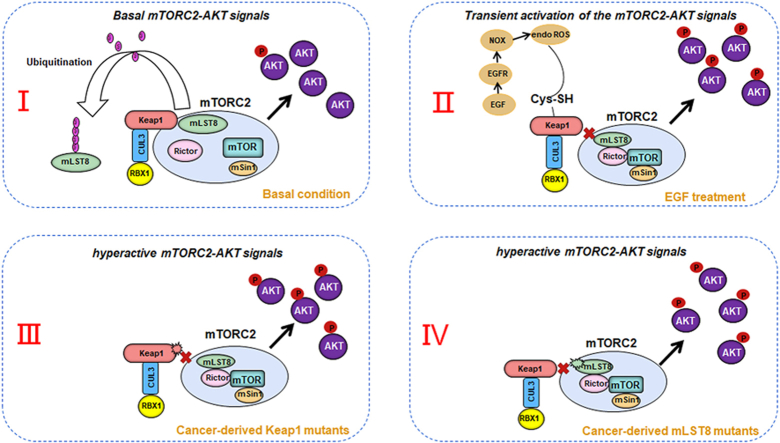
Abstract
The mechanistic target of the rapamycin (mTOR) pathway, which participates in the regulation of cellular growth and metabolism, is aberrantly regulated in various cancer types. The mTOR complex 2 (mTORC2), which consists of the core components mTOR, Rictor, mSin1, and mLST8, primarily responds to growth signals. However, the coordination between mTORC2 assembly and activity remains poorly understood. Keap1, a major sensor of oxidative stress in cells, functions as a substrate adaptor for Cullin 3-RING E3 ubiquitin ligase (CRL3) to promote proteasomal degradation of NF-E2-related factor 2 (NRF2), which is a transcription factor that protects cells against oxidative and electrophilic stress. In the present study, we demonstrate that Keap1 binds to mLST8 via a conserved ETGE motif. The CRL3Keap1 ubiquitin ligase complex promotes non-degradative ubiquitination of mLST8, thus reducing mTORC2 complex integrity and mTORC2-AKT activation. However, this effect can be prevented by oxidative/electrophilic stresses and growth factor signaling-induced reactive oxygen species (ROS) burst. Cancer-derived Keap1 or mLST8 mutations disrupt the Keap1-mLST8 interaction and allow mLST8 to evade Keap1-mediated ubiquitination, thereby enhancing mTORC2-AKT activation and promoting cell malignancy and remodeling cell metabolism. Our findings provide new insights into the molecular mechanisms of Keap1/mLST8 mutation-driven tumorigenesis by promoting mTORC2-AKT activation, which is independent of the canonical NRF2 pathway.
Keywords: mTORC2, AKT, Keap1, ROS, Ubiquitination, Lung cancer
Graphical abstract
Highlights
-
•
Keap1, a substrate adaptor for Cullin 3-RING E3 ubiquitin ligase (CRL3), is a novel mLST8-interacting protein.
-
•
CRL3Keap1 complex promotes non-degradative ubiquitination of mLST8, leading to reduced mTORC2 complex integrity and mTORC2-AKT activation.
-
•
Both H2O2 exposure and EGF stimulation decrease mLST8 ubiquitination, which correlate with increased mTORC2 activity.
-
•
Cancer-derived Keap1 or mLST8 mutations disrupt the Keap1-mLST8 interaction and allow mLST8 to evade Keap1-mediated ubiquitination.
-
•
Cancer-derived mLST8 mutations activate the mTORC2-AKT pathway, promoting cell malignancy and remodeling cell metabolism.
1. Introduction
The mechanistic target of rapamycin (mTOR) coordinates a diverse set of environmental cues such as growth factor signals and nutritional status to direct eukaryotic cell growth [1,2]. mTOR is an evolutionarily highly conserved serine/threonine protein kinase that belongs to the phosphatidylinositol kinase (PI3K)-related kinase family. mTOR is assembled into two large complexes, namely, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2), which are regulated independently by distinct binding partners. The two complexes share the mTOR kinase and mLST8 but also contain components unique to each complex (Raptor for mTORC1; Rictor and mSin1 for mTORC2). Although mLST8 was initially identified as an mTORC1 subunit, it is not required for mTORC1 activity but for mTORC2 activity [3,4]. mTORC1 orchestrates anabolic metabolism by regulating the key effectors of different biosynthetic and metabolic processes, while repressing catabolic processes such as autophagy. mTORC2 participates in physiological processes, such as cell survival, proliferation, polarity, and cytoskeletal dynamics by activating AGC kinases (e.g., AKT, SGK1, and PKC) [5]. The mTOR pathway is dysregulated in various human disorders such as cancer, obesity, and neurodegenerative diseases [2,5]. Although the regulation of mTORC1 has been widely studied, the dynamic regulation of mTORC2 activity under different environmental cues remains poorly understood [6].
