Abstract
Background
Glioblastoma (Gb) is the most common primary malignant tumor of brain with poor prognosis. Immune cells are the main factors affecting the prognosis of Gb, tumor-associated macrophages (TAMs) are the predominant infiltrating immune cell population in the immune microenvironment of Gb. Analyzing the relationship between magnetic resonance imaging (MRI) features and TAMs of Gb, and using imaging features to characterize the infiltration level of TAMs in tumor tissue may provide indicators for clinical decision-making and prognosis evaluation of Gb.
Methods
Data from 140 in patients with isocitrate dehydrogenase (IDH) wild-type Gb diagnosed via histopathology and molecular diagnosis in the Second Hospital of Lanzhou University from January 2018 to April 2022 were collected in this retrospective, cross-sectional study. MRI images were reviewed for lesion location, cyst, necrosis, hemorrhage, contrast-enhanced T1-weighted MRI signal intensity, average apparent diffusion coefficient (ADCmean), and minimum apparent diffusion coefficient (ADCmin). Immunohistochemical staining with anti-CD163 and anti-CD68 antibodies was employed for macrophage detection. The positive cell percentage was estimated in 9 microscopic fields at 400× magnification per whole-slide image with ImageJ software (National Institutes of Health). Additionally, the relationship between MRI features, molecular, states and the positive CD68 and CD163 expression was analyzed.
Results
Our study discovered that the mean or median values of CD68+ and CD163+ TAMs were 7.39% and 14.98%, respectively. There was an obvious correlation between CD163+ TAMs and CD68+ TAMs (r=0.497; P=0.000). CD68+ and CD163+ macrophage infiltration correlated with age at diagnosis in patients with Gb (CD68+: r=0.230, P=0.006; CD163+: r=0.172, P=0.042). The levels of Gb TAM infiltration in different tumor locations varied, with the temporal lobe having the highest CD163+ macrophage and CD68+ macrophage infiltration (18.58% and 9.46%, respectively). CD163+ macrophage infiltration was positively correlated with ADCmean (r=0.208; P=0.014). The infiltration of CD68+ macrophages differed significantly between groups with varying degrees of tumor enhancement (H =4.228; P=0.017). There was a significant difference in CD68+ TAMs and CD163+ TAMs between the wild-type and mutant-type telomerase reverse transcriptase (TERT) types (P=0.004 and P=0.031, respectively).
Conclusions
Age, location of the tumor, degree of tumor enhancement, ADC value, and TERT mutation status were associated with macrophage infiltration. These findings may serve as an effective tool for characterizing the tumor microenvironment in patients with Gb.
Keywords: Glioblastoma (Gb), tumor-associated macrophage (TAM), magnetic resonance imaging (MRI), tumor microenvironment
Introduction
In the 2021 World Health Organization (WHO) classification of central nervous system tumors, adult-type diffuse gliomas were classified according to isocitrate dehydrogenase (IDH)1/2 mutation or wild type and 1p/19q codeletion, which can be simplified into 3 main types: IDH-wild type glioblastoma (Gb; WHO grade 4), IDH-mutant astrocytoma (WHO grade 2–4), and IDH-mutant and 1p/19q codeletion oligodendroglioma (WHO grade 2–3) (1). Among these, IDH-wild type GB has the worst prognosis and shortest median survival (8 months) (2). The main reason for this is that Gb has an immunosuppressive and highly heterogeneous tumor microenvironment.
Tumor-associated macrophages (TAMs) are the predominant infiltrating immune cell population in the immune microenvironment of Gb, accounting for 30–40% of tumor tissues. In specific cases, the proportion of TAMs is as high as 50% (3,4). TAMs promote tumor cell proliferation, angiogenesis, and immune escape, resulting in treatment resistance and a poor prognosis in patients with Gb (5). An experimental animal study discovered that TAMs consisted of 85% bone marrow-derived monocytes or macrophages and 15% resident microglia, which were recruited to the tumor microenvironment early in tumor formation and localized to the surrounding blood vessels, thereby promoting tumor development (6). Under different stimuli, macrophages in the tumor microenvironment can be polarized to antitumor (M1) or tumor-promoting (M2) phenotypes. However, most macrophages recruited to the tumor microenvironment become the M2 subtype (7,8). M2 phenotypic macrophages promote tumor cell proliferation, tumor invasion, migration and angiogenesis by secreting extracellular matrix components. Tissue-resident memory T cells represent a large number of subsets of tumor-infecting lymphocytes. M2 phenotypic macrophages can inhibit T cells and induce T-cell anergy, help tumor immune escape, promote immunosuppression, and enhance drug resistance (9). M2 phenotypic macrophages can be further subdivided into different functional phenotypes such as M2a, M2b, and M2c, indicating that the function of TAMs changes with the changes in the cytokines in the tumor microenvironment, reflecting the high heterogeneity of Gb and considerable TAM plasticity (10,11). M2a and M2b subtypes play similar roles in immune regulation by stimulating Th2 response and participating in immune regulation. M2c subtypes are the main initiators of tumor growth (12).
Immunotherapy strategies targeting the Gb tumor microenvironment have become the focus of Gb treatment options, and TAM-targeted therapy has great potential in glioma treatment strategies (13). In particular, the infiltration of macrophages targeting the M2 phenotype opens the door for targeted therapy based on avoiding this phenotype or blocking its function (14,15). According to a large number of clinical trials, the targeted treatment of TAMs can be mainly divided into 3 strategies: preventing TAMs from being recruited to the tumor site, promoting TAM reprogramming to the M1 phenotype, and immune checkpoint blockade (16,17). Among them, inhibition of colony-stimulating factor-1 receptor (CSF-1R) has shown the ability to consume or reprogram TAMs in clinical mouse models (18), indicating that the inhibition of CSF-1R can effectively target TAMs (19). Immune checkpoint blocking can inhibit tumor progression by regulating the polarization state of TAMs. The combination of targeted TAMs and immune checkpoint inhibitors may enhance the antitumor effect of TILs (20). In addition, therapeutic radionuclides targeting CD163 and CD206 can reduce the number of M2 phenotype TAMs or reprogram the M2 phenotype TAMs to the M1 phenotype (21).
Diagnostic tools for detecting and quantifying TAMs include gene expression analysis, immunohistochemistry, and fluorescent magnetic nanoparticle labeling (22). CD68 has been widely reported as a specific marker of total TAM infiltration in tumors (23,24). Additionally, CD163, considered to be a reliable marker of M2 phenotype macrophages, has been used to evaluate the infiltration of M2 phenotype macrophages in various tumors (25,26). Structural magnetic resonance imaging (MRI) can be used to predict the disease progression or posttreatment recurrence of high-grade glioma (27). We also speculate that noninvasive imaging biomarkers are clinically valuable for evaluating and quantifying TAMs, as TAMs can be monitored noninvasively using imaging techniques. In particular, exploring imaging methods for M2-type TAMs is critical for monitoring tumor progression and assessing treatment efficacy (28). It may provide a new indicator for predicting tumor progression and poor prognosis and has the potential to guide the selection of individualized treatment options.
