Abstract
Over a period of 6 years (1989 to 1995), serum samples from 3,300 patients suspected to be infected by Coxiella burnetii were assayed for the presence of antibodies against antigen phase II of the microorganism by the indirect immunofluorescence antibody technique (IFAT). One hundred fifty-two cases were recorded, and blood samples from 17 patients were cultured for the isolation of the pathogen. By a centrifugation shell vial technique, eight strains were isolated from patients suffering from acute Q fever. The microorganism was detected in the cultures by IFAT, by Gimenez staining, and by the cytopathogenic effect on Vero and human embryonic lung (HEL) cells. PCR followed by restriction fragment length polymorphism analysis was used to confirm the diagnosis and identify the Coxiella burnetii strains within the cell cultures as well as to compare them with reference strains. In order to avoid time-consuming cultures, to achieve direct detection of Coxiella burnetii in clinical samples (blood, buffy coat, etc.), and to increase the specificity and sensitivity of the detection, nested PCR was performed. The first step of DNA extraction was performed with the QIAamp blood kit 250. For the second step of the PCR assays, the conditions of temperature and times of recycling were properly modified, and the microorganism was detected within 4 h. Our study demonstrates that Q fever is an endemic disease in Crete and that the diagnosis of Coxiella burnetii infection can be rapidly achieved by the detection of the microorganism in buffy coat samples by nested PCR. Although the presenting symptoms of the disease in this study differed from those in other studies, the Cretan strains do not differ genotypically from the reference strains (Nine Mile and Q212).
Coxiella burnetii, an obligate intracellular parasite with a worldwide distribution, is the causative agent of Q fever in humans and animals (1, 25).
The bacterium exists as a strict intracellular parasite when infecting its host but can also survive in the environment (1). Early assessments of the epidemiology of Q fever suggested that the diseases can be transmitted through contact with infected animals, blood transfusions, inhalation of infectious aerosols, the digestive tract, skin trauma, and sexual contact and, rarely, by a mother to the fetus (21, 24).
In humans, the infection has two forms, acute and chronic. The acute form is manifested by pneumonia, prolonged fever, granulomatous hepatitis, and, rarely, meningoencephalitis (23). The main clinical manifestation of chronic Q fever is endocarditis (1, 23, 24, 34).
In Greece in 1946, Caminopetros detected the microorganism in sera of German soldiers (2). Since 1950, only sporadic cases have been reported, and the microorganism was never cultured.
In a previous study on the Greek island of Crete, Tselentis et al. reported that the predominant clinical manifestations of the infection were fever and respiratory disease, whereas hepatitis occurred in only a minority of the infected patients (34). Reports from Australia (4), Great Britain (3, 8), the United States (10), Spain (17), France (5), and Canada (35) indicate that epidemiological and clinical features of the disease may vary from one area of the world to another. For example, in two Australian studies the prevailing clinical presentation was fever of unknown origin (16); in two studies from Nova Scotia (15) and Switzerland (6), the prevailing clinical presentation was pneumonia, while in a French study, hepatitis was the prevailing feature (5).
Since the reason for this clinical diversity between acute cases of Q fever is not known, the causative roles of strain differences cannot be excluded. However, no firm conclusions can be drawn because of the small number of C. burnetii strains isolated from patients suffering from acute Q fever (18–20, 36). Thus, isolation of C. burnetii strains from different geographic areas is needed.
Laboratory diagnosis of Q fever is mainly based on serological tests (29). The isolation of C. burnetii in cultures is time-consuming and hazardous and may give false-negative results. To overcome these problems, PCR and nested PCR techniques were developed (12, 29, 36). A number of C. burnetii strains originating from patients suffering from either chronic or acute Q fever have been isolated by a shell vial culture method. The method was successfully applied on valves, arterial prostheses, bone, skin biopsies, bone marrow, and blood (11, 18, 20, 29, 30).
The purpose of this study was (i) the isolation and molecular identification of clinical strains of C. burnetii in Greece, (ii) the comparison of our isolates with the reference strains by PCR-restriction fragment length polymorphism (RFLP), and the improvement of the methodology of rapid detection of C. burnetii in patient samples.