Kelch-like ECH-derived protein 1 (Keap1) is a substrate adaptor protein of the CRL3 ubiquitin ligase complex [7]. The best-characterized substrate of Keap1 is transcription factor NF-E2-related factor 2 (NRF2). NRF2 orchestrates an elaborate transcriptional program in response to environmental challenges caused by oxidants and electrophilic agents, allowing for adaptation and survival under stressed conditions. In unstressed cells, NRF2 is maintained at a low level through constitutive degradation via the proteasome system. Oxidative and electrophilic stresses trigger stabilization and activation of NRF2 by releasing NRF2 and Keap1 from the CUL3 scaffold. Stabilized NRF2 is then translocated into the nucleus, forms heterodimers with small Maf proteins, and drives the transcription of its cognate target genes, the protein products of which are involved in cellular antioxidant, detoxification, and metabolic pathways [[8], [9], [10]]. The Neh2 domain of NRF2 contains two degrons (DLG and ETGE motifs) that are specifically recognized by the KLECH domains of Keap1. The Keap1–NRF2 interaction surface is frequently affected by missense mutations in various types of cancers, underscoring the critical role of this pathway in cell physiology and homeostasis [11]. The Cancer Genome Atlas cataloged genomic alterations that affect the Keap1-NRF2 axis in 6.3% of patient samples across 226 studies, with substantial enrichment in lung, endometrial, liver, and bladder cancers [12]. These alterations lead to constitutive activation of NRF2-dependent gene transcription to promote many cancer hallmarks, including cellular resistance to oxidative stress, xenobiotic efflux, proliferation, and metabolic reprogramming [13]. Although the pathophysiological importance of Keap1 in redox sensing has been well established, focus on Keap1 beyond NRF2 repression is still rare.
In the present study, we aim to investigate new mTORC2 regulatory pathways by identifying mLST8-interacting partners, considering that mLST8 is functionally essential for mTORC2 assembly and activation [3,14]. We demonstrate that Keap1 binds to mLST8 and induces its non-degradative ubiquitination, thereby decreasing mTORC2 assembly and mTORC2-AKT activation. We propose that Keap1 acts as an upstream regulator of the mTORC2-AKT pathway by sensing cellular redox state, and that such regulation can be disrupted by cancer-derived gene mutations.
2. Materials and methods
2.1. Cell culture and lentivirus infection
293T, H1299, A549, H1437, H1703, HeLa, and MIHA cells were obtained from the American Type Culture Collection, EBC-1 cells were obtained from Japanese Collection of Research Bioresources Cell Bank. 293T, H1299, H1437, H1703, EBC-1 and HeLa cells were cultured in DMEM. MIHA and A549 cells were maintained in RPMI 1640. pCDH-based overexpression or pLKO-based knockdown lentiviral and packaging plasmids were transfected into 293T cells. The lentiviral particles were harvested 48 h after transfection. H1299 and A549 cells were infected with lentiviruses in the presence of polybrene (8 μg/ml) and selected using puromycin (1.5 μg/ml). The sequences of gene-specific shRNAs are summarized in Supplementary Table 1.
2.2. Antibodies and chemicals
The commercial antibodies and chemical reagents used in this study are summarized in Supplementary Tables 2 and 3
2.3. Plasmids and mutagenesis
Constructs of Keap1, mLST8, and their mutants were generated in our laboratory. The mLST8 and Keap1 mutants were established using the KOD-Plus-Mutagenesis Kit (TOYOBO), following the manufacturer's protocol.
2.4. Protein complexes purification
The affinity purification strategy used to isolate mLST8-containing protein complexes from H1299 cells was performed as previously described [15].
2.5. CRISPR-Cas9 mediated gene knockout (KO) stable cell generation
sgRNAs targeting KEAP1, MLST8, and NFE2L2 genes were designed using the online CRISPR design tool (http://crispr.mit.edu) and subcloned into the pX459 construct from Dr. Feng Zhang's lab. The sgRNA construct was transfected into 293T and H1299 cells for 24 h. The cells were then treated with puromycin (1 μg/ml) for 3 days. After two weeks, KO cells were validated by western blot and Sanger sequencing. The sgRNA sequences are summarized in Supplementary Table 1.
2.6. Western blot
Cell lysates or immunoprecipitates were subjected to SDS-PAGE and the proteins were transferred to nitrocellulose membranes (GE Healthcare). The membranes were blocked in Tris-buffered saline (pH 7.4) containing 5% non-fat milk and 0.1% Tween-20, washed twice in TBS containing 0.1% Tween-20, incubated with the primary antibody for 2 h, and then with the secondary antibody for 1 h at room temperature. The proteins of interest were visualized using ECL chemiluminescence system (Santa Cruz Biotechnology). The WB detection for each experiment was performed in triplicate, and band intensities of WB results were calculated by image J according to the manufacturer's instructions. The quantitative data are shown in Supplementary Figs. 7–13.
2.7. Doxycycline inducible expression
pGLV-FLAG-Keap1 was lentivirally infected into H1299 cells in the presence of polybrene (8 μg/ml) for 48 h, and then cells were selected in puromycin (1.5 μg/ml) for 3 days to generate stable cell lines. Inducible FLAG-Keap1 expression was achieved by adding Doxycycline (10 ng/ml) to the medium for 24 h.
