Abstract
There is a need to develop noninvasive methods for the diagnosis of Pneumocystis carinii pneumonia in patients unable to undergo bronchoscopy or induction sputum. Oral wash specimens are easily obtained, and P. ca- rinii nucleic acid can be amplified and demonstrated by PCR. In routine clinical use, easy sample processing and single-round PCR are needed to ensure rapid analysis and to reduce the risk of contamination. We developed a single-round Touchdown PCR (TD-PCR) protocol with the ability to detect PCR inhibition in the specimen. The TD-PCR was evaluated in a routine diagnostic laboratory and was compared to a previously described PCR protocol (mitochondrial RNA) run in a research laboratory. Both PCR methods amplified a sequence of the mitochondrial rRNA gene of P. carinii. Paired bronchoalveolar lavage (BAL) and oral wash specimens from 76 consecutive human immunodeficiency virus type 1-infected persons undergoing a diagnostic bronchoscopy were included. The TD-PCR procedure was quicker than the mitochondrial PCR procedure (<24 versus 48 h) and, compared to microscopy, had sensitivity, specificity, and positive and negative predictive values of 89, 94, 93, and 91%, respectively, for oral wash specimens and 100, 91, 90, and 100%, respectively, for BAL specimens. Our results suggest that oral wash specimens are a potential noninvasive method to obtain a diagnostic specimen during P. carinii pneumonia infection and that it can be applied in a routine diagnostic laboratory.
For years, diagnosis of Pneumocystis carinii pneumonia (PCP) has relied on microscopic visualization of P. carinii in specimens obtained from the lung either by bronchoalveolar lavage (BAL) or by induction of sputum. Currently, the “gold standard” for diagnosis of PCP involves colorimetric and immunofluorescent stains of BAL fluid, with sensitivity and specificity values of >95% (11, 16). In contrast, the sensitivity of conventionally stained induced-sputum specimens is variable (45 to 78%) (8, 16, 25). PCR-based assays have been shown to increase diagnostic sensitivity in induced-sputum specimens with small numbers of P. carinii organisms (6, 8, 21, 24–26, 34). However, less-invasive diagnostic procedures are needed for patients with severe respiratory distress or tendency to bleed, because both BAL and induced-sputum methods may prove difficult, contraindicated, or unpleasant for the patient.
PCR amplification of noninvasive serum or blood samples has shown conflicting results, with sensitivities ranging from 0 to 100% (3, 9, 12, 19, 28, 29, 32). Currently, the use of such specimens remains to be established (12, 19).
PCR detection of P. carinii in oral wash specimens represents an alternative noninvasive method. Oral wash specimens are obtained by having the patient rinse and gargle the mouth in sterile saline. By this method, Wakefield et al. detected P. carinii DNA in 78% of 18 human immunodeficiency virus (HIV)-positive patients with confirmed PCP and in none of 13 patients without PCP (35). Atzori et al. have reported a sensitivity of 90% using a nested PCR on oral wash specimens from 10 HIV-positive patients (2). We have recently used a similar method on specimens from 26 patients with hematological malignancies (14). We detected P. carinii in oral wash specimens of all 8 patients with verified PCP using a simple extraction method and a Touchdown PCR (TD-PCR). Oral wash specimens are easy to obtain even among severely ill patients unable to undergo more invasive procedures.
In order to use PCR in clinical diagnostic laboratories, easy sample extraction and, when possible, single-round PCR methods are needed to avoid the possibility of contamination by nested PCR. Furthermore, a large number of samples are needed to determine the diagnostic sensitivity and specificity of a new PCR with oral wash specimens for the diagnosis of PCP. We prospectively evaluated the use of two different PCR protocols on oral wash specimens for the diagnosis of PCP and compared them to PCR and microscopic examination of BAL. The principal aim of this study was to develop a single-round PCR applicable in routine laboratories which could generate a clinical usable answer within 1 working day.
MATERIALS AND METHODS
Patient specimens.
Paired samples of BAL and oral wash specimens from 76 HIV-infected patients undergoing diagnostic bronchoscopy due to respiratory symptoms were collected between January 1995 and March 1997 at the Department of Infectious Diseases, Hvidovre Hospital, Copenhagen, Denmark. Oral wash specimens were collected prior to bronchoscopy by patients rinsing and gargling their oral cavities with 10 ml of sterile saline for 1 min. All BAL specimens and 10 oral wash specimens obtained from patients with microscopic verified P. carinii organisms in BAL were stained with Giemsa and immunofluorescence stains (Genetic Systems, Seattle, Wash.) as previously described (16, 30). A case of PCP was defined by detection of P. carinii organisms in the BAL specimens by microscopy. From seven patients, more than one oral wash specimens (two to eight per patient) were collected during the treatment of PCP.
Control specimens.
A total of 60 paired BAL and oral wash specimens were collected from 41 immunosuppressed heart and lung transplant patients during routine control bronchoscopy after transplantation.
DNA extraction.
