Abstract
Correct floral development is the result of a sophisticated balance of molecular cues. Floral mutants provide insight into the main genetic determinants that integrate these cues, as well as providing opportunities to assess functional variation across species. In this study, we characterize the barley (Hordeum vulgare) multiovary mutants mov2.g and mov1, and propose causative gene sequences: a C2H2 zinc-finger gene HvSL1 and a B-class gene HvMADS16, respectively. In the absence of HvSL1, florets lack stamens but exhibit functional supernumerary carpels, resulting in multiple grains per floret. Deletion of HvMADS16 in mov1 causes homeotic conversion of lodicules and stamens into bract-like organs and carpels that contain non-functional ovules. Based on developmental, genetic, and molecular data, we propose a model by which stamen specification in barley is defined by HvSL1 acting upstream of HvMADS16. The present work identifies strong conservation of stamen formation pathways with other cereals, but also reveals intriguing species-specific differences. The findings lay the foundation for a better understanding of floral architecture in Triticeae, a key target for crop improvement.
Keywords: Barley, B-class, cereal, C2H2, flower development, MADS-box, multiovary, transcription factors, Triticeae
Analysis of barley multiovary mov1and mov2loci indicates that HvSL1 and HvMADS16 exhibit both unique and conserved roles in the specification and development of cereal florets.
Introduction
The ABC model of flower development postulates that each organ within a flower is specified by the combinatorial action of distinct gene classes, referred to as the A-, B-, C-, D-, and E-class genes (Schwarz-Sommer et al., 1990; Coen and Meyerowitz, 1991; Colombo et al., 1995). These genes act in concentric rings (whorls) of tissue within the floral meristem to establish organ identity. Generally, A+E-class genes control the development of sepals (or bract-like organs) in the outermost whorl (whorl 1), while the action of A+B+E-class genes in whorl 2 gives rise to petals or equivalent structures. Stamens in the third whorl are specified by B+C+E-class genes while, in the fourth innermost whorl, carpels require C+E-class genes, and D-class genes are essential for ovule development. Most genes within the ABC model encode transcription factors belonging to the MADS-box gene family (Coen and Meyerowitz, 1991; Pelaz et al., 2000; Favaro et al., 2003). Numerous studies have added to this model over time, creating a more comprehensive view of the complex genetic network leading to flower formation (Thomson and Wellmer, 2019). Importantly, dimers of MADS-box proteins can interact to form tetrameric complexes, described as the floral quartet model (Theissen, 2001). The floral tetramers can promote DNA looping to bring distal promoter regions closer and allow recruitment of transcription cofactors, chromatin-remodelling proteins, and other transcription factors to regulate the expression of downstream genes (Melzer and Theissen, 2009; Smaczniak et al., 2012).
Cys2/His2 (C2H2) zinc-finger transcription factors also play an important role in flower development by acting upstream and downstream of MADS-box genes, and by coordinating cell proliferation and differentiation during floral organogenesis (Lyu and Cao, 2018). For example in Arabidopsis, JAGGED (JAG) functions during petal primordia formation and redundantly acts with its paralogue NUBBIN (NUB) to control lateral growth and differentiation of stamens and carpels (Dinneny et al., 2004, 2006). In rice (Oryza sativa), the JAG orthologue STAMENLESS1 (SL1) specifies lodicule and stamen identity (Xiao et al., 2009).
Understanding floral development provides opportunities to manipulate floral structures for application in agriculture. In crop species, considerable effort has been devoted to understanding flower development in rice (Arora et al., 2007; Yoshida and Nagato, 2011) and maize (Zea mays) (Bommert et al., 2005; Bartlett et al., 2015; Ali et al., 2019; Abraham-Juárez et al., 2020). Less information is available for members of the Triticeae, such as wheat (Triticum aestivum) and barley (Hordeum vulgare), despite their importance in the food and feed industries. Recent functional studies (e.g. on HvMADS1, HvLFY, HvAP2, and HvMADS29; Li et al., 2021; Selva et al., 2021; Shoesmith et al., 2021), coupled with mining of genomic resources have started to address the diversity of ABC genes in barley (Callens et al., 2018; Kuijer et al., 2021). This has important implications for the Triticeae cereals, where manipulating floral structures to improve hybrid grain production is still a promising avenue for increased yield and crop improvement (Selva et al., 2020).
Diversity in floral structure can be obtained through specific targeting of regulatory genes (e.g. CRISPR/Cas9; Li et al., 2021; Selva et al., 2021), through the exploitation of natural genetic variation (e.g. in barley; Caldwell et al., 2004; Talamè et al., 2008; Druka et al., 2011; Szarejko et al., 2017; Szurman-Zubrzycka et al., 2018; Schreiber et al., 2019; and wheat; Krasileva et al., 2017), or through the induction of mutations by chemical mutagenesis (Knudsen et al., 2022). In terms of the latter, historical forward genetic screens have uncovered an array of floral phenotypes. Of these, pistillody is the conversion of stamens into additional pistils (Murai and Tsunewaki, 1993; Murai et al., 2002; Peng, 2003). Pistillody is one avenue to increase the number of seed-bearing units per plant (Yang and Tucker, 2021), as well as to create a male-sterile mother plant for cross-pollination in hybrid breeding (Selva et al., 2020). Studies in wheat mutants often show that pistillody correlates with changes in expression of genes belonging to the ABC model (Yamada et al., 2009; Wang et al., 2015; Yang et al., 2015). This is particularly true for B-, C-, and D-classes, which are directly implicated in stamen, carpel, and ovule development.
Until now, at least three pistillody loci have been reported in barley: multiovary1 (mov1), mov2, and mov5 (Franckowiak and Lundqvist, 2013). Early linkage and mapping attempts positioned the mov1 locus, proposed to encode a MADS-box gene, on chromosome 7H (Tazhin, 1980; Soule et al., 1996, 2000). The mov2 locus was mapped to chromosome 3H and also suggested to encode a member of the MADS-box gene family (Soule et al., 1996, 2000). The barley mov5 locus was recently characterized and shown to encode a FLORICAULA/LEAFY (HvLFY) transcription factor on chromosome 2H (Selva et al., 2021). Despite a general description of the mutant phenotype and rough mapping of the mov2 and mov1 loci, the causative gene sequences have yet to be reported.
To increase our knowledge of floral development in the Triticeae, this study aimed to map and characterize the barley mov1 and mov2 loci. We show that mov2 maps to a region on 3H encoding a C2H2 zinc-finger transcription factor, named here as HvSL1 based on homology to rice STAMENLESS1 (Xiao et al., 2009). Moreover, we show that a total of three genes on chromosome 7H, including the MADS-box B-class gene HvMADS16, are absent in mov1 plants. We investigate the interaction of HvSL1 with barley B-class genes, report the developmental changes and effect on ABC gene expression during floral development in mov1 and mov2.g, and propose a model for stamen specification in barley. Our results highlight strong conservation in the core determinants of stamen formation, despite intriguing differences in MADS-box regulation between cereals.
Materials and methods
Plant material and genotyping
Grains segregating for the mov2.g allele (mov2 locus) and for the mo6b allele (mov1 locus) in cv. Steptoe were kindly provided by Professor A. Kleinhofs. Germination was synchronized by cold treatment at 4 °C for 3 d in the dark. The germinated grains were then transplanted into pots and grown in the glasshouse at 23 °C/16 °C day/night temperatures and long days (~12 h). Phenotyping was done at the heading stage by visually inspecting the central florets of several spikes per plant. Florets were imaged with a Nikon SMZ25 Stereo Fluorescence Microscope equipped with DS-Ri1 colour-cooled digital camera.
Genotyping of the plants was performed by copy number analysis using TaqMan™ assay (Thermo Fisher Scientific, USA) designed for HvSL1 and HvMADS16 as genes of interest. The barley CONSTANS-like CO2 (HORVU6Hr1G072620) was used as the internal positive control (Kang et al., 1997). The sequences of primers and probes can be found in Supplementary Table S1. The reaction was set up as follows: 1× PrecisionFast™ qRT-PCR Master Mix with Low ROX (Primerdesign Ltd, UK), 200 nM of each primer, 100 nM of probe, 150 ng of template DNA, and water to a final volume of 10 μl. The reaction was performed with an initial activation step at 95 °C for 2 min, followed by 40 cycles at: 95 °C for 5 s and 60 °C for 20 s in a QuantStudio 6 Flex Real-Time PCR machine (Thermo Fisher Scientific, USA). The detectors used were FAM-BHQ1 and HEX-BHQ1, with ROX as internal passive reference.
The mov2 bi-parental mapping population was created by manually cross-pollinating mov2.g florets (cv. Steptoe) with wild-type pollen from cv. Morex. Heterozygosity of F1 plants was confirmed by KASP™ marker analysis across two known single nucleotide polymorphisms (SNPs), at positions chr3H_1006543 and chr3H_28805649, and by Sanger sequencing across a third SNP at position chr7H_557244517 according to the Morex reference assembly (Hv_IBSC_PGSB_v2). For fine-mapping of the mov2 locus, SNPs spanning the 3HS region were identified and developed as KASP™ markers. SNPs were identified from a published Steptoe×Morex dataset (Close et al., 2009) and from Steptoe leaf transcriptomic data mapped to the reference Hv_IBSC_PGSB_v2 Morex assembly. Primer Picker (LGC Genomics) and the LGC Genomics SNPline™ were used to design and prepare the KASP™ marker assays. Assays were performed using KASP™ Master mix as instructed by the manufacturer. Sequences of KASP™ markers can be found in Supplementary Table S2.
Nucleic acid extraction and genomic PCR
For genotyping, genomic DNA was extracted from freeze-dried 2-week-old plant material using a phenol–chloroform method as described by Rogowsky et al. (1991). PCR was performed using Q5® high-fidelity DNA polymerase (New England BioLabs, USA), following the manufacturer’s protocol in a final volume of 12.5 μl. All PCR products were separated by agarose gel electrophoresis, purified using the ISOLATE II PCR and Gel Kit (Bioline, Australia), and Sanger sequenced at the Australian Genome Resource Facility (AGRF). Primers used for PCR can be found in Supplementary Tables S3–S5.
RNA extraction, cDNA synthesis, and quantitative real-time PCR (qRT-PCR)
Ultra fine-pointed tweezers were used to manually dissect inflorescences at Waddington (W) developmental stages W2.0, W3.5, W4.0, and W6.0 (Waddington et al., 1983), corresponding to ~17, 20, 23, and 26 d post-germination in the growing conditions described above. Inflorescences were snap-frozen in liquid nitrogen, RNA extraction was performed using the ISOLATE II RNA Plant Kit (Bioline, Australia), followed by TURBO™ DNase treatment (Thermo Fisher Scientific, USA), and cDNA synthesis using the SuperScript™ IV First Strand Synthesis (Thermo Fisher Scientific, USA) as per the manufacturers’ instructions.