2.8. In vivo ubiquitination assays
293T cells were co-transfected with (His)6-tagged ubiquitin and the indicated constructs. After 24 h, cells were lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole (pH 8.0)). After sonication, the cell lysates were incubated with Ni–NTA beads (QIAGEN) for 3 h at room temperature. Subsequently, the pull-down products were washed once with buffer A, twice with buffer A/TI (buffer A: buffer TI = 1:3), and once with buffer TI (25 mM Tris-HCl and 20 mM imidazole; (pH 6.8)). Pull-down proteins and Whole cell lysates (WCL) were detected using Western blot.
2.9. Identification of the ubiquitin attachment sites by affinity purification and mass spectrometry
To prepare ubiquitinated mLST8 conjugates, FLAG-mLST8, HA-Ub, and Myc-Keap1 plasmids were transfected into 293T cells (5x100 mm dish). After 48 h, the cells were lysed in RIPA buffer, and FLAG-mLST8 was purified using anti-FLAG M2 beads before being resolved by 10% SDS-PAGE. After Coomassie Blue staining, the bands of the mLST8 conjugates were excised and subjected to mass spectrometry (Orbitrap Fusion Lumos, Thermo) analysis. The ubiquitinated peptides of mLST8 are summarized in Supplementary Table 4.
2.10. Proximity ligation assays (PLA)
The in situ interaction between mLST8 and mTOR in mLST8 KO H1299 cells stably overexpressing FLAG-mLST8-WT, -G302R, or -E303D mutants was detected using a Duolink® in situ detection Kit (Sigma). Briefly, H1299 cells were attached to 23-well slides and fixed in 4% paraformaldehyde. and permeabilized with 0.5% Triton x-100 for 30 min. H1299 cells were incubated with two primary antibodies (mTOR and FLAG), and then incubated with a pair of PLA probes composed of species-specific secondary antibodies conjugated to complementary oligonucleotides. After adding the hybridization solution and ligase, oligonucleotides formed a circle when proteins were very close. Finally, the polymerase and nucleotides participate in the formation of rolling-circle amplification, which is visualized by red fluorescence. The cells were imaged using a confocal laser scanning microscope (LSM880, Zeiss).
2.11. ROS measurement
Reactive oxygen was detected using the fluorescent probe DCFH-DA, which can oxidize non-fluorescent DCFH to generate fluorescent DCF, and the fluorescence of DCF was detected to measure the level of intracellular reactive oxygen species. Cells were seeded into 24-well plates and incubated for 24 h. DCFH-DA diluted in serum-free medium was added to the cells and incubated at 37 °C for 30 min. The cells were washed twice with PBS, and the fluorescence of each well was measured using a fluorescent microplate reader at an excitation wavelength of 480 nm and an emission wavelength of 530 nm.
2.12. CCK-8 cell growth assays
Cell growth was detected using the Cell Counting Kit-8 (CCK-8) (Beyotime), according to the manufacturer's instructions. The cells (1x103 cells/well) were seeded in 96-well plates. CCK-8 solution (10 μl) was added to the media and incubated for 2 h, and then cell counts were tested at the same time period for seven consecutive days. Cell growth was evaluated by measuring absorbance at 450 nm using a microplate absorbance reader (Bio-Rad).
2.13. Migration and invasion assays
Transwell chambers, with and without Matrigel coating, are generally used to determine cell migration and invasion. H1299 cells (1 × 105 cells/well) were seeded into the upper chamber of a 24-well Transwell plate containing serum-free medium, and 600 μl of DMEM containing 10% FBS was added to the lower chamber. After 48 h, the cells in the upper chamber were wiped off with a cotton swab, and the cells that migrated or invaded under the membrane were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet for 15 min, and examined under a microscope (IX73, Olympus) at nine different counts in the field of view.
2.14. Sphere-formation assays
Cells (1x103 cells/well) were mixed with Matrigel (BD) and plated in 24-well ultra-low-attachment plates (Corning). Fresh medium was added every three days. Floating spheres that grew in 1-2 weeks were quantified using a Nikon digital photo camera and ImageJ software.
2.15. BiFC assays
The full-length sequences of mLST8 and Keap1 were amplified and fused to the N-terminus of YFP (pBiFC-VN173 Vector) and the C-terminus of YFP (pBiFC-VC155 Vector), respectively. 293T cells were cultured in a 6-well plate, and 2 μg of pBiFC-VN173-mLST8 and pBiFC-VC155-Keap1 were co-transfected into 293T cells. Protein-VN alone and protein-VC alone served as negative controls. The cells were visualized 24 h after transfection (excitation and emission maxima at OD 494 and 507 nm, respectively) and imaged using an inverted fluorescence microscope (IX73, Olympus).
2.16. Xenograft tumor assays
3-4-week-old female nu/nu immunodeficient mice (SLAC Laboratory) were bred and maintained at our institutional pathogen-free mouse facilities. The cell suspension was concentrated to 5 × 106 cells/100 μl PBS and injected into the flanks of the nude mice. After 30 days, the mice were sacrificed and xenograft tumors were harvested, weighed, and photographed.
