Abstract
Since the outbreak of the COVID-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp) has become a main target for antiviral therapeutics due to its essential role in viral replication and transcription. Thus, nucleoside analogs structurally resemble the natural RdRp substrate and hold great potential as inhibitors. Until now, extensive experimental investigations have been performed to explore nucleoside analogs to inhibit the RdRp, and concerted efforts have been made to elucidate the underlying molecular mechanisms further. This review begins by discussing the nucleoside analogs that have demonstrated inhibition in the experiments. Second, we examine the current understanding of the molecular mechanisms underlying the action of nucleoside analogs on the SARS-CoV-2 RdRp. Recent findings in structural biology and computational research are presented through the classification of inhibitory mechanisms. This review summarizes previous experimental findings and mechanistic investigations of nucleoside analogs inhibiting SARS-CoV-2 RdRp. It would guide the rational design of antiviral medications and research into viral transcriptional mechanisms.
Keywords: SARS-CoV-2, RNA-dependent RNA polymerase, Nucleoside analog, Cryo-EM structure, Computational simulation
Graphical Abstract
1. Introduction
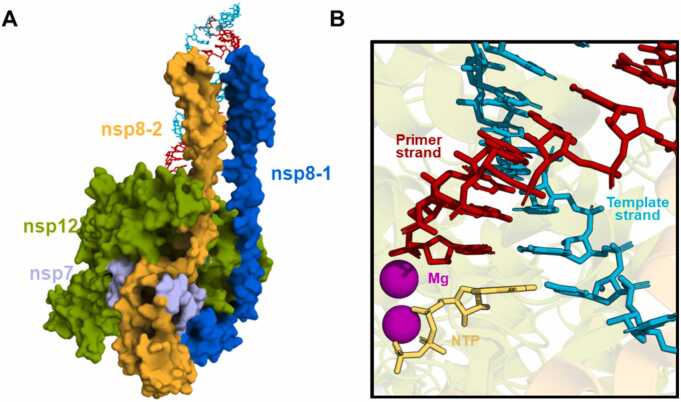
The SARS-CoV-2 virus is responsible for the COVID-19 pandemic. It is a new member of the betacoronavirus genus and possesses an approximately 30-kilobyte RNA genome [1], [2]. The RNA-dependent RNA polymerase complex (RdRp) is the minimum replicase required to catalyze the replication of the RNA genome. It contains non-structural protein 12 (nsp12) with nsp7 and nsp8 as cofactors [3], [4] (Fig. 1A). Considering the constant mutations of SARS-CoV-2 virus [5], the highly conserved and functionally essential enzyme RdRp has become a promising target for the development of antiviral drugs [6], [7], [8], [9], [10], [11].
Fig. 1.
The structure of the SARS-CoV-2 RdRp complex and the active site. (A) The structure of nsp12 bound with nsp7 and nsp8 is shown in surface representation (PDB ID: 7UOB). Nsp12, nsp8–1, nsp8–2, and nsp7 are colored green, blue, orange, and light purple, respectively. The primer and template strand of the double-stranded RNA (dsRNA) are displayed with red and cyan sticks, respectively. (B) The active site of RdRp complexed with NTP (shown in yellow). The primer strand and template strand are colored red and cyan, respectively. The two Mg2+ ions are shown in magenta spheres.
Nucleoside analogs structurally similar to natural nucleosides are promising inhibitors of viral replication and transcription. The nucleoside analog’s prodrug is typically metabolized into its 5’-triphosphate form in the cells. It can therefore compete with the natural substrate nucleoside triphosphate (NTP) for incorporation into the primer strand during the nucleotide addition cycle [12], [13], [14], [15], [16], [17], [18]. Such incorporation would result in the presence of nucleoside analogs at the primer strand or later at the template strand during the second round of RNA synthesis, allowing nucleoside analogs to exert inhibition via different mechanisms [17], [19], [20], [21], [22], [23], [24].
Until now, nucleoside analogs have been extensively explored as RdRp inhibitors [25], [26], [27]. In addition, concerted efforts have been made to comprehend the molecular details of inhibition, which would facilitate the mechanism-based drug design. In this review, we begin by introducing the nucleoside analogs that have experimentally demonstrated efficacy against SARS-CoV-2 by targeting the active site of RdRp. These analogs are classified according to the positions of their chemical modifications. The current understanding of the inhibitory mechanisms exerted by nucleoside analogs at the molecular level is then reviewed. For each mechanism, the previous structural biology and computational simulation findings are introduced. In conclusion, we discuss the perspectives based on our current knowledge.
2. Nucleoside analogs showing inhibitions on the RNA synthesis of SARS-CoV-2 RdRp
Nucleoside analogs are small molecules structurally similar to the natural RdRp substrate NTP, but with distinct chemical substitutions. Due to the structural similarity, RdRp may “mistakenly” recognize the analogs as the substrate and catalyze their incorporation into the primer strand. Thus, nucleoside analogs may appear in the RNA duplex and inhibit viral transcription and replication. In this section, we classify the analogs experimentally validated as RdRp inhibitors based on their chemical modification positions. When applicable, the findings of wet lab investigations and the respective computational results are discussed for each analog.
2.1. 1’-ribose modification
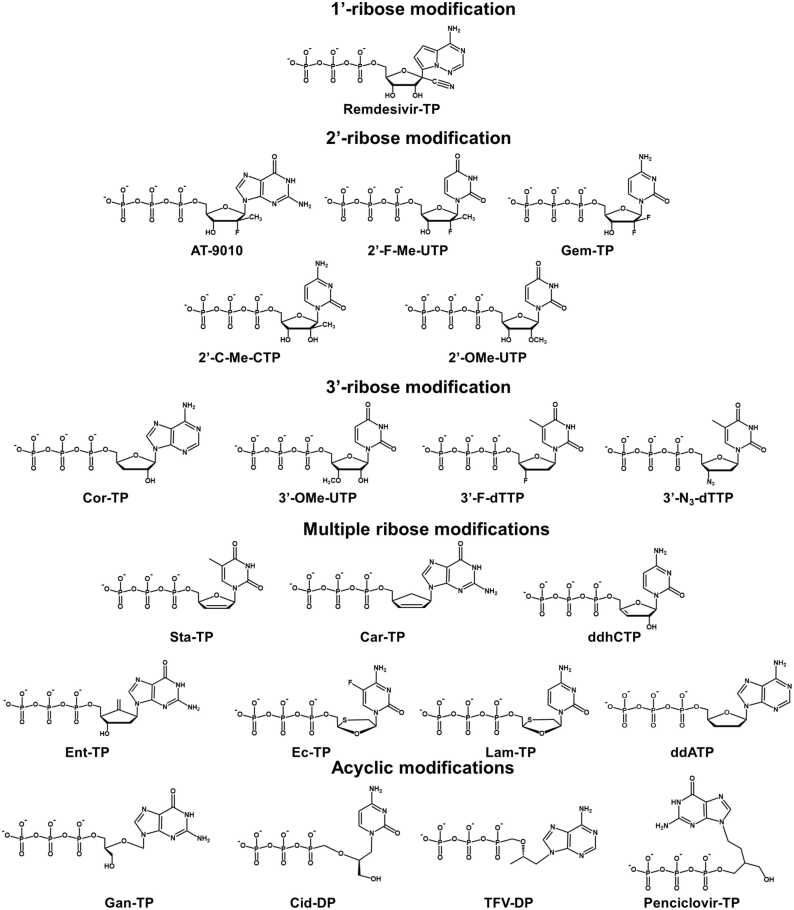
Remdesivir has been one of the most studied nucleoside analogs targeting the viral RdRp of SARS-CoV-2. The typical modification in its active form is a 1’-cyano ribose substitution, although its base has also been modified (Fig. 2). Multiple experiments have been performed to demonstrate its antiviral activity [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38]. One intriguing property of Remdesivir as an RdRp inhibitor is its higher incorporation efficiency than its counterpart natural substrate ATP. For example, Lu et al. have measured the K1/2 of Remdesivir against ATP and found that Remdesivir has a higher incorporation efficiency (0.03 µM) than ATP (0.04 µM) [39]. Moreover, Gordon et al. found the selectivity of ATP over Remdesivir-TP is 0.26–0.35 [40], [41], and Dangerfield et al. consistently show that the specificity (kcat/KM) of Remdesivir-TP is higher than that of ATP [42]. Besides the ensemble kinetic studies, the efficient incorporation of Remdesivir-TP was consistently observed in the single-molecular experiment [43]. Computational investigations also demonstrated that Remdesivir-TP is strongly bound with SARS-CoV-2 RdRp, for example, with a ΔΔG of −2.8∼−6.2 kcal/mol against ATP using free energy perturbation method [44], [45] or −13.6 kcal/mol using MM-GBSA protocol [46]. Molecular docking studies based on the static model also indicated more negative binding energy of Remdesivir-TP than that of ATP (for example, −7.6 kcal/mol versus −7.0 kcal/mol in Elfiky et al.’s study [47], and −9.3 kcal/mol versus −8.6 kcal/mol in Celik et al.’s work [48]). Further experimental assays indicate that after the efficient incorporation of Remdesivir into the primer strand, the inhibition does not take effect immediately but occurs at a later stage [20], [43]. Specifically, three additional substrates can still be added to the primer strand before the RNA synthesis stalls [19], [22], [23], [39], [40], [41], [49], [50]. However, such inhibition is profound only at a relatively low NTP concentration. It can be overcome when the NTP concentration increases [19], [22], [23], [39], [40], [43] and 90% full product can be generated with the NTP concentration as low as 10 µM [22]. Interestingly, Tchesnokov et al. found that inhibition can also happen when Remdesivir is embedded in the template strand, and such inhibition would be significantly (>90%) compromised when the NTP concentration reaches ∼100 µM [22]. Together, these studies have suggested a complex scenario of inhibition exerted by Remdesivir in SARS-CoV-2 RdRp. Further computational simulations have been performed to elucidate the underlying molecular mechanisms, which we will review in Section 3.