DNA was extracted from BAL by phenol-chloroform extraction or by Chelex extraction.
(i) Phenol-chloroform extraction.
A 2-ml volume of BAL or oral wash specimens was centrifuged at 3,000 × g for 15 min. The pellet was digested by proteinase K at 60°C for 2 h, followed by boiling for 10 min, and DNA was extracted by phenol-chloroform as previously described; 2 μl was used for PCR (36).
(ii) Chelex extraction.
A 2-ml volume of BAL or oral wash fluid was centrifuged at 20,000 × g for 15 min. The pellet was mixed with 300 μl of 20% (wt/vol) Chelex 100 (Bio-Rad, Richmond, Calif.) in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), vortexed for 1 min, and incubated at 95°C for 10 min (37). After centrifugation at 20,000 × g for 5 min, 2 μl of supernatant was used for PCR.
Plasmid DNA.
A plasmid containing the mitochondrial RNA (mtRNA) insert was generated by cloning PCR products into the pCRII vector with the TA cloning kit (Invitrogen, San Diego, Calif.). A 346-bp mtRNA DNA fragment was generated via PCR (GeneAmp PCR kit; Perkin-Elmer Cetus, Norwalk, Conn.) with the pAZ102-H and pAZ102E primers by using DNA extracted from a P. carinii sp. f. hominis-infected lung. Both mitochondrial PCR (mtPCR) and TD-PCR detected a dilution of 1:4,000 of the plasmid.
Internal process control for inhibition.
In order to detect the presence of Taq DNA polymerase inhibitors or subobtimal reaction conditions, an internal process control was constructed. Primers amplifying a part of the phage lambda genome were synthesized. The primers entailed the sequences of each of the P. carinii-specific primers added to the 5′ end of the corresponding lambda primer. PCR products thus containing the binding sites of the P. carinii primers were obtained by amplification of 1 ng of purified lambda DNA with an annealing temperature of 40°C. After gel purification of the amplicons, a 10-fold titration was performed. The dilution of the internal process control, which produced no increase in the detection limit of the P. carinii-positive control, was used in the assay.
DNA amplification.
Two PCR methods were evaluated, i.e., the PCR method previously described by Wakefield (mtPCR) and a TD-PCR. Both methods amplify a fragment of the P. carinii mitochondrial large-subunit rRNA gene by using the primers pAZ102-H and pAZ102E (36). In addition, a PCR amplifying the small-subunit rRNA of P. carinii with the primers pAZ112-10F and pAZ112-10R (15) was used as a confirmatory PCR. Finally, a previously described PCR detecting human beta globulin was used as a process control for inhibition in the mtRNA PCR (4).
TD-PCR.
All reactions were performed with thin-walled GeneAmp reaction tubes (Perkin-Elmer, Birkerød, Denmark) in a final volume of 100 μl containing 1× PCR buffer (Perkin-Elmer) (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 0.01% [wt/vol] gelatin) with 0.01% bovine serum albumin (Sigma Chemical Company, St. Louis, Mo.), 125 mM (each) dATP, dGTP, and dCTP, 250 mM dUTP, 4.5 mM MgCl2 (final concentration), 0.4 mM each primer, and an internal process control in order to control for inhibition. Taq DNA polymerase (2 U of AmpliTaq Gold; Perkin-Elmer) was used in each reaction. Thermocycling was performed in a Perkin-Elmer 9600 thermocycler. After an initial preheating step for 10 min at 95°C to achieve a hot start procedure (7), a TD procedure (10, 13) consisting of denaturation at 94°C for 15 s and annealing at 72 to 62°C for 30 s with a 1°C decrement per cycle was applied during the first 10 cycles. The subsequent cycles (no. 11 to 50) each consisted of steps 92°C for 15 s, 62°C for 30 s, and 72°C for 15 s. After the last cycle, an extension step of 72°C for 5 min was included. The amplified samples were subjected to agarose gel electrophoresis, resulting in detection of a 346-bp P. carinii amplicon and an internal process control amplicon with a size of 656 bp.
Confirmatory PCR.
To validate the PCR-positive results, a confirmatory PCR was done by amplifying the small-subunit rRNA gene (ssuPCR) of P. carinii (15). All PCR results detected by TD-PCR were confirmed by amplifying P. carinii small-subunit rRNA in a PCR assay under the same reaction conditions as those for the primary TD-PCR assay, except that 2.5 mM MgCl2 (final concentration) was used. After activation of AmpliTaq Gold, the cycling conditions consisted of 10 cycles of denaturation at 94°C for 15 s, annealing at 65 to 55°C for 30 s, with a 1°C decrement per cycle, and extension at 72°C for 60 s. The subsequent cycles, i.e., cycles 11 to 50, each consisted of steps at 92°C for 15 s, 55°C for 30 s, and 72°C for 60 s. After the last cycle, an extension step at 72°C for 5 min was included. The amplicon was 423 bp long and was visualized on an agarose gel.
mtPCR.
mtPCR was done according to the method described by Wakefield et al. (36). Amplification products were visualized on an agarose gel by oligoblotting with a BlueGENE detection system (BRL, Gaithersburg, Md.) (30). A previously described human beta globulin PCR was performed to validate the presence of DNA in the samples tested (4).