Therefore, this study aimed to clarify the association between MRI features and TAM infiltration, especially M2-type TAM infiltration, to predict macrophage infiltration based on preoperative MRI features. The findings are expected to be useful in treatment decision-making and prediction prognosis. We present this article in accordance with the STROBE reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-126/rc).
Methods
This study was performed in line with the principles of the Declaration of Helsinki (as revised in 2013) and approved by the Clinical Ethics Committee of Lanzhou University Second Hospital (No. 2020A-070). Individual consent for this retrospective analysis was waived.
Study population
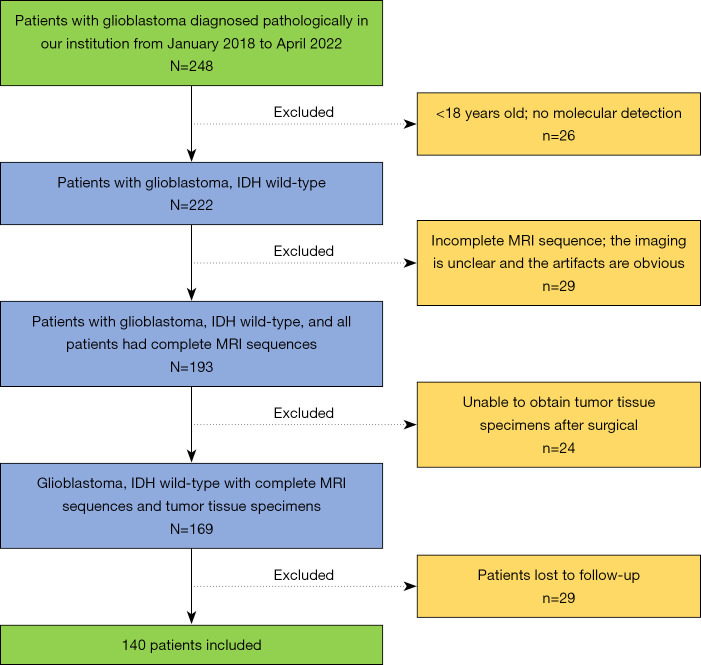
Data from 140 in patients with IDH-wildtype GB diagnosed via histopathology and molecular diagnosis at the Second Hospital of Lanzhou University from January 2018 to April 2022 were collected for this retrospective, cross-sectional study. The inclusion criteria were as follows: (I) Gb histopathological type, (II) IDH wild-type according to molecular detection, (III) complete clinicopathological information, (IV) MRI multisequence scanning [T1 weighted images (T1WI), T2 weighted images (T2WI), contrast enhancement-T1WI (CE-T1WI), diffusion weighted imaging (DWI)] completed before operation; (V) treatment via surgical resection. Meanwhile, the exclusion criteria were as follows: (I) patients younger than 18 years old; (II) IDH mutant type according to molecular detection; (III) preoperative MRI multisequence scanning images missing, unclear, or with obvious artifacts; (IV) inability to evaluate hematoxylin and eosin (HE) staining and immunohistochemical sections of tumor tissue specimens postoperatively; and (V) patients lost in survival follow-up. The inclusion and exclusion processes are shown in Figure 1. Ultimately, this study included 140 participants, comprising 82 males and 58 females, with an average age of 52.84 years.
Figure 1.
Flowchart of the patient selection process. IDH, isocitrate dehydrogenase; MRI, magnetic resonance imaging.
Molecular information included p53 (positive/negative), O6-methyl-guanine DNA methyltransferase (MGMT) methylation (positive/negative), and TERT (mutant/wild type). Overall survival (OS) was defined as the time from the date of surgery to the date of death or the last follow-up. Follow-up was performed via consultation of patients’ revisit records, assessment of MR images, telephone follow-up, and reference to the hospital information system and Picture Archiving and Communication System. The deadline for follow-up was October 31, 2022. The follow-up period of OS for surviving patients was at least 6 months after surgery and 49 months at most. Patients who were lost follow-up were excluded from the study.
MRI acquisition
MR images were obtained using the Siemens Verio 3.0 T (Siemens AG, Henkestrasse 127, D-91052, Erlangen, Germany) and Philips Achieva 3.0 T (Philips Medical Systems Nederland B.V., Veenpluis 4-6, 5684 PC, Amsterdam, The Netherlands) MR scanners. The scanning parameters for T1WI (gradient echo sequence) were repetition time (TR) 2,000 ms, echo time (TE) 9 ms, layer thickness 6.0 mm, layer spacing 1.5 mm, field of view (FOV) 230 mm × 230 mm, and matrix 320×340; the parameters for T2WI were TR 4,000 ms, TE 95 ms, FOV 230 mm × 230 mm, slice thickness 6.0 mm, interslice gap 1.5 mm, and matrix 384×284. Diffusion-weighted imaging used the echo planar-imaging sequence plus frequency selective fat-suppression technology under the following parameters: TR 3,300 ms, TE 64 ms, layer thickness 6 mm, layer spacing 1 mm, FOV 220 mm × 220 mm, and matrix 256×160. Diffusion gradients (b value =0, 1,000 s/mm2) were applied in the directions of the x-, y-, and z-axes. Enhanced scanning was performed using gadolinium-DTPA (Bayer Schering Pharma AG, Berlin, Germany) as the contrast agent, which was intravenously administered via a bolus injection of 0.1 mmol/kg at a flow rate of 3.0 mL/s.
MR image processing and analysis
All MRI studies were reviewed retrospectively and in consensus by 2 experienced radiologists (with 6 years of experience). MR images were reviewed for (I) lesion location (frontal lobe, temporal lobe, parietal lobe, occipital lobe, and other positions); (II) cyst, necrosis, or hemorrhage (yes or no); and (III) enhanced T1WI signal intensity (none, minimal/mild, marked/avid). According to the Visually Accessible Rembrandt Images (VASARI) MRI visual feature guide, the qualitative degree of contrast enhancement was defined as all or portions of the tumor with a significantly higher signal on the postcontrast T1WI than on precontrast T1W images. Mild or minimal enhancement indicated was defined as a barely discernable degree of enhancement relative to precontrast images, whereas marked or avid enhancement was defined as obvious tissue enhancement. The exact description can be found on the National Cancer Institute’s Cancer Imaging Archive (https://wiki.cancerimagingarchive.Net/display/Public/VASARI+Research+Project). (IV) Average apparent diffusion coefficient (ADCmean) and minimum ADC (ADCmin) were also reviewed on the MR images: the solid part of the tumor on the ADC map of multiple levels was selected and manually placed in the region of interest (ROI) with an area of 20–30 mm2. Subsequently, the average of the ADCmean and ADCmin values was calculated.
TAM quantitative analysis
One pathologist (with 7 years of experience in pathologic examination) retrospectively reviewed all tissue specimens (140 surgical specimens). All tumor paraffin-embedded specimens were resliced. Immunohistochemical staining was conducted using anti-CD163 antibody (1:1,200, Rosemont, IL, USA) and anti-CD68 antibody (1:200, MXB Biotechnologies, Fuzhou, China) for macrophage detection. In hematoxylin staining, the nucleus stained blue, and positive expression was brown. The TG TissueFAXS Plus digital pathological analysis system (TissueGnostics, Wien, Austria) and 3DHIESTECH digital slice scanning system (3DHISTECH, Budapest, Hungary) were used to scan CD68 and CD163 with whole-slide images (WSI). The WSI images were analyzed with TissueFAXS Viewer and CaseViewer software, and 9 ROIs were randomly segmented (Figure 2). Microsections were quantitatively evaluated using ImageJ software (version ImageJ2, National Institutes of Health, Bethesda, MD, USA). The positive cells were analyzed via the color threshold, and the respective coverage areas of CD68 and CD163 positive expression relative to the whole ROI image were calculated and are expressed as percentages (%). The final positive expression of CD68 and CD163 was considered to be the average of the expression in the 9 ROIs.