Fig. 2.
The chemical structures of the representative nucleoside analogs in the active form. The analogs are grouped by the type of modifications, and each group is labeled.
2.2. 2’-ribose modification
The natural substrate NTP of SARS-CoV-2 RdRp possesses a hydroxyl group at the 2’-ribose position, and analogs with 2’-substitution have shown promising inhibitory effects toward SARS-CoV-2 RdRp. For instance, AT-527 is the guanosine analog phosphoramidate prodrug, and a previous study has demonstrated that its free base form AT-511 can effectively inhibit the replication of SARS-CoV-2 in vitro with an EC90 of 0.47 µM and CC50 > 86 µM [51]. The active form of AT-527 after metabolism is named AT-9010, a GTP analog with a 2’-hydroxyl group and a 2’-hydrogen atom replaced with a fluorine atom and a methyl group, respectively (Fig. 2). Previous biochemical data has shown that AT-9010 can competitively incorporate into the primer strand against GTP with discrimination of ∼5-fold in vitro [17]. In addition, after incubation with the prodrug AT-527, the intracellular concentration of AT-9010 is comparable with that of GTP, which also shows its antiviral potential [17]. The triphosphate active form of Sofosbuvir (named 2’-F-Me-UTP, Fig. 2) possesses the same 2’-modifications as AT-9010 except that it is an analogy to UTP. Although computational studies have indicated a comparable binding free energy of 2’-F-Me-UTP relative to that of UTP [47], [52], several experimental studies have shown that the incorporation efficiency of 2’-F-Me-UTP is lower than that of UTP [17], [39], [40], [43]. Even so, polymerase extension assays have indicated it can be readily incorporated by SARS-CoV-2 RdRp and then effectively terminate the RNA synthesis [53]. However, the performance of Sofosbuvir in cell culture is controversial. Although Sofosbuvir inhibits more than half of SARS-CoV-2 replication in the human lung epithelial Calu-3 cells and human hepatoma lineage cells (HuH-7 cells) at a concentration of 10 μM [54], it exhibits no antiviral activity against SARS-CoV-2 in the A549-hACE2 cells (a human alveolar epithelial cell stably expressing hACE2) at concentrations up to 10 μM using high-through luciferase SARS-CoV-2 based neutralization assay [55]. Gemcitabine is a cytidine analog also with two fluorine substitutions on the 2’-ribose position (its active form named Gem-TP, Fig. 2). Its antiviral property has been validated previously using live virus infection in human respiratory cells [32]; however, primer extension assays suggest that it seldom exhibits inhibitory effect in SARS-CoV-2 RdRp [39]. Besides, primer extension assays have shown several extra 2’-substituted nucleoside analogs can also terminate RNA synthesis catalyzed by SARS-CoV-2, including 2’-C-Me-CTP and 2’-OMe-UTP [35], [39], [56] (Fig. 2). It is noted that the inhibition exerted by 2’-OMe-UTP is distinct as more than one 2’-OMe-UTP can be incorporated. However, incorporating the second 2’-OMe-UTP is compromised, suggesting the inhibition still takes effect although is delayed [35], [39]. Also, a recent work using primer extension assays has shown that 2’-OMe-G can exert inhibition when embedded in the template strand [57].
2.3. 3’- ribose modification
Nucleoside analogs with the 3’-modifications have also been proposed or designed, as the 3’-hydroxyl group serves as a nucleophile for an incoming NTP and is also the prerequisite for the NTP incorporation. In this regard, 3’-modifications endow the analogs with the inherent capability to inhibit the next NTP incorporation. For instance, the active form Cordycepin (named Cor-TP) is structurally similar to ATP while has replaced the 3’-hydroxyl group with a hydrogen atom (Fig. 2). The radioactive elongation assay has shown that Cordycepin could compete against all NTPs (at an NTP concentration of 10 µM) and terminate elongation with the Cordycepin’s concentration at 50 µM [49], consistent with the observation in a recent anti-SARS-CoV-2 assay [58]. Computational simulations have consistently proposed that Cordycepin is readily catalyzed by SARS-CoV-2 RdRp and incorporated into the RNA strand to exert termination [44], [59]. 3’-OMe-UTP has the methoxy substitution at the 3’-ribose position of UTP (Fig. 2), and its capability to terminate the RNA synthesis was shown by the extension reactions with SARS-CoV-2 RdRp [35]. The triphosphate forms of Alovudine and AZT (3’-F-dTTP and 3’-N3-dTTP, Fig. 2) both have 3’-ribose substitutions in comparison with dTTP (deoxythymidine triphosphate). In particular, a fluorine atom and an azide group have substituted the 3’-hydroxyl group for 3’-F-dTTP and 3’-N3-dTTP, respectively. Primer extension assays have also shown that the chain extension is immediately terminated after incorporating 3’-F-dTTP and 3’-N3-dTTP [53]. It is noted that the 3’-modification could usually be combined with other ribose modifications to render the nucleoside analogs inhibition capability and these analogs would be discussed in the next subsection.
2.4. Multiple ribose modifications
It is common that nucleoside analogs simultaneously modify two or even three ribose positions in comparison with their counterpart natural nucleotides. For example, the active form of Stavudine (named Sta-TP) is a thymidine analog, and it has distinct modifications on 2’- and 3’-ribose positions (Fig. 2). In specific, Sta-TP is a didehydro-dideoxynucleotide with a double bond between the two sp2 carbon atoms at 2’- and 3’-ribose positions. Previous extension reactions with SARS-CoV-2 RdRp have shown that such modifications could render Sta-TP capable of terminating the RNA synthesis after its incorporation [35]. The active form of Abacavir and Carbovir is named Car-TP (Fig. 2), and it is structurally similar to Sta-TP but has an extra substitution to render Car-TP a carbocyclic guanosine didehydro-dideoxynucleotide. Experimental assays have shown that Car-TP can also act as an effective inhibitor for SARS-CoV-2 [35], [53]. 3’-deoxy-3’,4’-didehydro-cytidine triphosphate (ddhCTP) has similar modifications as Sta-TP and Car-TP except that the double bond is formed between 3’- and 4’-ribose (Fig. 2). Previous study has shown it can be incorporated by SARS-CoV-2 RdRp in the single molecular experiment in vitro, however it seldom impacts the viral replication in cells [43]. The triphosphate form of Entecavir (Ent-TP) has modified 2’- and 4’-ribose simultaneously, with the 2’-hydroxyl group replaced by a hydrogen atom and the 4’-oxo replaced by an ethenyl group (Fig. 2). Previous primer extension assays have suggested that Ent-TP can exert the inhibition on SARS-CoV-2 RdRp [35]. In contrast, another cell-based assay using Entecavir in the non-triphosphorylation form shows limited inhibition [33]. The active forms of Emtricitabine and Lamivudine (Ec-TP and Lam-TP) both contain an oxathiolane ring with an unnatural (-)-β-L-stereochemical configuration, while Ec-TP has an extra fluorine substitution at the C5 position of cytosine’s base in comparison with Lam-TP (Fig. 2). Previous experimental studies using primer extension assays have suggested that their incorporations could also immediately inhibit the RNA synthesis in SARS-CoV-2 RdRp [53], [60]. However, a recent study has suggested that Emtricitabine shows no antiviral activity in cell cultures [61]. Didanosine simultaneously has the 2’- and 3’-ribose modifications by replacing the hydroxyl groups with the hydrogen atoms (its active form named ddATP, Fig. 2). Previous in vitro antiviral evaluation suggests that it can serve as a potential inhibitor for SARS-CoV-2 [62], corroborated by bioinformatics study and computational simulations [44], [63].