For all PCRs, two positive controls were incorporated (DNA extracted from a P. carinii-infected autopsy lung homogenate); one corresponded to the detection limit, and the second was 10-fold more concentrated. Furthermore, two negative controls were included; one was distilled water, and the other was distilled water subjected to pretreatment along with the clinical specimens. All specimens were tested in a blind fashion.
Data analysis.
For comparison of the sensitivities of different diagnostic tests, the χ2 test was used. A two-sided P value of <0.05 was considered significant.
RESULTS
Paired oral wash and BAL specimens from 76 HIV-positive patients who underwent diagnostic bronchoscopy due to respiratory symptoms were evaluated by two PCR methods (Table 1). Detection of P. carinii organisms in Giemsa- and immunofluorescence-stained BAL specimens was considered a gold standard for the diagnosis of PCP (Table 1). Only 10 of the PCR-positive oral wash specimens from patients with confirmed PCP were evaluated by microscopy using immunofluorescent stains, since all 10 were negative.
TABLE 1.
Specimens obtained from 76 HIV-positive patients and 41 control patients evaluated by two different PCR methodsa
| PCR method(s) | No. of HIV-positive patients | No. of patients with PCP (%)a | No. of control patients |
|---|---|---|---|
| TD-PCR and mtPCR | 32 | 14 (44) | |
| TD-PCR only | 29 | 14 (48) | 41b |
| mtPCR only | 15 | 10 (66) | |
| Total | 76 | 38 (50) | 41 |
PCP according to detection of P. carinii in BAL specimens by Giemsa and immunofluorescence staining.
None of the patients had microscopically verified or clinical PCP in 60 BAL specimens.
TD-PCR.
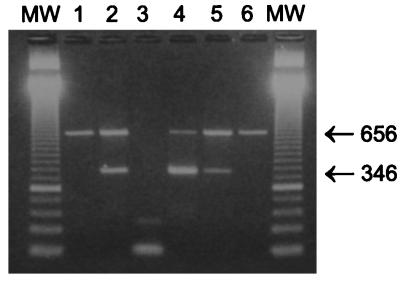
Sixty-one paired oral wash and BAL specimens were evaluated by TD-PCR in a laboratory handling routine PCRs for various pathogens. A lambda DNA internal process control, which was included to control for inhibition of the PCR, was added to the same reaction tube as the clinical specimen. Amplification products were visualized on agarose gels, as shown in Fig. 1, in which lanes 1 and 6 are true negative samples amplifying the internal process control DNA at 656 bp. Lanes 2, 4, and 5 are P. carinii-positive samples amplifying both internal process control and P. carinii DNA at 346 bp, whereas lane 3 is a false negative, since no internal process control is present due to inhibition of the PCR. In this way, inhibition was observed in 5 of 85 oral wash specimens and in 1 of 61 BAL specimens; however, in all cases inhibition could be circumvented by diluting the sample 1:2.
FIG. 1.
PCR results after DNA extraction by Chelex 100. PCR products were subjected to agarose gel electrophoresis and stained with ethidium bromide. MW, molecular weight marker. Lanes: 1 and 6, true-negative samples amplifying the internal process control at 656 bp; 2, 4, and 5, P. carinii-positive samples amplifying both the internal process control and P. carinii DNA at 346 bp; 3, false-negative sample, since no internal process control was present due to inhibition of the PCR.
A total of 25 oral wash specimens as well as 28 BAL specimens from 28 patients with confirmed PCP were PCR positive, as shown in Table 2. All results generated by the TD-PCR were confirmed by the confirmatory ssuPCR. When microscopy was considered a gold standard for the diagnosis of PCP, the sensitivity, specificity, and positive and negative predictive values of TD-PCR were 89, 94, 93, and 91, respectively, for oral wash specimens and 100, 91, 90, and 100%, respectively, for BAL specimens.
TABLE 2.
Detection of P. carinii by TD-PCR in paired oral wash and BAL specimens obtained from HIV-infected patients
| Specimen source | No. of specimens (%)a
|
|||
|---|---|---|---|---|
| PCR+/PCP+ | PCR−/PCP− | PCR−/PCP+ | PCR+/PCP− | |
| Oral wash | 25 (89) | 31 (94) | 3 | 2 |
| BAL | 28 (100) | 30 (91) | 0 | 3 |
The total number of patients was 61, of whom 28 were diagnosed with PCP according to detection of P. carinii in BAL specimens by Giemsa and immunofluorescence staining.