Figure 2.
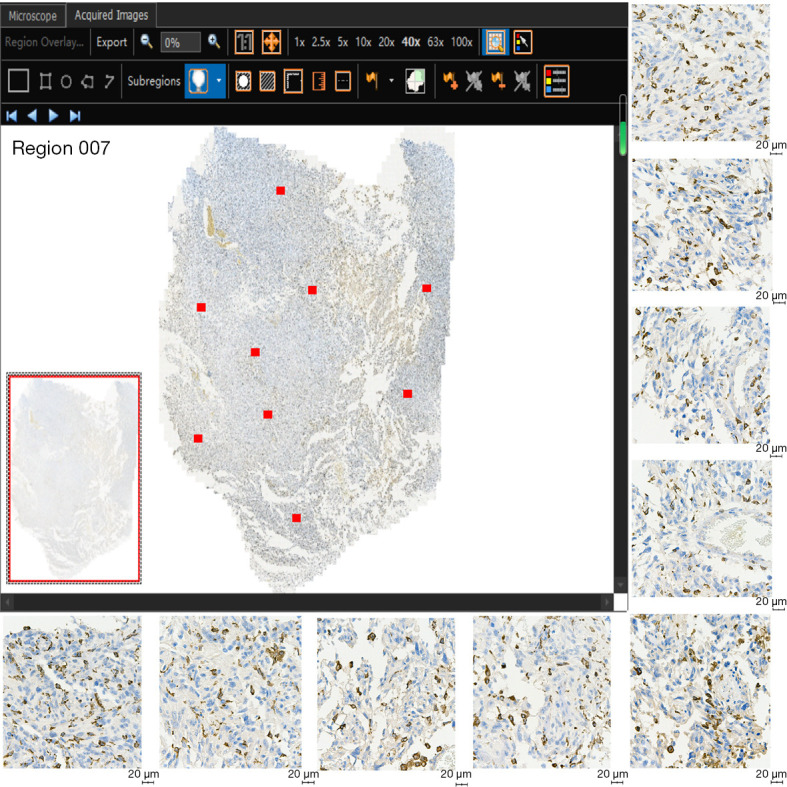
CD163 immunohistochemical staining of WSI was checked using TissueFAXS Viewer software, and 9 CD163 regions of interest were randomly segmented on the immunohistochemical images. Positive expression is indicated in brown (scale bar =20 µm). WSI, whole-slide images.
Statistical analysis
All statistical analyses were performed with SPSS 26.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism software (GraphPad Software, San Diego, CA, USA). All continuous variables were initially assessed for normality using the Kolmogorov-Smirnov test. Measurement and count data are expressed as the mean ± standard deviation (mean ± SD) and percentage. Skewed distributed data are represented as the median (first quartile, third quartile) [M (Q1, Q3)]. Spearman correlation was used for the correlation analysis between MRI continuous variables and TAMs. The r value was used to judge the strength of the correlation as follows: 0.00–0.19, very weak; 0.20–0.39, weak; 0.40–0.59, moderate; 0.60–0.79, strong; and 0.80–1.0, very strong (29). Groups were compared using a t-test. Differences in variables between groups were evaluated with the Mann-Whitney test and Kruskal-Wallis test for nonnormally distributed data. Statistical significance was set at as a 2-sided P value ≤0.05.
Results
Patient demographics
The mean age of the 140 patients included in the study was 52 years, with 82 male and 58 female patients included. The mean values of CD68+ and CD163+ macrophage infiltration were 7.39% and 14.98%, respectively. In the correlation analysis, there was an obvious correlation between CD163+ TAMs and CD68+ TAMs (r=0.497; P=0.000). Age was correlated with the infiltration of CD163+ (r=0.172, P=0.042) and CD68+ (r=0.230, P=0.006) macrophages, and the cell proliferation index Ki-67 was negatively correlated with CD163+ TAMs. Tables 1,2 present the correlation between Gb macrophage infiltration and the clinical and MRI features.
Table 1. Correlation between Gb CD68+ macrophage infiltration and clinical and MRI features.
| Characteristic | Total (N=140) | CD68 (%) | r | P |
|---|---|---|---|---|
| Gender | 0.718 | 0.474 | ||
| Male | 82 (58.57) | 6.84 (2.83, 14.72) | ||
| Female | 58 (41.43) | 9.32 (4.42, 14.22) | ||
| Age (year) | 52.84±11.44 | 7.39 (3.21, 14.33) | 0.230 | 0.006* |
| Ki-67 (%) | 40.00 (30.00, 50.00) | 7.39 (3.21, 14.33) | −0.137 | 0.106 |
| Lateralization of tumor | 0.136 | 0.873 | ||
| Left | 68 (48.57) | 7.17 (2.95, 14.55) | ||
| Right | 59 (42.14) | 8.85 (4.35, 14.38) | ||
| Bilateral | 13 (9.29) | 6.73 (3.93, 12.54) | ||
| Major location of the tumor | 2.559 | 0.041* | ||
| Frontal lobe | 46 (32.86) | 6.46 (2.42, 12.85) | ||
| Temporal lobe | 61 (43.57) | 9.46 (5.68, 17.17) | ||
| Parietal lobe | 10 (7.14) | 4.86 (2.70, 9.21) | ||
| Occipital lobe | 15 (10.71) | 6.54 (3.69, 14.17) | ||
| Other | 8 (5.71) | 4.42 (2.13, 9.26) | ||
| Necrosis/cystic | 0.122 | 0.903 | ||
| Yes | 125 (89.29) | 7.39 (3.41, 17.26) | ||
| No | 15 (10.71) | 11.30 (2.70, 17.26) | ||
| Hemorrhage | −1.502 | 0.135 | ||
| Yes | 38 (27.14) | 10.20 (4.46, 16.35) | ||
| No | 102 (72.86) | 6.78 (3.08, 13.87) | ||
| Degree of enhancement | 4.228 | 0.017* | ||
| None | 8 (5.71) | 2.06 (0.55, 6.46) | ||
| Mild/minimal | 24 (17.14) | 4.72 (1.08, 10.53) | ||
| Marked/avid | 108 (77.14) | 9.25 (4.66, 14.97) | ||
| ADCmean (SD) (×10−6 mm2/s) | 886.63±107.60 | 7.39 (3.21, 14.33) | 0.070 | 0.414 |
| ADCmin (SD) (×10−6 mm2/s) | 788.09±116.62 | 7.39 (3.21, 14.33) | 0.081 | 0.344 |
Data are shown as n (%) or mean ± SD or median (Q1, Q3). *, P<0.05, P values are statistically significant. Gb, glioblastoma; MRI, magnetic resonance imaging; ADC, apparent diffusion coefficient; SD, standard deviation.