2.5. Acyclic modifications
The abovementioned nucleoside analogs have ribose modifications on specific sites while maintaining the ribose’s cyclic architecture. There is an alternative class of analogs that have broken the ring moiety of ribose and are named acyclic nucleoside analogs. For example, Ganciclovir triphosphate (Gan-TP) and Cidofovir diphosphate (Cid-DP) are acyclic guanosine and cytidine nucleotides, respectively (Fig. 2). Polymerase extension assays have shown that incorporating Gan-TP can immediately terminate the RNA synthesis [35]. However, the termination can still occur while delayed after incorporating Cid-DP [35]. Tenofovir diphosphate (TFV-DP) is the triphosphate form of its prodrug, and it is an acyclic adenosine nucleotide (Fig. 2). Molecular docking study has indicated that the binding affinity of TFV-DP is comparable with that of ATP, suggesting it is a potential inhibitor [47]. Consistently, polymerase assay observed that no further extension is observed once one molecule of TFV-DP is incorporated [60], and such inhibition can take effect when different lengths of RNA primer are used [53]. However, cell-based studies have shown that Tenofovir has no or only limited inhibition of SARS-CoV-2 RdRp activity [33], [61]. Penciclovir is also an alternative acyclic guanosine nucleotide (its active form named Penciclovir-TP, Fig. 2), and in vitro SARS-CoV-2 assay has suggested that its inhibition occurs only at a high concentration [31]. Such limited inhibition is consistent with another cell-based SARS-CoV-2 RdRp assay [33], even though computational work has proposed Penciclovir as a potential inhibitor due to its comparable binding free energy with Remdesivir [64].
2.6. Base modifications
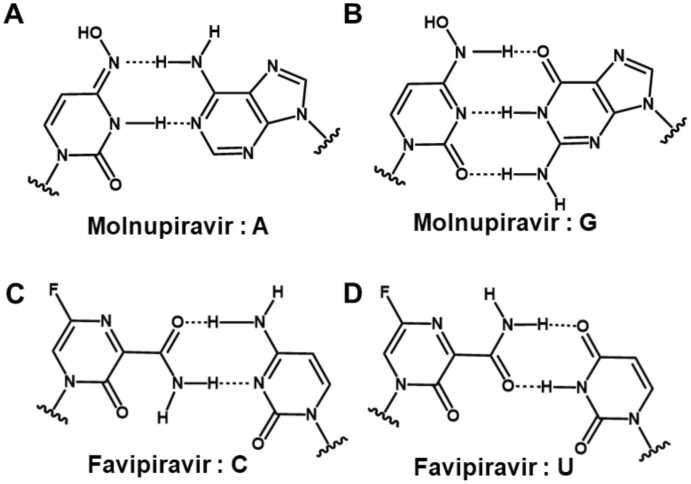
The nucleoside analogs with base modifications are less common than those with the ribose modifications, probably because they could usually impair the base pairing stability at the active site, thus impairing their incorporation efficiency into the primer strand. Even so, the experiments have explored several nucleoside analogs with the base modifications, and their efficacies to inhibit SARS-CoV-2 RdRp have been observed [32], [57], [65], [66]. For example, Schultz et al. conducted investigations on the antiviral activity of 18,000 drugs using live virus infection in human respiratory cells and found that several base-modified nucleosides analogs show antiviral activity, including 6-Thio-dG, 6-Thiopurine riboside as well as the well-studied Molnupiravir, etc [32]. The active form of Molnupiravir is β-d-N4-hydroxycytidine triphosphate (NHC-TP), and calculations of the binding free energies based on the static structural models suggest that it has higher binding affinity than the natural substrates, with ΔΔG= −21.8 and −10.1 kcal/mol for its enamine and oxime tautomer, respectively (Fig. 3A and B) [46]. RNA elongation assays have shown that NHC-TP can be catalyzed by SARS-CoV-2 RdRp and incorporated into the primer strand instead of CTP and UTP, although at lower incorporation efficiency. However, its incorporation will not stall the RdRp, and thus, it can appear in the template strand in the second round of RNA synthesis to incorporate either GTP or ATP (Figs. 3A and 3B). This would generate the mutated RNA products and lead to the lethal influence on viral replication [67]. The RNA mutagenesis exerted by Molnupiravir was also observed in another biochemical study [68], and its inhibition on SARS-CoV-2 was further validated by several experimental assays [33], [69], [70], [71] and even in animals [72], [73]. Favipiravir is a purine nucleic acid with base modifications. Its triphosphate form, Favipiravir-TP, can mimic both ATP and GTP to incorporate into the RNA strand (Fig. 3C-3D). Computational works show that Molnupiravir and Favipiravir have comparable or higher binding strength than their respective natural NTPs in SARS-CoV-2 RdRp [46], [48]. Inhibition or mutagenesis provoked by Favipiravir has been observed in SARS-CoV-2 RdRp [18], [74], although other experimental studies have shown that Favipiravir has only shown limited inhibition on SARS-CoV-2 [31], [33]. In addition to generating RNA mutations, the inhibition exerted by Favipiravir may be more complex. A previous study has shown that RNA extension will stall after several Favipiravir-TPs are consecutively incorporated [16]. Moreover, an alternative study has shown that incorporating the Favipiravir will make the further extension slower or less efficient [74].
Fig. 3.
The two base pairing patterns of Molnupiravir and Favipiravir. (A-B) Molnupiravir base paired with adenine and guanine. (C-D) Favipiravir base paired with cytosine and uridine.
3. Molecular mechanisms of nucleoside analogs inhibiting SARS-CoV-2 RdRp
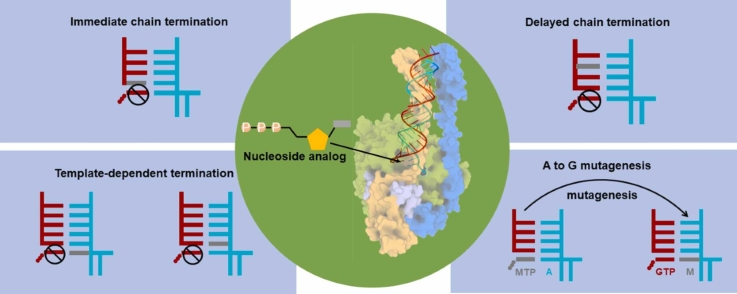
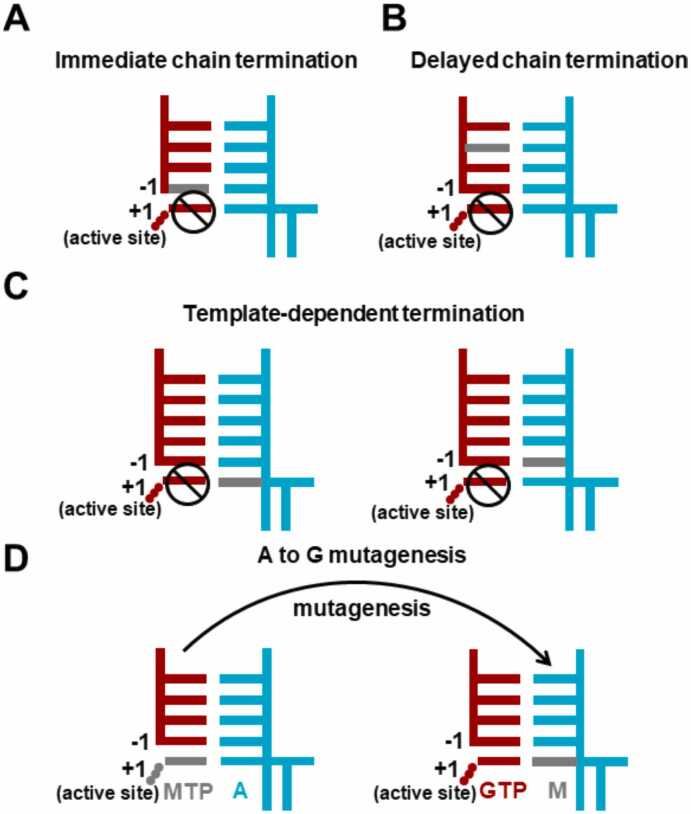
Although several nucleoside analogs have shown inhibitory efficacy against SARS-CoV-2 RdRp in the wet lab studies, the experimental assays alone cannot reveal the underlying molecular mechanisms, significantly impeding the further rational design of RdRp inhibitors. Under such circumstances, structural biology and computational simulations have emerged as promising methods and made significant progress in understanding the inhibitory mechanisms of nucleoside analogs on SARS-CoV-2 RdRp at the molecular level. The experimental structures have provided valuable insights into the transcriptional mechanisms of SARS-CoV-2 RdRp and lay the structural basis for exploring the inhibitory mechanisms of nucleoside analogs. Furthermore, computational simulations based on the experimental structures have been performed to elucidate the acting mechanism and design the nucleoside analogs, significantly deepening our knowledge about the inhibition that occurs in the SARS-CoV-2 RdRp. This section will review the current understanding of four common inhibitory mechanisms (Fig. 4), with the representative nucleoside analogs as examples.
