Abstract
Mesendodermal specification and cardiac differentiation are key issues for developmental biology and heart regeneration medicine. Previously, we demonstrated that FAM122A, a highly conserved housekeeping gene, is an endogenous inhibitor of protein phosphatase 2A (PP2A) and participates in multifaceted physiological and pathological processes. However, the in vivo function of FAM122A is largely unknown. In this study, we observed that Fam122 deletion resulted in embryonic lethality with severe defects of cardiovascular developments and significantly attenuated cardiac functions in conditional cardiac-specific knockout mice. More importantly, Fam122a deficiency impaired mesendodermal specification and cardiac differentiation from mouse embryonic stem cells but showed no influence on pluripotent identity. Mechanical investigation revealed that the impaired differentiation potential was caused by the dysregulation of histone modification and Wnt and Hippo signaling pathways through modulation of PP2A activity. These findings suggest that FAM122A is a novel and critical regulator in mesendodermal specification and cardiac differentiation. This research not only significantly extends our understanding of the regulatory network of mesendodermal/cardiac differentiation but also proposes the potential significance of FAM122A in cardiac regeneration.
Keywords: FAM122A, mESCs, mesendoderm, PP2A, cardiac differentiation
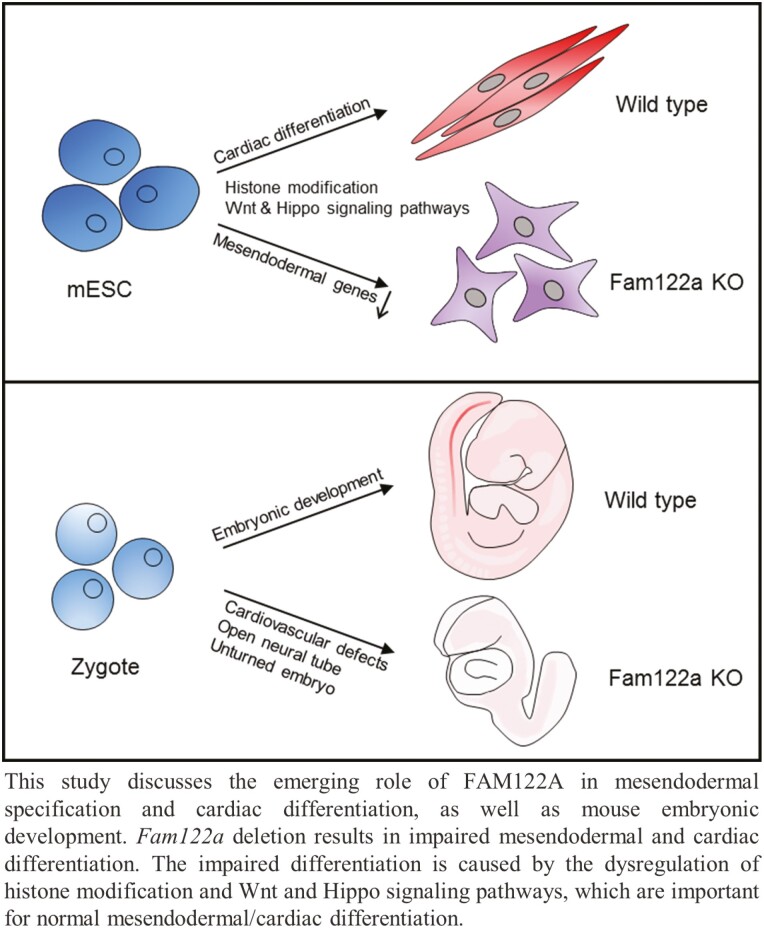
Graphical Abstract
Graphical Abstract.
Significance Statement.
Mesendodermal specification and cardiac differentiation are key issues for developmental biology and heart regeneration medicine. This study demonstrated for the first time that FAM122A, which was previously identified as a protein phosphatase 2A (PP2A) inhibitor, is a novel and critical regulator in mesendodermal/cardiac differentiation and early cardiac development. Impaired differentiation of Fam122a deletion was associated with the dysregulation of histone modification and Wnt and Hippo signaling pathways. These results reveal the essential role of FAM122A in cardiac differentiation and the possible potential significance in cardiac regeneration and linkage of PP2A and 3-germ layer development, which have not been characterized previously.
Introduction
Heart is the first and functional organ during embryonic development dedicated to the transportation of oxygen, nutrients, and wastes to and from the embryos, and it is vital to embryo survival. Cardiogenesis is a very complicated process involving cardiomyocyte (CM) differentiation, non-CM lineage specification, and a series of morphogenetic events.1,2 Briefly, it comprises pluripotent epiblast from inner cell mass, 3–germ layer cells or gastrulation stage (endoderm, mesoderm, and ectoderm), and subsequent differentiation along with cardiac mesoderm and cardiac progenitor specification, which includes myocardial and endocardial cells and smooth muscle cells. The cardiac precursor cells (CPCs) further migrate and fuse at the embryonic midline to form the linear primary heart tube and develop into functionally matured cells, finally transforming into a looped, multichambered, valved pump organ in a proper morphogenetic patterning.1,3
Mesendoderm (ME), referring to the primitive streak during gastrulation, has the potential to differentiate into mesoderm and endoderm, which further develop into a series of mesoderm and endoderm tissues, respectively, and organs, such as embryonic heart, vasculature, hematological system, muscle, lung, and liver.4 Thus, ME specification is an important and intermediate phase for heart development from pluripotent epiblast or embryonic stem (ES) cells during early embryonic development. ME formation is highly organized and spatiotemporally controlled via coordinated networks, including a number of essential transcription factors (TFs) specific for ME, epigenetic regulators, chromatin remodeling factors, and signaling pathways.5-7 However, the dynamic regulation and precise mechanism of ME specification are not well understood.7 Eomesodermin (Eomes) and brachyury (T), which are T-box TFs, are essential for ME specification between embryonic day (E) 6.5 and 7.5 in the mouse, and disruption of either one results in defects in cardiac mesoderm differentiation and embryonic lethality due to gastrulation defects.8-10 Eomes expression marks the earliest cardiac mesoderm and dictates the formation of cardiac precursors through regulating the master TF mesoderm posterior 1 (Mesp1).11 These critical TF expressions in ME are dependent on the activation of several important signaling pathways, including Wnt and transforming growth factor-β (TGF-β) signaling pathways.6,10 Wnt ligands bind to Frizzled receptors to activate β-catenin, which directly binds the upstream regulatory regions in most ME genes, including T, Eomes, and Mixl1.12,13 Besides Wnt signaling, TGF-β family member Nodal binds activin receptor to drive ME specification by Smad TF family members.14,15 β-catenin and Smad2/3 TFs collaboratively activate ME genes.5,16 In addition, Hippo signaling has recently been found to be involved in the modulation of this transcriptional program as a negative regulator.17 Upstream signals phosphorylate Yes-associated protein (YAP)/transcriptional co-activator with PDZ(PSD-95/Dlg/ZO-1)-binding motif (TAZ), leading to its restriction in cytoplasm by 14-3-3 binding and further proteasomal degradation. Non-phosphorylated YAP/TAZ translocates to the nucleus and binds with TEA domain transcription factors, which triggers the transcriptional activation of target genes.18 YAP exerts transcriptional repression in the regulation of ME gene by suppressing Wnt3.17 These signaling pathways and TFs have a crosstalk with histone modifications and chromatin remodeling to coordinately regulate ME specification and myocardial differentiation.5,6
Family with sequence similarity 122A (FAM122A) was first identified as an endogenous inhibitor of protein phosphatase 2A (PP2A),19,20 and its inhibitory effect in the regulation of replication stress was further confirmed by Li et al.21 Later, FAM122A was renamed as PP2A-Aα (PPP2R1A) and -B55A (PPP2R2A) interacting phosphatase regulator 1 (PABIR1) in human. FAM122A, a highly conserved housekeeping gene, localizes on chromosome 9q21.1 within the first intron of phosphatidylinositol 4-phosphate 5-kinase (PIP5K1B) gene and encodes 287 amino acids. FAM122A knockout (KO) suppresses the growth of hepatocellular carcinoma cells and acute myeloid leukemia cells in vitro and in vivo, with independence or dependence of PP2A inhibitory activity.22,23 In addition, FAM122A is essential for maintaining the self-renewal capability of hematological stem cells24 and required for the differentiation of erythroid cells;25 FAM122A also contributes to the maintenance of DNA stability in malignant tumor cells.26 These studies suggest that FAM122A has multifaceted functions under physiological or pathological circumstances. However, the in vivo function of FAM122A remains unknown.
In this study, we used Fam122a KO mice, conditional cardiac-specific Fam122a KO mice, and mouse ES cells (mESCs) to address the role of FAM122A in embryonic development and CM differentiation. We showed that Fam122a deficiency led to early embryonic lethality with severe defects of heart development, and impaired cardiac function was found in conditional KO mice. More importantly, we observed that FAM122A KO significantly disrupted ME specification and myocardial differentiation, which is cooperatively dysregulated by histone modifications and Wnt and Hippo signaling pathways.