Table 2. Correlation between Gb CD163+ macrophage infiltration and clinical and MRI features.
| Characteristic | Total (N=140) | CD163 (%) | r | P |
|---|---|---|---|---|
| Gender | −0.556 | 0.579 | ||
| Male | 82 (58.57) | 15.32±8.95 | ||
| Female | 58 (41.43) | 14.50±8.23 | ||
| Age (year) | 52.84±11.44 | 14.98±8.64 | 0.172 | 0.042* |
| Ki-67 (%) | 40.00 (30.00, 50.00) | 14.98±8.64 | −0.177 | 0.036* |
| Lateralization of tumor | 1.670 | 0.192 | ||
| Left | 68 (48.57) | 11.57 (6.69, 21.80) | ||
| Right | 59 (42.14) | 17.47 (10.30, 22.47) | ||
| Bilateral | 13 (9.29) | 11.32 (4.93, 19.88) | ||
| Major location of the tumor | 4.079 | 0.004* | ||
| Frontal lobe | 46 (32.86) | 10.84 (6.32, 20.88) | ||
| Temporal lobe | 61 (43.57) | 18.58 (11.54, 23.51) | ||
| Parietal lobe | 10 (7.14) | 11.45 (7.84, 25.23) | ||
| Occipital lobe | 15 (10.71) | 12.58 (10.81, 18.24) | ||
| Other | 8 (5.71) | 5.14 (2.24, 9.74) | ||
| Necrosis/cystic | −1.419 | 0.158 | ||
| Yes | 125 (89.29) | 15.17 (9.37, 22.35) | ||
| No | 15 (10.71) | 5.98 (2.75, 19.67) | ||
| Hemorrhage | −1.035 | 0.303 | ||
| Yes | 38 (27.14) | 17.08 (10.44, 2301) | ||
| No | 102 (72.86) | 13.17 (7.19, 21.34) | ||
| Degree of enhancement | 1.557 | 0.214 | ||
| None | 8 (5.71) | 10.71 (3.72, 15.95) | ||
| Mild/minimal | 24 (17.14) | 13.45 (6.50, 20.02) | ||
| Marked/avid | 108 (77.14) | 15.49 (8.88, 22.79) | ||
| ADCmean (SD) (×10−6 mm2/s) | 886.63±107.60 | 14.98±8.64 | 0.208 | 0.014* |
| ADCmin (SD) (×10−6 mm2/s) | 788.09±116.62 | 14.98±8.64 | 0.134 | 0.115 |
Data are shown as n (%) or mean ± SD or median (Q1, Q3). *, P<0.05, P values are statistically significant. Gb, glioblastoma; MRI, magnetic resonance imaging; ADC, apparent diffusion coefficient; SD, standard deviation.
Relationship between TAM infiltration and Gb survival
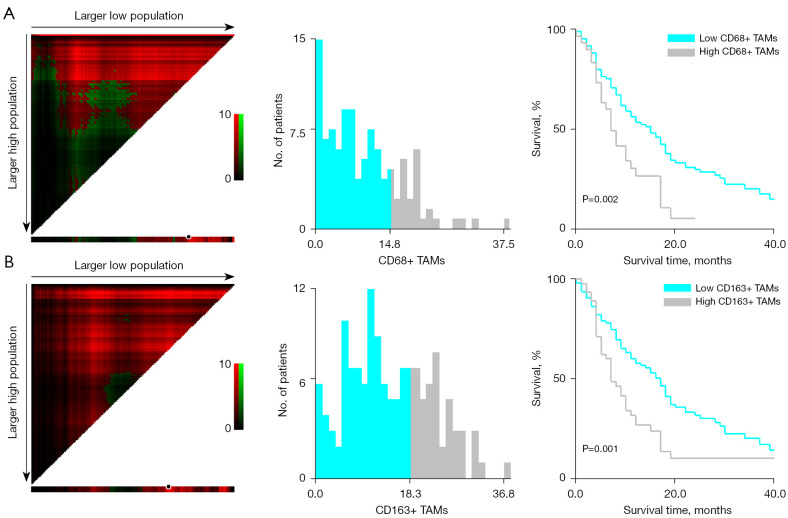
The median OS of the patient population in our study was 11 months. According to the outcome and OS of the patients at the end of follow-up, survival curves were drawn using the Kaplan-Meier method. CD68+ TAMs and CD163+ TAMs were classified into high and low expression with the critical values of 14.80% and 18.30%, respectively. The survival rate of patients with high expression of CD68+ TAMS and CD163+ TAMS decreased rapidly, the mortality was fast, and the survival time was relatively short; moreover, the high expression of CD68+ TAMs and CD163+ TAMs was negatively correlated with OS (P=0.002 and P=0.001, respectively; Figure 3).
Figure 3.
X-Tile software was used to determine correlations between macrophage expression and survival time. The image color represents the correlation intensity of each partition from low (dark/black) to high (bright green/red). Green represents a direct correlation, and red represents a reverse correlation between expression level and characteristic survival rate. (A) The best cutoff value of CD68+ TAMs was 14.80%. The relationship between high and low CD68+ TAM expression and survival time is shown (P=0.002). (B) The best cutoff value of CD163+ TAMs was 18.30%. The relationship between high and low CD163+ TAM expression and survival time is shown (P=0.001). TAMs, tumor-associated macrophages.