In this study, we report the isolation of eight strains of C. burnetii from Greek patients, the identification of these strains by PCR-RFLP with material from cell cultures, and the direct detection of the pathogen by nested PCR in buffy coat samples within 4 h.
MATERIALS AND METHODS
Our laboratory is the National Reference Centre of Parasitology, Zoonoses, and Geographical Medicine and a collaborating center of the World Health Organization. Over a period of 6 years (1989 to 1995), serum samples from 3,300 patients suspected to be infected by C. burnetii were assayed for the presence of antibodies against antigen phase II of the microorganism. Using the indirect immunofluorescence antibody technique (IFAT), we considered titers of immunoglobulin G (IgG) of ≥1/960 or titers of IgM of ≥1/400 and/or a fourfold increase of the titers between two assays as a strong indication of acute infection. A fever of ≥38°C, respiratory disease (dyspnea, expectoration, cough, and chest pain with associated X-ray abnormalities), hepatitis (a higher-than-twofold increase in serum glutamic oxalacetic transaminase and/or serum glutamic pyruvic transaminase levels), central nervous system involvement (neurological symptoms associated with normal or abnormal cerebrospinal fluid findings), and skin rash were considered cardinal manifestations of Q fever. The diagnosis was made according to clinical and serological criteria of the disease. One hundred fifty-two cases of Q fever were recorded.
Physicians were asked to provide buffy coat samples from patients who had not received C. burnetii-specific antibiotics at admission.
Samples.
Samples from 17 patients were assayed for the detection and identification of the microorganism. For blood cultures, a 5-ml sample of heparinized blood was obtained, and after sedimentation for 40 min, the supernatant monolayer was inoculated into the shell vials.
Isolation of C. burnetii.
Human embryonic lung (HEL) fibroblasts were grown in minimum essential medium with 10% fetal calf serum and then 1% glutamine. Shell vials (3 and 7 ml; Sterilin, Felthan, England) with 12-mm-diameter coverslips were seeded with 1 ml of medium containing 50,000 cells and incubated in a 5% CO2 incubator for 3 days to obtain a confluent monolayer. A portion of the buffy coat fraction of each sample (0.5 ml) was diluted with 1 volume of growth medium.
One milliliter of the mixture was placed in each shell vial. The shell vials were centrifuged at 700 × g for 1 h at 22°C. The inoculum was then removed, and 1 ml of growth medium was added to the cells. The shell vials were incubated in a 5% CO2 incubator at 37°C. At least three shell vials were inoculated per sample. The cytopathic effect of C. burnetii in HEL and Vero cells was also observed (20).
Immunofluorescence detection of C. burnetii.
The cell monolayers in the shell vials were examined for C. burnetii by IFAT on day 6 and again on day 12 if the first test was negative. For detection of C. burnetii, human serum samples collected by our laboratory (which display a high titer of immunofluorescent antibody to C. burnetii of >1/40,000), at a dilution of 1/100, and fluorescein-conjugated goat antiserum to human IgG (dilution, 1/200) (Kallestad, Austin, Tex.) were used. Specificity was evaluated by simultaneous staining of uninoculated cell monolayers and inoculated cultures with human serum negative for antibodies to C. burnetii.
Detection by PCR and nested PCR.
The DNA extraction from buffy coat was performed with the QIAamp blood kit 250 (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. Infected cells were used for PCR detection and RFLP identification of C. burnetii. Two hundred microliters of the cell suspension was incubated in the presence of 400 ng of proteinase K per ml (stock solution, 20 mg/ml in H2O) overnight at 56°C. Subsequently, proteinase K was inactivated by boiling for 10 min, and the solution was centrifuged at 2,000 rpm for 5 min at 22°C in a Beckman GS-6R centrifuge. The supernatant was kept at −20°C.
DNA amplification.