2.17. Metabolome analysis
Six sets of parallel cell samples were prepared for testing. The contents of glycometabolism of cell samples were quantified based on UHPLC-MS/MS platform. The cell samples containing 50% aqueous methanol collected by customer were added 20ul IS (4 μg/mL of FBP-13C6, 4 μg/mL of succinic acid-13C4, and 20 μg/mL Glucose-13C6) and processed by 5 cycles of 1 min ultra-sonication and 1 min interval in an ice-water bath and stood for 30 min at -20 °C. After centrifugation at 15000 rcf for 15 min at 4 °C. All supernatant was evaporated to dryness. The residues were reconstituted in 50 μL of 50% aqueous acetonitrile (1:1, v/v) prior to UHPLC-MS/MS analysis. The UHPLC-MS/MS analysis was performed on an Agilent 1290 Infinity II UHPLC system coupled to a 6470A Triple Quadrupole mass spectrometry (Agilent).
The raw data were processed by Agilent MassHunter Workstation Software (version B.08.00) by using the default parameters and assisting manual inspection to ensure the qualitative and quantitative accuracies of each compound. The peak areas of target compounds were integrated and output for quantitative calculation.
2.18. Oxygen consumption assays
Oxygen consumption rates (OCR) was measured under basal conditions in the presence of the mitochondrial inhibitors oligomycin (0.25 μmol/L, Calbiochem). Experiments were performed at 37 °C. OCR was calculated by the oligomycin-induced changes in comparison to basal rates. H1299 cells were seeded at a density of 20,000 cells in cell culture microplate. The total protein of each well was determined by Bradford assay and used as the reference to normalize the OCR.
2.19. NAD+/NADH measurement assays
After appropriate treatments, H1299 cells (1 × 106 cells/sample) were collected and intracellular NAD+/NADH levels were determined by using NAD+/NADH assay kit (Beyotime) with WST-8 according to the manufacturer's instructions. In brief, cells were lysed with 200 μl of cold lysis buffer. To measure total NAD+/NADH, 20 μl of cell lysates were added to a 96-well plate. The lysed cell suspension was incubated at 60 for 30 min and 20 μl was added to a 96-well plate. Subsequently, 90 μl of alcohol dehydrogenase was added and incubated at 37 °C for 10 min. Finally, 10 μl of chromogenic solution was added to the plate and the mixture was incubated at 37 °C for 30 min. Standard curve was generated and measured at the same time as the samples. The resulting color was assayed at 450 nm using a microplate absorbance reader.
2.20. Lactate production assays
Cells were seeded into six-well plates at a density of 1 × 106 cells in 2 ml DMEM medium per well and cultured overnight. According to the manual of the Lactate Acid Assay Kit (Solarbio), The resulting color was assayed was measured at 570 nm using a microplate absorbance reader.
2.21. Statistical analysis
The band intensities of the WB results were calculated using image J according to the manufacturer's instructions. Statistical analyses were performed using the GraphPad Prism software. All data are shown as the mean ± SD of experiments performed with at least three replicates. Differences between the two groups were analyzed using the One-way or the Two-way ANOVA tests. * represents p < 0.05; ** represents p < 0.01; *** represents p < 0.001.
3. Results
3.1. Keap1 is an mLST8 interacting protein
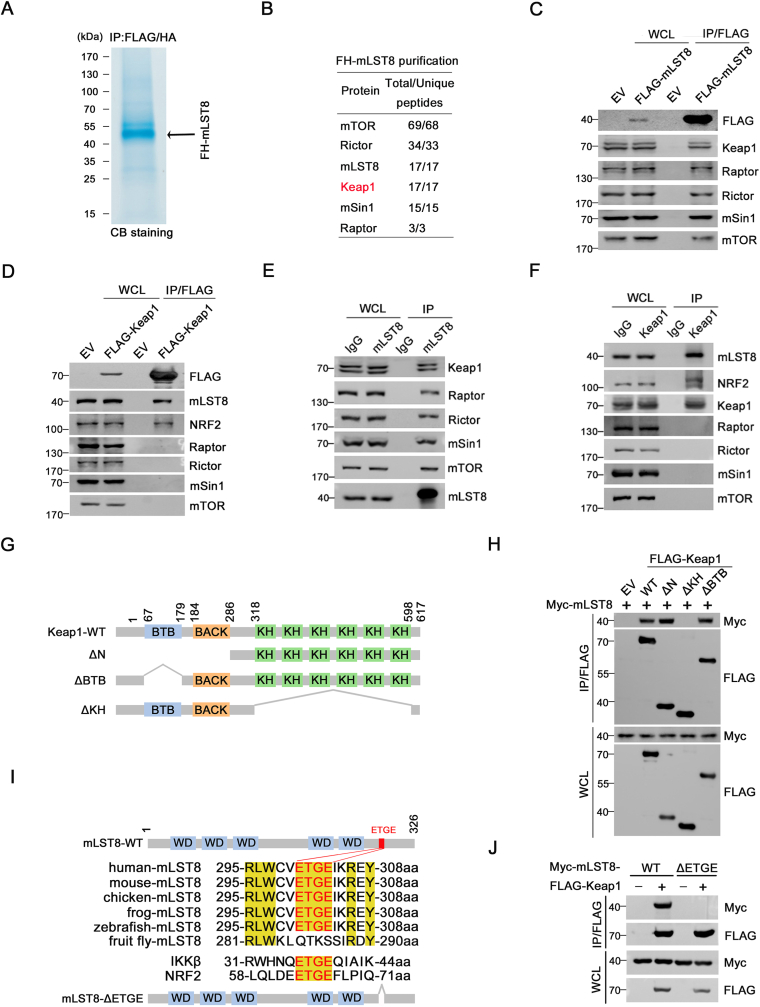
We isolated mLST8-associated protein complexes from non-small-cell lung carcinoma (NSCLC) H1299 cells that stably express FLAG-HA-mLST8 through tandem affinity purification and determined the proteins present in the complexes using mass spectrometry (MS, Fig. 1A). The validity of this approach was confirmed by the presence of multiple mTORC1/2 subunits. Unexpectedly, Keap1 was present in the complexes and ranked with high confidence in the interaction hit list (Fig. 1B). Notably, the potential interaction between mLST8 and Keap1 was also revealed via a large-scale proteome mapping analysis; however, the biological function of this interaction was not analyzed further [16]. Thus, we conducted further analyses of the pathophysiological significance of the mLST8-Keap1 interaction.