RNA from transfected protoplasts was extracted using a phenol–chloroform method. Briefly, samples were lysed in a 1.5 ml tube and homogenized in 500 μl of TRIzol (Thermo Fisher Scientific, USA) before adding 150 μl of chloroform. Samples were vortexed and centrifuged at maximum speed at 4 °C for 15 min. The aqueous phase was carefully transferred to a new 1.5 ml tube with 250 μl of isopropanol and incubated at 4 °C for 10 min before centrifuging at maximum speed at 4 °C for 20 min. The precipitated RNA was washed with 500 μl of 100% ethanol, allowed to air-dry, and resuspended in 20 μl of diethylpyrocarbonate (DEPC)-treated water.
qRT-PCR was carried out as described in Selva et al. (2021). Each time point is the result of three technical replicates and at least three biological replicates. Absolute quantification is reported as normalized transcript against the geometric means of the three least varying control genes as described in Vandesompele et al. (2002). Relative quantification in transfected protoplasts is expressed as calibrated normalized relative quantities (NRQ) as calculated by the qbase+ software (Biogazelle, Belgium; Hellemans et al., 2007). Primer information for the reference genes and genes of interest can be found in Supplementary Table S6.
CRISPR and plant transformation
Guide RNA (gRNA) design and cloning for HvSL1 CRISPR (clustered regularly interspaced palindromic repeats)-associated protein knockout was done following the procedure described by Ma et al. (2015) with vectors kindly provided by Professor Yao-Guang Liu (South China Agricultural University). Two gRNAs targeting positions +42 bp and +275 bp from the translational start site of HvSL1 were cloned in the same vector. Primer sequences used for cloning are listed in Supplementary Table S7.
Agrobacterium tumefaciens-mediated plant transformation (cv. Golden Promise) and genotyping of regenerant plants was performed as described previously (Selva et al., 2021).
In situ hybridization
Sense and antisense RNA probes for in situ hybridization were amplified using Q5® high-fidelity DNA polymerase (New England BioLabs, USA) with the T7 promoter extension to the 5ʹ end of primers (primers used can be found in Supplementary Table S8), transcribed, and digoxigenin (DIG) labelled. Inflorescences were prepared and in situ hybridization was performed as described in Selva et al. (2021)
Bi-molecular fluorescent complementation (BiFC)
The full-length coding sequences of HvMADS2, HvMADS4, HvMADS16, and ΔHvMADS16, containing only the MADS-domain (amino acids 1–65), were PCR-amplified with Q5® high-fidelity DNA polymerase (New England BioLabs, USA) and cloned in the BiFC vectors pSAT1-nEYFP-N1 (N-terminal fragment) and pSAT1-cEYFP-C1-B (C-terminal fragment) (Citovsky et al., 2006). Primers used for cloning are listed in Supplementary Table S9. A cyan fluorescent protein (CFP) co-transformation control and positive interaction control (OsERS1–nYFP and OsARC–cYFP) were included (Yang et al., 2018). All plasmids were checked by digestion and Sanger sequencing. Biolistic particle bombardment of onion epidermal cells was carried out as described in Selva et al. (2021). For each interaction test, four independent transfections were undertaken, including two technical replicates and at least two biological replicates. Interaction frequency was similar for each combination, showing a positive result, and was comparable with the positive control.
Light and electron microscopy
For light microscopy, wild-type, mov2, and mov1 carpels were fixed in FAA solution (50% ethanol, 5% glacial acetic acid, 10% formaldehyde, one drop of Tween-20) overnight, dehydrated in a 70–100% ethanol series, and embedded in LR white resin. Samples were sectioned using a Leica Rotary Microtome RM2265 at 1.5 μm. Slides were stained with 0.1% toluidine blue in 0.1% sodium tetraborate for 2 min and rinsed three times with water, dried, and mounted with DPX. After 72 h, slides were imaged with a Nikon Eclipse Ni-E optical microscope equipped with a DS-Ri1 colour cooled digital camera. Image analysis and processing were carried out with the NIS-Elements AR software.
For SEM, inflorescences were manually dissected and fixed overnight in 4% paraformaldehyde, 1.25% glutaraldehyde in phosphate-buffered saline (PBS), 4% sucrose, pH 7.2. Before processing, samples were washed three times in PBS and fixed in 2% OsO4 in PBS for 1 h. Samples were then dehydrated in a 50–100% ethanol series and dried with a critical point dryer. Dried samples were arranged on carbon tabs stuck to 12 mm aluminium stubs and coated with platinum. Samples were observed using a Hitachi FlexSem 1000 scanning electron microscope.
Protoplast isolation
Isolation of barley leaf protoplasts was performed as described by Yoo et al. (2007) with minor modifications. Briefly, the adaxial epidermal layer of leaves from 11-day-old barley seedlings (cv. Golden Promise) was manually peeled off and the leaf was cut into ~2 cm×0.5 cm strips using surgical blades. The leaf segments of ~10 plants were immediately transferred to a Petri dish containing 15 ml of 0.6 M mannitol for 30 min at room temperature to induce plasmolysis. After incubation, the leaf segments were transferred to another Petri dish containing 10 ml of freshly prepared enzyme solution [0.55 M mannitol, 40 mM MES-KOH at pH 5.7, 20 mM KCl, 2.0% cellulase R10 (Yakult Pharmaceutical, Japan), 0.75% macerozyme R10 (Yakult Pharmaceutical), 10 mM CaCl2, 0.1% BSA] and incubated for 3 h in the dark at 28 °C with gentle shaking (40–60 rpm). After enzymatic digestion, forceps were used to gently remove the remaining epidermis and leaf debris from the enzyme solution. An equal volume (10 ml) of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES-KOH at pH 5.7) was slowly added to the protoplasts and the solution was filtered with a 100 μM nylon mesh into a 50 ml round-bottom tube. The volume was adjusted by adding 5 ml of W5 solution. The filtered protoplasts were collected by centrifugation at 600 g for 3 min. The supernatant was replaced with 15 ml of fresh W5 and the protoplasts resuspended by gentle shaking. Protoplasts were allowed to pellet by gravity for 30 min in ice. After incubation, the supernatant was promptly removed and substituted with MMG solution (0.6 M mannitol, 15 mM MgCl2, and 4 mM MES-KOH at pH 5.7) at a concentration of 106 cells ml–1, determined by counting cells in 12 μl of a 1:10 dilution of protoplast solution with a haematocytometer.
PEG-mediated transfection of barley protoplasts
Polyethylene glycol (PEG)-mediated transfections were mostly carried out as described by Bai et al. (2014). Firstly, 200 μl of PEG–Ca2+ solution [40% (w/v) PEG 4000, 0.4 M mannitol, 0.1 M CaCl2] were pre-loaded in pipette tips. Secondly, 100 μl of protoplast solution (~105 cells) were added to 20 μg of each plasmid DNA in a 2.0 ml tube. The pre-loaded PEG–Ca2+ solution was immediately added to the protoplast–DNA mixture, mixed gently, and incubated for 15 min at room temperature in the dark. The transfection process was stopped by adding 840 μl of W5 and centrifuged at 600 g for 2 min. Cells were resuspended in 500 μlof W5 and transferred to multi-well plates previously coated with 5% (v/v) foetal bovine serum (FBS). Protoplasts were cultured at 28 °C for 40–48 h in the dark.
To test transfection efficiency, protoplasts were transfected with pUbi-YFP-rbcS in six independent transfections. At 40–48 h after transfection, the protoplasts were visualized under UV and bright light using a Nikon Eclipse Ni-E optical microscope equipped with a DS-Ri1 colour cooled digital camera.
Results
mov2.g florets have functional supernumerary carpels and produce multiple grains
In wild-type barley, floral organs are arranged in a defined pattern such that the outermost whorl contains the palea and lemma (whorl 1). These are followed by two lodicules in whorl 2 and three stamens in whorl 3, surrounding a single carpel at the centre of the floret in whorl 4 (Fig. 1A, B) (Brenchley, 1920).
Fig. 1.
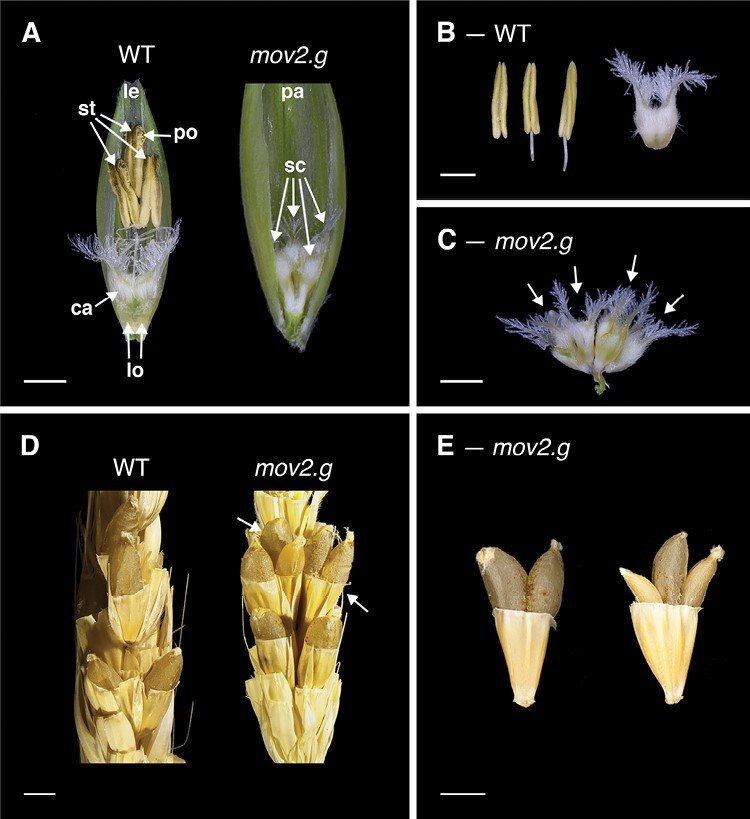
Florets, reproductive organs, and grains in the wild type (WT) and mov2.g. (A) Exposed WT and mov2.g florets. Palea or lemma have been removed to show internal floral organs. (B) Reproductive organs in WT florets consist of three stamens and one carpel. (C) Reproductive organs in mov2.g florets consist of supernumerary carpel-like structures (arrows). Scale bars: 1000 µm. (D) Manually cross-pollinated WT and mov2.g spikes. White arrows indicate multiple grains per floret. (E) Examples of multiple grains per floret produced by manually cross-pollinated mov2.g spikes. Scale bars: 2000 µm. Lemma (le), palea (pa), lodicule (lo), stamen (st), carpel (ca), supernumerary carpel-like structure (sc), pollen (po).