In order to perform DNA amplification of the Greek strains and compare them with the reference strains, Nine Mile and Q212, three different genomic primers were used. Additionally, in order to ascertain whether the Greek isolates contained plasmids, two additional types of primers which have been described to indicate acute or chronic Q fever were used (Table 1) (36). Primers QpH11 and QpH12 detect plasmids present during acute Q fever, whereas primers QpRS01 and QpRS02 detect plasmids present in chronic infection due to C. burnetii (36).
TABLE 1.
Primer sequences used in this studya
| Primer | Sequence (length [bp]) |
|---|---|
| Genomic | |
| C.B.1b | 5′-ACT CAA CGC ACT GGA ACC GC-3′ |
| C.B.2b | 5′-TAG CTG AAG CCA ATT CGC C-3′ (257 bp) |
| G4131b | 5′-CTG ATG TGT CAA GTA ATG TCG G-3′ |
| G4132b | 5′-GTT CAT GGT TAT GAT TCT GCG-3′ (183 bp) |
| 16S1c | 5′-CTC CTG GCG GCG AGA GTG GC-3′ |
| 16S2Nc | 5′-GTT AGC TTC GCT ACT AAG AAG GGA ACT TCC C-3′ (779 bp) |
| Plasmidic | |
| QpRS01d | 5′-CTCGTACCCAAAGACTATGAATATATCC-3′ |
| QpRS02d | 5′-CACATTGGGTATCGTACTGTCCCT-3′ (363 bp) |
| QpH11d | 5′-TGACAAATAGAATTTCTTCATTTTGATG-3′ |
| QpH12d | 5′-GCTTATTTTCTTCCTCGAATCTATGAAT-3′ (1,042 bp) |
| Hfrag1e | 5′-ATT GCT ATC ACT GAG GGT GAC G-3′ |
| Hfrag2e | 5′-CTG ACG AAG AAG CAG CAT TAG C-3′ (508 bp) |
| HF1e | 5′-TCC TAA ACA AGT GAT GGT CTC C-3′ |
| HF2e | 5′-TTC GCA GAA GTC AGC TAT GC-3′ (183 bp) |
Cycling conditions were as follows: 94°C for 5 min with 35 (total) cycles of denaturation (at 94°C for 30 min), annealing (at 55°C for 30 min), and extension (at 72°C for 30 min). After the 35 cycles, the PCR product was held for 10 min at 72°C.
Primers CB.1 and CB.2 were derived from the C. burnetii superoxide dismutase gene, and primers G4131 and G4132 were derived from a shotgun HindIII subcloning fragment (297 bp).
Primers 16S1 and 16S2N are based on DNA sequences of 16S rRNA.
Primers QpRS01 and QpRS02 are QpRS specific, and primers QpH11 and QpH12 are QpH1 and QpDG specific.
Primers Hfrag1, Hfrag2, HF1, and HF2 were used for nested PCR.
PCR was performed with 10 μl of supernatant from the proteinase-K-treated cell suspension in a total volume of 100 μl. The PCR mixture contained 1 μM of each primer, 200 μM of each deoxynucleoside triphosphate, 2.0 mM MgCl2, and 0.5 U of Taq polymerase (GIBCO BRL Life Technologies, Gaithersburg, Md.). The primers used, as well as the cycling conditions, are listed in Table 1.
For the direct PCR detection of C. burnetii in the buffy coat, a nested PCR assay was performed with primers Hfrag1 and Hfrag2 in the first PCR and primers HF1 and HF2 in the nested PCR. The conditions used are described in Table 1. The PCR products were separated on a 2% agarose gel and visualized by UV illumination.
Restriction endonuclease digestion.
The specificity of the amplification was evaluated by restriction analysis of the PCR products. The CB1 and CB2 and Hfrag1 and Hfrag2 products from infected cells were digested with the enzymes TaqI and AluI (New England Biolabs) as previously described (29, 36). The restriction fragments were examined by electrophoresis on a 3% low-melting-point agarose gel (GIBCO BRL), stained with ethidium bromide, and viewed by UV illumination. The restriction fragments were compared with those obtained with the reference strains, Nine Mile and Q212.
RESULTS
Isolation of C. burnetii.