Materials and Methods
Mouse Use and Maintenance
Fam122a KO mice were produced by East China Normal University (C57BL/6 and 129/SvJ mixed background). Fam122a gene was inactivated by targeting the unique exon. Supplementary Fig. S1A shows the schematics of targeted disruption of Fam122a. Fam122a floxed mice were produced by Shanghai Renyuan Biotechnology as described previously.24 Myh6-Cre mice were purchased from Shanghai Model Organisms Center, Inc., and Nkx2-5-Cre mice were bought from Jiangsu Gempharmatech Biotechnology. Fam122a conditional KO mice were on a C57BL/6 background. All animal experiments were carried out as approved by the Laboratory Animal Resource Center of Shanghai JiaoTong University School of Medicine.
Embryo Collection and Whole-Mount In Situ Hybridization
Whole-mount in situ hybridization assay was performed as described previously.27 For embryo collection, mice were maintained on a 12/12 h light/dark cycle. After mating, the morning of appearance of a vaginal plug was designated as embryonic day 0.5 (E0.5). At the determined embryonic day, mouse embryos were dissected in cold diethyl pyrocarbonate-treated phosphate-buffered saline + 0.1% Tween 20 (PBST). Then, the embryos were fixed with 4% paraformaldehyde (PFA) in PBST at 4 °C and dehydrated stepwise through a methanol/PBST series (25%, 50%, 75%, and 100% methanol). Embryos can be stored at −20 °C in 100% methanol. RNA probes were generated by in vitro transcription from linearized plasmids using a DIG RNA Labelling Kit (SP6/T7). Rehydrated embryos were incubated in 4:1 PBST:30% H2O2 for 1 h on ice and digested with 1 mL 10 µg mL−1 proteinase K in PBST in a 25 °C water bath. Digested embryos were fixed for 20 minutes at room temperature in 4% PFA/PBST and 0.2% glutaraldehyde and hybridized overnight at 65 °C. Embryos were pre-blocked in 0.5 mL Tris-buffered saline with 0.1% Tween 20 (TBST) + 10% heat-treated sheep serum and incubated with 1:2500 dilution of anti-DIG-alkaline phosphatase (AP) antibody at 4 °C overnight.
Then, the embryos were stained with BM Purple AP substrate, and the reaction was stopped by washing with 5 mm ethylenediaminetetraacetic acid (EDTA) in PBST. Embryos can be re-fixed and stored in 4% PFA/PBST.
Hematoxylin and Eosin Staining
Embryos were fixed in Bouin’s solution, and hearts from Fam122a conditional KO mice were fixed in 4% PFA and then processed for paraffin embedding. Sections (3.5 µm thick) were cut, dewaxed, rehydrated, and stained with hematoxylin and eosin (HE) solutions.
Genotyping
Mice were genotyped using a 2 mm piece of the toe or tail tip. The tissues were incubated overnight at 55 °C in 200 µL lysis buffer (50 mm NaCl, 10 mm Tris-HCl [pH 8.0], 5 mm EDTA, and 0.1% sodium dodecyl sulphate [SDS]) containing 75 µg proteinase K. The lysate can be used for polymerase chain reaction (PCR) directly or precipitated with isopropanol and dissolved in Tris-EDTA (TE) buffer. For a single embryo or yolk sac, DNA was extracted by KAPA Express Extract Kit following the manufacturer’s instruction. PCR primers are listed in Supplementary Table S1.
ESC Culture
R1/E (purchased from Cell Bank of Chinese Academy of Sciences) and TC1 (a gift from J. Kang)28 cells were maintained in Dulbecco’s Modified Eagle Medium (Basal Media) supplemented with 15% fetal bovine serum (Gibco), 1 × nonessential amino acids (Gibco), 1 × GlutaMAX (Gibco), 1 × sodium pyruvate (Gibco), 0.1 mm β-mercaptoethanol (Sigma), 1000 U mL−1 leukemia inhibitory factor (LIF) (Millipore), 100 U mL−1 penicillin, and 100 µg mL−1 streptomycin on mitomycin C (Selleck) inactivated mouse embryonic fibroblast feeder cells under 5% CO2 at 37 °C. ESCs were passaged every other day using 0.25% Trypsin at a split ratio of 1:5.
Generation of Fam122a KO mESCs
The sgRNAs targeted to the exon of Fam122a were inserted into the pX330 vector and are listed in Supplementary Table S1. The plasmids were electroporated into mESCs at 250 V and 500 µF in a 0.4 cm Gene Pulser cuvette (Bio-Rad). Then, mESCs were replated on feeder cells, and individual colonies were picked and expanded. The deletion of FAM122A was validated by Western blot.
AP Staining
ESC colonies were cultured on 12-well plates for 5 days, and AP staining was performed with the AP detection kit (Millipore) following the manufacturer’s instructions. Briefly, the cells were fixed with 4% PFA at room temperature for 2 minutes and rinsed with TBST. Then, the cells were stained with naphthol/Fast Red Violet solution (Fast Red Violet, naphthol AS-BI phosphate solution and water at a 2:1:1 ratio) in the dark at room temperature for 15 minutes, rinsed with TBST, covered with PBS, and imaged using a scanner.
Embryonic Body Formation and Cardiac Differentiation
The cardiac muscle cell differentiation protocol was performed as described previously.29 Briefly, mESCs were dissociated, and 300 cells in 30 µL culture medium without LIF were aggregated to form embryonic body (EB) by hanging drop for 2 days or 2.5 × 105 cell cultured with 5 mL medium in 60 mm bacterial culture dish in suspension for mass culture. EBs were harvested and transferred to 60 mm bacterial culture dish with 5 mL medium for 3 days. Then, a single EB was plated onto each well of 0.1% gelatin (Sigma)-coated 24-well microwell, and the medium was changed every other day. The beating EBs were counted, shot for videos, and used further analysis.
Immunostaining
For immunostaining, cells were fixed with 4% PFA at room temperature for 20 minutes and permeabilized with 0.1% Triton X-100 for 10 minutes. Then, the cells were blocked with 5% bovine serum albumin for 1 h and incubated with primary antibodies (anti-TNNT2, Abcam#ab8295; anti-SRY-box transcription factor 2 [SOX2], Abclonal#A0561) overnight at 4 °C and Alexa Fluor 488- or 594-conjugated secondary antibody (Abcam#ab150077, ab150116) at room temperature for 1 h. Finally, the slides were mounted in antifade mounting medium with 4ʹ,6-diamidino-2-phenylindole (Vectorshield) and imaged using a fluorescent or confocal microscope.
For D4 EBs, cells were permeabilized with 0.5% Triton X-100 for 15 minutes and incubated with primary antibody (T, Abcam#ab209665) overnight at 4 °C.
Quantitative Reverse Transcription (qRT)-PCR
Total RNA was extracted from cells using RNAiso (Takara). A total of 2 µg RNA was removed of genomic DNA and reverse transcribed into cDNA with FastKing-RT SuperMix (Tiangen). Diluted cDNA was used in qRT–PCR reaction with PowerUp SYBR Master Mix (ABI) on an Applied Biosystems QuantStudio 5. The relative expression level was calculated using the 2−ΔΔCt method, and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) gene served as the internal reference. Each experiment was performed in triplicate and repeated thrice. Supplementary Table S1 lists the primer sequences used in this study.
Western Blot
Proteins were extracted using 1 × SDS lysis buffer (50 mM Tris-HCl [pH 6.8], 100 mM DTT, 2% SDS, and 10% glycerol) and separated by SDS-polyacrylamide gel electrophoresis. Then the proteins were transferred to a nitrocellulose (NC) membrane (Millipore) and blocked with 5% skim milk in TBST at room temperature for 1 h. The membranes were incubated with primary antibody overnight at 4 °C and horseradish peroxidase (HRP)-linked secondary antibody (CST, 7074) at room temperature for 1 h. The signals were detected by reacting with chemiluminescent HRP substrate (Millipore) and visualized using a chemiluminescent detector (FUJIFILM). The primary antibodies included the following: anti-FAM122A (customized by Abclonal), anti–PIP5K1B (Abnova, H00008395-A01), HRP–conjugated anti-α-tubulin (Proteintech, HRP-66031), HRP-conjugated anti-GAPDH (Proteintech, HRP-60004), anti-octamer binding transcription factor 4 (OCT4; Abcam, ab181557), anti-SOX2 (Abcam, ab92494; Abclonal, A0561), anti-Homeobox protein NANOG (NANOG; Abcam, ab214549), anti–H3K4me3 (Abcam, ab8580), anti-H3K27me3 (Millipore, 07-449), anti-H3K27ac (Abcam, ab4729), anti-histone H3 (Abcam, ab1791), anti-p-YAP (CST, 13008), and anti-YAP (CST, 14074).
RNA-Seq Library Generation
Total RNA was isolated from mESCs at the indicated days using Trizol reagent (Thermo Fisher Scientific, 15596018) in accordance with the manufacturer’s protocol. Libraries were generated using NEBNext Ultra RNA Library Prep Kit (NEB, E7490) following the manual and purified using isopycnic Ampure XP beads (Beckman Coulter, A63881). Then, the libraries were sequenced using an Illumina Novaseq 6000 instrument with 150 bp reads and paired-end parameter.