In florets from mov2.g plants, the stamens are replaced by supernumerary carpels, rendering the plant unable to self-pollinate (Fig. 1A). The number of carpels within each floret typically varies between four and seven carpels or carpel-like structures (Supplementary Table S10). Visually, all carpels appear irregularly shaped, connected at the base, and of smaller size relative to a wild-type carpel (Fig. 1C). In contrast, development and morphology of the lemma, palea, and lodicules remains largely unaffected. Most notably, when mov2.g plants were used as a female recipient in manual cross-pollination, most florets were able to produce multiple grains with a maximum of three developing grains within a single floret (Fig. 1D, E). This is due to the ability of some mov2.g carpel structures to correctly differentiate fully functional female gametophytes, as was observed in transverse sections stained with toluidine blue (Supplementary Fig. S1). All grains from mov2.g plants were viable, and after germination gave rise to mature plants.
To better understand the developmental basis of the multiovary phenotype, SEM was used to compare wild-type (cv. Steptoe) and mov2.g inflorescences. Immature inflorescences were morphologically similar at the very early stages of development [i.e. floret primordium, at W3.0; Fig. 2; Waddington et al., 1983]. At stages W3.5–W3.75, both wild-type and mov2.g floral meristems initiated lateral dome-shaped protrusions, consistent with the appearance of stamen primordia from the floral meristem. Following this stage, the first morphological differences were identified. In the wild type, the stamen primordia differentiated into filament and anther tissues (W5.0–W8.5), while the meristematic tissue at the centre proliferated and terminally differentiated into a single ovule-containing carpel. As each wild-type floral meristem developed, a vertical symmetry along the central inflorescence rachis was maintained.
Fig. 2.
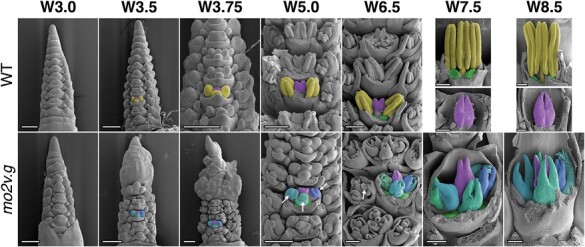
SEM of mov2.g developing inflorescences. In the wild type (WT), primordia giving rise to stamens are false-coloured in yellow, cells giving rise to the carpel in purple, and lodicules in green. In each panel from W3.5 onwards, the primordia are false-coloured in a single floret to enable comparison between genotypes. In mov2.g, the central carpel (purple) and lodicules (green) appear to be retained; cells giving rise to the additional carpel-like structures are false-coloured in blue (different shades). White arrows indicate ovule primordia. Waddington stage is indicated for each developmental time point; from stage W5.0, lemma and/or stamens have been removed to expose the carpels. Scale bars: 200 µm.
In contrast, in mov2.g, the lateral protrusions did not differentiate into stamens, but instead gave rise to organs that followed the characteristic morphogenesis of carpels (W5.0–W8.5) (Fig. 2). Interestingly, until W5.0, only three organ primordia were present in the third whorl, probably originating from converted stamen primordia. However, the number of carpels in whorl 3 dramatically increased by W6.5 and appeared to develop synchronously with the central carpel. These carpel-like organs initially developed an ovule primordium that was later surrounded by the growing carpel tissue. By stage W8.5, the ectopic carpels also differentiated stigmas bearing stigmatic papillae. Moreover, as floral organs developed, there was frequent loss in the vertical symmetry of florets along the central inflorescence rachis, and occasional fasciation of the growing tip, which was then reflected in a shorter and broader spike compared with the wild type (Supplementary Fig. S2). The inflorescence fasciation trait, although interesting, was not investigated further here.
mov2 encodes a C2H2 zinc-finger transcription factor
A previous study had mapped the barley mov2 locus to the telomeric region on the short arm of chromosome 3H (3HS) within an ~28 Mb genomic interval (Soule et al., 2000). This interval is too large to reliably identify the underlying causative gene(s). Therefore, a mov2.g (cv. Steptoe)×Morex bi-parental population was developed to map mov2 to a higher genetic resolution. The multiovary phenotype was not observed in F1 individuals, indicative of the mov2.g mutation being completely recessive. A total of 352 F2 plants were grown until maturity and used for recombination-based genetic mapping.
Seven KASP™ markers designed to span the previously published interval confirmed that mov2 mapped to the expected region on chromosome 3H. Following this, 10 additional KASP™ markers were designed to saturate the interval, reducing the critical mov2-containing region to ~1.9 Mb, based on flanking markers chr3H_9095799 and chr3H_11039299 (Supplementary Fig. S3). Further resolution of the locus was achieved with six additional KASP™ markers in 179 F3 individuals segregating for the multiovary phenotype. Comparison of phenotypic and genotypic segregation across these F3 plants reduced the critical interval to ~449 kb, between markers chr3H_9748112 and chr3H_10289104 (Supplementary Fig. S3). This region contains 20 annotated gene sequences, based on the Morex genome reference sequence (Supplementary Table S11).
Expression in floral tissues was assessed for each of the 20 annotated gene sequences to identify the likely causative gene, especially in tissue types affected in mov2.g florets. Barley expression datasets were obtained from a range of transcriptomes including 2-week-old seedlings (Zadoks stage Z12; Liu et al., 2019), developing inflorescences at stages W2.0, W3.5, and W8.0–W8.5 (Liu et al., 2019), developing pistils (W8.0–W10.0; Matros et al., 2021), and wild-type (cv. Steptoe) and mov2.g leaves (Z22; Zadoks et al., 1974). Overall, 10 of the 20 annotated gene sequences showed expression in either pistils or inflorescences. Of these, only HORVU3Hr1G003740 was identified to be uniquely expressed in reproductive tissues but not in vegetative tissues (leaf and seedling stage) or in mov2.g leaves (Supplementary Fig. S4). Sequence analysis indicated that HORVU3Hr1G003740 encodes a putative C2H2 zinc-finger transcription factor sharing 65.4% sequence identity with the rice protein STAMENLESS1 (OsSL1) (Supplementary Fig. S5; Supplementary Tables S12, S13). In rice, OsSL1 is known to play a crucial role in floral development, with loss of OsSL1 function leading to a multiovary phenotype (Xiao et al., 2009). Thus, HORVU3Hr1G003740 is hereafter referred to as HvSL1.
To validate the transcriptomic data, mRNA in situ hybridization was performed to confirm the HvSL1 spatial expression pattern in wild-type developing inflorescences. At stage W3.0, HvSL1 expression accumulated in the floret primordia, including the inflorescence tip, and was progressively found in the primordia of glumes, lemma, lodicules, stamens, carpels, and ovules (W3.5–W5.0) (Fig. 3). As the floral organs developed (W7.0), the signal persisted in the stamens, in the carpel, at the apical tip, and in the central vasculature of the lemma, but had decreased in the lodicules.
Fig. 3.
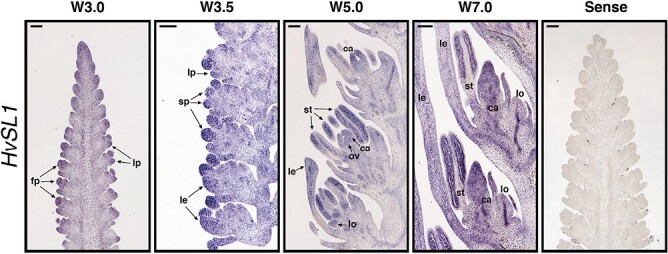
Spatial expression of HvSL1 during wild-type inflorescence development. Waddington stages correspond to lemma–floret primordia (W3.0), stamen primordia (W3.5), ovule primordium (W5.0), and stamen and carpel development (W7.0). Annotations and arrows indicate floral primordia (fp), lemma primordia (lp), stamen primordia (sp), lemma (le), lodicule (lo), stamen (st), carpel (ca), and ovule (ov). A sense HvSL1 probe was used as negative control to determine probe specificity. Scale bars: 100 µm.
HvSL1 is deleted in mov2.g plants and its absence causes multiovary
To determine whether the mov2.g phenotype is a consequence of mutation in HvSL1, PCR primers were designed to amplify the entire HvSL1 coding sequence. Comparisons between Morex, Steptoe, and mov2.g (cv. Steptoe) genomic DNA suggested that HvSL1 is absent in mov2.g plants but present in both Steptoe and Morex (Supplementary Fig. S6A). In contrast, predicted high-confidence neighbouring gene sequences were still present, indicating that HvSL1 could be the only deleted gene at the mov2 locus (Supplementary Table S11). For additional confirmation, the HvSL1-specific PCR was repeated on 36 critical recombinant F3 individuals from the mov2.g×Morex mapping population. For all samples, the absence of HvSL1 correlated with mutant phenotype expression. Furthermore, HvSL1 expression by qRT-PCR in developing inflorescences indicated that HvSL1 expression was completely absent in mov2.g samples (Supplementary Fig. S6B), whereas HvSL1 transcript abundance increased in the wild type between stages W2.0 and W6.0 (double ridge—carpel development), with the biggest increase between W4.5 (carpel primordium) and W6.0.
To provide additional support for HvSL1 being the causative gene underlying the mov2.g phenotype, Hvsl1 knockout plants were generated by CRISPR/Cas9 editing. Transformation of immature barley embryos (cv. Golden Promise) led to the generation of five T0 lines—HvSL1-1, HvSL1-3, HvSL1-5, HvSL1-6, and HvSL1-10—that phenocopied mov2.g plants. Specifically, HvSL1-6 was found to be a homo-allelic edit while HvSL1-1, HvSL1-3, and HvSL1-5 were all bi-allelic and HvSL1-10 was hetero-allelic (Supplementary Fig. S7). In all cases, edits occurred at the canonical cut site and resulted in loss-of-function frameshift mutations. The resulting phenotypes differed among lines, reflecting the phenotypic variation also observed in mov2.g plants. While all retained correct lodicule development, there was variation in the degree of stamen to carpel conversion, as well as in the number and complexity of supernumerary carpel-like structures (Fig. 4).
Fig. 4.