Fig. 4.
Diagrams demonstrating the four common inhibitory mechanisms. In the diagram, the primer and template strand are colored red and cyan, respectively. The analog is highlighted in grey.
3.1. Immediate chain termination
Immediate chain termination requires the nucleoside analog to effectively compete with the natural substrate NTP to incorporate into the primer strand. Its incorporation would then immediately inhibit the addition of the next nucleotide (Fig. 4A), and thus, it is named an “immediate” chain terminator.
AT-9010 is structurally similar to GTP but has the 2’-substituions (Fig. 2). The cryo-EM structure containing AT-9010-TP at the active site (+1 site) and AT-9010-MP at −1 site of the primer strand has been reported (PDB ID: 7ED5) [17]. This structure has shed light on the molecular mechanism for its capability as an immediate chain terminator. In particular, the presence of AT-9010-MP at the −1 site has suggested that it can be incorporated into the primer strand and smoothly translocate to the −1 site, satisfying the prerequisite of the immediate chain terminator. Moreover, as shown by the structure, the 2’-methyl substitution of the incorporated AT-9010 at the −1 site would create a repulsive hydrophobic-polar contact with the ribose 4’-oxygen atom of the AT-9010-TP at the active site. The substrate, thus, is not correctly poised for catalysis and abolishing the chain extension [17], providing the molecular mechanism of AT-9010 as an immediate chain terminator.
Sofosbuvir is the uridine analog, and its active form 2’-F-Me-UTP has similar ribose modifications as the AT-9010 (Fig. 2). Recent experiments have demonstrated that its incorporation can also immediately terminate the next nucleotide addition and thus act as an immediate terminator [53]. A computational study by MD simulations based on the cryo-EM structure of SARS-CoV-2 RdRp has further elucidated its inhibitory mechanism. Specifically, the simulations have shown that 2’-F-Me-UTP can maintain the catalytically active conformation at the active site (+1 site) of SARS-CoV-2 RdRp [52]. Moreover, they have found that after its incorporation and translocation to the −1 site, the 2’-methyl group of Sofosbuvir would clash with the base of the next substrate NTP, thus disrupting the stability of NTP at the active site and exerting the “immediate” termination as observed in the experiments [53].
3.2. Delayed chain termination
Delayed chain termination implies that the nucleoside analog does not immediately impair the nucleotide addition after its incorporation while inhibiting the RNA synthesis after consecutive incorporations of several natural NTPs. Therefore, the inhibition occurs when the nucleoside analog has moved to an upstream site of the primer strand and causes a “delayed” termination (Fig. 4B), in sharp contrast to the “immediate” termination discussed above.
Remdesivir is one typical delayed chain terminator that has been extensively studied, and several RdRp structures containing Remdesivir have been resolved. For example, cryo-EM structure has shown that Remdesivir-TP can pair well with uridine-MP at the active site (PDBID: 7UO4) and further revealed that the higher affinity of Remdesivir relative to ATP is gained by the extra polar interaction between the 1’-cyano group and the protein residues T687, N691, and S759 [15]. This is consistent with the biochemical experiments showing that the incorporation efficiency of Remdesivir-TP is significantly reduced upon S759A mutation [15], [75]. Computational studies have complemented the structural investigations to suggest the high incorporation efficiency of Remdesivir-TP. For instance, a previous computational work using umbrella sampling has investigated the insertion of Remdesivir-TP into the active site and found that the Remdesivir-TP has lower free energy than ATP [76]. Several MD simulation works have consistently shown that Remdesivir-TP can maintain the catalytically active conformation for incorporation into the primer strand [77], [78], further rationalizing the high incorporation efficiency of Remdesivir-TP. In addition, the cryo-EM structures of SARS-CoV-2 RdRp after Remdesivir’s incorporation have also been resolved. For example, the RdRp structure in the pre-translocation state has been determined with Remdesivir-monophosphate (RMP) embedded at the −1 site of the primer strand (PDB ID: 7C2K) [24]. In this structure, the GMP is present at the 3’-terminal of the primer strand (at +1 site), and it is well stacked with the RMP and base paired with the CMP of the template strand. Such a conformation has again corroborated that Remdesivir does not act as an immediate terminator. Instead, it would exert inhibition when it moves to the upstream site of the primer strand. This has been observed in the resolved cryo-EM structures of SARS-CoV-2 RdRp with Remdesivir at the −3 site of the primer strand (PDB IDs: 7B3C in the pre-translocation state and 7B3B in the post-translocation state) [23], and rationalized by structural modeling showing Remdesivir at −4 site clashed with S861. Consistently, several computational works have further elucidated that its translocation from the −3 site to the −4 site is inhibited due to the steric clash between the 1’-cyano group of Remdesivir and S861 [77], [79], [80], [81]. Moreover, previous computational works have also suggested that the interactions between the 1’-cyano group of Remdesivir and a neighboring salt bridge formed by Asp865 and Lys593 would further halt the translocation of Remdesivir from −3 to −4 site [77].
Besides Remdesivir, previous work has also suggested that the 2’-OMe-UTP could cause a delayed chain termination [35], [39]. Recently, computational work has provided the molecular details of the inhibition exerted by 2’-OMe-UMP, although it suggests that it is more accurate to name 2’-OMe-UMP as a partial chain terminator [52]. In this computational work, they have found that the presence of 2’-OMe-UMP at the −1 site of the primer strand would create a steric clash between its 2’-OMe group and the base of the next NTP at the active site (+1 site), which would weaken the base stacking between + 1 and −1 site and thus reduces the binding affinity of the next substrate as well as impairs the primer extension. However, they also found that the critical distance for catalysis (the distance between the O3’ atom of the 2’-OMe-UMP at the −1 site and the Pα atom of the 2’-OMe-UTP at the +1 site) is well maintained as that of the wildtype dsRNA with natural NTP. This also explains the experimental observations that primer extension is only partially terminated [35], [39].
3.3. Template-dependent termination
Template-dependent termination characterizes the situation that the nucleoside analog terminates the RNA synthesis when it is present at the template strand (Fig. 4C). Such kind of terminator has been found in other viral RdRp [21], [82], such as Ribavirin against HCV RdRp. For SARS-CoV-2 RdRp, biochemical experiments have demonstrated that Remdesivir can act as a template-dependent terminator [21], [22]. Specifically, it has been found that Remdesivir embedded at the template strand would inhibit the incorporation of its complementary NTP and the next NTP. Interestingly, the first inhibition is reduced by the V557L mutation, while the second one is seldom affected [22]. Our recent computational works have provided molecular insights into such template-dependent mechanisms and the drug resistance gained by V557L mutation [83], [84]. We found that the inhibitions do not take effect by directly destroying the NTP incorporation at the active site. Instead, the translocation of Remdesivir along the template strand is inhibited; thus, the active site is occupied and unavailable for the arrival of NTP. Specifically, protein residue V557 would form a steric clash with the 1’-cyano group of Remdesivir and thus hampers its translocation from the + 2 site to the + 1 site along the template strand [83]. However, upon V557L mutation, the obstacle on the translocation pathway is relieved due to the side-chain’s rotation of V557L, explaining the reduced inhibition upon V557L mutation [83].
Moreover, our work suggests that the interaction between the 1’-cyano group of Remdesivir and two conserved protein residues, S682 and G683, on motif B has further impeded the translocation of Remdesivir from the + 1 site to the −1 site along the template strand [84]. Specifically, G683 would destabilize the post-translocation state with Remdesivir at the −1 site of the template strand, and S682 would kinetically hamper the translocation by sterically clashing with the Remdesivir. In addition to Remdesivir, a recent work using primer extension assays has found that 3-MeU and 1-MeG present at the template strand can inhibit the NTP incorporation opposite to the nucleoside analogs, and 2’-OMe-G at the template strand can inhibit the next NTP incorporation [57]. However, how these analogs induce the RdRp stalling awaits further exploration.