Discrepant results between PCR and microscopy were found in three BAL and two oral wash specimens from a total of three patients without microscopy-verified PCP (Table 3). One patient was on a regimen of inhaled-pentamidine prophylaxis, had clinical PCP, and received therapy but died after 10 days; no autopsy was performed. The second patient did not receive prophylaxis prior to bronchoscopy, and had a CD4 cell count of 39 cells/μl and Mycobacterium tuberculosis was isolated from the BAL fluid. After bronchoscopy, the patient started an antituberculosis medication regimen and PCP prophylaxis but died 1.5 months later. Autopsy verified the diagnosis of pulmonary tuberculosis but not of PCP. The third patient had a CD4 cell count of 182 cells/μl and was started on antiretrovirus therapy and PCP prophylaxis 4 days prior to bronchoscopy. No lung diagnosis could be made based on routine examination of the BAL specimens, and the patient had not developed PCP at 4 months of follow-up while still receiving PCP prophylaxis.
TABLE 3.
TD-PCR-positive specimens without microscopically verified P. carinii in BAL
| Patient no. | PCR result (BAL/oral wash) | CD4 cell count/μl | Lung diagnosis | Prophylaxis against PCP
|
Development of PCP (time of follow-up) | |
|---|---|---|---|---|---|---|
| Prior to BAL | After BAL | |||||
| 1 | +/− | 5 | PCPa | Yes | No | Dead (day 10 after BAL) |
| 2 | +/+ | 39 | Tuberculosis | No | Yes | Dead (day 40 after BAL, no PCP) |
| 3 | +/+ | 182 | ?b | Yes (4 days) | Yes | No PCP (after 6 mo) |
Received pentamidine intravenously because of suspicion of PCP; no P. carinii organisms detected in BAL specimens.
?, no specific pathogen was identified.
From seven HIV-positive patients, serial oral wash specimens were obtained from the day on which each patient was started on anti-PCP therapy (Table 4). Thirty-one specimens from these seven patients were evaluated by TD-PCR. Seven specimens obtained from three patients were negative, and five of these were obtained from the same patient 2 or more days after therapy was initiated (Table 4). For two of the patients, positive results of the PCR test were obtained after one negative test. Positive results could be detected up to day 12 after the introduction of therapy.
TABLE 4.
Detection of P. carinii by TD-PCR in serially collected oral wash specimens from seven patients with confirmed PCP
| Patient no. | Detection of P. carinii DNA in oral wash specimens at the following days after initiation of anti-PCP treatment:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 12 | |
| 1 | + | + | + | + | + | + | ||||
| 2 | + | + | + | |||||||
| 3 | + | + | + | + | − | |||||
| 4 | − | + | + | + | + | |||||
| 5 | + | + | ||||||||
| 6 | + | + | ||||||||
| 7 | + | + | − | − | − | − | + | − | ||
As a control, 60 paired BAL and oral wash specimens from 41 immunosuppressed heart and lung transplant patients were evaluated by this method. In one patient, oral wash and BAL specimens were positive by TD-PCR as well as by the confirmatory ssuPCR, but routine stains were negative for P. carinii. Specimens were obtained from this patient 2 weeks after a double lung transplant; when the patient was receiving high-dose steroids due to rejection of the transplant as well as bactrim prophylaxis, the patient did not develop clinical PCP.
mtPCR.
Prior to the development of TD-PCR, 47 HIV-positive patients were evaluated by the more-laborious phenol- chloroform extraction method and a previously described PCR performed in a research laboratory (36). P. carinii DNA was detected in oral wash specimens obtained from 71% (17 of 24) of the patients and in 96% (23 of 24) of the BAL specimens from patients with confirmed PCP. No P. carinii DNA was detected in any of the oral wash or BAL specimens from patients without PCP. The sensitivity, specificity, and positive and negative predictive values of mtPCR were 71, 100, 100, and 77%, respectively, for oral wash specimens and 96, 100, 100, and 100%, respectively, for BAL specimens. A human beta globulin PCR was negative in four of seven oral wash specimens from patients with confirmed PCP. In total, inhibition was present in nine oral wash specimens and in one BAL specimen, generating false-negative results.
Comparison of the two PCR methods could be made for 32 paired original specimens (Table 1) which were available after the specimens had been processed for mtPCR. For both TD-PCR and mtPCR, the detection limit was the same dilution of a plasmid containing the mtPCR amplicon of P. carinii (1:4,000). Agreement between the two PCR methods was found among 28 paired specimens. In four patients, discrepancies between mtPCR and TD-PCR were observed. In three patients with PCP, oral wash specimens were TD-PCR positive but mtPCR negative. For one patient without microscopic or clinical PCP, TD-PCR was positive only for the BAL specimen, whereas the mtPCR result was negative. Among 14 patients with confirmed PCP (Table 1), TD-PCR and mtRNA detected P. carinii DNA in oral wash specimens from 79% (11 of 14) and from 57% (8 of 14), respectively (P = 0.22).
DISCUSSION
In this study, PCR detection of P. carinii DNA in oral wash and BAL fluid specimens was compared to microscopic detection of P. carinii in BAL fluid. An improved single-round PCR method with the ability to detect PCR inhibition in the specimen was developed.