Three cultures were obtained from each patient prior to any administration of antibiotics. We were able to isolate C. burnetii in blood cultures from eight of 17 patients suffering from Q fever. All patients except one presented with fever and pneumonia. The last patient had had fever for 10 days upon admission. Clinical, serological, therapeutic, and culture data from these eight patients are presented in Table 2.
TABLE 2.
Clinical, serological, therapeutic, and culture data for eight Q fever patients with blood cultures positive for C. burnetii
| Patient no. | Diagnosis at admission (duration of fever [days]) | Treatment | Phase II antibody titer for:
|
Blood culture (day)c | ||
|---|---|---|---|---|---|---|
| IgM | IgG | IgA | ||||
| CB1 | Fevera (10) | Vibramycin | 3,200 | 3,840 | 200 | + (10) |
| CB2 | Pneumoniab + fever (8) | Vibramycin | 800 | 7,680 | 800 | + (12) |
| CB3 | Pneumonia + fever (6) | Vibramycin | 6,400 | 30,700 | 6,900 | + (11) |
| CB4 | Pneumonia + fever (9) | Macrolide | 51,200 | 61,440 | 12,800 | + (8) |
| CB5 | Pneumonia + fever (10) | Macrolide | 102,400 | 122,880 | 200 | + (6) |
| CB6 | Pneumonia + fever (6) | Macrolide | 3,200 | 7,680 | 800 | + (10) |
| CB7 | Pneumonia + fever (8) | Vibramycin | 6,400 | 30,720 | 12,800 | + (8) |
| CB8 | Pneumonia + fever (5) | Macrolide | 800 | 3,840 | 200 | + (12) |
Body temperature greater than or equal to 38°C.
Dyspnea, cough, expectoration, hemoptysis, or chest pain associated with chest radiographic abnormalities.
+, positive shell vial cultures.
The microorganism was detected between days 6 and 12 by IFAT, Gimenez staining, and PCR (7). The subcultures derived from the initial shell vial were considered heavily infected after 15 to 21 days of incubation. At this point, the voluminous vacuolar formations in the cell cytoplasm, due to the cytopathic effect of C. burnetii, were prominent (20).
PCR identification.
C. burnetii-specific sequences were amplified by PCR from DNA derived from infected Vero or HEL cells. For further characterization of the isolates, PCR was carried out with infected cells with the genomic primers CB1 and CB2 and the plasmidic primers QpH11 and QpH12, which were initially designed as primers specific for acute infection (29, 36). Our results showed clearly that the specific PCR product appeared in all our samples and the Nine Mile strain but not in the Q212 strain (36). On the other hand, the primers QpRS01 and QpRS02, which were referred to as amplifying only chronic infection from specific sequences, did not give any amplification with our samples and the Nine Mile strain but produced a PCR-specific product with the Q212 strain (36).
The PCR methods were not sensitive enough to detect C. burnetii directly in the clinical samples (buffy coat, blood, etc.).
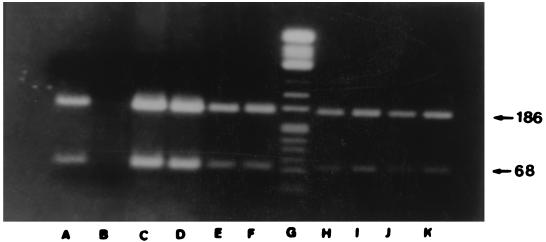
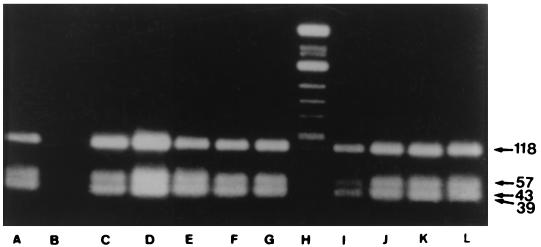
The assay specificity has been previously evaluated by restricted digestion of the PCR products (29). PCR products obtained (with primers CB1 and CB2) from our samples and the reference strains were subsequently digested with AluI, resulting in the generation of fragments of 186, 68, and 3 bp (Fig. 1), whereas cleavage with TaqI gave fragments of 118, 57, 43, and 39 bp (Fig. 2). The product of amplification with primers Hfrag1 and Hfrag2 in infected cells was digested with TaqI, and the sizes of the generated fragments were identical to those of the fragments derived from the reference strains, Nine Mile and Q212 (36).