Fig. 1.
Identification of Keap1 as a novel mLST8-interacting protein.
(A, B) Tandem affinity purification of mLST8-containing protein complexes was conducted from H1299 cells stably overexpressing FLAG-HA-mLST8. Derived proteins were separated by SDS-PAGE and visualized by CB staining (A). The number of total/unique peptides identified by mass spectrometry analysis is shown in the table (B). (C, D) 293T cells were transfected with the indicated plasmids. WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by Western blot (WB) with the indicated antibodies.
(E) H1299 WCL were prepared and subjected to co-IP with IgG or anti-mLST8 antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies. (F) H1299 WCL were prepared and subjected to co-IP with IgG or anti-Keap1 antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(G) Schematic representation of Keap1 deletion mutants.
(H) 293T cells were transfected with the indicated plasmids. WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(I) Alignment of ETGE motif sequence of mLST8 and its homologs across different species. IKKβ and NRF2 are known Keap1 substrates. Schematic representation of two cancer-derived mLST8 mutants.
(J) 293T cells were transfected with the indicated plasmids. WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
We first verified the binding of Keap1 to mLST8 by co-immunoprecipitation (co-IP) assays. FLAG-mLST8 immunoprecipitated endogenous Keap1 in 293T cells (Fig. 1C). Reciprocally, FLAG-Keap1 immunoprecipitated endogenous mLST8 in 293T cells, whereas Keap1 did not interact with other mTORC1/2 subunits, including mTOR, Raptor, Rictor, and mSin1 (Fig. 1D). Furthermore, we detected an endogenous Keap1-mLST8 interaction in cancer cell lines (H1299, HeLa) and immortalized human hepatocytes (MIHA, Fig. 1E, F, Supplementary Fig. 1A). mLST8 did not interact with the other CRL3 adaptors examined (Supplementary Fig. 1B). Keap1 did not interact with S6K1 or AKT1, which are downstream kinases of mTORC1 and mTORC2, respectively (Supplementary Fig. 1C). Immunofluorescence analysis showed that mLST8 and Keap1 were colocalized in the cytoplasmic compartments (Supplementary Fig. 1D). We performed BiFC assays for the direct visualization of protein–protein interactions at high spatial resolution in living cells. These results confirmed the occurrence of Keap1–mLST8 interaction in living cells (Supplementary Figs. 1E and F).
Keap1 contains a BTB domain and multiple KELCH domains responsible for substrate binding (Fig. 1G). Keap1's KELCH domains, but not the BTB domain, are essential for interaction with mLST8 (Fig. 1H). Finally, we determined the protein sequence in mLST8 that is responsible for Keap1-binding. Keap1 binds to NRF2 by recognizing ETGE and DLG motifs [13]. A perfectly matched ETGE motif is present in the mLST8 protein sequence. This motif is highly conserved between mLST8 orthologs from zebrafish to humans but not in fruit flies (Fig. 1I). Deletion of this ETGE motif completely abolished the Keap1–mLST8 interaction (Fig. 1J). Therefore, Keap1 specifically interacts with mLST8 in cells.