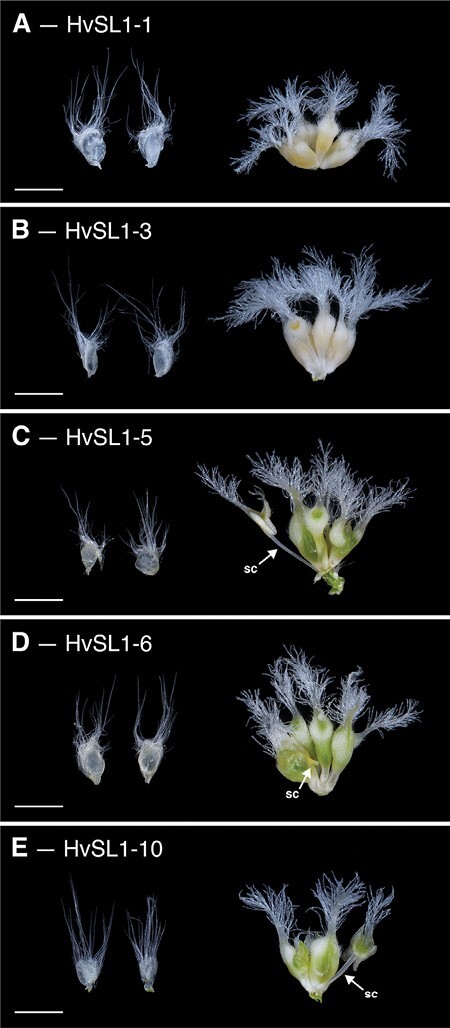
Phenotype of CRISPR Hvsl1 knockout plants. All knockout plants contained two lodicules and supernumerary carpels. (A) HvSL1-1, (B) HvSL1-3, (C) HvSL1-5, (D) HvSL1-6, and (E) HvSL1-10. Partial stamen conversions (sc) are annotated. Scale bars: 100 µm.
HvSL1 influences HvMADS16 expression but not that of other B-class genes
Since the early 1990s, flower development has always been described in the framework of the ABC model, whereby specific classes of genes are needed for the correct specification of each floral organ (Coen and Meyerowitz, 1991). The abnormal development of stamens and carpels in mov2.g is consistent with altered activity of B-class genes. Using a homology-based search, three B-class genes were found in barley, consisting of two GLO-like homologues (referred to here as HvMADS2 and HvMADS4) and a single DEF-like homologue (referred to as HvMADS16), confirming already published data (Callens et al., 2018). These genes were evaluated in further detail.
B-class proteins can act in either homodimeric or heterodimeric complexes involving DEF and GLO proteins (Winter et al., 2002). To assess interactions among the barley B-class proteins, BiFC was conducted in onion epidermal cells. Fluorescence signal was observed only in cells co-expressing HvMADS16 with HvMADS2 or HvMADS4 (Supplementary Fig. S8). For these combinations, fluorescence was predominantly confined to the nucleus. No signal was observed when the two GLO-like genes HvMADS2 and HvMADS4 were co-expressed in the same cells, or with a truncated version of HvMADS16 (ΔHvMADS16-nYFP), containing only the MADS-domain. Likewise, no signal could be detected when homodimeric combinations for each B-class gene were tested (Supplementary Fig. S8).
To determine whether heterodimerization is consistent with co-located gene expression, the expression patterns of the B-class genes were assessed by in situ hybridization in developing inflorescences and florets (Fig. 5). In the wild type (cv. Steptoe), HvMADS2 was found to be expressed in most floral tissues (glumes, lodicules, stamen primordia, and carpel) in the early stages (W3.5–W6.0), but the signal became more specific as development progressed (Fig. 5A). In stamens, expression was initially localized mainly in the locules, but was later restricted to the filament (W7.0) (Fig. 5B). HvMADS4 showed a similar pattern; however, it was also observed to accumulate in the ovule primordium (W4.5–W6.0). At maturity, both HvMADS2 and HvMADS4 were detected at the apex of the carpel, as well as in the lodicules. Expression of HvMADS16 was first detected in the stamen and lodicule primordia (W3.5) (Fig. 5C). As the stamens develop, HvMADS16 signal became weaker, while remaining strong in the lodicules (W6.0–W7.0). No expression was observed in the carpel or ovule at any stage and no signal was detected with the sense probes.
Fig. 5.
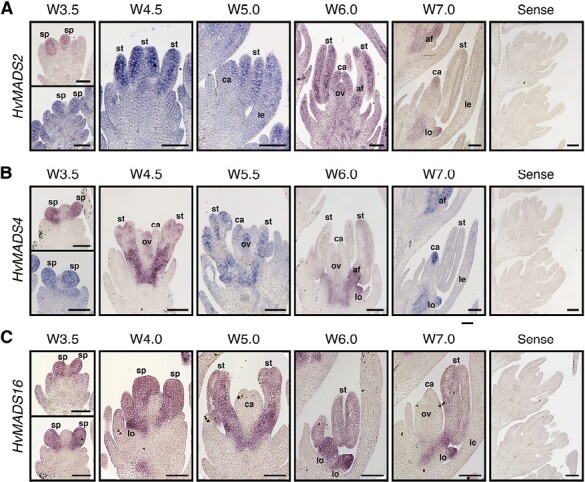
Expression pattern as assessed by in situ hybridization for barley B-class genes. (A) HvMADS2, (B) HvMADS4, and (C) HvMADS16. Annotations indicate stamen primordia (sp), lemma (le), lodicule (lo), stamen (st), anther filament (af), carpel (ca), and ovule (ov). Scale bars: 100 µm.
The overlapping expression domain of HvSL1 and B-class genes, together with the specific mov2.g phenotype, suggests that these genes may act in a common pathway. Moreover, studies conducted in rice suggest that OsSL1 acts as an upstream positive regulator of OsMADS16 transcription (Xiao et al., 2009). Expression assays in isolated barley protoplasts were performed to investigate a potential interaction between HvSL1 and barley B-class genes (Supplementary Fig. S9). Independent transfection experiments showed that protoplasts carrying a p35S:HvSL1 construct showed higher HvSL1 expression relative to protoplasts transfected with a mock solution (Supplementary Fig. S9B). In addition, a qRT-PCR assay indicated that endogenous HvMADS16 transcript abundance was significantly increased in transfected protoplasts compared with the mock, while HvMADS2 and HvMADS4 expression remained unchanged (Supplementary Fig. S9C). This suggests that HvSL1 may also act as a positive regulator of HvMADS16 in barley.
The B-class gene HvMADS16 is absent in mov1 plants
To further dissect the relationship between HvSL1 and HvMADS16, we aimed to create a Hvmads16 mutant using CRISPR/Cas9. Despite multiple attempts with different guides, edited plants were not recovered. To overcome this, we again investigated historical induced mutant resources. At least three multiovary loci have been reported to date (Franckowiak and Lundqvist, 2013). Of these, the mov1 locus (Tazhin, 1980; Soule et al., 2000) bears a striking resemblance to the rice Osmads16/superwoman1 mutant (Nagasawa et al., 2003). Under our growth conditions, mov1 florets consistently showed direct homeotic transformation of lodicules into leaf-like organs and of stamens into carpels (Fig. 6). In each instance, the palea, lemma, and central carpel were retained. Unlike mov2.g/Hvsl1 mutants, the four carpel-like organs were sterile as the female gametophyte failed to differentiate correctly (Supplementary Fig. S10). Furthermore, mov1 florets did not produce any grains when manually cross-pollinated with wild-type pollen.
Fig. 6.
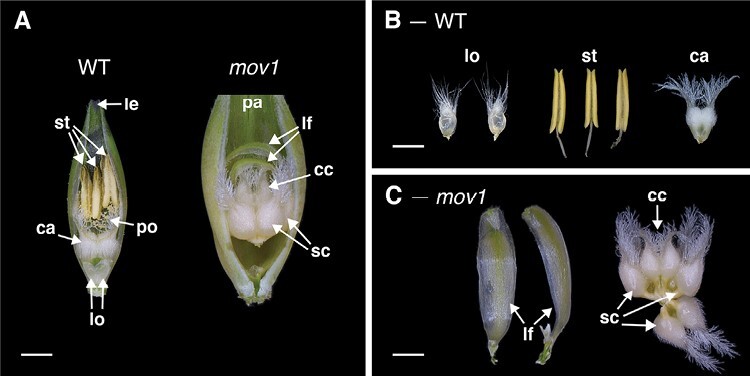
Florets and floral organs in the wild type (WT) and mov1. (A) Exposed WT and mov1 florets. Palea or lemma have been removed to show internal floral organs. (B) The floral organs in a WT floret consist of two lodicules, three stamens, and one carpel. (C) In mov1 florets, the lodicules are converted into bract-like organs, and stamens are converted into additional carpels. Scale bars: 1000 µm. Lemma (le), palea (pa), lodicule (lo), stamen (st), carpel (ca), supernumerary carpel-like structure (sc), central carpel (cc), leaf-like organ (lf), pollen (po).
SEM images of wild-type (cv. Steptoe) and mov1 developing inflorescences were compared to determine the effect of mov1 on floral organ development (Fig. 7). The earliest observable difference was seen immediately preceding the appearance of stamen primordia at W3.0, whereby a crease appeared in the basal floral meristems of mov1 inflorescences. At stage W3.5–W3.75, instead of developing dome-shaped stamen primordia, the meristems in mov1 divided into irregularly shaped protrusions which arranged into discernible multiple concentric creases by stage W5.0. As development progressed (W6.5–W7.0), each crease developed into a carpel, eventually leading to the four-carpel structure visible in the mature mov1 floret. Occasionally, it was observed that a single floral meristem could give rise to two distinct florets (Fig. 7; Supplementray Fig. S11).
Fig. 7.
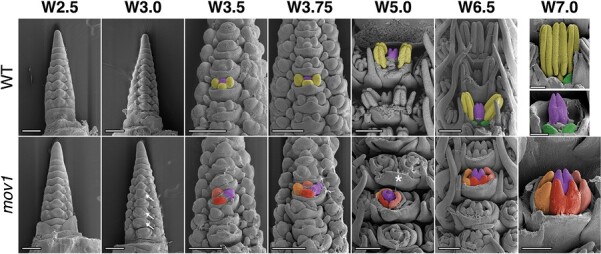
SEM of mov1 inflorescence development. In the wild type (WT), primordia giving rise to stamens are false-coloured in yellow, cells giving rise to the carpel in purple, and lodicules in green. In each panel from W3.5 onwards, the primordia are false-coloured in a single floret to enable comparison between genotypes. In mov1, the central carpel (purple) is retained while the stamens are converted into additional carpels. Cells giving rise to the additional carpels are false-coloured in different shades of red/orange. White arrows in mov1 (W3.0) indicate creases in the floral meristems. The white star at W5.0 indicates separation of a single floral meristem into two distinct florets; for greater detail, see Supplementary Fig. S11. Waddington stage is indicated for each developmental time point; from W5.0, lemma and/or stamens have been removed to expose the carpels. Scale bars: 200 µm.