3.4. Mutagenesis
Nucleoside analogs can also cause RNA mutagenesis to impair the genes. Such mutagenic nucleoside analog usually possesses base modifications, allowing itself to have different base pairing patterns. Therefore, the incorporation of such nucleoside analog into the primer strand would result in its presence at the template strand for the RNA synthesis at the later stage (Fig. 4D). Its two tautomer forms would then induce the viral mutations and finally lead to the lethal effect on the viral genes. In this subsection, we have reviewed the cryo-EM structures of SARS-CoV-2 RdRp with the mutagenic nucleoside analogs.
Molnupiravir (the prodrug of NHC) is one representative mutagenic nucleoside analog, and it has been marketed for treating COVID-19 [67], [85]. Two structures with the monophosphate form of NHC (named “NMP”) embedded at the −1 site of the template strand have been resolved [67]. The two structures share high similarity except that NMP can form three hydrogen bonds with GMP in one structure (PDBID: 7OZV) while forming two hydrogen bonds with AMP in another (PDBID: 7OZU), providing the structural basis to understand how Molnupiravir synthesizes the mutated RNA.
Favipiravir has also been suggested to act as a mutagenic nucleoside analog [18], [74], [86], [87], [88], [89], [90], as its triphosphate form could simulate both GTP and ATP to incorporate into the primer strand. SARS-CoV-2 RdRp structures with Favipiravir-TP at the active site have been resolved (PDB ID: 7AAP [16] and 7CTT [18]). In the 7AAP structure, the Favipiravir-TP is not well posed, as its triphosphate moiety rotated and misaligned with the nucleophile 3’-OH group of the nucleotide at the −1 site. The researchers thus anticipate such a structural distortion explains the inefficient incorporation of Favipiravir-TP into the primer strand as observed in their primer extension assay [16]. However, the 7CTT structure shows that the α-phosphate of Favipiravir-TP at the + 1 site is in the vicinity (<3.5 Å) of the 3’-OH group of the nucleotide at the −1 site, which suggests Favipiravir-TP could also adopt an appropriate conformation for incorporation at the active site. Although the conformations of Favipiravir-TP show discrepancies in the two RdRp structures, both structures have shown that Favipiravir-TP can form hydrogen bonds with CMP, which may explain why it can still be incorporated and cause mutagenesis. The incorporation of Favipiravir-TP into the primer strand has been captured by a cryo-EM structure of SARS-CoV-2 RdRp in the pre-translocation state (PDB ID: 7DFG) [91]. In this structure, Favipiravir-MP is covalently bound at the + 1 site of the primer strand and forms complex interactions with the protein residues. Besides, this structure shows that Favipiravir-MP forms two hydrogen bonds with UMP, thus complementing the structures with Favipiravir-TP:CMP pair to provide the structural basis for understanding the RNA mutation caused by Favipiravir.
4. Summary and outlook
The RdRp complex is one of the widely studied drug targets for SARS-CoV-2, and great efforts have been devoted to exploring its inhibitors. Nucleoside analogs are structurally similar to the natural substrate NTPs of the RdRp complex, and have been extensively investigated for their efficacy in inhibiting viral replication. In this review, we have summarized the nucleoside analogs proposed as potential RdRp inhibitors, especially those the experimental assays have validated. Furthermore, to provide molecular insights into the inhibition, experimental RdRp structures with the presence of analogs have been reviewed, and the corresponding computational studies elucidating the inhibitory mechanisms at the molecular level have been discussed.
Most analogs investigated by the experimental assays contain chemical modifications on 1’, 2’, or 3’-ribose positions, but those with the 4’-ribose modifications were seldom evaluated. Although none of the three 4’-substituted carbocyclic uridine analogs exhibits significant antiviral activities toward SARS-CoV-2 in a recent study [92], it is worth further investigations to explore the potential of analogs with 4’-substitution. One promising example is that 4’-fluorouridine has been identified as an effective inhibitor to block SARS-CoV-2 replication, and the in vitro assay shows that multiple incorporations of 4’-fluorouridine would trigger the polymerase stalling [93]. Therefore, 4’-ribose is an alternative site to consider or combine with other modifications in future drug screening and rational design efforts.
So far, most studies have focused on the binding affinity of nucleoside analogs relative to the cognate natural substrate in SARS-CoV-2 RdRp to suggest the potential inhibition against SARS-CoV-2 RdRp. However, it is essential to investigate if nucleoside analogs can selectively inhibit the viral RdRp rather than the human RNA polymerase to evaluate the overall inhibitory effect. One previous study has investigated the selective inhibition of the nucleoside analog Remdesivir in SARS-CoV-2 RdRp against the human DNA-dependent RNA polymerase (Pol II) by comparing the relative binding affinity (ΔΔG) of Remdesivir [81]. By performing alchemical free energy simulations, they have found that the ΔΔGATP->RTP in SARS-CoV-2 RdRp is around −6 kcal/mol, suggesting that RTP has a higher affinity than ATP. On the contrary, the ΔΔGATP->RTP in human RNA Pol II is + 1 kcal/mol, implicating that Remdesivir is less competitive than ATP and rarely incorporated into the nascent RNA strand in Pol II. Therefore, the calculations of ΔΔG rationalized the selective inhibition exerted by Remdesivir in SARS-CoV-2 RdRp. This is further consolidated by the different protein environments of the active site in two polymerases. Specifically, the cyano group of RTP can form hydrogen bonds with T687 and N691 in the active site of SARS-CoV-2 RdRp. On the contrary, P462 in the corresponding position in human RNA Pol II could not provide the hydrogen bonds to stabilize the cyano group of RTP, thereby reducing its binding affinity. Overall, this study has provided an example to investigate the selective inhibition of nucleoside analogs in SARS-CoV-2 RdRp against the human RNA Pol II. Considering the selectivity against human RNA Pol II is vital for designing nucleotide analogs or evaluating the inhibitory effect, future studies are suggested to investigate and compare the binding affinity of nucleotide analogs in viral polymerase and human RNA Pol II to assess the efficacy of nucleotide analogs more comprehensively.
It is also interesting to discuss the interplay between the structural modifications on the nucleoside analogs and the different SARS-CoV-2 variants. Although RdRp is relatively conserved compared to the surface proteins, mutations were found in the RdRp of each variant [37]. As RdRp is an important drug target, studies have been conducted to investigate the effect of the naturally occurring mutation on the nucleoside analogs’ antiviral activity. For instance, Salpini et al. have found that the frequently occurring mutation P323L can strengthen the binding affinity of the two acyclic nucleoside analogs Penciclovir and Tenofovir, while decreasing that of Remdesivir and Emtricitabine, which are nucleoside analogs with 1’-ribose modification and multiple ribose modifications, respectively [94]. In addition, Cho et al. have shown that P323L, A529V, and G671S (in Omicron subvariant) mutations would not influence the IC50 of Remdesivir or Molnupiravir (with base modification) [95]. Moreover, Pitts et al. have shown that P323L (among all variants), G671S (in the Delta variant), and F694Y (a highly prevalent substitution in early Omicron isolates) mutations have little effect on the EC50 of Remdesivir and GS-441524 by measuring EC50 [37]. In addition to the mutations occurring during the evolution of SARS-CoV-2, some mutations can appear when the virus is passaged or consecutively exposed in the presence of nucleoside analogs. Specifically, Gandhi et al. reported that E802D mutation was produced in a case of an immunocompromised patient infected with COVID-19 during the usage of Remdesivir, and such mutation increases the IC50 of Remdesivir by ∼6 folds [96]. As E802 forms an electrostatic network with D804 and K807 adjacent to the nascent RNA strand, they speculate that E802D mutation may distort the active site in a way that either enables the enzyme to exclude Remdesivir or alleviates the steric clash from S861, thereby allowing the enzyme to escape Remdesivir-mediated chain termination [96]. In addition, Stevens et al. have observed S759A and V792I mutations in the GS-441524 lineages when passaging the WA-1 clinical isolates in Vero E6 cells in the presence of GS-441524. They found that V792I gains drug resistance to Remdesivir by reducing the UTP concentration required to bypass the template-dependent inhibition, and S759A impairs the binding of Remdesivir by 10 folds because such mutation disrupts its hydrogen bond with the 1’-cyano group of Remdesivir [75]. Consistently, Malone et al. have shown that the selectivity of ATP over Remdesivir-TP is ∼5 upon S759 mutation. In contrast, the selectivity is ∼0.5 in the wildtype RdRp, indicating that S759A is resistant to Remdesivir [15]. They also resolved the cryo-EM structure of RdRp with Remdesivir-TP in the active site, consolidating that S759 can stabilize the binding of Remdesivir-TP by forming a hydrogen bond with the 1’-cyano group. The mutation breaks the hydrogen bond and thus reduces the binding of Remdesivir-TP. In summary, although several RdRp mutations have been reported during the evolution of the virus or in the constant exposure to the nucleoside analogs, further studies are expected to understand how the nucleoside analogs respond to the naturally occurring mutations or cause the drug-resistant mutations.