FIG. 1.
Restriction endonuclease profile analysis of the 257-bp amplification products of eight C. burnetii isolates digested with AluI. Lanes: A, strain Nine Mile; B, reagent control; C to F, four C. burnetii isolates; G, molecular size markers (ΦX174 cleaved with HinfI); H to K, four C. burnetii isolates.
FIG. 2.
Restriction endonuclease profile analysis of the 257-bp amplification products of eight C. burnetii isolates digested with TaqI. Lanes: A, strain Nine Mile; B, reagent control; C, strain Q212; D to G, four C. burnetii isolates; H, molecular size markers (ΦX174 cleaved with HinfI); I to L, four C. burnetii isolates.
Nested PCR detection.
Detection of C. burnetii by nested PCR (Fig. 3 and 4) succeeded only with buffy coat samples by the DNA extraction method with the QIAamp blood kit. When whole blood was used, we were unable to detect a positive signal, even in the samples from which C. burnetii had been previously isolated. The nested PCR test for the detection of C. burnetii was modified from the one previously described (36) in three ways. (i) Extraction of DNA was done by us with the QIAamp blood kit instead of by the traditional boiling method (36), (ii) the temperatures were changed, and (iii) the times of recycling were modified (Table 1). Thus, the overall duration of nested PCR was 4 h.
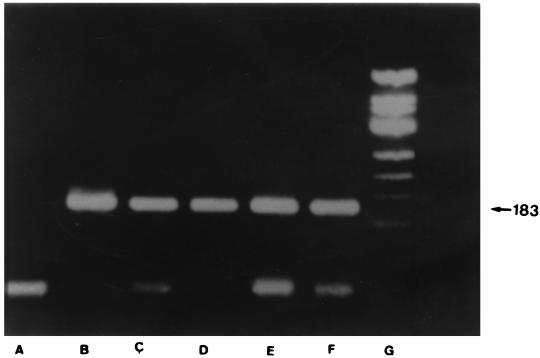
FIG. 3.
Direct detection of C. burnetii in buffy coat by nested PCR. Amplification products (183 bp in size) from four buffy coat samples. An agarose gel electrophoretogram of amplified DNA after 35 cycles of amplification. Lanes: A, reagent control; B to E, four buffy coat samples; F, strain Nine Mile; G, molecular size markers (ΦX174 cleaved with HaeIII).
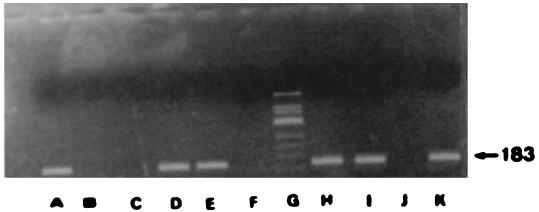
FIG. 4.
Direct detection of C. burnetii in buffy coat by nested PCR. Amplification products (183 bp in size) from four buffy coat samples. An agarose gel electrophoretogram of amplified DNA after 35 cycles of amplification. Lanes: A, strain Nine Mile; B, reagent control; C, buffy coat negative control; D, strain Q212; E, buffy coat sample; F, reagent control; G, molecular size markers (ΦX174/HinfI); H and I, buffy coat samples; J, buffy coat negative control; K, buffy coat sample.
C. burnetii was also isolated from the same samples by culture. The product obtained by the nested PCR had a size of 183 bp (Fig. 3).