To determine whether mutation of a B-class gene contributes to the mov1 phenotype, a PCR-based strategy, consisting of PCR amplification followed by Sanger sequencing of the amplicon, was used to survey all B-class genes for presence/absence, and structural and sequence variants in mov1 plants relative to the wild type (cv. Steptoe). HvMADS2 and HvMADS4 did not show differences between genotypes when tested for amplicon size by PCR or polymorphisms by sequencing. In contrast, HvMADS16 was not detected in mov1 mutants (Supplementary Fig. S12A). Genotyping by copy number analysis combined with phenotyping demonstrated that absence of HvMADS16 co-segregated perfectly with the mov1 phenotype and that mov1 segregates as a single recessive Mendelian locus (3:1, wild type:multiovary) (Supplementary Table S14). These findings indicate that mov1 lacks HvMADS16.
To define the size of the deletion surrounding HvMADS16, a PCR-based approach was used to test the presence/absence of neighbouring gene sequences based on annotations from the barley reference Morex genome Hv_IBSC_PGSB_v2. Overall, the mov1 mutant appeared to be missing a region of ~0.95 Mb relative to the wild type (Supplementary Fig. S12C). According to the reference sequence, this region is predicted to include three gene sequences: HORVU7Hr1G091190 (40S ribosomal protein), HORVU7Hr1G091200 (undescribed protein); and HvMADS16 (Supplementary Table S15). Consistent with a role for HvMADS16 in inflorescence specification, transcripts were predominantly detected in developing wild-type inflorescences when tested by qRT-PCR across a cv. Steptoe tissue series (Supplementary Fig. S12B). On the other hand, transcripts for HORVU7Hr1G091200 could not be detected in any of the tissues examined. Furthermore, a BLASTx query against NCBI non-redundant protein databases found no significant similarity in other species, suggesting tht HORVU7Hr1G091200 is a likely pseudogene. The presence of HORVU7Hr1G091190 encoding a 40S ribosomal protein could not be uniquely assayed due to the highly repetitive nature of its sequence. However, publicly available RNA-seq data (Colmsee et al., 2015) indicate that HORVU7Hr1G091190 is not expressed in a range of barley tissues including the inflorescence. Considering the specific homeotic conversion of floral organs in whorls 2 and 3, the absence of HvMADS16 in mov1 mutant plants and the role of B-class genes in other plant species, HvMADS16 appears to be the most likely causal agent for mov1.
HvSL1 and HvMADS16 differentially affect expression of floral regulators
To establish how the HvSL1 (mov2.g) and HvMADS16 (mov1) deletions might influence molecular pathways underlying stamen and carpel formation, qRT-PCR was performed on developing inflorescences from the wild type, mov2.g, and mov1 at stages W2.0–W6.0 (Fig. 8), which encompass the specification and differentiation of the reproductive organs.
Fig. 8.
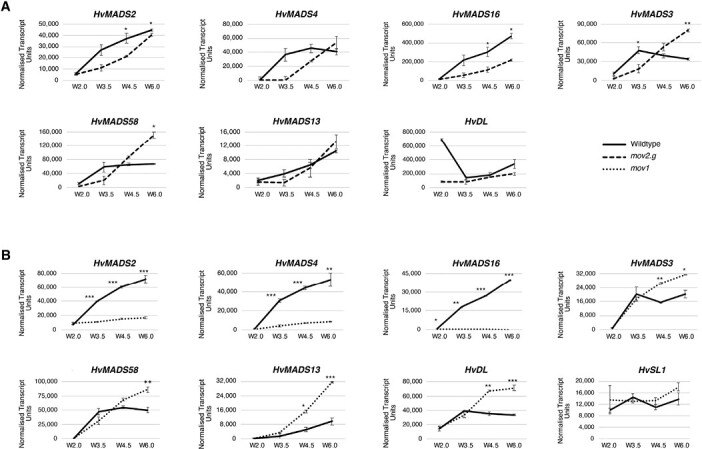
Transcript abundance of floral genes assessed by qRT-PCR in mov2.g and mov1 developing inflorescences. Gene expression of B-class genes (HvMADS2, HvMADS4, HvMADS16), C-class genes (HvMADS3, HvMADS58), D-class gene HvMADS13, and carpel genes HvDL and HvSL1 was assayed in (A) mov2.g and (B) mov1 inflorescences at stages W2.0 (double ridge), W3.5 (stamen primordia), W4.5 (carpel primordium), and W6.0 (stamen and carpel development). Error bars represent ±SE. For each time point, two-tailed t-test P-values ≤0.05 (*), ≤0.005 (**), and ≤0.001 (***) are shown for differences between the wild type and mov1. For each sample, n=3 independent biological replicates.
When comparing transcript abundance for the three B-class genes in mov2.g (Fig. 8A), the most pronounced difference was observed for HvMADS16, which was significantly reduced in the mutant. However, despite an overall reduction of transcript abundance in mov2.g, expression still increased with development. A similar trend was observed for HvMADS2, while for HvMADS4, transcript abundance remained lower in mov2.g only until stamen primordia specification (stage W3.5). After stage W3.5, transcript abundance for HvMADS4 steadily increased to match wild-type levels by stage W6.0. For C-class HvMADS3 and HvMADS58 MADS-box genes, transcript abundance in mov2.g showed a delayed accumulation relative to the wild type, followed by an increase eventually exceeding wild-type levels. Conversely, transcript abundance of the ovule-specific D-class gene HvMADS13 and the barley orthologue of the rice carpel-specific YABBY transcription factor HvDL (DROOPING LEAF) remained largely unaffected in mov2.g samples.
In mov1 (Fig. 8B), HvSL1 expression did not significantly differ from that in the wild type. In contrast, expression of the B-class genes HvMADS2 and HvMADS4 was significantly lower in mov1 from stage W3.5 onwards, and HvMADS16 expression was absent. Conversely, expression of the C-class genes (HvMADS3 and HvMADS58), a D-class gene (HvMADS13), and the carpel-specific HvDL gene was significantly increased in mov1 compared with the wild type following stamen primordia specification (W3.5). To determine the spatio-temporal expression pattern during floret development, in situ hybridization of selected genes was performed in wild-type (cv. Steptoe) and mov1 mature inflorescences (Fig. 9). As expected, HvMADS16 expression was undetectable in mov1 inflorescences in any floral organ, whereas it was expressed in wild-type lodicules and stamens. The expression patterns of HvMADS3 (C-class) and HvMADS13 (D-class) genes were found to be very similar: in wild-type florets, expression was confined to the ovule. In mov1, both HvMADS3 and HvMADS13 were also expressed in the primordia of ectopic ovules. Notably, HvDL expression in the wild type was restricted to the carpels and to the abaxial side of the lemma but was absent in lodicules or stamens. In mov1 florets, HvDL expression remained in the lemma and was present in the central and ectopic carpels. For all genes tested, sense probes gave no observable signal (Supplementary Fig. S13).
Fig. 9.
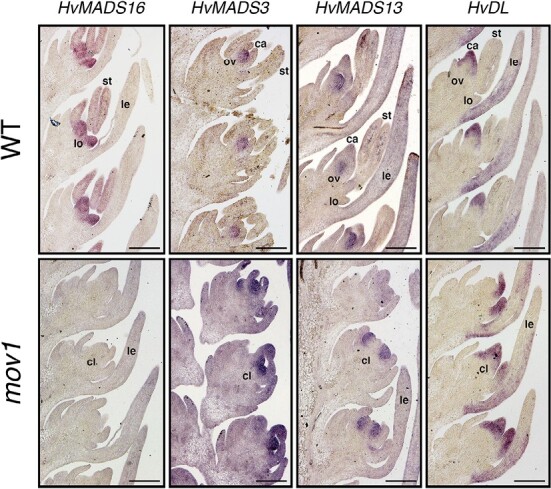
Spatial expression of floral homeotic genes in wild-type (WT) and mov1 inflorescences. Expression patterns as detected by in situ hybridization are shown in WT and mov1 inflorescences at stage W6.0 for genes HvMADS16, HvMADS3, HvMADS13, and HvDL. Lemma (le), lodicule (lo), stamen (st), carpel (ca), ovule (ov), and carpel-like structure (cl). Scale bars: 200 µm.
Discussion
Deletion of the zinc-finger transcription factor HvSL1 underlies the mov2.g multiovary phenotype
The present study demonstrates that HORVU3Hr1G003740 on chromosome 3H underlies the barley mov2.g locus. HORVU3Hr1G003740, referred to here as HvSL1, encodes a putative C2H2 zinc-finger transcription factor and is the likely orthologue of rice STAMENLESS1 (OsSL1) (Xiao et al., 2009). Indeed, the mov2.g multiovary phenotype closely resembles that of sl1 mutants in rice. Both barley and rice mutants exhibit a varied number of irregularly shaped carpels at the expense of stamens, and ectopic organs are present predominantly in whorl 3, while whorl 4 remains less affected and typically retains a single carpel (Xiao et al., 2009). Despite this, notable phenotypic differences are present between mov2.g and sl1. Unlike rice sl1, a high rate of abnormal or converted lodicules was not observed in mov2.g plants. Likewise, palea/lemma-shaped organs or undifferentiated tissue were not observed among the ectopic carpels in whorl 3 of Hvsl1. In addition, inflorescence meristem fasciation appears to be a unique feature of mov2.g, although this requires confirmation by further study.
Moreover, the most prominent feature of mov2.g spikes is the ability to produce multiple grains per floret upon cross-pollination (Fig. 1D, E). This is in accord with initial reports of barley multiovary mo mutants (Moh and Nilan, 1953; Kamra and Nilan, 1959), proposed to be allelic to mov2.g (Franckowiak and Lundqvist, 2013). However, production of multiple grains per floret has not been reported for rice sl1.
More pronounced functional differences are present between HvSL1 and the respective Arabidopsis orthologues, JAG and NUB. Both JAG and NUB appear to be involved in correct stamen and carpel development; however, lack of function in these genes does not alter organ identity, but rather organ morphogenesis (Dinneny et al., 2006). Furthermore, the role of JAG and NUB as shape determinants is not only restricted to the reproductive organs but also affects the outer floral whorls as well as vegetative tissues, indicating a more general role in lateral organ shape (Ohno et al., 2004). This broad comparison indicates that the function of barley HvSL1 is restricted to controlling specific floral organ identity in the Triticeae relative to both closely related monocots (rice) and the more distant dicots (Arabidopsis). Given that mov2.g plants only show conversion of whorl 3 organs (stamens) into supernumerary carpels (Fig. 1), we propose that HvSL1 is necessary for correct stamen development.