RNA-Seq Data Processing
Sequencing reads were mapped to mm10 reference genome using STAR (v.2.5.2b).30 The transcription levels of annotated genes (FPKM, fragments per kilobase of transcript per million mapped reads) were quantified using HTSeq-count (v.0.6.0).31 Differentially expressed gene (DEG) analysis was performed using DESeq232 and filtered by adjusted P < .05 and fold-change >2. The overlapping DEGs between Fam122a KO-1 and Fam122a KO-3 were used for Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and gene set enrichment analysis (GSEA).
Flow Cytometry Analysis
D4 EBs were disassociated with Accutase (Biolegend) and washed with PBS. Then 1 × 106 cells were incubated with anti–FLK-1-PE (Biolegend, 121905) and anti–PDGFR-α-APC (Biolegend, 135907) at 4 °C for 30 minutes and washed with PBS. Cells were sorted using a Beckman CytoFlex Sand analyzed with FlowJo software (v.7.6).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays of mESCs and EBs were performed using a ChIP-IT Express Kit (Active Motif, 53008) following the manufacturer’s instructions. Briefly, the cells were cross-linked with 1% formaldehyde at room temperature for 10 minutes and quenched with 0.25 m glycine for 5 minutes. The cell pellet was incubated in lysis buffer on ice for 30 minutes and stroked to release nuclei. Chromatin was sheared to 150-500 bp fragments with a sonicator (Qsonica Q700) in Shearing Buffer. Immunoprecipitation was performed with Protein G Magnetic Beads and primary antibodies (anti-H3K4me3, Abcam#ab8580; anti-H3K27me3, Millipore#07-449; anti-H3K27Ac, Abcam#ab4729) by rotation at 4 °C overnight. Beads were washed thrice with ChIP Buffer 1 and twice with ChIP Buffer 2. Chromatin was eluted by rotation at room temperature for 15 minutes and reversed cross-linked at 65 °C for 4 h with Reverse Cross-linking Buffer. Then, the samples were incubated with RNase A at 37 °C for 15 minutes and Proteinase K at 42 °C for 1.5 h. DNA was purified using NucleoSpin Extract II Kit (MN, 740609).
ChIP-Seq Library Generation
Libraries were generated using NEBNext Ultra DNA Library Prep Kit for Illumina (NEB, E7370L) following the manufacturer’s protocol. Libraries were quantified by Qubit2.0, and the qualities were determined using Agilent 2100. Then, the libraries were sequenced using an Illumina Novaseq 6000 instrument with 150 bp reads and paired-end parameter.
ChIP-Seq Data Processing
Reads were removed with poor quality calls and mapped to the mouse mm10 reference genome using Bowtie2 (v.2.4.2).33 Multiple mapped reads and PCR duplicates were further removed using Sambamba (v.0.6.8)34 with the parameter: −F “[XS] == null and not unmapped and not duplicate”. Genome coverage bigwig files were generated by deepTools (v.3.5.0)35 bamCoverage with the parameters “--normalizeUsing RPKM --binSize 20” and visualized using IGV (v.2.9.4).36 Heatmaps were created by deepTools computeMatrix and plot Heatmap.
Echocardiography
Echocardiography was performed using a digital small-animal ultrasound system (VisualSonics, Vevo 2100 or Vevo 3100). Male mice (12-16 weeks old) were constantly anesthetized with 2% isoflurane and placed in the supine position on a heated platform. The mouse chest skin was depilated with hair removal cream and applied with acoustic gel. Two-dimensional long- or short-axis B-Mode and M-Mode views were obtained. Ejection fraction (EF), fractional shortening (FS), cardiac output (CO), left ventricular internal diameter in systole (LVIDs), and interventricular septum (IVS) were calculated.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism 6. All data in this study are presented as means ± SD from 3 independent experiments. The P values were calculated using unpaired 2-tailed Student’s t test.
Data Availability
All RNA-seq and ChIP-seq data in this study have been deposited to the Gene Expression Omnibus with the accession number GSE211341.
Results
Fam122a Deletion Results in Embryonic Lethality and Defects of Cardiovascular Development
To investigate the potential function of FAM122A in vivo, we applied CRISPR/Cas9 technique to generate Fam122a gene KO mice (Supplementary Fig. S1A, Materials and Methods). Heterozygous Fam122a+/− mice with 29 bp deletion were intercrossed to produce Fam122a deletion homozygous mice (Fam122a−/−). Among a cohort of 210 progenies, 69 (32.86%) were wild-type (WT) for Fam122a+/+, and 141 (67.14%) were heterozygous for Fam122a+/−. These data are consistent with the predicted 1:2 Mendelian ratios (Supplementary Fig. S1B) for WT over heterozygous mice. No viable homozygous Fam122a−/− mouse was obtained, which suggests that the deletion of Fam122a gene led to embryonic lethality.
To determine the time of embryonic lethality in Fam122a−/− mice, we extracted genomic DNA from embryos harvested at different stages in pregnant Fam122a+/− mice. PCR-based analysis was used to determine the genotype of these embryos (Supplementary Fig. S1C). Western blot further confirmed that FAM122A protein decreased in Fam122a+/− and was absent in Fam122a–/– embryos, whereas no difference was observed in the PIP5K1B protein level (Supplementary Fig. S1D), which suggests that Fam122a gene was successfully and specifically ablated. Fam122a-null embryos decreased gradually from E9.5 to 11.5 but were absent on E12.5 due to reabsorption (Supplementary Fig. S1E). Fam122a–null embryos on E9.5 were easily distinguished from their WT littermates by severe growth retardation and multiple structural malformations, including smaller size, open anterior neural folds, defects of somatic formation and turning, enlarged pericardial cavity or pericardial effusion, together with the absence of large vessel formation in yolk sac, the latter being signs of cardiovascular defects (Supplementary Fig. S1G). Fam122a-deficient embryos on E8.5 also showed partial developmental delays, such as smaller head folds and reduced extra-embryonic tissues, compared with the WT ones (Supplementary Fig. S1F). Histological analysis further confirmed the morphological findings in E9.5 Fam122a-deficient embryos, particular the profound cardiac abnormalities, including defects of atrial and ventricular chambers with thinner myocardial walls (Supplementary Fig. S1H). Heterozygous FAM122A+/– mice were normal and fertile. These results strongly suggest that the loss of Fam122a leads to embryonic lethality with severe defects of cardiovascular development. Thus, in this study, we mainly focused on whether FAM122A has a role in cardiac differentiation and development.
Fam122a Deletion Impairs CM Differentiation in mESCs
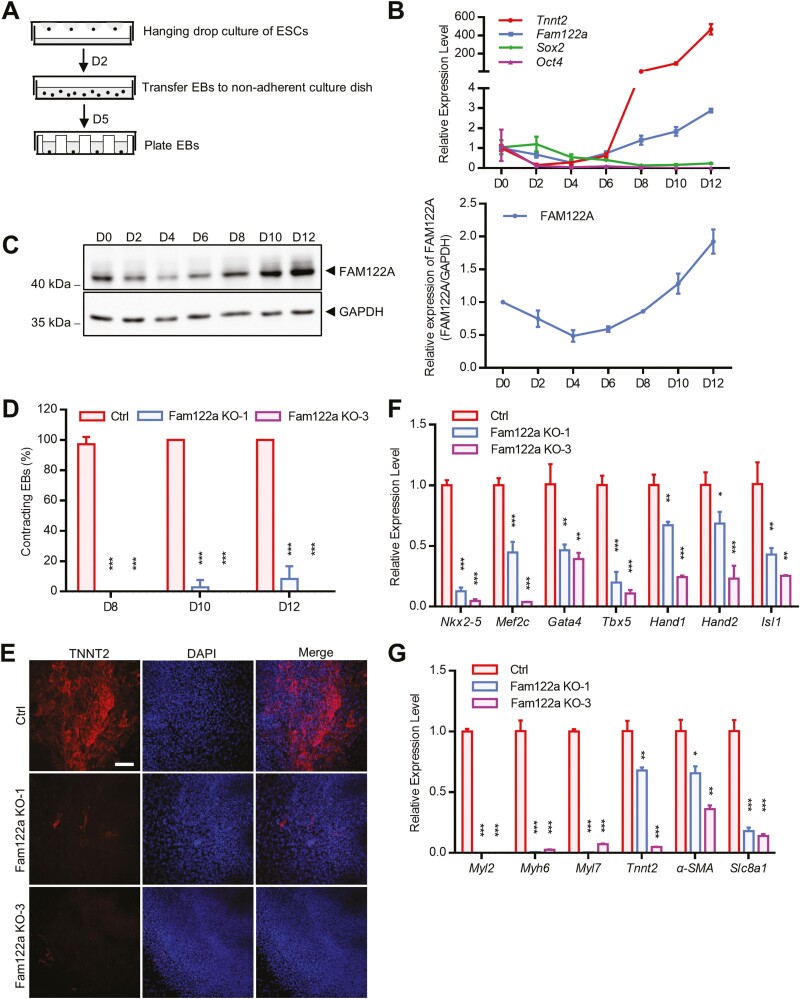
In situ hybridization analysis showed that Fam122a was expressed broadly without tissue specification and upregulated from E7.5 to E9.5 (Supplementary Fig. S2A). To evaluate the possible role of FAM122A in cardiac differentiation, we induced mESC differentiation by hanging drop EBs in LIF-free medium, which allowed the stepwise differentiation of mESCs toward the cardiogenic lineage, generating ME precursor cells (MEPCs), CPC, and ultimately differentiated CMs (Fig. 1A).6 We found that Fam122a gene and protein levels declined mildly at the initial 4 days and rose rapidly after 6 days of differentiation with the upregulation of Tnnt2 (a tropomyosin-binding subunit of cardiac troponin complex, an indicator of CMs37; Fig. 1B, 1C), which suggests that FAM122A may participate in the differentiation process. CRISPR/Cas9-mediated Fam122a KO mESCs were generated by 2 sgRNAs, designated as Fam122a KO-1 and Fam122a KO-3. Deletion of FAM122A in mESCs R1/E, which was confirmed by Western blot (Supplementary Fig. S2B), significantly reduced the percentages of beating EBs and the expressions of CPC (Nkx2-5, Gata4, Tbx5, and Isl138) and CM (Myl2, Myh6, Tnnt2, and α-SMA28) marker genes and TNNT2 protein on day 7 (D7) and D10 of EB differentiation (Fig. 1D–1G; Supplementary Fig. S2G; videos S1–S3), although FAM122A KO did not evidently change the morphology of ESC colonies (Supplementary Fig. S2C). Similar differentiation defects were found in another Fam122a deletion mESC TC1 (Supplementary Fig. S3D–S3F). Fam122a deletion did not affect the expression levels of stem cell-correlated genes (Oct4, Sox2, and Nanog; Supplementary Figs. S2D, S2E, S3A–S3C) and the activity of AP39 (Supplementary Fig. S2F). Thus, FAM122A may be essential for CM differentiation, but it shows no influence on stem cell identity.