3.2. Keap1 promotes the non-degradative ubiquitination of mLST8
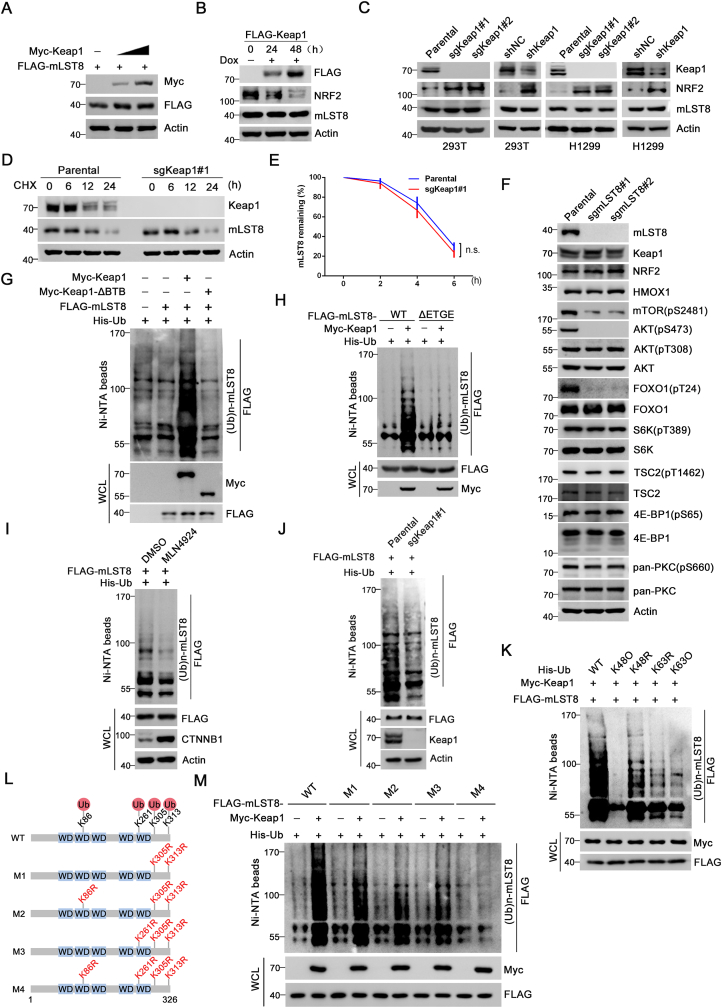
We explored whether Keap1 targets mLST8 for degradation, similarly to NRF2. However, Keap1 overexpression did not alter the protein levels of ectopically coexpressed mLST8 (Fig. 2A). To examine the effect of Keap1 on endogenous mLST8, we generated Tet-on-inducible H1299 cells conditionally expressing FLAG-Keap1. Although the induction of exogenous Keap1 led to a time-dependent reduction in NRF2 protein levels, no protein changes in mLST8 was observed (Fig. 2B). Keap1 depletion by CRISPR/Cas9-mediated KO or short hairpin RNA (shRNA)-mediated knockdown (KD) in H1299 and 293T cells markedly upregulated NRF2 protein levels, but did not alter mLST8 protein levels (Fig. 2C, Supplementary Fig. 2A). Moreover, protein turnover of mLST8 was not affected by Keap1 ablation (Fig. 2D, E).
Fig. 2.
Keap1 promotes the non-degradative ubiquitination of mLST8.
(A) H1299 cells were transfected with FLAG-mLST8 and increasing amount of Myc-Keap1. WCL were prepared for WB with the indicated antibodies.
(B) H1299 cells stably overexpressing tet-on inducible FLAG-Keap1 were treated with doxycycline (Dox) (10 nM) at the indicated times and WCL were prepared for WB with the indicated antibodies.
(C) Keap1 KO cell lines were generated through CRISPR/Cas9 methods. WCL from parental and Keap1 KO 293T (or H1299) cells were prepared for WB with the indicated antibodies. WCL from H1299 cells infected with lentivirus expressing Keap1-specific shRNA or negative control (shNC) were prepared for WB with the indicated antibodies. (D, E) WB analysis of the indicated proteins in WCL of parental and Keap1 KO H1299 cells treated with cycloheximide (CHX, 50 μg/ml) and harvested at different time points. At each time point, the intensity of mLST8 was normalized to the intensity of Actin and then to the value at 0 h (E). P values are calculated by the Two-way ANOVA test. n.s: not significant.
(F) mLST8 KO cell lines were generated through CRISPR/Cas9 methods. WCL from parental and Keap1 KO H1299 cells were prepared for WB with the indicated antibodies.
(G, H) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids.
(I) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids and treated with DMSO or MLN4924(10 nM) for 24 h
(J) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from parental or Keap1 KO H1299 cells transfected with the indicated plasmids.
(K) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids.
(L) Schematic representation of mLST8 domain architecture and ubiquitin attachment sites.
(M) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids.