Macrophage infiltration was associated with MRI characteristics of Gb
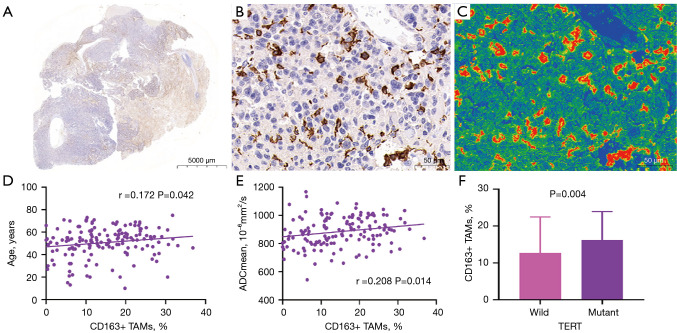
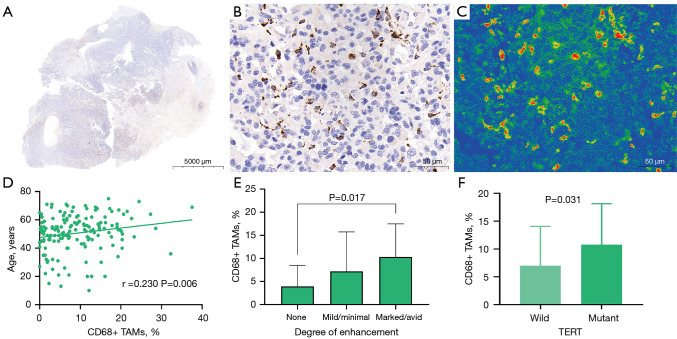
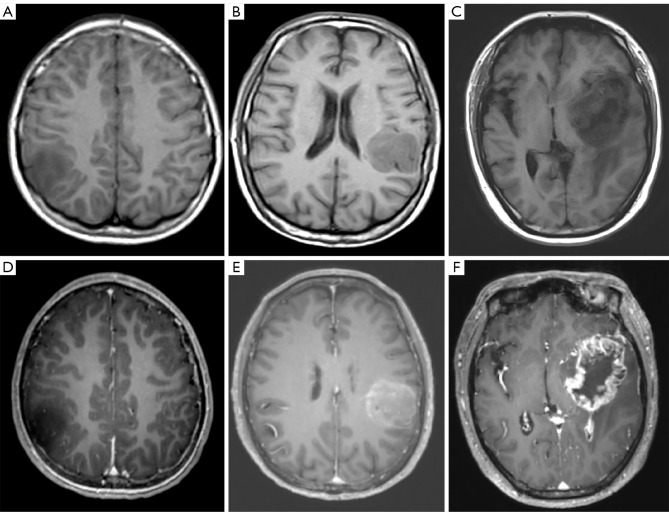
We divided the main locations of the tumor into frontal lobe, temporal lobe, parietal lobe, occipital lobe, and other parts and discovered a difference in macrophage infiltration between different tumor locations. Tumors in the temporal lobe had the highest CD163+ macrophage and CD68+ macrophage infiltration (18.58% and 9.46%, respectively). ADCmean was significantly correlated with CD163+ macrophage infiltration (r=0.208; P=0.014); the CD163 immunohistochemical images of Gb, correlation scatter plot, and boxplots are shown in Figure 4. The infiltration of CD68+ macrophages differed significantly between groups with different degrees of tumor enhancement (H =4.228; P=0.017). The CD68 immunohistochemical images of Gb, the scatter plot of the correlation between age and CD68+ TAMs, and the boxplots of CD68+ TAMs according to degree of enhancement and TERT status are shown in Figure 5. Figure 6 presents illustrative examples of MR images enhancement.
Figure 4.
CD163 immunohistochemical staining and correlation between CD163+ expression and clinical imaging features. (A) CD163 immunohistochemical whole-slide images of Gb tissue samples. (B) The 400× image of the region of interest of CD163 (scale bar =50 µm). (C) The 400× magnification pseudo-color image of the region of interest of CD163 (scale bar =50 µm). (D) Scatter plot showing the association of age with CD163+ TAMs. (E) Scatter plot showing the association of ADCmean with CD163+ TAMs. (F) Boxplot showing the differences in CD163+ TAMs between the mutant-type and wild-type TERT groups. TAMs, tumor-associated macrophages; ADCmean, average apparent diffusion coefficient; TERT, telomerase reverse transcriptase.
Figure 5.
CD68 immunohistochemical staining and correlation between CD163+ expression and clinical imaging features. (A) CD68 immunohistochemical whole-slide images of Gb tissue samples. (B) The 400× magnification image of the region of interest of CD68 (scale bar =50 µm). (C) The 400× magnification pseudo-color image of the region of interest of CD68 (scale bar =50 µm). (D) Scatter plot showing the association of age with CD68+ TAMs. (E) Boxplot showing the differences in CD68+ TAMs between groups with different degrees of tumor enhancement. (F) Boxplot showing the differences in CD68+ TAMs between the mutant-type and wild-type TERT groups. Gb, glioblastoma; TAMs, tumor-associated macrophages; ADCmean, average apparent diffusion coefficient; TERT, telomerase reverse transcriptase.
Figure 6.
Examples of magnetic resonance images with different degrees of enhancement of Gb. (A-C) T1WI. (D-F) Contrast-enhanced T1WI. (A,D) No enhancement. (B,E) Mild or minimal enhancement. (C,F) Marked or avid enhancement. Gb, glioblastoma; T1WI, T1-weighted imaging.
Correlation between macrophage infiltration and molecular status of Gb
There were significant differences in CD68+ TAMs and CD163+ TAMs between wild-type and mutant TERT (P=0.004 and P=0.031, respectively). The median proportions of CD163+ and CD68+ macrophage infiltration in patients with TERT-mutant Gb were 15.75% and 10.89%, respectively. The median proportions of CD163+ and CD68+ macrophage infiltration in patients with wild-type TERT were 11.24% and 5.21%, respectively. Differences in macrophage infiltration between groups are shown in Table 3.
Table 3. Macrophage infiltration associations with molecular features of Gb.
| Characteristic | Total (N=140) | CD68 (%) | CD163 (%) | |||||
|---|---|---|---|---|---|---|---|---|
| Value | r | P | Value | r | P | |||
| P53 | −2.209 | 0.029* | −1.263 | 0.209 | ||||
| Positive | 84 (60.00) | 9.44 (4.69, 14.74) | 15.73±9.01 | |||||
| Negative | 56 (40.00) | 5.82 (1.67, 11.92) | 13.85±7.99 | |||||
| MGMT methylation | −0.974 | 0.332 | −1.260 | 0.210 | ||||
| Positive | 70 (50.00) | 10.03 (3.17, 14.63) | 15.90±8.61 | |||||
| Negative | 70 (50.00) | 7.04 (3.21, 13.14) | 14.06±8.62 | |||||
| TERT | −2.969 | 0.004* | −2.201 | 0.031* | ||||
| Mutant | 90 (64.29) | 10.89 (4.70, 14.71) | 15.75 (10.62, 22.47) | |||||
| Wild | 50 (35.71) | 5.21 (1.42, 9.39) | 11.24 (4.38, 20.99) | |||||
Data are shown as n (%) or mean ± SD or median (Q1, Q3). *, P<0.05, P values are statistically significant. Gb, glioblastoma; MGMT, O6-methyl-guanine DNA methyl-transferase; TERT, telomerase reverse transcriptase.
Discussion
This study investigated the association of Gb MRI features with TAM infiltration in the tumor microenvironment and attempted to predict Gb macrophage infiltration using MRI features. TAMs and M2-TAMs were labelled as CD68 and CD163, respectively. The results revealed that age, tumor enhancement degree, major location of the tumor, ADC value, and TERT status were correlated with TAM infiltration.
Immunohistochemistry was used to detect and visualize tissue proteins for patient stratification (30). In this study, to quantify macrophages in the tumor microenvironment, we used immunohistochemical panmacrophage marker CD68 and M2 macrophage marker CD163 to analyze TAMs and M2-TAMs, respectively (31). CD163 is a membrane protein and a well-known marker for identifying monocytes or macrophages (32,33). In Gb, CD163+ TAMs are a promising immunotherapy target (34). In our exploratory study, we selected the 9 microscopic fields to ensure representative coverage of the overall tumor macrophage quantification; the median value of CD163-positive cells in Gb was approximately 14.98%, and the median value of CD68+ cells was 7.39%. In the correlation analysis, there was an obvious correlation between CD163+ TAMs and CD68+ TAMs (r=0.497; P=0.000).