DISCUSSION
C. burnetii is an obligate intracellular parasite that was isolated initially in animals (guinea pigs) and embryonated chicken eggs and later in cell cultures (Vero and L929). These modes of isolation are time-consuming, hazardous, and restricted to specialized laboratories. A less-hazardous technique for the isolation of C. burnetii was proposed by Raoult et al., who used a simplified shell vial culture system (20). Although this rapid culture radically accelerates the identification of C. burnetii, the process is still too lengthy to be employed in everyday clinical practice. The shell vial technique proved to be very efficient and able to yield large quantities of C. burnetii for further studies of strain identification and antibiotic susceptibility (22, 26). Although the genome of C. burnetii is still thought to be highly conserved, previous studies have shown that C. burnetii isolates can be differentiated by RFLP (9, 32) and/or plasmid DNA content (27, 36). The first C. burnetii plasmid, QpH1, was isolated and described by Samuel et al. (28). This low-copy-number plasmid (13) was obtained from tick isolate Nine Mile, the prototype strain of acute Q fever. Another plasmid, QpRS, also described by Samuel et al. (27), was obtained from a goat placenta C. burnetii isolate and found to be common to most of the chronic Q fever isolates (14).
PCR-RFLP is useful for detection and identification of C. burnetii in early shell vial cultures, for diagnosis of both acute and chronic infections, and for detection of the bacteria in certain clinical specimens (heart valves) (29).
The classification of C. burnetii strains into acute and chronic isolates by PCR is still preliminary. It is not yet known whether the virulence potential of C. burnetii is encoded by plasmids or genomic sequences or dependent on host factors as well (31, 33). We used both primers (genomic and plasmidic) in order to confirm the above differentiation.
All our patients presented the clinical manifestations of acute Q fever infection. Follow-up analysis of IgM and IgG showed a decline in antibody titers. No other clinical manifestations (e.g., cardiac or chronic hepatic involvement, etc.) were present during a 2- to 3-year follow-up period.
Our strains were detected with the primers QpH11 and QpH12, derived from plasmids of strains associated with acute Q fever, but not with primers derived from strains associated with chronic Q fever.
Recently, a nested PCR approach was used for the highly sensitive and specific direct detection of C. burnetii in clinical samples collected from animals and humans, with primers based on conserved plasmid sequences (33, 36).
However, this technique also proved inconvenient, since the procedure times are not yet appropriate for the everyday clinical practice and inhibitors of the reaction can falsify the results. Our method, which involved changing the DNA extraction procedures and optimizing the temperature and time conditions, improved the time required for the PCR procedure, making results available within 4 h.
This study presents the first successful attempt to isolate C. burnetii in Greece from patients suffering from the acute form of the infection.
In conclusion, we have successfully isolated C. burnetii in Greece from eight Greek patients by the shell vial assay. Isolates were detected and identified by molecular biology techniques. Optimization of nested PCR conditions allowed direct detection of C. burnetii within 4 h. The strains isolated did not differ from the standard reference strains.
REFERENCES