Absence of HvMADS16 underlies the mov1 multiovary mutation
The current study positioned the mov1 locus at higher resolution on chromosome 7H and significantly narrowed the critical interval to only three putative genes. Among these, the MADS-box transcription factor gene HvMADS16 emerges as the most likely candidate for mov1. Indeed, the mov1 phenotype is remarkably similar to characterized B-class mutants such as Arabidopsis apetala3 (ap3) (Jack et al., 1992), rice superwoman1 (spw1) (Nagasawa et al., 2003), and maize silky1-5 (si1-5) (Ambrose et al., 2000) which all show transformation of petals/lodicules and stamens into bract-like and carpel-like organs, respectively, and involve DEF-like genes. Additional molecular evidence supports our hypothesis that altered HvMADS16 function underlies the mov1 phenotype. Besides being physically absent in mov1 (Supplementary Fig. S12A), HvMADS16 is preferentially expressed in inflorescences (Supplementary Fig. S12B). Within wild-type developing florets, HvMADS16 expression is specifically confined to lodicules and stamens (Fig. 5C), similarly to SPW1 expression in rice (Nagasawa et al., 2003). The specific expression pattern is also consistent with the observation that mov1 only shows a major phenotype in floral development, but not in any other aspect of plant growth.
Another interesting mov1 phenotype is complete female sterility, as was observed for the orthologous rice mutant spw1 (Nagasawa et al., 2003). Histological analysis suggested that the ovules in all four mov1 carpels are unable to produce a fully differentiated female gametophyte (Supplementary Fig. S10). Instead, the ovule-like structures are filled with undifferentiated cell layers. This phenotype is distinct from mov2.g, which often produced multiple fertile ovules, but is similar to that observed in wheat pistillody lines (Yamada et al., 2009) and in rice spw1 (Nagasawa et al., 2003), suggestive of a potentially conserved role for HvMADS16 in ovule development. Interestingly, this contrasts with the Arabidopsis orthologue ap3 mutant that produces viable seeds (Jack et al., 1992), and with maize si1-5 in which only the transformed carpels in the tassels (the male florets) were reported to be sterile (Ambrose et al., 2000). It also contrasts with the lack of HvMADS16 expression in ovules, and is suggestive of an indirect non-cell-autonomous effect, which might be considered in future studies.
HvSL1 and HvMADS16 functionally overlap during floral specification
As revealed by SEM analysis, phenotypic differences in mov2 and mov1 inflorescences appear very early in floret development (Figs 2 and 7). This is consistent with HvSL1 and HvMADS16 being expressed from double ridge (W2.0) and gradually increasing expression until differentiation of the reproductive organs (W6.0) in wild-type inflorescences (Supplementary Figs S6B, 8).
Studies in rice have suggested that OsSL1 may act as a positive upstream regulator of OsMADS16 (Xiao et al., 2009). Considering that HvSL1 expression temporally precedes that of HvMADS16 and that their overlapping expression domains overlap, we hypothesized that HvSL1 might regulate B-class genes in barley, particularly HvMADS16. Indeed, the presence of HvSL1 appears to positively influence endogenous HvMADS16 expression in transfected protoplasts (Supplementary Fig. S9C). Moreover, expression of HvMADS16 is down-regulated in mov2.g even before stamen primordia initiation, whereas the expression profile of HvSL1 does not significantly differ from that of the wild type in mov1 plants (Fig. 8). These results suggest a conserved regulatory network with rice in controlling stamen identity. Further study is required to establish whether HvSL1 directly regulates HvMADS16 or acts through an intermediate.
Absence of HvSL1 and HvMADS16 disrupts the expression of B-class genes
The absence of HvMADS16 and HvSL1 in mov1 and mov2.g influenced the expression of the transcription factors driving floral development. As expected, there is no expression of HvMADS16 in mov1 (Fig. 8B). Expression of the other B-class genes (HvMADS2 and HvMADS4) in mov1 is significantly reduced from a very early stage onwards, suggesting a tight regulatory relationship between these three genes (Fig. 8B). Evidence supporting this hypothesis comes from the overlapping expression of B-class genes in barley lodicules and stamens (Fig. 5), and the obligate interaction of HvMADS16 (DEF-like) with either GLO-like protein (HvMADS2 and HvMADS4) (Supplementary Fig. S8). These findings are consistent with reports in other species whereby obligate DEF–GLO heterodimers can activate their own expression and initiate a positive autoregulatory feedback loop (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994; McGonigle et al., 1996; Bartlett et al., 2015).
The reduced expression of all three B-class genes HvMADS2, HvMADS4, and HvMADS16 in mov2.g inflorescences is consistent with the lack of stamens (Fig. 8A). Nevertheless, expression of these genes is not completely abolished and is sufficient to drive normal lodicule development in whorl 2. Indeed, transcript abundance of both HvMADS2 and HvMADS4 reaches wild-type levels by stage W6.0. The low levels of HvMADS2 and HvMADS4 at earlier stages could indicate a slower initiation of the positive autoregulatory feedback loop typical of B-class genes, due to the low HvMADS16 levels.
HvSL1 and HvMADS16 have distinct effects on C- and D-class genes
The expression of C-class genes (HvMADS3 and HvMADS58), D-class genes (HvMADS13), and HvDL increased in mov1 inflorescences (Fig. 8B). In wild-type barley, these genes are expressed in the fourth whorl and are predicted to specify carpel (HvMADS58 and HvDL) and ovule (HvMADS3 and HvMADS13) development. The HvDL expression pattern is consistent with that of its rice orthologue OsDL (Nagasawa et al., 2003), and partly with that of the maize co-orthologues drl1 and drl2 (Strable and Vollbrecht, 2019). Whereas transcript of maize drl genes accumulated in the carpel primordia and lemma as well as in the palea, glumes, and cryptic bracts (Strable and Vollbrecht, 2019), expression of rice OsDL was shown to be confined to the carpel and to the central vein of the lemma (Ohmori et al., 2011). Similarly, HvDL expression in both the wild type and mov1 was also restricted to the central carpel, ectopic carpels, and the base of the lemma (Fig. 9), overall suggesting a conserved function in carpel morphogenesis among grasses. Likewise, HvMADS13 signal in wild-type barley ovules is similar to that of OsMADS13 in rice (Lopez-Dee et al., 1999), whereas the specific expression pattern of HvMADS3 in ovules highlights a potential subfunctionalization of C-class genes in the grasses. Both the rice (OsMADS3) and maize (ZMM2) HvMADS3 orthologues have been reported to be strongly expressed in stamen primordia (Dreni et al., 2011) and anthers (Mena et al., 1996), respectively. OsMADS3 plays an important role in specifying stamen identity, as well as in repressing lodicule formation (Yamaguchi et al., 2006). The ovule-specific HvMADS3 expression (Fig. 9), and the increased transcript abundance detected in mov1 (Fig. 8B) argue against a conserved role for HvMADS3 in stamen specification in barley.
In mov1 inflorescences, the increased transcript levels of both HvMADS3 and HvMADS13 can be explained by the additional ovule structures observed by histochemical analysis (Supplementary Fig. S10) and corresponds well to the spatial expression of both genes in the mutant (Fig. 9). However, expression of these ovule identity genes is apparently insufficient to drive correct ovule formation in mov1, suggesting that their activity may be compromised, or that ovule fertility defects are manifested later during gametogenesis. We speculate that the increased transcript levels of DL-, C-, and D-class genes in mov1 can be explained by a reduced repressive action of B-class genes on carpel- and ovule-promoting genes, together with the expansion of these genes to the third whorl, resulting in florets with carpels in two consecutive whorls (whorl 3 and whorl 4).
In mov2.g, the increased abundance of carpel- and ovule-specific C-class genes (HvMADS3 and HvMADS58) (Fig. 8A) is also consistent with the presence of supernumerary carpels (Fig. 1). Despite this, expression levels of the D-class gene HvMADS13 and the carpel-specific HvDL gene remain unaffected in mov2.g with respect to the wild type (Fig. 8A). This contrasts with mov1 and confirms that an increase in ovule number alone does not always relate to increased HvMADS13 and HvDL expression. A possible explanation is that these two genes may play a minor role in the formation of additional ovules and carpels in mov2.g compared with other genes. Alternatively, HvDL in mov2.g may have slower expression dynamics. Indeed, most of the investigated genes seem to have a delayed response in mov2.g compared with the wild type.
A model for barley stamen specification
Although the ABC model was initially proposed as a universal model to explain flower development, differences have been found across plant species, even among members of the same family, as is the case between rice and barley (Poaceae). For example, some MADS-box genes have acquired grass-specific functions as seen for the A-class genes in rice which specify palea and lodicules (reviewed in Callens et al., 2018). Other classes underwent partial subfunctionalization, as is the case of the C-class genes in rice (Kang et al., 1995; Yamaguchi et al., 2006; Kuijer et al., 2021) and maize (Ali et al., 2019), or the D-lineage in Brachypodium distachyon (Wei et al., 2013).
In barley, an expression atlas throughout inflorescence development of 34 MIKc MADS genes highlighted how some transcription patterns strongly deviate from the predicted model (Kuijer et al., 2021). Indeed, several barley MADS-box genes have acquired roles in addition to floral specification. This is the case for HvMADS14 (A-class) which controls vernalization-induced flowering, acting as the VERNALIZATION1 equivalent in other temperate cereals (Trevaskis et al., 2003). Another example is HvMADS1 (E-class), which has been recently reported to direct plant thermomorphogenesis (Li et al., 2021). Conversely, the core B-class function, particularly stamen development, appears to be extremely conserved among domesticated grasses.
Based on the results obtained in this study, together with previous reports in barley and other plant species, we formulate a testable model for barley stamen specification (Fig. 10). In this model, HvSL1 acts predominantly in whorl 3 and promotes HvMADS16 in the wild type (Fig. 10A). The B-class genes HvMADS16, HvMADS2, and HvMADS4 are expressed in floral whorls 2 and 3 and form DEF–GLO heterodimers. Based on the floral quartet model, the heterodimers associate in higher order protein complexes with A- and E-class genes, leading to the formation of lodicules in whorl 2. In whorl 3, instead, the floral quartet complex forms between B-, C-, and E-class genes, promoting stamen development while concomitantly repressing carpel formation.
Fig. 10.