Keap1-interacting proteins, such as DPP3 and SQSTM1/p62, sequester Keap1, thus increasing the stabilization and activation of NRF2 [17,18]. Thus, we investigated the effect of mLST8 on NRF2 protein levels. We ablated mLST8 expression by KO and observed that the levels of AKT (pS473) and AKT (pS473)-dependent phosphorylation of FOXO1 (a phosphorylated substrate of AKT) decreased, whereas the levels of AKT (pT308), S6K (pT389), and 4E-BP1(pS65) remained unchanged (Fig. 2F, Supplementary Fig. 2B). This finding supported the notion that mLST8 is essential for mTORC2 but not for PDK1 and mTORC1 signaling. However, mLST8 KO had no effect on the protein levels of Keap1, NRF2, and HMOX1 (a transcriptional target of NRF2) (Fig. 2F). In contrast to p62, mLST8 overexpression did not elevate NRF2 and HMOX1 protein levels (Supplementary Fig. 3A). These results indicate that mLST8 does not regulate NRF2 protein stability.
mLST8 was robustly polyubiquitinated by the co-expression of Keap1-WT but not by the Keap1-ΔBTB mutant (which is deficient for CUL3 binding) or the other CRL3 adaptors examined (Fig. 2G, Supplementary Fig. 3B). Deletion of the ETGE motif completely abolished the Keap1-mediated mLST8 ubiquitination (Fig. 2H). MLN4924, a CRL inhibitor, decreased Keap1-mediated mLST8 ubiquitination (Fig. 2I). Accordingly, Keap1 KO decreased endogenous mLST8 ubiquitination (Fig. 2J). In contrast, Keap1 did not affect ubiquitination levels of AKT1 and S6K1 (Supplementary Fig. 3C). The E3 ubiquitin ligase TRAF2 promotes mLST8 ubiquitination by recognizing a TRAF2-binding motif ([P/S/T/A]x([Q/E]) in mLST8 [19]. However, the mLST8 mutant (PEAA), which cannot bind TRAF2, was still ubiquitinated by Keap1 to a similar extent as mLST8-WT, indicating that Keap1-mediated mLST8 ubiquitination is independent of TRAF2 (Supplementary Fig. 3D).
CRL3-Keap1 complex primarily catalyzes a mixture of K48 and K63 polyubiquitin chains on its substrates [20]. We performed in vivo ubiquitination assays using two ubiquitin (Ub) mutants with a single K to R mutation at K48 or K63. We also included two Ub mutants that contain only intact K48 or K63, and six other lysine residues were mutated to arginine (K48O or K63O). Co-expression of the K48O-Ub mutant nearly abolished Keap1-mediated mLST8 ubiquitination, whereas co-expression of the K48R, K63R, or K63O-Ub mutant reduced Keap1-mediated mLST8 ubiquitination to various degrees (Fig. 2K). Other types of Ub linkage mutants (K6, K11, K27, K29, and K33) were also tested; however, it was found that these Ub linkage types were not involved in Keap1-mediated mLST8 ubiquitination (Supplementary Figs. 3E and F). We also utilized linkage-specific K27/K29/48/63-Ub antibodies to demonstrate that ubiquitinated mLST8 contains K48-, and K63-Ub linkages, but K27-, K29-Ub linkages (Supplementary Fig. 3G). Therefore, Keap1 promotes the synthesis of mixed K48/63 linkage polyubiquitin chains on mLST8. Next, we identified the ubiquitin attachment sites on mLST8. TRAF2 ubiquitinated mLST8 at K305 and K313 [19]. However, Keap1-mediated mLST8 ubiquitination was only moderately reduced when K305/K313 was mutated, indicating that other lysine residues in mLST8 were also modified (Fig. 2L, M). We co-transfected Keap1, mLST8, and Ub plasmids into 293T cells and immunoprecipitated Ub-mLST8 conjugates were analyzed by MS, revealing that mLST8 was ubiquitinated at two additional lysine residues, K86 and K261 (Supplementary Table 1). We constructed a panel of combinatorial K to R mLST8 mutants to evaluate whether K86 and K261 were truly targeted by Keap1. The results confirmed that these mutants had similar Keap1-binding affinities as mLST8-WT (Supplementary Fig. 3H). Additional K→R mutations of K86 or K261 in the mLST8-K305/K313R mutant markedly reduced Keap1-mediated mLST8 ubiquitination, and the quadruple K→R mutant of mLST8 was resistant to Keap1-mediated ubiquitination, indicating that four lysine residues are ubiquitin attachment sites targeted by Keap1 (Fig. 2L, M). Overall, these results indicate that Keap1 promotes the non-degradative ubiquitination of mLST8.