Wakefield et al. first reported the use of PCR for oral wash specimens in detecting P. carinii (35). Among 18 patients with PCP, 14 could be diagnosed by oral wash specimens by using both PCR and oligoblotting. PCR was negative for all 13 patients without verified PCP. With the ITS primers and a nested PCR, preliminary data have indicated that P. carinii DNA could be detected in oral wash specimens from 9 of 10 patients with PCP (2). We have recently evaluated oral wash specimens for the diagnosis of PCP in 26 patients with hematological malignancies, detecting P. carinii DNA in 8 patients with verified PCP and in none of 18 patients without PCP (14). These studies were done in research laboratories and with a relatively small number of PCP patients; no control groups were included, and the definitive diagnosis was not systematically assessed (2, 14, 35).
Most reports evaluating PCR detection of P. carinii DNA have been done with BAL and induced-sputum specimens. Different gene targets and both single-round and nested PCRs have been evaluated. Lu and colleagues evaluated six different PCR methods and found that nested PCR was more sensitive than single-round PCR when 30 BAL specimens from PCP patients were evaluated (20). BAL has a high sensitivity in diagnosing PCP with routine stains (30), and recent reports have found that PCR detects P. carinii DNA in BAL specimens obtained from patients without clinical disease, suggesting colonization or subclinical infection (23, 26, 27). Therefore, a PCR-positive result obtained for a BAL specimen may not necessarily correlate with clinical disease caused by P. carinii (5, 18, 25–27, 31, 38). In order to introduce PCR into the routine diagnosis of PCP, the method needs to be at least as sensitive as conventional stains and a positive PCR result must correlate with clinical disease. If PCR-based methods are to be used in the routine diagnosis of PCP, simple sample extraction procedures and short procedure times, as well as the inclusion of internal inhibitor controls, are needed to detect false-negative results (1, 23, 33). Although nested PCR has been reported to increase the sensitivity of PCP detection (20, 38), in our view, single-round PCR is preferable to nested PCR in a routine laboratory to reduce the risk of contamination.
To properly evaluate the diagnostic use of oral wash specimens in detecting P. carinii, we collected paired oral wash and BAL specimens from 76 HIV-positive patients who underwent bronchoscopy due to respiratory symptoms. By using the mtPCR on oral wash specimens from 47 patients, it was possible to obtain a sensitivity of 71%, which was comparable to the 78% originally reported (35). Subsequently, we optimized the PCR methodology and had the specimens analyzed in a routine laboratory. We modified the previously described mtPCR to a more user-friendly single-round TD-PCR and evaluated it for 61 HIV-positive patients (Table 1). Specimens were extracted by Chelex 100, which is easy to do in a routine diagnostic laboratory because it requires few handling procedures and takes less than 20 min. We used the same primers as those in the mtRNA PCR but used a TD-PCR procedure to increase sensitivity in clinical specimens. There was no difference in the detection limits of a purified plasmid DNA between the two PCR methods. However, the advantage of the TD-PCR is that the procedure can detect specific DNA in clinical specimens containing a variety of different DNAs with a higher sensitivity (13).
PCR inhibition caused by the presence of PCR inhibitors in respiratory-tract specimens can be a problem; in this way, PCR inhibition has been described for 36% of 102 induced-sputum specimens, for 5% of 83 BAL specimens, and for 50% of 6 sputum samples (23). Therefore, the use of an internal process control is particularly important in respiratory-tract specimens. We found PCR inhibition in 19% of the oral wash specimens and in 2% of the BAL specimens using proteinase K extraction and mtPCR. With 2 μl of the Chelex-extracted specimen and by TD-PCR, 6% of 85 oral wash specimens inhibited the PCR. However, after dilution of the specimen, the inhibition problem was eliminated as previously described (23).
By TD-PCR, a final result could be generated within 6 h. A negative result could be inferred for all lanes containing only lambda DNA, and a presumptive positive result could be inferred for lanes with a band at 346 bp, suggesting that P. carinii cells were present in the sample (Fig. 1). A confirmatory result generated by a second PCR could be done within 24 h. By TD-PCR, the sensitivity and specificity in oral wash specimens were found to be 89 and 94%, respectively, for diagnosis of 28 patients with PCP (Table 2). With three patients, a discrepancy between direct microscopy of BAL specimens and the PCR result was observed (Table 3). For two patients, P. carinii DNA was detected in both oral wash and BAL specimens but no P. carinii organisms could be detected by direct microscopy performed by two independent laboratories blinded to the findings of the other. All three patients may have been colonized as previously described (27), and only few P. carinii organisms were present in the BAL specimens; hence, they were not detected by the microscopists. Contamination in these specimens was unlikely, since the results were confirmed by the confirmatory PCR amplifying a different gene target and also after reextraction of the original specimen.