Figure 1.
Cardiac differentiation defect in Fam122a knockout mESCs. (A) Schematic of mESCs differentiation assay. The mESCs are trypsinized, aggregated to EBs by hanging drop and subsequently plated to differentiation. (B) qRT–PCR analysis of Tnnt2, Fam122a, Sox2, and Oct4 expression. (C) Western blot analysis of FAM122A expression during mESCs differentiation (left), and quantification of 3 experiments (right). (D) Percentages of contracting EBs determined from D8 to D12 of mESCs differentiation. (E) TNNT2 immunostaining on D10 of mESCs differentiation. Scale bar = 100 μm. (F) qRT-PCR analysis of cardiac progenitor marker genes on D7 of mESCs differentiation. (G) Expression analysis of cardiomyocyte marker genes on D10 of mESCs differentiation. In (D), (F) and (G), data represent means ± SD from n = 3 independent experiments, and P values were calculated by 2-tailed unpaired t test (* P < .05, ** P < .01, and *** P < .001).
FAM122A Regulates a Core Network of Genes that Drive Cardiac Differentiation
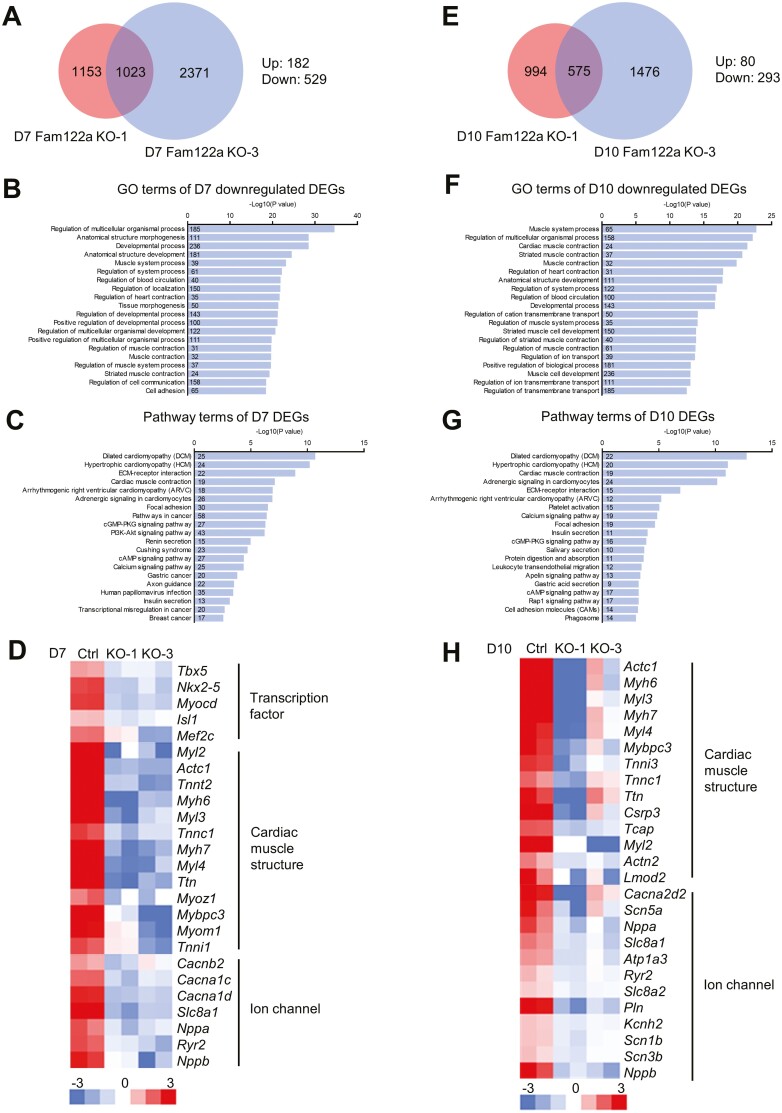
To further explore the mechanism of FAM122A on CM differentiation, we performed RNA sequencing for global gene expressions in WT and 2 Fam122a KO mESCs during different differentiation stages. The overlapping DEGs (2-fold cutoff) in Fam122a KO-1 and Fam122a KO-3 vs. WT totaled 1023 on D7 of differentiation, and most of them were downregulated (Fig. 2A). GO and pathway analysis showed that Fam122a deletion-modulated genes were mainly enriched in multicellular organismal and developmental process and different cardiomyopathies and heart functions, including dilated cardiomyopathy, hypertrophic cardiomyopathy, cardiac muscle contraction, arrhythmogenic right ventricular cardiomyopathy, and adrenergic signaling in CMs (Fig. 2B, 2C). The downregulation of multiple CPC-related genes, including several TFs, and cardiac muscle- and iron channel-related genes were observed in Fam122a-deleted mESCs on D7 of differentiation (Fig. 2D). Consistently, the RNA-seq result on D10 differentiation also showed that Fam122a deletion suppressed the terminal CM differentiation genes (Fig. 2E–2H).
Figure 2.
RNA-seq analysis of D7 and D10 Fam122a knockout differentiated mESCs. (A) Venn diagram showing the overlapped DEGs in D7 KO mESCs. (B) Gene ontology analysis of the D7 downregulated DEGs. (C) KEGG pathway analysis of the D7 DEGs. (D) Heatmap showing the representative D7 downregulated cardiac genes. (E) Venn diagram showing the overlapped DEGs in D10 KO mESCs. (F) Gene ontology analysis of the D10 downregulated DEGs. (G) KEGG pathway analysis of the D10 DEGs. (H) Heatmap showing the representative D10 downregulated cardiac genes. Numbers in bars show the gene numbers of the corresponding gene ontology or pathway categories (B, C, F, and G).