In the survival analysis, CD163+ TAMs and CD68+ TAMs were classified into high and low expression, and their critical values were 18.30% and 14.80%, respectively. Significant differences in TAM expression were observed on the survival curve (CD163+: P=0.001; CD68+ TAMs: P=0.002). The high expression of TAMs indicated a short survival time in patients with Gb. In the study of Idoate Gastearena et al. (35), the median value of CD163+ TAMs was 10.48%. Based on this cutoff value, there was a negative correlation between CD163+ TAM infiltration and OS. Another study based on a large samples from the Chinese Glioma Genome Atlas (CGGA) and The Cancer Genome Atlas (TCGA) datasets confirmed that patients with Gb with higher expression of CD68+ TAMs had a significantly shorter survival time and that CD68-targeted therapy could improve the prognosis of patients with Gbs (23). Other TAM markers, such as high expression of CD204, are associated with poor prognosis, and these markers increase as the degree of malignancy of gliomas increases and contribute to the formation of the precancerous microenvironment (33). The expression of allograft inflammatory factor 1 (IBA-1) is related to a longer survival time in patients with glioma (36,37). CD206 is one of the 3 targets of positron emission tomography tracers for the specific expression of M2 phenotypic macrophages (CD206, CD163, and arginase) (38). Its expression level is significantly increased in grade I–IV gliomas. Moreover, progression-free survival and OS in patients with high expression of CD206 were found to be significantly lower than those in patients with a low expression of CD206 (39). Therefore, our study and other recent studies agree that high levels of TAM infiltration can distinguish patients with Gb with poor prognosis (31,40). This further supports the necessity of evaluating the level of TAM infiltration using noninvasive imaging indicators.
Many studies have focused on multiple grades of glioma and demonstrated that the infiltration of tumor immune cells in the tumor immune microenvironment increases with the grade (41). For instance, the higher the tumor grade in brain tumors, the higher the TAM concentration, which is associated with poor prognosis (42). Our study on IDH wild-type Gb according to the latest WHO classification, showed that the expression of CD68+ and CD163+ cells was positively correlated with the age at Gb diagnosis; that is, the older the age at Gb diagnosis, the higher the level of macrophage infiltration. Additionally, more than 60% of gliomas occur in the supratentorial area, including the frontal, temporal, parietal, and occipital lobes. Studies have shown that different brain regions have different susceptibilities to Gb progression (43). Our study revealed that Gbs growing in the temporal lobe had the highest macrophage infiltration, whereas Gbs located in the frontal lobes had low macrophage infiltration; the median proportion of CD68+ TAMs in the temporal lobe was 9.46%, and that of CD163+ TAMs was 18.58%. The level of macrophage infiltration in different cerebral lobes showed a certain difference. This conclusion is in agreement with that of Schulze Heuling et al. (44), who reported that frontal glioma tumors had higher numbers of tumor cells than did the tumors in other brain lobes, indicating that the microenvironment of frontal gliomas contains relatively fewer macrophages. According to these results, we can speculate that Gb in the temporal lobe is more malignant, the prognosis may be worse, and the tumor should be removed as completely as possible during surgery. We also analyzed the TAM infiltration in tumors with different P53, MGMT, and TERT statuses. Patients with TERT-mutant Gb had higher TAMs infiltration. Therefore, patients with IDH wild-type Gb and mutant-TERT Gb might have a worse prognosis than may those with wild-type TERT Gb (45), and this conclusion is further confirmed by the highly invasive nature of TAMs.
Imaging features are macroscopic manifestations of tumor biological heterogeneity, and combining imaging and molecular features can improve prognosis prediction. We believe that it is necessary to explore biomarkers to monitor the immune properties of the tumor microenvironment based with noninvasive imaging methods. In recent years, only a few studies have used MRI to characterize the tumor microenvironment (46). One of these studies was based on MRI features, including tumor margins (regular or irregular), cystic component, radiation necrosis, peritumoral edema, multifocality, and hemorrhage, illustrating that conventional MRI features can predict TAM content in different Gb molecular subtypes (47). A recent study reported the correlation between structural MRI features and the ratio of recruited macrophages and resident microglia, with their results suggesting that the infiltration of blood-borne macrophages has an effect on some of the MRI parameters of Gb (48).
Considering that the ADC value represents the cell density, we quantitatively analyzed the ADCmean and the ADCmin values based on the ADC map and discovered that ADCmean was positively correlated with macrophage infiltration, indicating that the higher the macrophage infiltration is, the higher the ADC value. This is consistent with the notion that water diffusion is restricted at high tumor cell ratios, suggesting that the tumor immune environment affects ADC values. An earlier study showed that CD68+ TAM infiltration is closely related to neovascularization in gliomas (49). Aghighi et al. further confirmed that enhanced MRI signals were related to the presence of CD68+ TAMs and CD163+ TAMs (22). Moreover, in our study, the expression of CD68+ TAMs on contrast-enhanced T1WI varied according to the degree of enhancement (H =4. 228; P=0.017). Given the correlation between preoperative MRI features and macrophage infiltration in the tumor microenvironment, the detection of these MRI parameters may be an effective tool for characterizing the tumor microenvironment.
Other advanced technologies such as nanoparticle MRI can be used to track macrophages and assess immune responses in cancer (22). MR susceptibility imaging has potential as a noninvasive method for quantifying and phenotyping TAMs, with gradient echo relaxation (R2*) measurements correlating with both CD68 and CD86+ cell counts (50). Preoperative radiomics signatures reflect the tumor microenvironment, and related research suggests that a radiomics prediction model based on preoperative T2WI-MRI can assess tumor-infiltrating macrophages in patients with glioma (51). These results support the possibility of characterizing the heterogeneity of the Gb microenvironment based on advanced MRI techniques and radiomics methods. However, these methods require further research and confirmation.
This study had several limitations. First, our sample size was relatively small. Second, our study was based on conventional MRI features. However, conventional MRI features remain clinically available and reproducible. Finally, our results only showed the correlation between imaging features and the level of TAM infiltration. On the basis of this study, the next stage of our research will be focused on the use of MRI radiomics for predicting TAM infiltration.
Conclusions
In this preliminary study, we used CD68 and CD163 to label Gb panmacrophage infiltration and M2-type macrophage infiltration, respectively, with the aim of determining the association between preoperative conventional MRI features and TAMs. We discovered that age, location of the tumor, degree of enhancement, ADC value, and TERT status were associated with macrophage infiltration. Our findings may serve as an informative reference for characterizing the tumor microenvironment in patients with Gb and may further assist in the survival stratification and prognosis prediction of these patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82071872), the Science and Technology Program of Gansu Province (No. 21YF5FA123, 21JR11RA105), and the China International Medical Foundation (No. Z-2014-07-2101).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was performed in line with the principles of the Declaration of Helsinki (as revised in 2013) and approved by the Clinical Ethics Committee of Lanzhou University Second Hospital (No. 2020A-070). Individual consent for this retrospective analysis was waived.
Footnotes
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-126/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-126/coif). All authors report that this work was supported by the National Natural Science Foundation of China (No. 82071872), the Science and Technology Program of Gansu Province (No. 21YF5FA123, 21JR11RA105), and the China International Medical Foundation (No. Z-2014-07-2101). The authors have no other conflicts of interest to declare.