For seven patients, serial oral wash specimens were available. P. carinii DNA could be detected in the majority of these patients up to day 12 after initiation of anti-PCP therapy. Systematically collected oral wash specimens are needed to evaluate the use of oral wash specimens in monitoring therapy. In oral wash specimens, the load of P. carinii organisms is low, since no organisms were identified by immunofluorescence in 10 PCR-positive oral wash specimens obtained from patients with verified PCP. However, when a patient has PCP, small amounts of P. carinii DNA appear to be present in the oropharynges and probably all specimens obtained from the oropharynx and nasopharynx are potentially useful in detection of PCP by PCR. This will especially be an advantage in diagnosing PCP among hematological patients often unable to sustain invasive diagnostic procedures because of bleeding tendencies (14). PCR diagnosis of PCP may be of benefit to children (17); however, the exact value of oral wash specimens in diagnosing PCP in this patient category needs to be established.
To further verify the specificity of the test, 60 oral wash and BAL specimens obtained from control bronchoscopy of 41 severely immunosuppressed heart and lung transplant patients were evaluated by TD-PCR. Only one oral wash specimen and one BAL specimen obtained from the same patient were positive. This patient was receiving a high dose of a steroid known to dispose for development of PCP and bactrim prophylaxis at the same time, which probably prevented development of clinical PCP. Paired oral wash and BAL specimens were obtained 60 days later from this patient, and P. carinii was not detected either by microscopic evaluation of BAL or by PCR. These results do not support the high prevalence of PCR-positive BAL specimens found in patients without clinical PCP (23, 27). This is not caused by a low prevalence of PCP in general, as illustrated by the findings among AIDS patients in Denmark (22), but may reflect differences in populations submitted to bronchoscopy or differences in diagnostic techniques.
In conclusion, our findings indicate that it is possible to apply the PCR technique in diagnosing PCP with oral wash specimens in a routine diagnostic laboratory. Oral wash specimens are easy to obtain, and they do not require special equipment or specially trained staff. The sample extraction method and PCR conditions used are the same as those for other respirator pathogens, which may allow testing for more than one microorganism simultaneously. Although investigation of a larger number of patients with PCP would have been preferable, our results indicate that oral wash specimens are a potentially useful noninvasive method for diagnosing PCP by PCR. This method may be especially useful for patients unable to sustain more-invasive diagnostic procedures. In addition, the possibility of detecting P. carinii DNA in oral wash specimens may facilitate study of the genotyping, epidemiology, and response to therapy of infection with P. carinii.
ACKNOWLEDGMENTS
We thank Birthe Dohn, Aase Mayer, and Lena Hansen for excellent technical assistance.
This study was supported in part by the AIDS Foundation and ECA Bio-Med 1 no. PL 941118 and by the Danish Medical Research Council (grant 9601812). Jannik Helweg-Larsen was supported by the Danish Medical Research Council (grant 9702462).
REFERENCES
- 1.Andersen A B, Thybo S, Fausset G P, Stoker N G. Polymerase chain reaction for detection of Mycobacterium tuberculosis in sputum. Eur J Clin Microbiol Infect Dis. 1993;12:922–927. doi: 10.1007/BF01992166. [DOI] [PubMed] [Google Scholar]
- 2.Atzori C, Agostoni F, Gubertini G, Cargnel A. Diagnosis of PCP by ITSs nested PCR on noninvasive oropharyngeal samples. J Eukaryot Microbiol. 1996;43:41S. doi: 10.1111/j.1550-7408.1996.tb04977.x. [DOI] [PubMed] [Google Scholar]
- 3.Atzori C, Lu J-J, Jiang B, Bartlett M S, Orlando G, Queener S F, Smith J W, Cargnel A, Lee C-H. Diagnosis of Pneumocystis carinii pneumonia in AIDS patients by using polymerase chain reactions on serum specimens. J Infect Dis. 1995;172:1623–1626. doi: 10.1093/infdis/172.6.1623. [DOI] [PubMed] [Google Scholar]
- 4.Bauer H M, Ting Y, Green C E, Chambers J C, Tashiro C J, Chimera J, Reingold A, Manos M M. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA. 1991;265:472–477. [PubMed] [Google Scholar]
- 5.Calderón E, Regordán C, Medrano F J, Ollero M, Varela J M. Pneumocystis carinii infection in patients with chronic bronchial disease. Lancet. 1996;347:977. doi: 10.1016/s0140-6736(96)91468-3. [DOI] [PubMed] [Google Scholar]