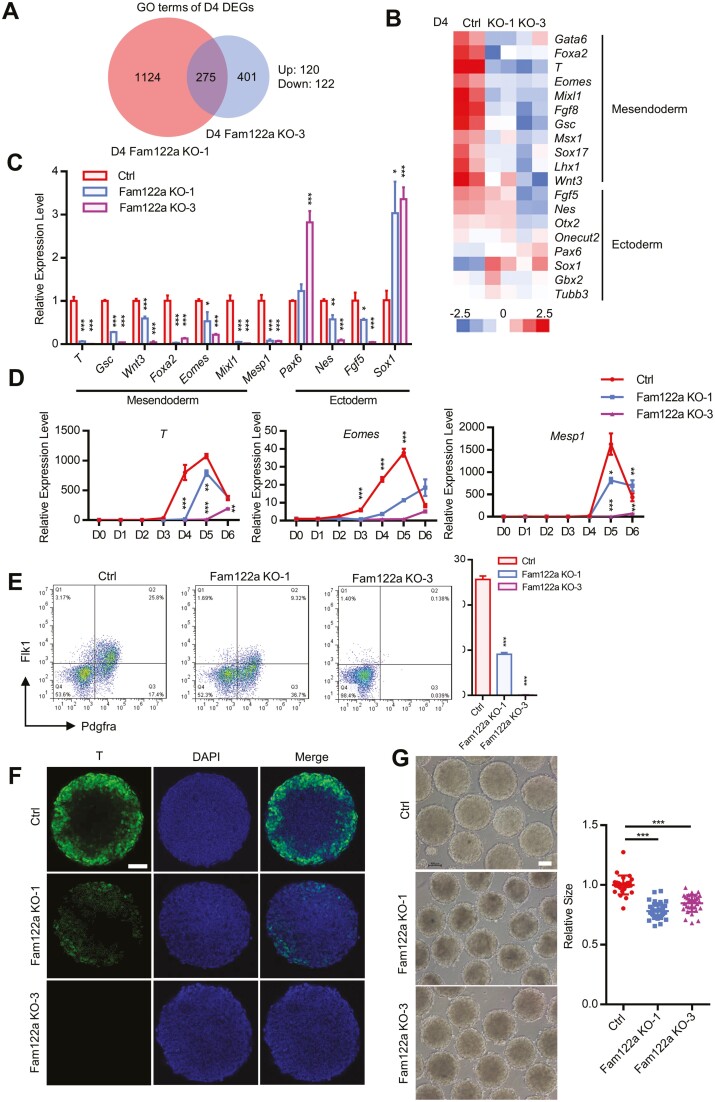
In vitro cardiac differentiation, EBs in suspension formed by pluripotent ESCs may recapitulate the signaling and transcriptional events for the specification of 3-germ layers, which is mirrored in the gastrulation process in vivo.12 Eomes, T, Mesp1, and Mixl1 are critical TFs, and depletion of these factors individually can significantly impair ME specification and cardiac differentiation.10,11,40 Intriguingly, RNA-seq result on D4 EBs showed the reduced expression of 122 genes in Fam122a KO vs. WT, whereas 120 genes were upregulated (Fig. 3A). GO analysis further revealed that Fam122a deletion led to the significant downregulation of ME genes, including T, Eomes, Mixl1, Wnt3, Gsc, and Foxa2 (Fig. 3B; Supplementary Fig. S3G), which was further confirmed by qPCR (Fig. 3C) and time course analysis (Fig. 3D). Fam122a deletion also reduced the immunofluorescence staining for T protein (Fig. 3F) and the percentages of Flk1+/Pdgfra+ cardiac MEPCs (Fig. 3E).41 These results strongly suggest that ME differentiation was blocked in Fam122a KO mESCs. In addition, Fam122a-deleted mESCs demonstrated smaller sizes of EBs on D4 of differentiation (Fig. 3G). Fam122a deletion also reduced the expression of partial ectoderm-related genes Nes and Fgf5 (Fig. 3C). Collectively, FAM122A is critical for ME specification of mESCs.
Figure 3.
Mesendodermal differentiation defect in D4 Fam122a knockout EBs. (A) Venn diagram showing the overlapped RNA-seq DEGs in D4 KO EBs. (B) Heatmap showing the representative D4 DEGs of 3-germ layer markers. (C) qRT-PCR analysis of 3-germ layer marker genes on D4 of EB differentiation. (D) Expression analysis of T, Eomes, and Mesp1 determined from D0 to D6 of EB differentiation. (E) Fluorescence-activated cell sorting (FACS) analysis of the percentage of Flk1+/Pdgfra+ cells differentiated from D4 EBs (left) and quantification (right). (F) T immunostaining of control and Fam122a KO EBs on D4. Scale bar = 50 μm. (G) Images of D4 EBs in control and Fam122a KO mESCs (left), and quantification of EBs size (right). Scale bar = 100 μm. In (C), (D), (E), and (G), data represent means ± SD from n = 3 independent experiments, and P values were calculated by 2-tailed unpaired t test (* P < .05, ** P < .01, and *** P < .001).
Fam122a Deletion Reduces the Binding Activities of H3K4me3 and H3K27ac on Mesendodermal Genes
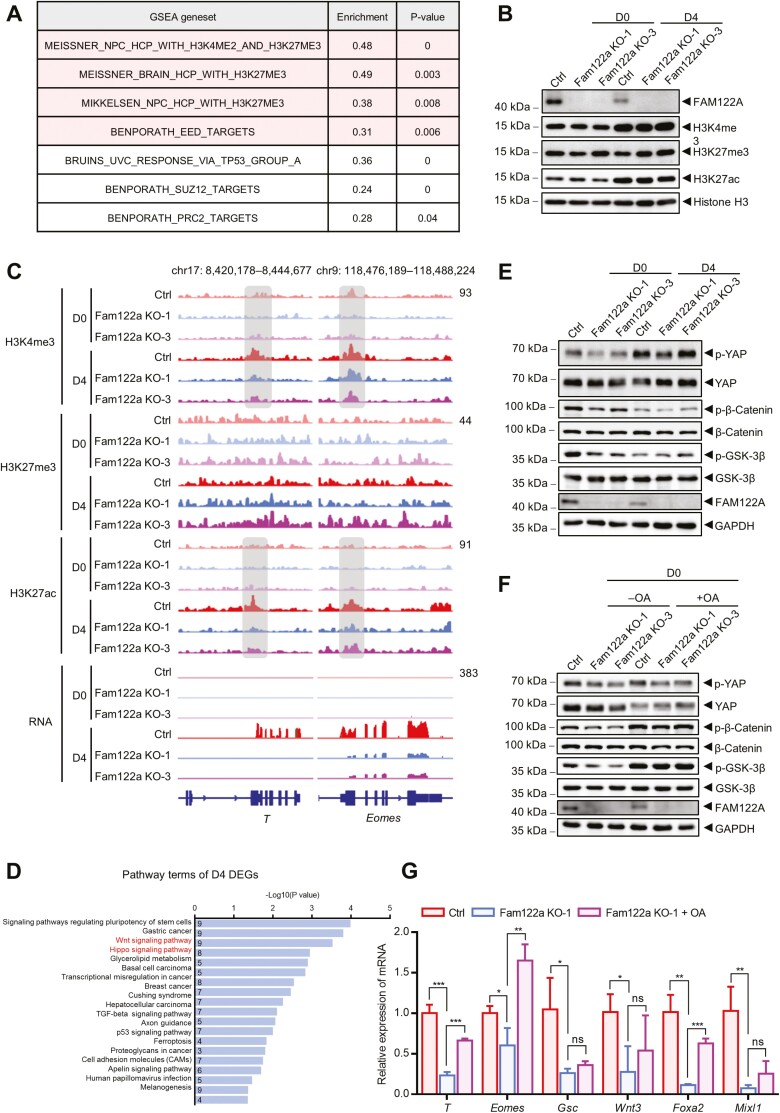
Substantial ME genes were suppressed in Fam122a depleted mESC-formed EBs on D4, which suggests that FAM122A may directly or indirectly regulate gene expression as a TF, by modulating transcriptional activity as a cofactor or by changing the chromatin structure as a modifier. GSEA of Fam122a KO and WT mESCs showed that DEGs were significantly enriched in H3K4me2 and H3K27me3 datasets (Fig. 4A), which indicates that FAM122A may modulate histone post-translational modifications. Therefore, we performed ChIP-seqs of H3K4me3 and H3K27ac (transcriptional activation markers) and H3K27me3 (a transcriptional repression marker).5,42 We also compared their protein levels in Fam122a KO and WT ESCs on D0 and D4 EBs. Heatmaps showed that the enrichments of H3K4me3 and H3K27ac were significantly enhanced in D4 EBs compared with D0 (Supplementary Fig. S4A–S4C), which paralleled the shift of transcriptional repression to activation following differentiation induction. Fam122a deletion decreased H3K4me3 binding on D0 and increased H3K27me3 binding on D4 EBs, whereas H3K27ac showed no remarkable change. The changes in binding capacity were accompanied by the increase in H3K27me3 protein in D4 EBs compared with D0, but no significant alteration was observed in H3K4me3 and H3K27ac proteins (Fig. 4B). More importantly, Fam122a depletion significantly decreased the occupancy of H3K4me3 and H3K27ac in the promoters of T and Eomes genes in D4 EBs but did not change the binding of H3K27me3 in these genes on D0 and D4 ESCs (Fig. 4C). The reduced H3K4me3 and H3K27ac binding on ME genes were consistent with our observation that Fam122a depletion disrupted ME gene expression and specification. In addition, decreased expression of ectoderm gene Fgf5 upon Fam122a deletion was accompanied by reduced H3K27ac binding on its promoter in D4 EBs, whereas enhanced expression of Sox1 was paralleled by the increased binding activity within H3K4me3 and H3K27ac loci (Supplementary Fig. S4D). These results implied that Fam122a deletion induces the suppression of ME genes through histone modification.
Figure 4.
Wnt and Hippo signalings as well as histone modification dysregulated in Fam122a knockout mESCs. (A) GSEA of D0 Fam122a KO-1 DEGs. Gene sets changed in both KO mESCs are colored in red background (for color figure refer to online version). (B) Western blot analysis of histone modification proteins in D0 and D4 control and Fam122a knockout mESCs. (C) Genome browser views of H3K4me3, H3K27me3, and H3K27ac ChIP-seq signals as well as RNA expression in D0 and D4 indicated mESCs at mesendodermal genes T and Eomes. (D) KEGG pathway analysis of D4 DEGs. Numbers in bars show the gene numbers of the corresponding pathway categories. (E) Western blot analysis of p-YAP, p-β-catenin, and p-GSK-3β in D0 and D4 mESCs. (F) Western blot analysis of p-YAP, p-β-catenin, and p-GSK-3β in D0 indicated mESCs treated with or without 50 nm OA for 6 h. (G) qRT-PCR analysis of mesendodermal genes in D4 control and Fam122a KO EBs treated with or without 20 nm OA for the first 2 days. Data represent means ± SD from n = 3 independent experiments, and P values were calculated by 2-tailed unpaired t test (* P < .05, ** P < .01, and *** P < .001).