References
- 1.Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 2021;23:1231-51. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014-2018. Neuro Oncol 2021;23:iii1-iii105. 10.1093/neuonc/noab200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pires-Afonso Y, Niclou SP, Michelucci A. Revealing and Harnessing Tumour-Associated Microglia/Macrophage Heterogeneity in Glioblastoma. Int J Mol Sci 2020;21:689. 10.3390/ijms21030689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci 2016;19:20-7. 10.1038/nn.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther 2021;6:75. 10.1038/s41392-021-00484-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Feng X, Herting CJ, Garcia VA, Nie K, Pong WW, Rasmussen R, Dwivedi B, Seby S, Wolf SA, Gutmann DH, Hambardzumyan D. Cellular and Molecular Identity of Tumor-Associated Macrophages in Glioblastoma. Cancer Res 2017;77:2266-78. 10.1158/0008-5472.CAN-16-2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur J Cancer 2006;42:717-27. 10.1016/j.ejca.2006.01.003 [DOI] [PubMed] [Google Scholar]
- 8.Kemmerer CL, Schittenhelm J, Dubois E, Neumann L, Häsler LM, Lambert M, Renovanz M, Kaeser SA, Tabatabai G, Ziemann U, Naumann U, Kowarik MC. Cerebrospinal fluid cytokine levels are associated with macrophage infiltration into tumor tissues of glioma patients. BMC Cancer 2021;21:1108. 10.1186/s12885-021-08825-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, He L, Zhu J, Zhang P, Liang S. Targeting tumor-associated macrophages for cancer treatment. Cell Biosci 2022;12:85. 10.1186/s13578-022-00823-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szulzewsky F, Pelz A, Feng X, Synowitz M, Markovic D, Langmann T, Holtman IR, Wang X, Eggen BJ, Boddeke HW, Hambardzumyan D, Wolf SA, Kettenmann H. Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One 2015;10:e0116644. 10.1371/journal.pone.0116644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Xu Y, Sun Q, Zhou X, Ma W, Wu J, Zhuang J, Sun C. New insights from the single-cell level: Tumor associated macrophages heterogeneity and personalized therapy. Biomed Pharmacother 2022;153:113343. 10.1016/j.biopha.2022.113343 [DOI] [PubMed] [Google Scholar]
- 12.Bianconi A, Aruta G, Rizzo F, Salvati LF, Zeppa P, Garbossa D, Cofano F. Systematic Review on Tumor Microenvironment in Glial Neoplasm: From Understanding Pathogenesis to Future Therapeutic Perspectives. Int J Mol Sci 2022;23:4166. 10.3390/ijms23084166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang B, Zhang H, Gu L, Ye B, Jian Z, Stary C, Xiong X. Advances in Immunotherapy for Glioblastoma Multiforme. J Immunol Res 2017;2017:3597613. 10.1155/2017/3597613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong W, Li C, Kong G, Wan B, Wang S, Fan J. Glioblastoma: two immune subtypes under the surface of the cold tumor. Aging (Albany NY) 2022;14:4357-75. 10.18632/aging.204067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Bossche WBL, Kleijn A, Teunissen CE, Voerman JSA, Teodosio C, Noske DP, van Dongen JJM, Dirven CMF, Lamfers MLM. Oncolytic virotherapy in glioblastoma patients induces a tumor macrophage phenotypic shift leading to an altered glioblastoma microenvironment. Neuro Oncol 2018;20:1494-504. 10.1093/neuonc/noy082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Zhong H, Liang X, Ni S. Targeting tumor-associated macrophages for the immunotherapy of glioblastoma: Navigating the clinical and translational landscape. Front Immunol 2022;13:1024921. 10.3389/fimmu.2022.1024921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y, Ye Z, Song F, He Y, Liu J. The Role of TAMs in Tumor Microenvironment and New Research Progress. Stem Cells Int 2022;2022:5775696. 10.1155/2022/5775696 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V, Setty M, Leslie CS, Oei Y, Pedraza A, Zhang J, Brennan CW, Sutton JC, Holland EC, Daniel D, Joyce JA. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med 2013;19:1264-72. 10.1038/nm.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morisse MC, Jouannet S, Dominguez-Villar M, Sanson M, Idbaih A. Interactions between tumor-associated macrophages and tumor cells in glioblastoma: unraveling promising targeted therapies. Expert Rev Neurother 2018;18:729-37. 10.1080/14737175.2018.1510321 [DOI] [PubMed] [Google Scholar]
- 20.Tomaszewski W, Sanchez-Perez L, Gajewski TF, Sampson JH. Brain Tumor Microenvironment and Host State: Implications for Immunotherapy. Clin Cancer Res 2019;25:4202-10. 10.1158/1078-0432.CCR-18-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee S, Sonanini D, Maurer A, Daldrup-Link HE. The yin and yang of imaging tumor associated macrophages with PET and MRI. Theranostics 2019;9:7730-48. 10.7150/thno.37306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aghighi M, Theruvath AJ, Pareek A, Pisani LL, Alford R, Muehe AM, Sethi TK, Holdsworth SJ, Hazard FK, Gratzinger D, Luna-Fineman S, Advani R, Spunt SL, Daldrup-Link HE. Magnetic Resonance Imaging of Tumor-Associated Macrophages: Clinical Translation. Clin Cancer Res 2018;24:4110-8. 10.1158/1078-0432.CCR-18-0673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Zhang C, Zhang Z, Han B, Shen Z, Li L, Liu S, Zhao X, Ye F, Zhang Y. Specific clinical and immune features of CD68 in glioma via 1,024 samples. Cancer Manag Res 2018;10:6409-19. 10.2147/CMAR.S183293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XL, Jiang JT, Wu CP. Prognostic significance of tumor-associated macrophage infiltration in gastric cancer: a meta-analysis. Genet Mol Res 2016. [DOI] [PubMed] [Google Scholar]
- 25.Nam SJ, Go H, Paik JH, Kim TM, Heo DS, Kim CW, Jeon YK. An increase of M2 macrophages predicts poor prognosis in patients with diffuse large B-cell lymphoma treated with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone. Leuk Lymphoma 2014;55:2466-76. 10.3109/10428194.2013.879713 [DOI] [PubMed] [Google Scholar]
- 26.Vidyarthi A, Agnihotri T, Khan N, Singh S, Tewari MK, Radotra BD, Chatterjee D, Agrewala JN. Predominance of M2 macrophages in gliomas leads to the suppression of local and systemic immunity. Cancer Immunol Immunother 2019;68:1995-2004. 10.1007/s00262-019-02423-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Chen L, Song Y, Dai G, Duan L, Luo Y, Wang G, Xiao Q, Li G, Bai S. Predictive value of magnetic resonance imaging radiomics-based machine learning for disease progression in patients with high-grade glioma. Quant Imaging Med Surg 2023;13:224-36. 10.21037/qims-22-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Ren F, Wang T, Jiang Z, Sun Q, Li Z. Targeted Immunoimaging of Tumor-Associated Macrophages in Orthotopic Glioblastoma by the NIR-IIb Nanoprobes. Small 2022;18:e2202201. 10.1002/smll.202202201 [DOI] [PubMed] [Google Scholar]