- 6.Cartwright C P, Nelson N A, Gill V J. Development and evaluation of a rapid and simple procedure for detection of Pneumocystis carinii by PCR. J Clin Microbiol. 1994;32:1634–1638. doi: 10.1128/jcm.32.7.1634-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou Q, Russel M, Birch D E, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–1723. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouaid C, Roux P, Lavard I, Poirot J L, Housset B. Use of polymerase chain reaction technique on induced-sputum samples for the diagnosis of Pneumocystis carinii pneumonia in HIV-infected patients. Am J Clin Pathol. 1995;104:72–75. doi: 10.1093/ajcp/104.1.72. [DOI] [PubMed] [Google Scholar]
- 9.Contini C, Romani R, Manganaro M, Sorice F, Delia S. Tissue culture isolation of Pneumocystis carinii from peripheral blood mononuclear cells of AIDS patients with PCP. AIDS. 1993;7:1137–1138. doi: 10.1097/00002030-199308000-00026. [DOI] [PubMed] [Google Scholar]
- 10.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elvin K, Björkman A, Linder E, Heurlin N, Hjerpe A. Pneumocystis carinii pneumonia: detection of parasites in sputum and bronchoalveolar lavage fluid by monoclonal antibodies. Br Med J. 1988;297:381–384. doi: 10.1136/bmj.297.6645.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans R, Joss A W L, Pennington T H, Ho-Yen D O. The use of a nested polymerase chain reaction for detecting Pneumocystis carinii from lung and blood in rat and human infection. J Med Microbiol. 1995;42:209–213. doi: 10.1099/00222615-42-3-209. [DOI] [PubMed] [Google Scholar]
- 13.Hecker K H, Roux K H. High and low annealing temperatures increase both specificity and yield in Touchdown and stepdown PCR. BioTechniques. 1996;20:478–485. doi: 10.2144/19962003478. [DOI] [PubMed] [Google Scholar]
- 14.Helweg-Larsen J, Jensen J S, Lundgren B. Non-invasive diagnosis of Pneumocystis carinii pneumonia in haematological patients using PCR on oral washes. Lancet. 1997;350:1365. doi: 10.1111/j.1550-7408.1997.tb05779.x. [DOI] [PubMed] [Google Scholar]
- 15.Hunter J A C, Wakefield A E. Genetic divergence at the small subunit ribosomal RNA gene among isolates of Pneumocystis carinii from five mammalian host species. J Eukaryot Microbiol. 1996;43:24S–25S. doi: 10.1111/j.1550-7408.1996.tb04962.x. [DOI] [PubMed] [Google Scholar]
- 16.Kovacs J A, Ng V L, Masur H, Leoung G, Hadley W K, Evans G, Lane F P, H C, Ognibene F P, Shelhamer J, Parillo J E, Gill V J. Diagnosis of Pneumocystis carinii pneumonia: improved detection of in induced sputum with use of monoclonal antibodies. N Engl J Med. 1988;318:583–593. doi: 10.1056/NEJM198803103181001. [DOI] [PubMed] [Google Scholar]
- 17.Leibovitz E, Pollack H, Rigaud M, Kaul A, Persaud D, Gallo L, Papellas J, Krasinski W, Borkowsky W. Polymerase chain reaction is more sensitive than standard cytologic stains in detecting Pneumocystis carinii in bronchoalveolar lavage from human immunodeficiency virus type 1-infected infants and children with pneumonia. Pediatr Infect Dis J. 1995;14:714–716. [PubMed] [Google Scholar]
- 18.Leigh T R, Kangro H O, Gazzard B G, Jefferies D J, Collins J V. DNA amplification by polymerase chain reaction to detect subclinical Pneumocystis carinii colonization in HIV-positive and HIV-negative homosexuals with and without respiratory symptoms. Respir Med. 1993;87:525–529. doi: 10.1016/0954-6111(93)90008-n. [DOI] [PubMed] [Google Scholar]
- 19.Lipschik G Y, Gill V J, Lundgren J D, Andrawis V A, Nelson N A, Nielsen J O, Ognibene F P, Kovacs J A. Improved diagnosis of Pneumocystis carinii infection by polymerase chain reaction on induced sputum and blood. Lancet. 1992;340:203–206. doi: 10.1016/0140-6736(92)90469-j. [DOI] [PubMed] [Google Scholar]
- 20.Lu J-J, Chen C-H, Bartlett M S, Smith J W, Lee C-H. Comparison of six different PCR methods for detection of Pneumocystis carinii. J Clin Microbiol. 1997;33:2785–2788. doi: 10.1128/jcm.33.10.2785-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundgren B, Lundgren J D, Nielsen T, Mathiesen L, Nielsen J O, Kovacs J A. Antibody responses to a major Pneumocystis carinii antigen in human immunodeficiency virus-infected patients with and without P. carinii pneumonia. J Infect Dis. 1992;165:1151–1155. doi: 10.1093/infdis/165.6.1151. [DOI] [PubMed] [Google Scholar]
- 22.Lundgren J D, Barton S E, Lazzarin A, Danner S, Goebel F D, Pehrson P, Mulcahy F, Kosmidis J, Pedersen C, Phillips A N. Factors associated with the occurrence of Pneumocystis carinii pneumonia in 5025 European AIDS patients. AIDS in Europe Study Group. Clin Infect Dis. 1995;21:106–113. doi: 10.1093/clinids/21.1.106. [DOI] [PubMed] [Google Scholar]