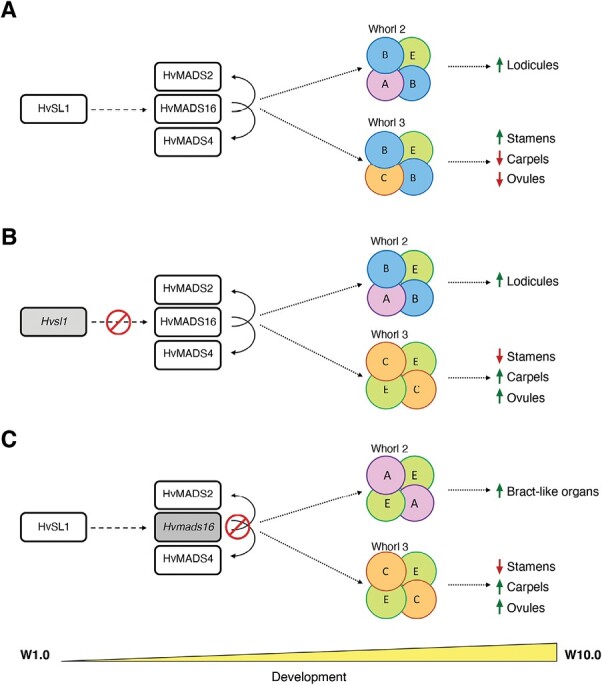
Model for barley stamen specification. (A) As inflorescence development progresses in the wild type, genes of the ABC model combine in floral quartets to specify lodicules in whorl 2 and stamens in whorl 3, with putative repression of carpel and ovule-specific genes. The absence of (B) HvSL1 in mov2.g plants and (C) of HvMADS16 in mov1 plants affects the balance and composition of the floral quartets that form in these whorls, leading to an altered specification of the resulting floral organs. Within the floral quartets, B-class genes act as obligate GLO (HvMADS2 and HvMADS4)/DEF (HvMADS16) heterodimers. Dashed arrows indicate direct or indirect interaction, solid arrows indicate direct interactions found in this study, while dotted arrows indicate a process or consequence. For each mutant, the missing protein is indicated in grey, and the red symbol indicates the affected interaction.
The lack of HvSL1 in mov2.g plants results in a shift in this balance (Fig. 10B). At a molecular level, we speculate that B-class activity in mov2.g florets is predominantly maintained in whorl 2 where enough HvMADS16 is present to develop normal lodicules. At the same time, the reduced levels of HvMADS16 in whorl 3 are insufficient to successfully form functional B-class heterodimers. Consequently, the predominant quaternary complexes forming in whorl 3 are between C- and E-class genes, which promote carpel formation. Concomitant expansion of ovule-promoting genes such as HvMADS3 to whorl 3 may then explain multiple carpels having a functional female gametophyte.
In mov1, HvMADS16 is absent. The remaining B-class GLO-like proteins HvMADS2 and HvMADS4 do not interact and therefore cannot compensate for the absence of HvMADS16. Thus, the only protein complexes that can form in whorl 2 are between A- and E-class genes, and between C- and E-class genes in whorl 3. As a result, bract-like organs develop in whorl 2 and carpel formation is promoted in whorl 3, leading to the multiovary phenotype observed in mov1 (Fig. 10C).
In conclusion, we have characterized two barley multiovary loci, mov2 and mov1, and identified likely causative genes, namely a C2H2 zinc-finger transcription factor gene termed here HvSL1, and the B-class gene HvMADS16. Although both mutants exhibit a similar multiovary phenotype, the difference in fertility is remarkable: while mov1 is completely sterile, mov2 can produce multiple grains per floret. This indicates that the presence of multiovary is not necessarily indicative of fully functional reproductive development in barley. Studying the molecular changes underlying these differences will improve our understanding of floral development at a species-specific level and expand the tools available for modification of cereal florets for breeding.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. Primer sequences and Taqman probes used for copy number analysis to genotype mov2.g and mov1 plants.
Table S2. Sequence of KASP™ markers on chromosome 3H used to map the mov2 locus.
Table S3. PCR primer sequences for testing the presence of genes upstream and downstream from HvSL1 on chromosome 3H.
Table S4. PCR primer sequences for testing the presence of barley B-class genes.
Table S5. PCR primer sequences for testing the presence of genes upstream and downstream from HvMADS16 on chromosome 7H.
Table S6. qRT-PCR primer sequences.
Table S7. Primer sequences for HvSL1 CRISPR knockout.
Table S8. Primer sequence for cloning of in situ hybridization antisense and sense probes.
Table S9. Primer sequences for BiFC cloning.
Table S10. Floral organ frequency in the wild type and mov2.g.
Table S11. Annotated genes present in the mapped mov2 critical interval between flanking markers chr3H_9748112 and chr3H_10289104.
Table S12. BLASTp results using barley HvSL1 as query against the rice genome.
Table S13. BLASTp results using rice OsSL1 as query against the barley genome.
Table S14. Observed segregation ratios of mov1 phenotype in heterozygous growing material.
Table S15. Genes on chromosome 7H tested by PCR.
Fig. S1. Histological sections of mov2.g carpels.
Fig. S2. Spike morphology in wild-type and mov2.g plants.
Fig. S3. Mapping of the mov2 locus in a mov2.g×Morex bi-parental population.
Fig. S4. Heatmap of gene expression for genes in the critical mov2 interval.
Fig. S5. HvSL1 and OsSL1 gene models and alignment.
Fig. S6. HvSL1 deletion in mov2.g.
Fig. S7. CRISPR design and analysis of Hvsl1 knockout plants.
Fig. S8. BiFC assays showing interaction between barley B-class genes.
Fig. S9. Transfection efficiency and transcript abundance in barley protoplasts.
Fig. S10. Histological sections of mov1 carpels.
Fig. S11. Details of mov1 inflorescence development.
Fig. S12. Characterization of the mov1 deletion and HvMADS16.
Fig. S13. In situ hybridization with sense probes on wild-type and mov1 inflorescences.
Acknowledgements
The authors are grateful to Dr Margaret Pallotta and Mr Chao Ma for technical advice, and to the Adelaide Microscopy staff at the University of Adelaide.
Contributor Information
Caterina Selva, Waite Research Institute, School of Agriculture Food and Wine, University of Adelaide, Waite Campus, Urrbrae 5064, South Australia, Australia.
Xiujuan Yang, Waite Research Institute, School of Agriculture Food and Wine, University of Adelaide, Waite Campus, Urrbrae 5064, South Australia, Australia.
Neil J Shirley, Waite Research Institute, School of Agriculture Food and Wine, University of Adelaide, Waite Campus, Urrbrae 5064, South Australia, Australia.
Ryan Whitford, Waite Research Institute, School of Agriculture Food and Wine, University of Adelaide, Waite Campus, Urrbrae 5064, South Australia, Australia.
Ute Baumann, Waite Research Institute, School of Agriculture Food and Wine, University of Adelaide, Waite Campus, Urrbrae 5064, South Australia, Australia.
Matthew R Tucker, Waite Research Institute, School of Agriculture Food and Wine, University of Adelaide, Waite Campus, Urrbrae 5064, South Australia, Australia.
Madelaine Bartlett, University of Massachusetts Amherst, USA.
Author contributions
CS, RW, UB, and MRT: conceptualization; RW, UB, and MRT: supervision; CS: performing the experiments; NJS: performing qRT-PCR and assisting in identifying KASP™ markers for mapping; XY: assisted with in situ hybridization and provided technical advice on cloning; CS: writing the manuscript; all authors edited and reviewed the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Funding
This research was supported by funding from the Australian Research Council (MRT; DP210103491), the Australian Centre for Plant Functional Genomics (RW and UB; ACPFG), the University of Adelaide and the Australian Government who provided a Research Training Program Stipend PhD scholarship to CS.
Data availability
All data supporting the findings of this study can be found within the paper and its supplementary data published online. They are also available from the corresponding author, Matthew R. Tucker, upon request.