It is worth noting that the current investigations of the nucleoside analogs as inhibitors have mainly focused on terminating the nucleotide addition in RdRp. However, the efficacy of the nucleoside analogs is also challenged by the exoribonuclease (ExoN) domain in the nsp14 [35], [56], [65], [97], [98], [99], [100]. The ExoN domain only exists in coronaviruses and a few other virus families of the Nidovirales order [101], [102], [103], [104], and it helps to preserve the fidelity of RNA replication by excising the mismatched nucleotide from the 3’-terminal of the RNA strand [105], [106]. Such a function of ExoN also brings challenges to the design of nucleoside analog for the treatment of SARS-CoV-2, as the analog could be backtracked to the ExoN domain [107], [108] and cleaved from the primer strand [71], [109], [110], [111]. Therefore, a complete exploration of the nucleoside analog against SARS-CoV-2 would not only consider its inhibition on the RdRp but also examine if it can evade the excision by ExoN. Fortunately, the structures nsp14-nsp10 have been reported and shed light on the mismatch recognition and fidelity control by SARS-CoV-2 [101], [110], [112]. Further experimental and computational investigations of the nucleoside analogs’ performance on the cleavage site of the nsp14 would be helpful to provide a full evaluation of its inhibitory effect, as well as orientate the drug design to optimize its performance simultaneously on the RdRp and ExoN [71], [77], [113].
Overall, we have systematically reviewed the nucleoside analogs that show inhibition against SARS-CoV-2 RdRp and discussed the underlying molecular mechanisms. We hope this work will not only provide a comprehensive knowledge of nucleoside analogs inhibiting the SARS-CoV-2 RdRp but also be conducive to the further investigations of inhibitory mechanisms, orientating the mechanism-driven and structure-based drug design by optimizing the modifications on the nucleoside analogs.
CRediT authorship contribution statement
Tiantian Xu: Writing – review & editing. Lu Zhang: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge the financial support from the National Natural Science Foundation of China (22273107) and the Chinese Academy of Sciences (JCTD-2022-12) to L.Z. We thank Prof. Peter Pak-Hang Cheung from the Chinese University of Hong Kong for editing the manuscript.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci. 2014;111(37):E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Tracking SARS-CoV-2 variants. 2023. 〈https://cov-lineages.org/lineage_list.html〉.
- 6.Wu J., Chen Z., Han X., Chen Q., Wang Y., et al. SARS-CoV-2 RNA-dependent RNA polymerase as a target for high-throughput drug screening. Future Virol. 2023 doi: 10.2217/fvl-2021-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naidu S.A.G., Mustafa G., Clemens R.A., Naidu A.S. Plant-Derived Natural Non-Nucleoside Analog Inhibitors (NNAIs) against RNA-Dependent RNA Polymerase Complex (nsp7/nsp8/nsp12) of SARS-CoV-2. J Diet. 2021;Suppl:1–30. doi: 10.1080/19390211.2021.2006387. [DOI] [PubMed] [Google Scholar]
- 8.Jin Z., Wang H., Duan Y., Yang H. The main protease and RNA-dependent RNA polymerase are two prime targets for SARS-CoV-2. Biochem Biophys Res Commun. 2021;538:63–71. doi: 10.1016/j.bbrc.2020.10.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J., Song W., Huang H., Sun Q. Pharmacological therapeutics targeting RNA-Dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. J Clin Med. 2020;9(4):1131. doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., et al. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discov. 2020;25(10):1141–1151. doi: 10.1177/2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S., Attar F., Bloukh S.H., Sharifi M., Nabi F., et al. A review on the interaction of nucleoside analogues with SARS-CoV-2 RNA dependent RNA polymerase. Int J Biol Macromol. 2021;181:605–611. doi: 10.1016/j.ijbiomac.2021.03.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu B., Gong P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc Natl Acad Sci USA. 2016;113(28):E4005–E4014. doi: 10.1073/pnas.1602591113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamyatkin D.F., Parra F., Martin Alonso J.M., Harki D.A., Peterson B.R., et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem. 2008;283(12):7705–7712. doi: 10.1074/jbc.M709563200. [DOI] [PubMed] [Google Scholar]
- 14.Gong P. Within and beyond the nucleotide addition cycle of viral RNA-dependent RNA polymerases. Front Mol Biosci. 2022;8 doi: 10.3389/fmolb.2021.822218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malone B.F.F., Perry J.K.K., Olinares P.D.B., Lee H.W.W., Chen J., et al. Structural basis for substrate selection by the SARS-CoV-2 replicase. Nature. 2023;614:781–787. doi: 10.1038/s41586-022-05664-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naydenova K., Muir K.W., Wu L.-F., Zhang Z., Coscia F., et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc Natl Acad Sci. 2021;118(7) doi: 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon A., Fattorini V., Sama B., Selisko B., Feracci M., et al. A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase. Nat Commun. 2022;13(1):621. doi: 10.1038/s41467-022-28113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng Q., Peng R., Yuan B., Wang M., Zhao J., et al. Structural basis of SARS-CoV-2 polymerase inhibition by favipiravir. Innov (Camb) 2021;2(1) doi: 10.1016/j.xinn.2021.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol Cell. 2021;81(7):1548–1552. doi: 10.1016/j.molcel.2021.01.035. e1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin W., Mao C., Luan X., Shen D.-D., Shen Q., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon C.J., Lee H.W., Tchesnokov E.P., Perry J.K., Feng J.Y., et al. Efficient incorporation and template-dependent polymerase inhibition are major determinants for the broad-spectrum antiviral activity of remdesivir. J Biol Chem. 2022;298(2) doi: 10.1016/j.jbc.2021.101529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., et al. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J Biol Chem. 2020;295(47):16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat Commun. 2021;12(1):279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Wu J., Wang H., Gao Y., Liu Q., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182(2):417–428. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson K.A., Dangerfield T. Mechanisms of inhibition of viral RNA replication by nucleotide analogs. Enzymes. 2021;49:39–62. doi: 10.1016/bs.enz.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J Med Virol. 2021;93(1):300–310. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y., Yin W., Xu H.E. RNA-dependent RNA polymerase: structure, mechanism, and drug discovery for COVID-19. Biochem Biophys Res Commun. 2021;538:47–53. doi: 10.1016/j.bbrc.2020.08.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., et al. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585:273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tempestilli M., Caputi P., Avataneo V., Notari S., Forini O., et al. Pharmacokinetics of remdesivir and GS-441524 in two critically ill patients who recovered from COVID-19. J Antimicrob Chemother. 2020;75(10):2977–2980. doi: 10.1093/jac/dkaa239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaw Z.R., Kim D.H., Wei L.-J. Remdesivir for the treatment of Covid-19-preliminary report. N Engl J Med. 2020;383(10):993. doi: 10.1056/NEJMc2022236. [DOI] [PubMed] [Google Scholar]
- 31.Wang M., Cao R., Zhang L., Yang X., Liu J., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz D.C., Johnson R.M., Ayyanathan K., Miller J., Whig K., et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature. 2022;604(7904):134–140. doi: 10.1038/s41586-022-04482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J., Guo S., Yi D., Li Q., Ma L., et al. A cell-based assay to discover inhibitors of SARS-CoV-2 RNA dependent RNA polymerase. Antivir Res. 2021;190 doi: 10.1016/j.antiviral.2021.105078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schafer A., Martinez D.R., Won J.J., Meganck R.M., Moreira F.R., et al. Therapeutic treatment with an oral prodrug of the remdesivir parental nucleoside is protective against SARS-CoV-2 pathogenesis in mice. Sci Transl Med. 2022;14(643):eabm3410. doi: 10.1126/scitranslmed.abm3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jockusch S., Tao C., Li X., Anderson T.K., Chien M., et al. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir Res. 2020;180 doi: 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiba S., Kiso M., Nakajima N., Iida S., Maemura T., et al. Co-administration of favipiravir and the remdesivir Metabolite GS-441524 effectively reduces SARS-CoV-2 replication in the lungs of the syrian hamster model. mBio. 2022;13(1) doi: 10.1128/mbio.03044-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitts J., Li J., Perry J.K., Du Pont V., Riola N., et al. Remdesivir and GS-441524 retain antiviral activity against delta, omicron, and other emergent SARS-CoV-2 variants. Antimicrob Agents Chemother. 