- 23.Mathis A, Weber R, Kuster H, Speich R. Simplified sample processing combined with a sensitive one-tube nested PCR assay for detection of Pneumocystis carinii in respiratory specimens. J Clin Microbiol. 1997;35:1691–1695. doi: 10.1128/jcm.35.7.1691-1695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer C L, Palmer C J. Evaluation of PCR, nested PCR, and fluorescent antibodies for detection of Giardia and Cryptosporidium species in wastewater. Appl Environ Microbiol. 1996;62:2081–2085. doi: 10.1128/aem.62.6.2081-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsson M, Elvin K, Löfdahl S, Linder E. Detection of Pneumocystis carinii DNA in sputum and bronchoalveolar lavage samples by polymerase chain reaction. J Clin Microbiol. 1993;31:221–226. doi: 10.1128/jcm.31.2.221-226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabodonirina M, Raffenot D, Cotte L, Boibieux A, Mayencon M, Bayle G, Persat F, Rabatel F, Trepo C, Payramond D, Piens M-A. Rapid detection of Pneumocystis carinii in bronchoalveolar lavage specimen from human immunodeficiency virus-infected patients: use of a simple DNA extraction procedure and nested PCR. J Clin Microbiol. 1997;35:2748–2751. doi: 10.1128/jcm.35.11.2748-2751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribes J A, Limper A H, Espy M J, Smith T F. PCR detection of Pneumocystis carinii in bronchoalveolar lavage specimens: analysis of sensitivity and specificity. J Clin Microbiol. 1997;35:830–835. doi: 10.1128/jcm.35.4.830-835.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux P, Lavrard I, Chouaid C, Denis M, Olivier J L, Nigou M, Miltgen M. Usefulness of PCR for detection of Pneumocystis carinii DNA. J Clin Microbiol. 1994;32:2324–2326. doi: 10.1128/jcm.32.9.2324-2326.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schluger N, Godwin T, Sepkowitz K, Armstrong D, Bernard E, Rifkin M, Cerami A, Bucala R. Application of DNA amplification to pneumocystosis: presence of serum Pneumocystis carinii DNA during human and experimentally induced Pneumocystis carinii pneumonia. J Exp Med. 1992;176:1327–1333. doi: 10.1084/jem.176.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sk¢t J, Lerche A, Kolmos H J, Lundgren J D. Pneumocystis carinii in bronchoalveolar lavage and induced sputum: detection with nested polymerase chain reaction. Scand J Infect Dis. 1995;27:363–367. doi: 10.3109/00365549509032732. [DOI] [PubMed] [Google Scholar]
- 31.Tamborini E, Mencarini P, De Luca A, Maiuro G, Ventura G, Siracusano A, Ortona E, Vicari G. Diagnosis of Pneumocystis carinii pneumonia: specificity and sensitivity of polymerase chain reaction in comparison with immunofluorescence in bronchoalveolar lavage specimens. J Med Microbiol. 1993;38:449–453. doi: 10.1099/00222615-38-6-449. [DOI] [PubMed] [Google Scholar]
- 32.Tamborini E, Mencarini P, Visconti E, Zolfo M, De Luca A, Siracusano A, Ortona E, Wakefield A E. Detection of Pneumocystis carinii DNA in blood by PCR is not of value for diagnosis of P. carinii pneumonia. J Clin Microbiol. 1996;34:1568–1588. doi: 10.1128/jcm.34.6.1586-1588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ursi J P, Ursi D, Ieven M, Pattyn S R. Utility of an internal control for the polymerase chain reaction—application to detection of mycoplasma pneumonia in clinical specimens. APMIS. 1992;100:635–639. doi: 10.1111/j.1699-0463.1992.tb03978.x. [DOI] [PubMed] [Google Scholar]
- 34.Wakefield A E, Guliver L, Miller R F, Hopkin J M. DNA amplification on induced sputum samples for diagnosis of Pneumocystis carinii pneumonia. Lancet. 1991;337:1378–1379. doi: 10.1016/0140-6736(91)93062-e. [DOI] [PubMed] [Google Scholar]
- 35.Wakefield A E, Miller R F, Guiver L A, Hopkin J M. Oropharyngeal samples for detection of Pneumocystis carinii by DNA amplification. Q J Med. 1993;86:401–406. [PubMed] [Google Scholar]
- 36.Wakefield A E, Pixley F J, Banerji S, Sinclair K, Millar R F, Maxon E R, Hopkin J M. Detection of Pneumocystis carinii with DNA amplification. Lancet. 1990;336:451–453. doi: 10.1016/0140-6736(90)92008-6. [DOI] [PubMed] [Google Scholar]
- 37.Walsh P S, Metzger D A, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques. 1991;10:506–513. [PubMed] [Google Scholar]
- 38.Weig M, Klinker H, Wilhem M, Lemmer K, Gross U. Correlation of Pneumocystis carinii PCR with clinical diagnosis in immunocompromised patients. Lancet. 1996;347:1266. doi: 10.1016/s0140-6736(96)90787-4. [DOI] [PubMed] [Google Scholar]