 10.1038/s41422-020-0346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore E, Clavijo PE, Davis R, Cash H, Van Waes C, Kim Y, et al. Established T cell-inflamed tumors rejected after adaptive resistance was reversed by combination STING activation and PD-1 pathway blockade. Cancer Immunol Res (2016) 4(12):1061–71. doi: 10.1158/2326-6066.CIR-16-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dorta-Estremera S, Hegde VL, Slay RB, Sun R, Yanamandra AV, Nicholas C, et al. Targeting interferon signaling and CTLA-4 enhance the therapeutic efficacy of anti-PD-1 immunotherapy in preclinical model of HPV(+) oral cancer. J Immunother Cancer (2019) 7(1):252. doi: 10.1186/s40425-019-0728-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su T, Zhang Y, Valerie K, Wang XY, Lin S, Zhu G. STING activation in cancer immunotherapy. Theranostics (2019) 9(25):7759–71. doi: 10.7150/thno.37574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Flood BA, Higgs EF, Li SY, Luke JJ, Gajewski TF. STING pathway agonism as a cancer therapeutic. Immunol Rev (2019) 290(1):24–38. doi: 10.1111/imr.12765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhuo XX, Dai H, Yu S. The cGAS-STING pathway-related gene signature can predict patient prognosis and immunotherapy responses in prostate adenocarcinoma. Medicine (2022) 101(50):e31290. doi: 10.1097/Md.0000000000031290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C, Wang J, Dong C, Lim D, Feng Z. Development of a risk model to predict prognosis in breast cancer based on cGAS-STING-related genes. Front Genet (2023) 14:1121018. doi: 10.3389/fgene.2023.1121018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Q, Fu Y, Chen XB, Li L, Wu HY, Liu YX, et al. Prognostic perspectives of STING and PD-L1 expression and correlation with the prognosis of epstein-barr virus-associated gastric cancers. Gut Liver (2022) 16(6):875–91. doi: 10.5009/gnl210359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marletta S, Calio A, Bogina G, Rizzo M, Brunelli M, Pedron S, et al. STING is a prognostic factor related to tumor necrosis, sarcomatoid dedifferentiation, and distant metastasis in clear cell renal cell carcinoma. Virchows Archiv (2023) 483(1):87–96. doi: 10.1007/s00428-023-03549-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen SY, Chen SY, Feng WJ, Li ZT, Luo YX, Zhu XD. A STING-related prognostic score predicts high-risk patients of colorectal cancer and provides insights into immunotherapy. Ann Trans Med (2021) 9(1):14. doi: 10.21037/atm-20-2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang ZC, Zhang L, Li ZL, He J, Cai TT, Yang DJ, et al. Expression of STING and MIF in tumor infiltration lymphocytes as prognostic factors in patients with ESCC. Int J Clin Exp Pathol (2017) 10(9):10066. [PMC free article] [PubMed] [Google Scholar]
- 34. Wu Z, Lin Y, Liu LM, Hou YL, Qin WT, Zhang L, et al. Identification of Cytosolic DNA Sensor cGAS-STING as Immune-Related Risk Factor in Renal Carcinoma following Pan-Cancer Analysis. J Immunol Res (2022) 2022:7978042. doi: 10.1155/2022/7978042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials (2007) 8:16. doi: 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. ChonL HJ, Kim H, Noh JH, Yang HN, Lee WS, Kong SJ, et al. STING signaling is a potential immunotherapeutic target in colorectal cancer. J Cancer (2019) 10(20):4932–8. doi: 10.7150/jca.32806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kol A, Lubbers JM, Terwindt ALJ, Workel HH, Plat A, Wisman GBA, et al. Combined STING levels and CD103+T cell infiltration have significant prognostic implications for patients with cervical cancer. Oncoimmunology (2021) 10(1):1936391. doi: 10.1080/2162402x.2021.1936391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lohinai Z, Dora D, Caldwell C, Rivard CJ, Suda K, Yu H, et al. Loss of STING expression is prognostic in non-small cell lung cancer. J Surg Oncol (2022) 125(6):1042–52. doi: 10.1002/jso.26804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parkes EE, Humphries MP, Gilmore E, Sidi FA, Bingham V, Phyu SM, et al. The clinical and molecular significance associated with STING signaling in breast cancer. NPJ Breast Cancer (2021) 7(1):81. doi: 10.1038/s41523-021-00283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song SS, Peng PK, Tang ZQ, Zhao JJ, Wu WC, Li HJ, et al. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep (2017) 7:39858. doi: 10.1038/srep39858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y, Zhai Q, Feng X, Chen D, Lu Y, Hu J, et al. Cancer cell-intrinsic STING is associated with CD8+T-cell infiltration and might serve as a potential immunotherapeutic target in hepatocellular carcinoma. Clin Trans Oncol (2021) 23(7):1314–24. doi: 10.1007/s12094-020-02519-z [DOI] [PubMed] [Google Scholar]
- 42. Zhong GS, Peng CC, Chen YA, Li JY, Yang R, Wu MN, et al. Expression of STING and PD-L1 in colorectal cancer and their correlation with clinical prognosis. Int J Clin Exp Pathol (2018) 11(3):1256–64. [PMC free article] [PubMed] [Google Scholar]
- 43. Biesaga B, Smolarczyk R, Mucha-Malecka A, Czapla J, Rys J, Malecki K. Prognostic significance of STING immunoexpression in relation to HPV16 infection in patients with squamous cell carcinomas of oral cavity and oropharynx. Biomedicines (2022) 10(10):2538. doi: 10.3390/biomedicines10102538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discovery (2020) 10(1):26–39. doi: 10.1158/2159-8290.CD-19-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun (2015) 6:8971. doi: 10.1038/ncomms9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu J, Sanchez-Rivera FJ, Wang Z, Johnson GN, Ho YJ, Ganesh K, et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature (2023) 616(7958):806–13. doi: 10.1038/s41586-023-05880-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ahn J, Barber GN. STING signaling and host defense against microbial infection. Exp Mol Med (2019) 51(12):1–10. doi: 10.1038/s12276-019-0333-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science (2015) 347(6227):aaa2630. doi: 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.