- 1.Aitken I D, Bogel K, Cracea E, Edlinger E, Houwers D, Krauss H, Rady M, Rehacek J, Schiefer H G, Schmeer N, Tarasevich I V, Tringali G. Q fever in Europe: curent aspects of aetiology, epidemiology, human infection diagnosis and therapy. Infection. 1987;15:323–327. doi: 10.1007/BF01647731. [DOI] [PubMed] [Google Scholar]
- 2.Caminopetros J. La fièvre Q en Grèce. Le lait source de l’infection pour l’homme et les animaux. Ann Parasitol Hum Comp. 1948;23:107–108. [PubMed] [Google Scholar]
- 3.Coyle P V, Connolly J H, Adgey A A J. Q fever endocarditis in Northern Ireland. Lancet. 1983;i:411. doi: 10.1016/s0140-6736(83)91520-9. [DOI] [PubMed] [Google Scholar]
- 4.Derrick E H. The course of infection with Coxiella burnetii. Med J Aust. 1973;1:1051–1057. [PubMed] [Google Scholar]
- 5.Dupont H T, Raoult D, Brouquil P, Janbon F, Peyramond D, Weiller P J, Chicheportiche C, Nezri M, Poirier R. Epidemiologic features and clinical presentation of acute Q fever in hospitalized patients: 323 French cases. Am J Med. 1992;93:427–434. doi: 10.1016/0002-9343(92)90173-9. [DOI] [PubMed] [Google Scholar]
- 6.Dupuis G, Peter O, Pedroni D. Aspects cliniques observés lors d’une épidémie de 415 cas de fièvre Q. Schweiz Med Wochenschr. 1985;115:814–818. [PubMed] [Google Scholar]
- 7.Gimenez D F. Staining rickettsiae in yolk-sac cultures. Stain Technol. 1964;39:135–140. doi: 10.3109/10520296409061219. [DOI] [PubMed] [Google Scholar]
- 8.Haldane E V, Marrie T J, Faulkner R S, Lee S H S, Cooper J H, MacPherson D D, Montague T J. Endocarditis due to Q fever in Nova Scotia: experience with five patients in 1981–1982. J Infect Dis. 1983;148:978–985. doi: 10.1093/infdis/148.6.978. [DOI] [PubMed] [Google Scholar]
- 9.Heinzen R A, Stiegler G L, Whitting L L, Schmitt A S, Mallavia L P, Frazier M E. Use of pulsed gel electrophoresis to differentiate Coxiella burnetii strains. Ann N Y Acad Sci. 1990;590:504–513. doi: 10.1111/j.1749-6632.1990.tb42260.x. [DOI] [PubMed] [Google Scholar]
- 10.Kimbrough R C, III, Ormsbee R A, Peacock M, Rogers W R, Bennetts R W, Raaf J, Krause A, Gardner C. Q fever endocarditis in the United States. Ann Intern Med. 1979;91:400–402. doi: 10.7326/0003-4819-91-3-400. [DOI] [PubMed] [Google Scholar]
- 11.Levy P V, Drancourt M, Etienne J, Auvergnat J C, Beytout J, Sainty J M, Goldstein F, Raoult D. Comparison of different antibiotic regimens for therapy of 32 cases of Q fever endocarditis. Antimicrob Agents Chemother. 1991;35:533–537. doi: 10.1128/aac.35.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mallavia L P, Whiting L L, Minnick M F, Heinzen R, Reschke D, Foreman M, Baca O G, Frazier M E. Strategy for detection and differentiation of Coxiella burnetii strains using the polymerase chain reaction. Ann N Y Acad Sci. 1990;590:572–581. doi: 10.1111/j.1749-6632.1990.tb42268.x. [DOI] [PubMed] [Google Scholar]
- 13.Mallavia L P, Samuel J E, Kahn M L, Thomashow L S, Frazier M E. Coxiella burnetii plasmid DNA. In: Leive L, Schlessinger D, editors. Microbiology—1984. Washington, D.C: American Society for Microbiology; 1984. pp. 293–296. [Google Scholar]
- 14.Mallavia L P, Samuel E J. Genetic diversity of Coxiella burnetii strains. In: Moulder J, editor. Intracellular parasitism. Boca Raton, Fla: CRC Press; 1989. pp. 117–126. [Google Scholar]
- 15.Marrie T J. Q fever, 1979–1987, Nova Scotia. Can Dis Wkly Rep. 1988;14:69–70. [PubMed] [Google Scholar]
- 16.Marrie T J. Epidemiology of Q fever. In: Marrie T J, editor. Q fever. The disease. Boca Raton, Fla: CRC Press; 1990. pp. 49–70. [Google Scholar]
- 17.Montejo Baranda M, Corral Carranceja J, Aquirre Errasti C. Q fever in the Basque Country: 1981–1984. Rev Infect Dis. 1985;7:700–701. doi: 10.1093/clinids/7.5.700. [DOI] [PubMed] [Google Scholar]