To test whether FAM122A modulates the transcriptional activity of ME genes by interacting with any essential enzymes for histone modification, we performed immunoprecipitation and mass spectrometry (IP-MS) to screen interacting protein profiles in Flag-FAM122A-overexpressing mESCs and 293T cells. However, we found no interaction of FAM122A with histone modifiers, such as methyltransferase, acetyltransferase, demethylase, or deacetylase (data not shown), which suggests that FAM122A regulating ME genes and histone modification is not directly modulated by histone modifiers.
Fam122a Deletion Aberrantly Regulates Wnt and Hippo Signaling Pathways
To further determine the underlined causes of Fam122a deletion impairing the ME genes and differentiation, we closely analyzed the DEGs in D4 differentiated EBs with or without Fam122a depletion. KEGG analysis showed that these DEGs were mainly enriched in the regulation of pluripotent stem cell fate and Wnt and Hippo signaling pathways (Fig. 4D), with both signaling pathways having an effect on histone modification.43-46 Wnt signaling activation is critical for ME gene expression and specification,16,47 whereas the effector of Hippo signaling YAP directly antagonizes ME gene activation as a co-repressor.17,48 Considering that FAM122A has been identified originally as an endogenous inhibitor of PP2A, we examined the phosphorylation levels of PP2A substrates in Wnt (β-catenin and GSK-3β) and Hippo (YAP) signaling.49-54 The results showed that the phosphorylations of YAP, β-catenin, and GSK-3β were significantly reduced in D0 Fam122a KO mESCs compared with those of WT cells (Fig. 4E). However, these effects were not significant in D4 EBs. The addition of okadaic acid (OA, a PP2A inhibitor) not only eliminated the reduced phosphorylation of YAP, β-catenin, and GSK-3β by Fam122a deletion but also partially rescued Fam122a deletion-induced decrease in the expression of ME genes (Fig. 4F, 4G).
Fam122a Depletion Impairs Cardiac Development and Function in Conditional Cardiac KO Mice
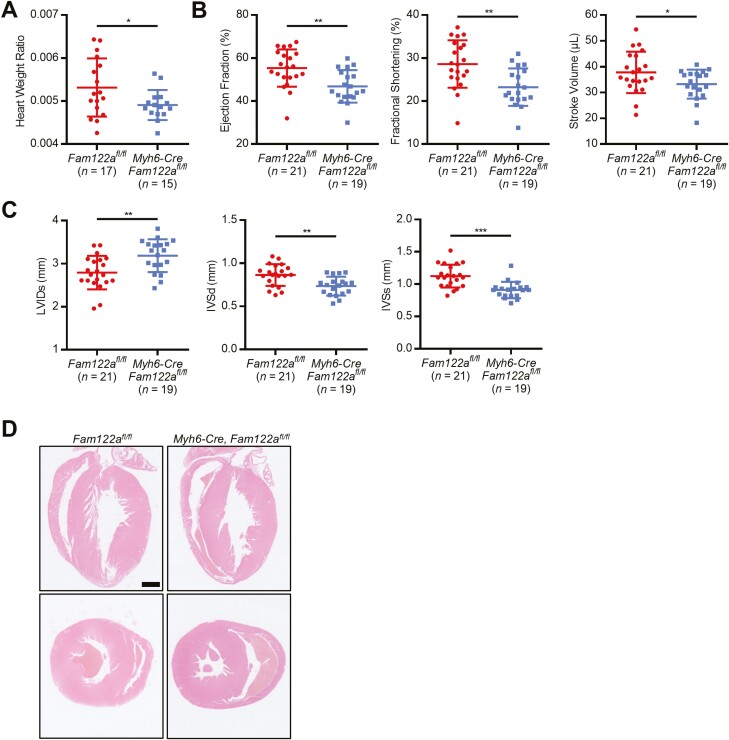
To strengthen the critical role of FAM122A in cardiac development and function, we crossed Fam122afl/fl mice (Supplementary Fig. S5A, S5B) with Myh6-Cre transgenic mice55 to generate mice where Fam122a is genetically and specifically inactivated in CMs of the heart tube on E8.5. Western blot confirmed the KO effect of FAM122A (Supplementary Fig. S5C), and echocardiography analysis showed that Fam122a deletion significantly impaired cardiac functions, as determined by the decreases in heart weight ratio, percentages of EF and FS, and stroke volume (SV) in Myh6-Cre Fam122afl/fl mice compared with Fam122afl/fl ones (Fig. 5A, 5B). Moreover, we observed that LVIDs increased, whereas IVS thickness in diastole (IVSd) and in systole (IVSs) reduced in Myh6-Cre Fam122afl/fl mice (Fig. 5C, 5D). Similar effects of Fam122a deletion on cardiac dysfunction were found in other conditional cardiac-specific Fam122a KO mice (Nkx2-5-Cre Fam122afl/fl) by using Nkx2-5-Cre mice, with Nkx2-5-Cre-mediated recombination occurring on E7.5 cardiac progenitor cells during cardiac crescent,56 which can elucidate the function of FAM122A during the earliest stages of cardiogenesis (Supplementary Fig. S5D–S5H). Notably, Fam122a deletion can significantly decrease the body and heart weights of Nkx2-5-Cre Fam122afl/fl mice but not Myh6-Cre Fam122afl/fl mice. Thus, Fam122a deletion at earlier cardiac development may result in more severe effects on heart development and function.
Figure 5.
Cardiac function attenuated in Myh6-Cre Fam122a CKO mice. (A) Heart weight ratio analysis in Fam122aflox/flox (n = 17) and Fam122a CKO (n = 15) mice. (B) Echocardiography analyses for the EF, FS, and SV in Fam122aflox/flox (n = 21) and Fam122a CKO (n = 19) mice. (C) Echocardiography analyses for the LVID and IVS in diastole or systole in Fam122aflox/flox (n = 21) and Fam122a CKO (n = 19) mice. (D) H&E-stained sagittal or transverse sections of heart in Fam122aflox/flox and Fam122a CKO mice. Scale bar = 1 mm. In (A), (B), and (C), data represent means ± SD and P values were calculated by 2-tailed unpaired t test (* P < .05, ** P < .01, and *** P < .001).
Discussion
ME is an important event in early embryonic development. In-depth understanding of the regulatory networks of ME specification from ESCs in vitro or pluripotent epiblast in vivo may provide important insights into early embryonic patterning and a possible guidance for ES applications in regenerative medicine, especially in heart regeneration field.57,58 Therefore, studies should find new target molecules in the regulation or manipulation of ES cells differentiating into ME, followed stepwise by differentiation into CMs. In our study, we have shown that FAM122A is critical for ME specification and CM differentiation from mESC in vitro culture system, as demonstrated by Fam122a deletion not only significantly impairing the expressions of MEPC-, CPC-, and CM-related genes but also reducing the percentages of spontaneously contracting EBs and Flk1+/Pdgfra+ cardiac progenitors and TNNT2 protein. However, Fam122a deletion does not influence the stem cell identity.
The role of FAM122A in vivo study showed that Fam122a deletion led to early embryonic lethality with significantly severe defects in the cardiovascular system, including the deficiency in the normal structure of 4-chamber embryonic heart on E9.5 shown by HE staining and pericardial bulge revealed by anatomic microscopy. This finding supports the essential role of FAM122A in early embryonic and heart development. Two-conditional cardiac-specific Fam122a KO mice also displayed abnormal heart development and functions, as demonstrated by the decrease in cardiac functional indexes (EF, FS, and SV) and heart/weight ratio and altered structural parameters (LVIDs, IVSd, and IVSs). However, these conditional Fam122a KO mice did not reproduce severe defects of heart development and embryonic lethality, which occurred in total Fam122a KO mice. Noticeably, the specific depletion of Fam122a in cardiac progenitors by Nkx2-5-Cre on E7.5 did not reveal striking cardiac defects, which suggests that FAM122A may affect considerably earlier developmental stages, such as ME, the dysfunction of which may further disrupt cardiac development and cardiomyocytic differentiation. Therefore, ME-specific Cre mice, such as Mesp1-Cre, or other ME drivers (Brachyury or Mixl1) may be used to further investigate the in vivo effect of Fam122a deletion on ME specification and confirm whether the in vivo effect is similar to the in vitro effect in mESCs (loss of ME gene expression upon Fam122a depletion). Considering that nascent mesoderm toward cardiac fates also relies on the signaling from proper endoderm development,59 the effect of endodermal Cre (Foxa2 or Gsc)–driven deletion of Fam122a must be examined to exclude the possibility of cardiac defects upon Fam122a deletion resulting from impaired signaling from the overlaying foregut endoderm. These animal model should be analyzed in the future to confirm whether ME specification is disrupted upon Fam122a KO in vivo.