- 29.Li F, Sun H, Li Y, Bai X, Dong X, Zhao N, Meng J, Sun B, Zhang D. High expression of eIF4E is associated with tumor macrophage infiltration and leads to poor prognosis in breast cancer. BMC Cancer 2021;21:1305. 10.1186/s12885-021-09010-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson RA, Adams RA, McArt DG, Salto-Tellez M, Jasani B, Hamilton PW. Epidermal growth factor receptor immunohistochemistry: new opportunities in metastatic colorectal cancer. J Transl Med 2015;13:217. 10.1186/s12967-015-0531-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim AR, Choi KS, Kim MS, Kim KM, Kang H, Kim S, Chowdhury T, Yu HJ, Lee CE, Lee JH, Lee ST, Won JK, Kim JW, Kim YH, Kim TM, Park SH, Choi SH, Shin EC, Park CK. Absolute quantification of tumor-infiltrating immune cells in high-grade glioma identifies prognostic and radiomics values. Cancer Immunol Immunother 2021;70:1995-2008. 10.1007/s00262-020-02836-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Zhang C, Maimela NR, Yang L, Zhang Z, Ping Y, Huang L, Zhang Y. Molecular and clinical characterization of CD163 expression via large-scale analysis in glioma. Oncoimmunology 2019;8:1601478. 10.1080/2162402X.2019.1601478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol 2008;216:15-24. 10.1002/path.2370 [DOI] [PubMed] [Google Scholar]
- 34.Zhu C, Kros JM, Cheng C, Mustafa D. The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro Oncol 2017;19:1435-46. 10.1093/neuonc/nox081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idoate Gastearena MA, López-Janeiro Á, Lecumberri Aznarez A, Arana-Iñiguez I, Guillén-Grima F. A Quantitative Digital Analysis of Tissue Immune Components Reveals an Immunosuppressive and Anergic Immune Response with Relevant Prognostic Significance in Glioblastoma. Biomedicines 2022;10:1753. 10.3390/biomedicines10071753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J, Chen CC, Li M. Macrophages/Microglia in the Glioblastoma Tumor Microenvironment. Int J Mol Sci 2021;22:5775. 10.3390/ijms22115775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sørensen MD, Dahlrot RH, Boldt HB, Hansen S, Kristensen BW. Tumour-associated microglia/macrophages predict poor prognosis in high-grade gliomas and correlate with an aggressive tumour subtype. Neuropathol Appl Neurobiol 2018;44:185-206. 10.1111/nan.12428 [DOI] [PubMed] [Google Scholar]
- 38.Fernandes B, Feltes PK, Luft C, Nazario LR, Jeckel CMM, Antunes IF, Elsinga PH, de Vries EFJ. Potential PET tracers for imaging of tumor-associated macrophages. EJNMMI Radiopharm Chem 2022;7:11. 10.1186/s41181-022-00163-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding P, Wang W, Wang J, Yang Z, Xue L. Expression of tumor-associated macrophage in progression of human glioma. Cell Biochem Biophys 2014;70:1625-31. 10.1007/s12013-014-0105-3 [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Shi Y, Wu M, Liu J, Wu H, Xu C, Chen L. Hypoxia-induced polypoid giant cancer cells in glioma promote the transformation of tumor-associated macrophages to a tumor-supportive phenotype. CNS Neurosci Ther 2022;28:1326-38. 10.1111/cns.13892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Wang G, Wang W, Pan J, Zhou H, Han X, Su L, Ma Z, Hou L, Xue X. A Focal Adhesion-Related Gene Signature Predicts Prognosis in Glioma and Correlates With Radiation Response and Immune Microenvironment. Front Oncol 2021;11:698278. 10.3389/fonc.2021.698278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prosniak M, Harshyne LA, Andrews DW, Kenyon LC, Bedelbaeva K, Apanasovich TV, Heber-Katz E, Curtis MT, Cotzia P, Hooper DC. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res 2013;19:3776-86. 10.1158/1078-0432.CCR-12-1940 [DOI] [PubMed] [Google Scholar]
- 43.Mughal AA, Zhang L, Fayzullin A, Server A, Li Y, Wu Y, Glass R, Meling T, Langmoen IA, Leergaard TB, Vik-Mo EO. Patterns of Invasive Growth in Malignant Gliomas-The Hippocampus Emerges as an Invasion-Spared Brain Region. Neoplasia 2018;20:643-56. 10.1016/j.neo.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulze Heuling E, Knab F, Radke J, Eskilsson E, Martinez-Ledesma E, Koch A, Czabanka M, Dieterich C, Verhaak RG, Harms C, Euskirchen P. Prognostic Relevance of Tumor Purity and Interaction with MGMT Methylation in Glioblastoma. Mol Cancer Res 2017;15:532-40. 10.1158/1541-7786.MCR-16-0322 [DOI] [PubMed] [Google Scholar]
- 45.Hewer E, Phour J, Gutt-Will M, Schucht P, Dettmer MS, Vassella E. TERT Promoter Mutation Analysis to Distinguish Glioma From Gliosis. J Neuropathol Exp Neurol 2020;79:430-6. 10.1093/jnen/nlaa004 [DOI] [PubMed] [Google Scholar]
- 46.Nabavizadeh SA, Ware JB, Wolf RL. Emerging Techniques in Imaging of Glioma Microenvironment. Top Magn Reson Imaging 2020;29:103-14. 10.1097/RMR.0000000000000232 [DOI] [PubMed] [Google Scholar]
- 47.Zhou J, Reddy MV, Wilson BKJ, Blair DA, Taha A, Frampton CM, Eiholzer RA, Gan PYC, Ziad F, Thotathil Z, Kirs S, Hung NA, Royds JA, Slatter TL. MR Imaging Characteristics Associate with Tumor-Associated Macrophages in Glioblastoma and Provide an Improved Signature for Survival Prognostication. AJNR Am J Neuroradiol 2018;39:252-9. 10.3174/ajnr.A5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salvalaggio A, Silvestri E, Sansone G, Pinton L, Magri S, Briani C, Anglani M, Lombardi G, Zagonel V, Della Puppa A, Mandruzzato S, Corbetta M, Bertoldo A. Magnetic Resonance Imaging Correlates of Immune Microenvironment in Glioblastoma. Front Oncol 2022;12:823812. 10.3389/fonc.2022.823812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishie A, Ono M, Shono T, Fukushi J, Otsubo M, Onoue H, Ito Y, Inamura T, Ikezaki K, Fukui M, Iwaki T, Kuwano M. Macrophage infiltration and heme oxygenase-1 expression correlate with angiogenesis in human gliomas. Clin Cancer Res 1999;5:1107-13. [PubMed] [Google Scholar]
- 50.Nazem A, Guiry SC, Pourfathi M, Ware JB, Anderson H, Iyer SK, Moon BF, Fan Y, Witschey WR, Rizi R, Bagley SJ, Desai A, O'Rourke DM, Brem S, Nasrallah M, Nabavizadeh A. MR susceptibility imaging for detection of tumor-associated macrophages in glioblastoma. J Neurooncol 2022;156:645-53. 10.1007/s11060-022-03947-3 [DOI] [PubMed] [Google Scholar]
- 51.Li G, Li L, Li Y, Qian Z, Wu F, He Y, Jiang H, Li R, Wang D, Zhai Y, Wang Z, Jiang T, Zhang J, Zhang W. An MRI radiomics approach to predict survival and tumour-infiltrating macrophages in gliomas. Brain 2022;145:1151-61. 10.1093/brain/awab340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as