2022;66(6) doi: 10.1128/aac.00222-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai X., Sun H., Wu S., Li Y., Wang L., et al. Identifying small-molecule inhibitors of SARS-CoV-2 RNA-dependent RNA polymerase by establishing a fluorometric assay. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.844749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu G., Zhang X., Zheng W., Sun J., Hua L., et al. Development of a simple in vitro assay to identify and evaluate nucleotide analogs against SARS-CoV-2 RNA-Dependent RNA polymerase. Antimicrob Agents Chemother. 2021;65(1):e01508–e01520. doi: 10.1128/AAC.01508-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23(12) doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seifert M., Bera S.C., van Nies P., Kirchdoerfer R.N., Shannon A., et al. Inhibition of SARS-CoV-2 polymerase by nucleotide analogs from a single-molecule perspective. Elife. 2021;10 doi: 10.7554/eLife.70968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Zhang D., Gao X., Wang X., Zhang L. 2'- and 3'-Ribose modifications of nucleotide analogues establish the structural basis to inhibit the viral replication of SARS-CoV-2. J Phys Chem Lett. 2022;13(18):4111–4118. doi: 10.1021/acs.jpclett.2c00087. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase. J Phys Chem B. 2020;124(32):6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 46.Prussia A.J., Chennamadhavuni S. Biostructural models for the binding of nucleoside analogs to SARS-CoV-2 RNA-dependent RNA polymerase. J Chem Inf Model. 2021;61(3):1402–1411. doi: 10.1021/acs.jcim.0c01277. [DOI] [PubMed] [Google Scholar]
- 47.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Celik I., Erol M., Duzgun Z. In silico evaluation of potential inhibitory activity of remdesivir, favipiravir, ribavirin and galidesivir active forms on SARS-CoV-2 RNA polymerase. Mol Divers. 2022;26(1):279–292. doi: 10.1007/s11030-021-10215-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vatandaslar H. A systematic study on the optimal nucleotide analogue concentration and rate limiting nucleotide of the SARS-CoV-2 RNA-Dependent RNA polymerase. Int J Mol Sci. 2022;23(15):8302. doi: 10.3390/ijms23158302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J., Wang H., Liu Q., Li R., Gao Y., et al. Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex. Cell Rep. 2021;37(4) doi: 10.1016/j.celrep.2021.109882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Good S.S., Westover J., Jung K.H., Zhou X.-J., Moussa A., et al. AT-527, a double prodrug of a guanosine nucleotide analog, is a potent inhibitor of SARS-CoV-2 in vitro and a promising oral antiviral for treatment of COVID-19. Antimicrob Agents Chemother. 2021;65(4):e02479–02420. doi: 10.1128/AAC.02479-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan C., Goonetilleke E.C., Unarta I.C., Huang X. Incorporation efficiency and inhibition mechanism of 2 '-substituted nucleotide analogs against SARS-CoV-2 RNA-dependent RNA polymerase. Phys Chem Chem Phys. 2021;23(36):20117–20128. doi: 10.1039/d1cp03049c. [DOI] [PubMed] [Google Scholar]
- 53.Chien M., Anderson T.K., Jockusch S., Tao C., Li X., et al. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J Proteome Res. 2020;19(11):4690–4697. doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sacramento C., Fintelman-Rodrigues N., Temerozo J., de Paula Dias Da Silva A., da Silva Gomes Dias S., et al. J Antimicrob Chemother. 2020;Vol. 76:1874–1885. doi: 10.1093/jac/dkab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun. 2020;11(1):5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones A.N., Mourao A., Czarna A., Matsuda A., Fino R., et al. Characterization of SARS-CoV-2 replication complex elongation and proofreading activity. Sci Rep. 2022;12(1):9593. doi: 10.1038/s41598-022-13380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petushkov I., Esyunina D., Kulbachinskiy A. Effects of natural RNA modifications on the activity of SARS-CoV-2 RNA-dependent RNA polymerase. FEBS J. 2023;290(1):80–92. doi: 10.1111/febs.16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rabie A.M. Potent inhibitory activities of the adenosine analogue cordycepin on SARS-CoV-2 replication. Acs Omega. 2022;7(3):2960–2969. doi: 10.1021/acsomega.1c05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bibi S., Hasan M.M., Wang Y.-B., Papadakos S.P., Yu H. Cordycepin as a promising inhibitor of SARS-CoV-2 RNA dependent RNA polymerase (RdRp) Curr Med Chem. 2022;29(1):152–162. doi: 10.2174/0929867328666210820114025. [DOI] [PubMed] [Google Scholar]
- 60.Jockusch S., Tao C., Li X., Anderson T., Chien M., et al. Triphosphates of the two components in DESCOVY and TRUVADA are inhibitors of the SARS-CoV-2 polymerase. bioRxiv. 2020 doi: 10.1101/2020.1104.1103.022939. doi: 10.1101/2020.1104.1103.022939. [DOI] [Google Scholar]
- 61.Feng J.Y., Du Pont V., Babusis D., Gordon C.J., Tchesnokov E.P., et al. The Nucleoside/Nucleotide Analogs Tenofovir and Emtricitabine Are Inactive against SARS-CoV-2. Molecules. 2022;27(13):4212. doi: 10.3390/molecules27134212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabie A.M. Efficacious preclinical repurposing of the nucleoside analogue didanosine against COVID-19 polymerase and exonuclease. ACS Omega. 2022;7(25):21385–21396. doi: 10.1021/acsomega.1c07095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alakwaa F.M. RepurposinG Didanosine as A Potential Treatment for COVID-19 using single-Cell RNA sequencing data. mSystems. 2020;5(2):e00297–00220. doi: 10.1128/mSystems.00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dey S.K., Saini M., Dhembla C., Bhatt S., Rajesh A.S., et al. Suramin, penciclovir, and anidulafungin exhibit potential in the treatment of COVID-19 via binding to nsp12 of SARS-CoV-2. J Biomol Struct Dyn. 2022;40(24):14067–14083. doi: 10.1080/07391102.2021.2000498. [DOI] [PubMed] [Google Scholar]
- 65.Abdalla M., Rabie A.M. Dual computational and biological assessment of some promising nucleoside analogs against the COVID-19-Omicron variant. Comput Biol Chem. 2022;104 doi: 10.1016/j.compbiolchem.2022.107768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bennett R.P., Postnikova E.N., Eaton B.P., Cai Y., Yu S., et al. Sangivamycin is highly effective against SARS-CoV-2 in vitro and has favorable drug properties. JCI Insight. 2022;7(1) doi: 10.1172/jci.insight.153165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kabinger F., Stiller C., Schmitzoval J., Dienemann C., Kokic G., et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat Struct Mol Biol. 2021;28(9):740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J Biol Chem. 2021;297(1) doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stegmann K.M., Dickmanns A., Heinen N., Blaurock C., Karrasch T., et al. Inhibitors of dihydroorotate dehydrogenase cooperate with molnupiravir and N4-hydroxycytidine to suppress SARS-CoV-2 replication. iScience. 2022;25(5) doi: 10.1016/j.isci.2022.104293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X., Sacramento C.Q., Jockusch S., Chaves O.A., Tao C., et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun Biol. 2022;5(1):154. doi: 10.1038/s42003-022-03101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenke K., Hansen F., Schwarz B., Feldmann F., Haddock E., et al. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat Commun. 2021;12(1):2295. doi: 10.1038/s41467-021-22580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenke K., Okumura A., Lewis M.C., Feldmann F., Meade-White K., et al. Molnupiravir inhibits SARS-CoV-2 variants including Omicron in the hamster model. JCI Insight. 2022;7(13) doi: 10.1172/jci.insight.160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon A., Selisko B., Le N.-T.-T., Huchting J., Touret F., et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat Commun. 2020;11(1):4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stevens L.J., Pruijssers A.J., Lee H.W., Gordon C.J., Tchesnokov E.P., et al. Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci Transl Med. 2022;14(656):eabo0718. doi: 10.1126/scitranslmed.abo0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Romero M.E., Long C.H., La Rocco D., Keerthi A.M., Xu D.J., et al. Probing remdesivir nucleotide analogue insertion to SARS-CoV-2 RNA dependent RNA polymerase in viral replication. Mol Syst Des Eng. 2021;6(11):888–902. doi: 10.1039/D1ME00088H. [DOI] [Google Scholar]
- 77.Zhang L., Zhang D., Wang X., Yuan C., Li Y., et al. 1'-Ribose cyano substitution allows Remdesivir to effectively inhibit nucleotide addition and proofreading during SARS-CoV-2 viral RNA replication. Phys Chem Chem Phys. 2021;23(10):5852–5863. doi: 10.1039/d0cp05948j. [DOI] [PubMed] [Google Scholar]