Currently, the understanding of the regulation networks of ME specification is limited, and its dynamic alteration is largely unclear. ME formation is highly organized and spatiotemporally controlled via the integrated interactions of TFs, epigenetic regulators, chromatin remodeling factors, and signaling pathways.5,7 Substantial ME genes are suppressed upon Fam122a deletion, which includes the essential T-box TFs (Eomes and T) for ME differentiation. The GSEA dataset from DEGs in D4 EBs suggests that Fam122a deletion may affect histone modification of H3K4me2 and H3K27me3, which are closely related to ME specification.60,61 ChIP-seq data regarding several histone modifications showed that Fam122a deletion decreased H3K4me3 binding on D0 and increased H3K27me3 binding in D4 EBs in a genome-wide manner, whereas no significant alteration was observed in H3K27ac. The altered histone modification pattern was accompanied by the upregulation of H3K27me3 protein in D4 EBs, which implies that FAM122A may regulate histone modification markers with different patterns before and after ME. More importantly, Fam122a depletion significantly decreased the association of H3K4me3 and H3K27ac with the promoters of T and Eomes genes in D4 EBs, which is consistent with the inhibitory effect of ME differentiation upon Fam122a depletion. However, IP-MS results depicted that no histone post-translational modifier interacted with FAM122A, which suggests that FAM122A may indirectly modulate histone modification by other unidentified mechanism or signaling pathway. In general, a default path of ectoderm differentiation exists when ME differentiation is absent10; however, our results suggest that Fam122a deletion may upregulate or downregulate ectoderm genes, and therefore, FAM122A participates in the regulation of ectoderm genes with a complexed mechanism.
FAM122A has no classical DNA binding motif and did not directly bind to the promoters of Eomes or T examined by ChIP-seq with Flag-FAM122A mESCs (data not shown), which suggests that FAM122A may modulate the expression of ME genes as a co-regulator or by influencing the chromatin structure. To address this question, scientists may apply ATAC-seq62 to elucidate the effect of FAM122A modulation on chromatin architecture directly. Previously, we observed that FAM122A acts as a co-repressor of GATA-1 (an essential TF for erythroid differentiation) to suppress the terminal differentiation of erythroids25; therefore, FAM122A possibly interacts with essential ME-related TFs to participate in the regulation of ME gene activation, which needs to be checked dynamically in different stages of differentiation with IP-MS.
Mounting pieces of evidence show that TGF-β and Wnt signaling are critical for ME specification, and Wnt/Hippo signals are involved in the regulation of histone modification. Our results showed that DEGs between Fam122a KO and WT in D4 EBs are enriched in Wnt and Hippo signaling. YAP, an essential effector in Hippo signaling, has been recently reported to act as a negative regulator of ME differentiation. We observed that Fam122a deletion significantly decreased the phosphorylation levels of GSK-3β, β-catenin, and YAP, which suggests that Fam122a silencing dysregulates Wnt and YAP signaling pathways, thus affecting histone modification and impairing ME specification. OA can eliminate Fam122a KO–reduced phosphorylation of GSK-3β, β-Catenin, and YAP proteins, but it partially reverses the decreased effect of ME gene expression by Fam122a deletion, which strongly suggests that FAM122A regulates ME differentiation as a PP2A inhibitor. Zheng et al have identified a new dual-enzyme complex called INTAC for transcription regulation, and it is composed of PP2A core enzyme and multi-subunit RNA endonuclease integrator.63 In their model, PP2A dephosphorylates the C-terminal domain of RNA polymerase II to attenuate transcription. Therefore, whether FAM122A modulates ME gene expression through PP2A and/or INTAC warrants investigation.
Conclusion
Collectively, the results reveal that FAM122A, as possible PP2A inhibitor, is a critical regulator for ME specification and CM differentiation from mESCs through the dysregulation of histone modification and Wnt and Hippo signaling pathways. Moreover, Fam122a deletion leads to embryonic lethality with severe cardiovascular developmental defects and impairs cardiac function. The potential effects of FAM122A on stem cell differentiation into CMs not only provide new insights into its role in the development of 3-germ layers but can also contribute to clinical/therapeutic advancement in cardiac regenerative medicine in the future.
Supplementary Material
Acknowledgments
We thank Shanghai Frontiers Science Center of Cellular Homeostasis and Human Diseases for providing the experimental platforms and instruments.
Contributor Information
Yun-Sheng Yang, Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education and Chinese Academy of Medical Sciences Research Unit (2019RU043, Stress and Tumor), Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai, People’s Republic of China.
Man-Hua Liu, Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education and Chinese Academy of Medical Sciences Research Unit (2019RU043, Stress and Tumor), Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai, People’s Republic of China.
Zhao-Wen Yan, Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education and Chinese Academy of Medical Sciences Research Unit (2019RU043, Stress and Tumor), Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai, People’s Republic of China.
Guo-Qiang Chen, Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education and Chinese Academy of Medical Sciences Research Unit (2019RU043, Stress and Tumor), Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai, People’s Republic of China.
Ying Huang, Department of Pathophysiology, Key Laboratory of Cell Differentiation and Apoptosis of Chinese Ministry of Education and Chinese Academy of Medical Sciences Research Unit (2019RU043, Stress and Tumor), Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai, People’s Republic of China.
Funding
This study was supported by the grant from Chinese National Natural Science Foundation (32070720, 82270244, H.Y.), Shanghai Jiao-Tong University, Research Units of Stress and Tumor (2019RU043, C.G.Q.), and Chinese Academy of Medical Sciences.
Conflict of Interest
The authors declare no potential conflicts of interests.
Author Contributions
Y.Y.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; M.L.: provision of study material; Z.Y.: collection of data; G.C.: final revision and approval of manuscript; Y.H.: conception and design, administrative support, manuscript writing, final approval of manuscript.
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or Supplementary Materials. Additional data related to this paper may be requested from the authors.
References
- 1. Santini MP, Forte E, Harvey RP, Kovacic JC. Developmental origin and lineage plasticity of endogenous cardiac stem cells. Development. 2016;143(8):1242-1258. https://doi.org/ 10.1242/dev.111591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Savolainen SM, Foley JF, Elmore SA. Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol Pathol. 2009;37(4):395-414. https://doi.org/ 10.1177/0192623309335060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sivakumar A, Kurpios NA. Transcriptional regulation of cell shape during organ morphogenesis. J Cell Biol. 2018;217(9):2987-3005. https://doi.org/ 10.1083/jcb.201612115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tada S, Era T, Furusawa C, et al. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132(19):4363-4374. https://doi.org/ 10.1242/dev.02005 [DOI] [PubMed] [Google Scholar]
- 5. Wang L, Chen YG. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J Mol Biol. 2016;428(7):1409-1422. https://doi.org/ 10.1016/j.jmb.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 6. Alexanian M, Maric D, Jenkinson SP, et al. A transcribed enhancer dictates mesendoderm specification in pluripotency. Nat Commun. 2017;8(1):1806. https://doi.org/ 10.1038/s41467-017-01804-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jansen C, Paraiso KD, Zhou JJ, et al. Uncovering the mesendoderm gene regulatory network through multi-omic data integration. Cell Rep. 2022;38(7):110364. https://doi.org/ 10.1016/j.celrep.2022.110364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beddington RS, Rashbass P, Wilson V. Brachyury—a gene affecting mouse gastrulation and early organogenesis. Dev Suppl. 1992;116(Supplement):157-165. 10.1242/dev.116.Supplement.157 [DOI] [PubMed] [Google Scholar]
- 9. Wilson V, Manson L, Skarnes WC, Beddington RS. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development. 1995;121(3):877-886. https://doi.org/ 10.1242/dev.121.3.877 [DOI] [PubMed] [Google Scholar]
- 10. Tosic J, Kim GJ, Pavlovic M, et al. Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat Cell Biol. 2019;21(12):1518-1531. https://doi.org/ 10.1038/s41556-019-0423-1 [DOI] [PubMed] [Google Scholar]
- 11. Costello I, Pimeisl IM, Dräger S, et al. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol. 2011;13(9):1084-1091. https://doi.org/ 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Q, Zou Y, Nowotschin S, et al. The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell. 2017;20(1):70-86. https://doi.org/ 10.1016/j.stem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao M, Tang Y, Zhou Y, Zhang J. Deciphering role of Wnt signalling in cardiac mesoderm and cardiomyocyte differentiation from human iPSCs: four-dimensional control of Wnt pathway for hiPSC-CMs differentiation. Sci Rep. 2019;9(1):19389. https://doi.org/ 10.1038/s41598-019-55620-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brennan J, Lu CC, Norris DP, et al. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411(6840):965-969. https://doi.org/ 10.1038/35082103 [DOI] [PubMed] [Google Scholar]
- 15. Pauklin S, Vallier L. Activin/Nodal signalling in stem cells. Development. 2015;142(4):607-619. https://doi.org/ 10.1242/dev.091769 [DOI] [PubMed] [Google Scholar]