- 18.Mühlemann K, Matter L, Meyer B, Schopfer K. Isolation of Coxiella burnetii from heart valves of patients treated for Q fever endocarditis. J Clin Microbiol. 1995;33:428–431. doi: 10.1128/jcm.33.2.428-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musso D, Raoult D. Coxiella burnetii blood cultures from acute and chronic Q fever patients. J Clin Microbiol. 1995;33:3129–3132. doi: 10.1128/jcm.33.12.3129-3132.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raoult D, Vestris G, Enea M. Isolation of 16 strains of Coxiella burnetii from patients by using a sensitive centrifugation cell culture system and establishment of the strains in HEL cells. J Clin Microbiol. 1990;28:2482–2484. doi: 10.1128/jcm.28.11.2482-2484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raoult D, Stein A. Q fever during pregnancy—a risk for women, fetuses, and obstetricians. N Engl J Med. 1994;330:371. doi: 10.1056/nejm199402033300518. [DOI] [PubMed] [Google Scholar]
- 22.Raoult D, Torres H, Drancourt M. Shell-vial assay: evaluation of a new technique for determining antibiotic susceptibility, tested in 13 isolates of C. burnetii. Antimicrob Agents Chemother. 1991;35:2070–2077. doi: 10.1128/aac.35.10.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raoult D, Marrie T. Q fever. Clin Infect Dis. 1995;20:489–496. doi: 10.1093/clinids/20.3.489. [DOI] [PubMed] [Google Scholar]
- 24.Raoult D. Q fever: still a query after all these years. J Med Microbiol. 1996;44:77–78. doi: 10.1099/00222615-44-2-77. [DOI] [PubMed] [Google Scholar]
- 25.Raoult D. La fièvre Q: infection à Coxiella burnetii. Encycl Medico-Chir (Paris) 1988;8077:1–10. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Samuel J E, Frazier M E, Mallavia L P. Correlation of plasmid type and disease caused by Coxiella burnetii. Infect Immun. 1985;49:775–779. doi: 10.1128/iai.49.3.775-779.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuel J E, Frazier M E, Kahn M L, Thomashow L S, Mallavia L P. Isolation and characterization of a plasmid from phase I Coxiella burnetii. Infect Immun. 1983;41:488–493. doi: 10.1128/iai.41.2.488-493.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein A, Raoult D. Detection of Coxiella burnetii by DNA amplification using polymerase chain reaction. J Clin Microbiol. 1992;30:2462–2466. doi: 10.1128/jcm.30.9.2462-2466.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein A, Raoult D. Phenotypic and genotypic heterogeneity of 8 new human Coxiella burnetii isolates. Acta Virol. 1992;36:7–12. [PubMed] [Google Scholar]
- 31.Stein A, Raoult D. Lack of pathotype specific gene in human Coxiella burnetii isolates. Microb Pathog. 1993;15:177–185. doi: 10.1006/mpat.1993.1068. [DOI] [PubMed] [Google Scholar]
- 32.Thiele D, Willems H, Kopf G, Krauss H. Polymorphism in DNA restriction patterns of Coxiella burnetii isolates investigated by pulsed field electrophoresis and image analysis. Eur J Epidemiol. 1993;9:419–425. doi: 10.1007/BF00157400. [DOI] [PubMed] [Google Scholar]
- 33.Thiele D, Willems H. Is plasmid based differentiation of Coxiella burnetii in ‘acute’ and ‘chronic’ isolates still valid? Eur J Epidemiol. 1994;10:427–434. doi: 10.1007/BF01719667. [DOI] [PubMed] [Google Scholar]
- 34.Tselentis Y, Gikas A, Kofteridis D, Kyriakakis E, Lydataki N, Bouros D, Tsaparas N. Q fever in the Greek island of Crete: epidemiologic, clinical, and therapeutic data from 98 cases. Clin Infect Dis. 1995;20:1311–1316. doi: 10.1093/clinids/20.5.1311. [DOI] [PubMed] [Google Scholar]
- 35.Vellend H, Salit I E, Spence L. Q fever—Ontario. Can Dis Wkly Rep. 1982;8:171. [Google Scholar]
- 36.Willems H, Thiele D, Krauss H. Plasmid based differentiation and detection of Coxiella burnetii in clinical samples. Eur J Epidemiol. 1993;9:411–418. doi: 10.1007/BF00157399. [DOI] [PubMed] [Google Scholar]