References
- Abraham-Juárez MJ, Schrager-Lavelle A, Man J, Whipple C, Handakumbura P, Babbitt C, Bartlett M.. 2020. Evolutionary variation in MADS box dimerization affects floral development and protein abundance in maize. The Plant Cell 32, 3408–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali K, Raza Q, Atif RM, Aslam U, Ajmal M, Chung G.. 2019. Genetic and molecular control of floral organ identity in cereals. International Journal of Molecular Sciences 20, 20743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ.. 2000. Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Molecular Cell 5, 569–579. [DOI] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S.. 2007. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Han N, Wu J, Yang Y, Wang J, Zhu M, Bian H.. 2014. A transient gene expression system using barley protoplasts to evaluate microRNAs for post-transcriptional regulation of their target genes. Plant Cell, Tissue and Organ Culture 119, 211–219. [Google Scholar]
- Franckowiak JD, Lundqvist U.. 2013. Barley Genetics Newsletter 43, 48–223. https://wheat.pw.usda.gov/ggpages/bgn/43/index.html [Google Scholar]
- Bartlett ME, Williams SK, Taylor Z, DeBlasio S, Goldshmidt A, Hall DH, Schmidt RJ, Jackson DP, Whipple CJ.. 2015. The maize PI/GLO ortholog Zmm16/sterile tassel silky ear1 interacts with the zygomorphy and sex determination pathways in flower development. The Plant Cell 27, 3081–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Satoh-Nagasawa N, Jackson D, Hirano H.. 2005. Genetics and evolution of inflorescence and flower development in grasses. Plant and Cell Physiology 46, 69–78. [DOI] [PubMed] [Google Scholar]
- Brenchley WE. 1920. The development of the flower and grain of barley. Journal of the Institute of Brewing 26, 615–632. [Google Scholar]
- Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R.. 2004. A structured mutant population for forward and reverse genetics in barley (Hordeum vulgare L.). The Plant Journal 40, 143–150. [DOI] [PubMed] [Google Scholar]
- Callens C, Tucker MR, Zhang D, Wilson ZA.. 2018. Dissecting the role of MADS-box genes in monocot floral development and diversity. Journal of Experimental Botany 69, 2435–2459. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee L-Y, Vyas S, Glick E, Chen M-H, Vainstein A, Gafni Y, Gelvin SB, Tzfira T.. 2006. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. Journal of Molecular Biology 362, 1120–1131. [DOI] [PubMed] [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, et al. 2009. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10, 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM.. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, Scholz U, Mascher M.. 2015. BARLEX—the barley draft genome explorer. Molecular Plant 8, 964–966. [DOI] [PubMed] [Google Scholar]
- Colombo L, Franken J, Koetje E, van Went J, Dons HJ, Angenent GC, van Tunen AJ.. 1995. The petunia MADS box gene FBP11 determines ovule identity. The Plant Cell 7, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF.. 2006. NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D.. 2004. The role of JAGGED in shaping lateral organs. Development 131, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D Kater MM.. 2011. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. The Plant Cell 23, 2850–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, et al. 2011. Genetic dissection of barley morphology and development. Plant Physiology 155, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro R, Pinyopich A, Battaglia R, Kooiker M, Borghi L, Ditta G, Yanofsky MF, Kater MM, Colombo L.. 2003. MADS-box protein complexes control carpel and ovule development in Arabidopsis. The Plant Cell 15, 2603–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM.. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes & Development 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J.. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM.. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Kamra OP, Nilan RA.. 1959. Multi-ovary in barley floral anatomy and embryo-sac development. Journal of Heredity 50, 159–166. [Google Scholar]
- Kang H-G, Jang S, Chung J-E, Cho Y-G, An G.. 1997. Characterization of two rice MADS box genes that control flowering time. Molecules & Cells 7, 5569. [PubMed] [Google Scholar]
- Kang H-G, Noh Y-S, Chung Y-Y, Costa MA, An K, An G.. 1995. Phenotypic alterations of petal and sepal by ectopic expression of a rice MADS box gene in tobacco. Plant Molecular Biology 29, 1–10. [DOI] [PubMed] [Google Scholar]
- Knudsen S, Wendt T, Dockter C, et al. 2022. FIND-IT: accelerated trait development for a green evolution. Science Advances 8, eabq2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Vasquez-Gross HA, Howell T, et al. 2017. Uncovering hidden variation in polyploid wheat. Proceedings of the National Academy of Sciences, USA 114, E913–E921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijer HN, Shirley NJ, Khor SF, Shi J, Schwerdt J, Zhang D, Li G, Burton RA.. 2021. Transcript profiling of MIKCc MADS-box genes reveals conserved and novel roles in barley inflorescence development. Frontiers in Plant Science 12, 705286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Kuijer HN, Yang X, et al. 2021. MADS1 maintains barley spike morphology at high ambient temperatures. Nature Plants 7, 1093–1107. [DOI] [PubMed] [Google Scholar]
- Liu H, Li G, Yang X, Kuijer HNJ, Liang W, Zhang D.. 2019. Transcriptome profiling reveals phase-specific gene expression in the developing barley inflorescence. The Crop Journal 8, 71–86. [Google Scholar]
- Lopez-Dee ZP, Wittich P, Pè EM, Rigola D, Del Buono I, Gorla MS, Kater MM, Colombo L.. 1999. OsMADS13, a novel rice MADS-box gene expressed during ovule development. Developmental Genetics 25, 237–244. [DOI] [PubMed] [Google Scholar]
- Lyu T, Cao J.. 2018. Cys2/His2 Zinc-Finger proteins in transcriptional regulation of flower development. International Journal of Molecular Sciences 19, 2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Matros A, Houston K, Tucker MR, et al. 2021. Genome-wide association study reveals the genetic complexity of fructan accumulation patterns in barley grain. Journal of Experimental Botany 72, 2383–2402. [DOI] [PubMed] [Google Scholar]
- McGonigle B, Bouhidel K, Irish VF.. 1996. Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes & Development 10, 1812–1821. [DOI] [PubMed] [Google Scholar]
- Melzer R, Theissen G.. 2009. Reconstitution of ‘floral quartets’ in vitro involving class B and class E floral homeotic proteins. Nucleic Acids Research 37, 2723–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ.. 1996. Diversification of C-function activity in maize flower development. Science 274, 1537–1540. [DOI] [PubMed] [Google Scholar]
- Moh CC, Nilan RA.. 1953. Multi-ovary in barley: a mutant induced by atomic bomb irradiation. Journal of Heredity 44, 183–184. [Google Scholar]
- Murai K, Takumi S, Koga H, Ogihara Y.. 2002. Pistillody, homeotic transformation of stamens into pistil-like structures, caused by nuclear–cytoplasm interaction in wheat. The Plant Journal 29, 169–181. [DOI] [PubMed] [Google Scholar]
- Murai K, Tsunewaki K.. 1993. Photoperiod-sensitive cytoplasmic male sterility in wheat with Aegilops crassa cytoplasm. Euphytica 67, 41–48. [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y.. 2003. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. [DOI] [PubMed] [Google Scholar]
- Ohmori Y, Toriba T, Nakamura H, Ichikawa H, Hirano H-Y.. 2011. Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice. The Plant Journal 65, 77–86. [DOI] [PubMed] [Google Scholar]
- Ohno CK, Reddy GV, Heisler MGB, Meyerowitz EM.. 2004. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF.. 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Peng Z-S. 2003. A new mutation in wheat producing three pistils in a floret. Journal of Agronomy and Crop Science 189, 270–272. [Google Scholar]
- Rogowsky PM, Guidet FL, Langridge P, Shepherd KW, Koebner RM.. 1991. Isolation and characterization of wheat–rye recombinants involving chromosome arm 1DS of wheat. Theoretical and Applied Genetics 82, 537–544. [DOI] [PubMed] [Google Scholar]
- Schreiber M, Barakate A, Uzrek N, Macaulay M, Sourdille A, Morris J, Hedley PE, Ramsay L, Waugh R.. 2019. A highly mutagenised barley (cv. Golden Promise) TILLING population coupled with strategies for screening-by-sequencing. Plant Methods 15, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lönnig W-E, Saedler H, Sommer H.. 1992. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. The EMBO Journal 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H.. 1990. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. [DOI] [PubMed] [Google Scholar]
- Selva C, Riboni M, Baumann U, Würschum T, Whitford R, Tucker MR.. 2020. Hybrid breeding in wheat: how shaping floral biology can offer new perspectives. Functional Plant Biology 47, 675–694. [DOI] [PubMed] [Google Scholar]
- Selva C, Shirley N, Houston K, Whitford R, Baumann U, Li G, Tucker MR.. 2021. HvLEAFY controls the early stages of floral organ specification and inhibits the formation of multiple ovaries in barley. The Plant Journal 108, 509–527. [DOI] [PubMed] [Google Scholar]
- Shoesmith JR, Solomon CU, Yang X, et al. 2021. APETALA2 functions as a temporal factor together with BLADE-ON-PETIOLE2 and MADS29 to control flower and grain development in barley. Development 148, dev194894. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muiño JM, et al. 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences, USA 109, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule JD, Kudrna DA, Kleinhofs A.. 2000. Isolation, mapping, and characterization of two barley multiovary mutants. Journal of Heredity 91, 483–487. [DOI] [PubMed] [Google Scholar]
- Soule JD, Skodova I, Kudrna D, Kilian A, Kleinhofs A.. 1996. Molecular and genetic characterization of barley flower development mutants. Barley Genetics Newsletter 25, 76–80. [Google Scholar]
- Strable J, Vollbrecht E.. 2019. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate floret development and floral meristem determinacy. Development 146, dev171181. [DOI] [PubMed] [Google Scholar]
- Szarejko I, Szurman-Zubrzycka M, Nawrot M, Marzec M, Gruszka D, Kurowska M, Chmielewska B, Zbieszczyk J, Jelonek J, Maluszynski M.. 2017. Creation of a TILLING population in barley after chemical mutagenesis with sodium azide and MNU. In: Jankowicz-Cieslak J, Tai TH, Kumlehn J, Till BJ, eds. Biotechnologies for plant mutation breeding: protocols. Cham: Springer International Publishing, 91–111. [Google Scholar]
- Szurman-Zubrzycka ME, Zbieszczyk J, Marzec M, et al. 2018. HorTILLUS—a rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (Hordeum vulgare L.). Frontiers in Plant Science 9, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamè V, Bovina R, Sanguineti MC, Tuberosa R, Lundqvist U, Salvi S.. 2008. TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotechnology Journal 6, 477–485. [DOI] [PubMed] [Google Scholar]
- Tazhin OT. 1980. The linkage of the genes mo5* and n in barley. Barley Genetics Newsletter 10, 69–72. [Google Scholar]
- Theissen G. 2001. Development of floral organ identity: stories from the MADS house. Current Opinion in Plant Biology 4, 75–85. [DOI] [PubMed] [Google Scholar]
- Thomson B, Wellmer F.. 2019. Molecular regulation of flower development. In: Grossniklaus U, ed. Plant development and evolution. Amsterdam: Elsevier Inc., 185–210. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES.. 2003. MADS box genes control vernalization-induced flowering in cereals. Proceedings of the National Academy of Sciences, USA 100, 13099–13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F.. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington SR, Cartwright PM, Wall PC.. 1983. A quantitative scale of spike initial and pistil development in barley and wheat. Annals of Botany 51, 119–130. [Google Scholar]
- Wang QH, Yang ZJ, Wei SH, Jiang ZY, Yang YF, Hu ZS, Sun QX, Peng ZS.. 2015. Molecular cloning, characterization and expression analysis of WAG-1 in the pistillody line of common wheat. Genetics and Molecular Research 14, 12455–12465. [DOI] [PubMed] [Google Scholar]
- Wei B, Liu D, Guo J, Leseberg CH, Zhang W, Mao L.. 2013. Functional divergence of two duplicated D-lineage MADS-box genes BdMADS2 and BdMADS4 from Brachypodium distachyon. Journal of Plant Physiology 170, 424–431. [DOI] [PubMed] [Google Scholar]
- Winter KU, Weiser C, Kaufmann K, Bohne A, Kirchner C, Kanno A, Saedler H, Theissen G.. 2002. Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Molecular Biology and Evolution 19, 587–596. [DOI] [PubMed] [Google Scholar]
- Xiao H, Tang J, Li Y, et al. 2009. STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. The Plant Journal 59, 789–801. [DOI] [PubMed] [Google Scholar]
- Yamada K, Saraike T, Shitsukawa N, Hirabayashi C, Takumi S, Murai K.. 2009. Class D and Bsister MADS-box genes are associated with ectopic ovule formation in the pistil-like stamens of alloplasmic wheat (Triticum aestivum L.). Plant Molecular Biology 71, 1–14. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY.. 2006. Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. The Plant Cell 18, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li G, Tian Y, Song Y, Liang W, Zhang D.. 2018. A rice glutamyl-tRNA synthetase modulates early anther cell division and patterning. Plant Physiology 177, 728–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tucker MR.. 2021. Establishing a regulatory blueprint for ovule number and function during plant development. Current Opinion in Plant Biology 63, 102095. [DOI] [PubMed] [Google Scholar]
- Yang Z, Peng Z, Wei S, Liao M, Yu Y, Jang Z.. 2015. Pistillody mutant reveals key insights into stamen and pistil development in wheat (Triticum aestivum L.). BMC Genomics 16, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J.. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y.. 2011. Flower development in rice. Journal of Experimental Botany 62, 4719–4730. [DOI] [PubMed] [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF.. 1974. A decimal code for the growth stages of cereals. Weed Research 14, 415–421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study can be found within the paper and its supplementary data published online. They are also available from the corresponding author, Matthew R. Tucker, upon request.