- 16. Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2(1):60-71. https://doi.org/ 10.1016/j.stem.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Estaras C, Hsu HT, Huang L, Jones KA. YAP repression of the WNT3 gene controls hESC differentiation along the cardiac mesoderm lineage. Genes Dev. 2017;31(22):2250-2263. https://doi.org/ 10.1101/gad.307512.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Q, Li L, Zhao B, Guan KL. The hippo pathway in heart development, regeneration, and diseases. Circ Res. 2015;116(8):1431-1447. https://doi.org/ 10.1161/CIRCRESAHA.116.303311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan L, Liu MH, Guo M, et al. FAM122A, a new endogenous inhibitor of protein phosphatase 2A. Oncotarget. 2016;7(39):63887-63900. https://doi.org/ 10.18632/oncotarget.11698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan F, Zhao J, Liu Y, et al. Identifying the SUMO1 modification of FAM122A leading to the degradation of PP2A-Calpha by ubiquitin-proteasome system. Biochem Biophys Res Commun. 2018;500(3):676-681. https://doi.org/ 10.1016/j.bbrc.2018.04.135 [DOI] [PubMed] [Google Scholar]
- 21. Li F, Kozono D, Deraska P, et al. CHK1 inhibitor blocks phosphorylation of FAM122A and promotes replication stress. Mol Cell. 2020;80(3):410-422.e6. https://doi.org/ 10.1016/j.molcel.2020.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Y, Shi WY, He W, et al. FAM122A supports the growth of hepatocellular carcinoma cells and its deletion enhances Doxorubicin-induced cytotoxicity. Exp Cell Res. 2020;387(1):111714. https://doi.org/ 10.1016/j.yexcr.2019.111714 [DOI] [PubMed] [Google Scholar]
- 23. Liu MH, Chen J, Yang YS, et al. FAM122A promotes acute myeloid leukemia cell growth through inhibiting PP2A activity and sustaining MYC expression. Haematologica. 2021;106(3):903-907. https://doi.org/ 10.3324/haematol.2020.251462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu MH, Zhang XC, Chen J, et al. FAM122A is required for hematopoietic stem cell function. Leukemia. 2021;35(7):2130-2134. https://doi.org/ 10.1038/s41375-020-01099-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J, Zhou Q, Liu MH, et al. FAM122A inhibits erythroid differentiation through GATA1. Stem Cell Rep. 2020;15(3):721-734. https://doi.org/ 10.1016/j.stemcr.2020.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang YQ, Yang YS, Chen J, et al. FAM122A maintains DNA stability possibly through the regulation of topoisomerase IIalpha expression. Exp Cell Res. 2020;396(1):112242. https://doi.org/ 10.1016/j.yexcr.2020.112242 [DOI] [PubMed] [Google Scholar]
- 27. Wei Q, Manley NR, Condie BG. Whole mount in situ hybridization of E8.5 to E11.5 mouse embryos. J Vis Exp. 2011;(56):e2797. 10.3791/2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo X, Xu Y, Wang Z, et al. A Linc1405/Eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell. 2018;22(6):893-908.e6. https://doi.org/ 10.1016/j.stem.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 29. Wobus AM, Guan K, Yang HT, Boheler KR. Embryonic stem cells as a model to study cardiac, skeletal muscle, and vascular smooth muscle cell differentiation. Methods Mol Biol. 2002;185:127-156. https://doi.org/ 10.1385/1-59259-241-4:127 [DOI] [PubMed] [Google Scholar]
- 30. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. https://doi.org/ 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166-169. https://doi.org/ 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357-359. https://doi.org/ 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31(12):2032-2034. 10.1093/bioinformatics/btv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramirez F, Ryan DP, Grüning B, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1);W160-165. https://doi.org/ 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178-192. https://doi.org/ 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farah CS, Reinach FC. The troponin complex and regulation of muscle contraction. FASEB J. 1995;9(9):755-767. https://doi.org/ 10.1096/fasebj.9.9.7601340 [DOI] [PubMed] [Google Scholar]
- 38. Barreto S, Hamel L, Schiatti T, Yang Y, George V. Cardiac progenitor cells from stem cells: learning from genetics and biomaterials. Cells. 2019;8(12):1536. 10.3390/cells8121536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stefkova K, Prochazkova J, Pachernik J. Alkaline phosphatase in stem cells. Stem Cells Int. 2015;2015:628368. 10.1155/2015/628368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hart AH, Hartley L, Sourris K, et al. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development. 2002;129(15):3597-3608. https://doi.org/ 10.1242/dev.129.15.3597 [DOI] [PubMed] [Google Scholar]
- 41. Hong SP, Song S, Cho SW, et al. Generation of PDGFRalpha(+) cardioblasts from pluripotent stem cells. Sci Rep. 2017;7(1):41840. https://doi.org/ 10.1038/srep41840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen CY, Cheng YY, Yen CY, Hsieh PC. Mechanisms of pluripotency maintenance in mouse embryonic stem cells. Cell Mol Life Sci. 2017;74(10):1805-1817. https://doi.org/ 10.1007/s00018-016-2438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma A, Mir R, Galande S. Epigenetic regulation of the Wnt/beta-catenin signaling pathway in cancer. Front Genet. 2021;12:681053. 10.3389/fgene.2021.681053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu X, Li Z, Song Y, et al. AURKA induces EMT by regulating histone modification through Wnt/beta-catenin and PI3K/Akt signaling pathway in gastric cancer. Oncotarget. 2016;7(22):33152-331643. https://doi.org/ 10.18632/oncotarget.8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oh H, Slattery M, Ma L, et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 2014;8(2):449-459. https://doi.org/ 10.1016/j.celrep.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qing Y, Yin F, Wang W, et al. The Hippo effector Yorkie activates transcription by interacting with a histone methyltransferase complex through Ncoa6. Elife. 2014;3:e02564. 10.7554/eLife.02564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arkell RM, Fossat N, Tam PP. Wnt signalling in mouse gastrulation and anterior development: new players in the pathway and signal output. Curr Opin Genet Dev. 2013;23(4):454-460. https://doi.org/ 10.1016/j.gde.2013.03.001 [DOI] [PubMed] [Google Scholar]
- 48. Mia MM, Singh MK. The hippo signaling pathway in cardiac development and diseases. Front Cell Dev Biol. 2019;7:211. https://doi.org/ 10.3389/fcell.2019.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jiang X, Hu J, Wu Z, et al. Protein phosphatase 2A mediates YAP activation in endothelial cells upon VEGF stimulation and matrix stiffness. Front Cell Dev Biol. 2021;9:675562. 10.3389/fcell.2021.675562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brewer CM, Nelson BR, Wakenight P, et al. Adaptations in Hippo-Yap signaling and myofibroblast fate underlie scar-free ear appendage wound healing in spiny mice. Dev Cell. 2021;56(19):2722-2740.e2726. https://doi.org/ 10.1016/j.devcel.2021.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiao Z, Wen L, Zeng D, et al. Protein phosphatase 2A inhibiting beta-catenin phosphorylation contributes critically to the anti-renal interstitial fibrotic effect of norcantharidin. Inflammation. 2020;43(3):878-891. https://doi.org/ 10.1007/s10753-019-01173-0 [DOI] [PubMed] [Google Scholar]
- 52. Wu MY, Xie X, Xu ZK, et al. PP2A inhibitors suppress migration and growth of PANC-1 pancreatic cancer cells through inhibition on the Wnt/beta-catenin pathway by phosphorylation and degradation of beta-catenin. Oncol Rep. 2014;32(2):513-522. 10.3892/or.2014.3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chu DD, Tan J, Xie S, et al. GSK-3 beta is dephosphorylated by PP2A in a Leu309 methylation-independent manner. J Alzheimers Dis. 2016;49(2):365-375. 10.3233/Jad-150497 [DOI] [PubMed] [Google Scholar]
- 54. Mitra A, Menezes ME, Pannell LK, et al. DNAJB6 chaperones PP2A mediated dephosphorylation of GSK3beta to downregulate beta-catenin transcription target, osteopontin. Oncogene. 2012;31(41):4472-4483. https://doi.org/ 10.1038/onc.2011.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang X, Yan L, Kou S, et al. Generation and characterization of a Myh6-driven Cre knockin mouse line. Transgenic Res. 2021;30(6):821-835. https://doi.org/ 10.1007/s11248-021-00285-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31(4):176-180. https://doi.org/ 10.1002/gene.10022 [DOI] [PubMed] [Google Scholar]
- 57. Bertero A, Murry CE. Hallmarks of cardiac regeneration. Nat Rev Cardiol. 2018;15(10):579-580. 10.1038/s41569-018-0079-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Choi WY, Poss KD. Cardiac regeneration. Curr Top Dev Biol. 2012;100:319-344. https://doi.org/ 10.1016/B978-0-12-387786-4.00010-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang L, Nomura-Kitabayashi A, Sultana N, et al. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev Biol. 2014;390(1):68-79. 10.1016/j.ydbio.2014.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xie W, Schultz MD, Lister R, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134-1148. 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xie R, Everett LJ, Lim HW, et al. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12(2):224-237. 10.1016/j.stem.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun Y, Miao N, Sun T. Detect accessible chromatin using ATAC-sequencing, from principle to applications. Hereditas. 2019;156(1):29. 10.1186/s41065-019-0105-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zheng H, Qi Y, Hu S, et al. Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase. Science. 2020;370(6520):eabb5872. 10.1126/science.abb5872 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq and ChIP-seq data in this study have been deposited to the Gene Expression Omnibus with the accession number GSE211341.
All data needed to evaluate the conclusions in the paper are present in the paper and/or Supplementary Materials. Additional data related to this paper may be requested from the authors.