- 78.Wang J., Shi Y., Reiss K., Maschietto F., Lolis E., et al. Structural insights into binding of remdesivir triphosphate within the replication-transcription complex of SARS-CoV-2. Biochemistry. 2022;61(18):1966–1973. doi: 10.1021/acs.biochem.2c00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Naseem-Khan S., Berger M.B., Leddin E.M., Maghsoud Y., Cisneros G.A. Impact of remdesivir incorporation along the primer strand on SARS-CoV-2 RNA-Dependent RNA polymerase. J Chem Inf Model. 2022;62(10):2456–2465. doi: 10.1021/acs.jcim.2c00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi Y., Wang J., Batista V.S. Translocation pause of remdesivir-containing primer/template RNA duplex within SARS-CoV-2's RNA polymerase complexes. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.999291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aranda J., Wieczor M., Terrazas M., Brun-Heath I., Orozco M. Mechanism of reaction of RNA-dependent RNA polymerase from SARS-CoV-2. Chem Catal. 2022;2(5):1084–1099. doi: 10.1016/j.checat.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maag D., Castro C., Hong Z., Cameron C.E. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J Biol Chem. 2001;276(49):46094–46098. doi: 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
- 83.Luo X., Wang X., Yao Y., Gao X., Zhang L. Unveiling the "Template-Dependent" inhibition on the viral transcription of SARS-CoV-2. J Phys Chem Lett. 2022;13(31):7197–7205. doi: 10.1021/acs.jpclett.2c01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo X., Xu T., Gao X., Zhang L. Alternative role of Motif B in template dependent polymerase inhibition. Chin J Chem Phys. 2022;35(3):407–412. doi: 10.1063/1674-0068/cjcp2203053. [DOI] [Google Scholar]
- 85.Zhou S., Hill C.S., Sarkar S., Tse L.V., Woodburn B.M.D., et al. beta-D-N-4-hydroxycytidine Inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J Infect Dis. 2021;224(3):415–419. doi: 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baranovich T., Wong S.-S., Armstrong J., Marjuki H., Webby R.J., et al. T-705 (Favipiravir) induces lethal mutagenesis in influenza A H1N1 viruses in vitro. J Virol. 2013;87(7):3741–3751. doi: 10.1128/JVI.02346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin Z., Smith L.K., Rajwanshi V.K., Kim B., Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) ribofuranosyl 5 '-Triphosphate towards influenza a virus polymerase. PLoS One. 2013;8(7) doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arias A., Thorne L., Goodfellow I. Favipiravir elicits antiviral mutagenesis during virus replication in vivo. Elife. 2014;3 doi: 10.7554/eLife.03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vanderlinden E., Vrancken B., Van Houdt J., Rajwanshi V.K., Gillemot S., et al. Distinct effects of T-705 (Favipiravir) and ribavirin on influenza virus replication and viral RNA synthesis. Antimicrob Agents Chemother. 2016;60(11):6679–6691. doi: 10.1128/AAC.01156-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Driouich J.-S., Cochin M., Lingas G., Moureau G., Touret F., et al. Favipiravir and severe acute respiratory syndrome coronavirus 2 in hamster model. Nat Commun. 2020;12:1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yin W., Luan X., Li Z., Xie Y., Zhou Z., et al. Structural basis for repurpose and design of nucleoside drugs for treating COVID-19. bioRxiv. 2020 doi: 10.1101/2020.1111.1101.363812. https://doi.org/10.1101/2020.11.01.363812. [DOI] [Google Scholar]
- 92.Biteau N.G., Amichai S.A., Azadi N., De R., Downs-Bowen J., et al. Synthesis of 4'-substituted carbocyclic uracil derivatives and their monophosphate prodrugs as potential antiviral agents. Viruses. 2023;15(2):544. doi: 10.3390/v15020544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sourimant J., Lieber C.M., Aggarwal M., Cox R.M., Wolf J.D., et al. 4 '-Fluorouridine is an oral antiviral that blocks respiratory syncytial virus and SARS-CoV-2 replication. Science. 2022;375(6577):161–167. doi: 10.1126/science.abj5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Salpini R., Alkhatib M., Costa G., Piermatteo L., Ambrosio F.A., et al. Key genetic elements, single and in clusters, underlying geographically dependent SARS-CoV-2 genetic adaptation and their impact on binding affinity for drugs and immune control. J Antimicrob Chemother. 2021;76(2):396–412. doi: 10.1093/jac/dkaa444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cho J., Shin Y., Yang J.-S., Kim J.W., Kim K.-C., et al. Evaluation of antiviral drugs against newly emerged SARS-CoV-2 Omicron subvariants. Antivir Res. 2023:214. doi: 10.1016/j.antiviral.2023.105609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gandhi S., Klein J., Robertson A., Pena-Hernandez M.A., Lin M.J., et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. medRxiv Prepr Serv Health Sci. 2021 doi: 10.1101/2021.11.08.21266069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pourfarjam Y., Ma Z., Kim I.-K. ATP enhances the error-prone ribonucleotide incorporation by the SARS-CoV-2 RNA polymerase. Biochem Biophys Res Commun. 2022;625:53–59. doi: 10.1016/j.bbrc.2022.07.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rabie A.M., Abdalla M. A series of adenosine analogs as the first efficacious anti-SARS-CoV-2 drugs against the B.1.1.529.4 lineage: a preclinical repurposing research study. ChemistrySelect. 2022;7(46) doi: 10.1002/slct.202201912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moeller N.H., Passow K.T., Harki D.A., Aihara H. SARS-CoV-2 nsp14 exoribonuclease removes the natural antiviral 3'-Deoxy-3',4'-didehydro-cytidine Nucleotide from RNA. Viruses. 2022;14(8):1790. doi: 10.3390/v14081790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chinthapatla R., Sotoudegan M., Srivastava P., Anderson T.K., Moustafa I.M., et al. Interfering with nucleotide excision by the coronavirus 3'-to-5' exoribonuclease. Nucleic Acids Res. 2023;51(1):315–336. doi: 10.1093/nar/gkac1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu C., Shi W., Becker S.T., Schatz D.G., Liu B., et al. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science. 2021;373(6559):1142–1146. doi: 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117(1):17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., et al. Coronavirus RNA Proofreading: molecular basis and therapeutic targeting. Mol Cell. 2020;79(5):710–727. doi: 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ogando N.S., Zevenhoven-Dobbe J.C., van der Meer Y., Bredenbeek P.J., Posthuma C.C., et al. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J Virol. 2020;94(23) doi: 10.1128/JVI.01246-20. e01246-01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Baddock H.T., Brolih S., Yosaatmadja Y., Ratnaweera M., Bielinski M., et al. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022;50(3):1484–1500. doi: 10.1093/nar/gkab1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yin X., Popa H., Stapon A., Bouda E., Garcia-Diaz M. Fidelity of ribonucleotide incorporation by the SARS-CoV-2 replication complex. J Mol Biol. 2023;435(5) doi: 10.1016/j.jmb.2023.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Malone B., Chen J., Wang Q., Llewellyn E., Choi Y.J., et al. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc Natl Acad Sci. 2021;118(19) doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen J., Wang Q., Malone B., Llewellyn E., Pechersky Y., et al. Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex. Nat Struct Mol Biol. 2022;29(3):250–260. doi: 10.1038/s41594-022-00734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang X., Tao C., Morozova I., Kalachikov S., Li X., et al. Identifying structural features of nucleotide analogues to overcome SARS-CoV-2 exonuclease activity. Viruses. 2022;14(7):1413. doi: 10.3390/v14071413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moeller N.H., Shi K., Demir Ö., Belica C., Banerjee S., et al. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc Natl Acad Sci. 2022;119(9) doi: 10.1073/pnas.2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jockusch S., Tao C., Li X., Chien M., Kumar S., et al. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci Rep. 2020;10(1):16577. doi: 10.1038/s41598-020-73641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yan L., Yang Y., Li M., Zhang Y., Zheng L., et al. Coupling of N7-methyltransferase and 3 '-5 ' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell. 2021;184(13):3474–3485. doi: 10.1016/j.cell.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khater S., Kumar P., Dasgupta N., Das G., Ray S., et al. Combining SARS-CoV-2 proofreading exonuclease and RNA-Dependent RNA polymerase inhibitors as a strategy to combat COVID-19: a high-throughput in silico screening. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.647693. [DOI] [PMC free article] [PubMed] [Google Scholar]