3.3. Keap1 reduces mTORC2 integrity and mTORC2-AKT activation
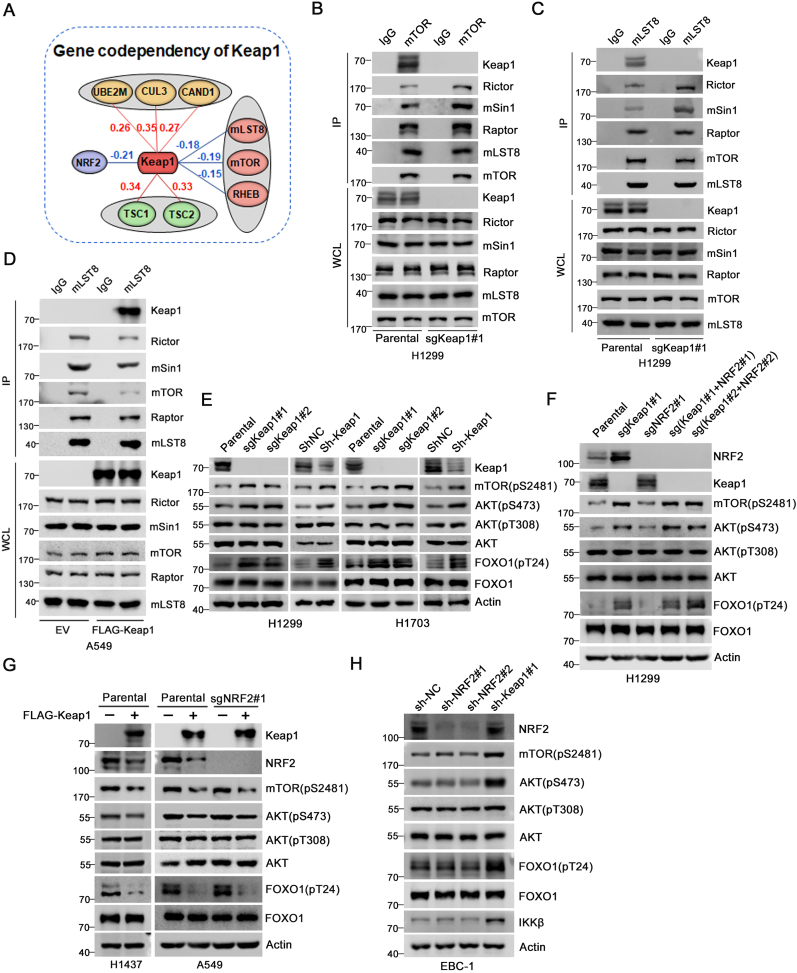
We next investigated whether the Keap1-mLST8 interaction has a functional effect on the mTORC2 pathway. First, the genetic co-dependency between Keap1 and other proteins was analyzed using Broad's 21Q2 DepMap dataset, which was derived from large-scale loss-of-function sgRNA screens for vulnerabilities in 990 cancer cell lines [21]. This dataset can be used to identify genes with similar functions or pathways [22,23]. Among the top 100 co-dependent genes of Keap1, CRL3-Keap1 complex components, such as CUL3, CAND1, and UBE2M, were among the strongest positive hits, whereas NRF2 was among the strongest negative hits. Additionally, Keap1 showed a strong positive correlation with TSC1/2, but a negative correlation with mLST8, mTOR, and RHEB, indicating that Keap1 is closely linked to the mTOR pathway (Fig. 3A).
Fig. 3.
Keap1 negatively regulates mTORC2 complex integrity and mTORC2-AKT activation.
(A) Representative gene co-dependency map for Keap1 from Broad's 21Q2 DepMap dataset.
(B) WCL from parental and Keap1 KO H1299 cells were prepared and subjected to co-IP with IgG or anti-mTOR antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(C) WCL from parental and Keap1 KO H1299 cells were prepared and subjected to co-IP with IgG or anti-mLST8 antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(D) WCL from A549 cells stably overexpressing EV or FLAG-Keap1 were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(E) WB analysis of the indicated proteins in WCL from parental or Keap1 KO H1299 and H1703 cells. Western blot of the indicated proteins in WCL from H1299 and H1703 cells infected with lentivirus expressing Keap1-specific shRNA or shNC.
(F) WB analysis of the indicated proteins in WCL from parental, Keap1 KO, NRF2 KO, and Keap1/NRF2 double KO H1299 cells.
(G) WB analysis of the indicated proteins in WCL from parental H1437 and A549 cells or NRF2 KO A549 cells stably overexpressing EV or FLAG-Keap1.
(H) WB analysis of the indicated proteins in WCL from EBC-1 cells infected with lentivirus expressing Keap1-specific and NRF2-specific shRNA or shNC.
We investigated the potential regulation of mTORC2 activity by Keap1 using five NSCLC cell lines with distinct genetic backgrounds. H1299 and H1703 cells harbor wild-type Keap1 and NRF2. A549 cells harbor an inactive Keap1G333C mutant, and Keap1 protein expression was nearly undetectable [24,25]. H1437 cells show no detectable Keap1 mRNA, due to bi-allelic loss of the Keap1 gene [26]. EBC-1 cells harbor an activating NRF2D77V mutant [27] (Supplementary Figs. 4A and B). We found that Keap1 KO in H1299 cells led to an increase in mTOR-immunoprecipitated mTORC2 subunits Rictor, and mSin1 but without affecting the mTORC1-specific subunit Raptor (Fig. 3B). Similarly, Keap1 KO in H1299 cells led to an increase in mLST8-immunoprecipitated mTORC2 subunits (Fig. 3C). The re-introduction of Keap1-WT into A549 cells led to a decrease in mTORC2 but not mTORC1 complex integrity (Fig. 3D). When Keap1 expression was depleted in H1299 or H1703 cells, mTOR (pS2481), AKT (pS473), and FOXO1 (pT24) levels increased, but phospho-AKT (T308) levels remained unchanged (Fig. 3E, Supplementary Figs. 2A and 5A).
To rule out the possibility that the elevation of mTORC2-AKT activity by Keap1 depletion was indirectly caused by NRF2 stabilization, we compared the AKT (pS473) levels in Keap1 KO, NRF2 KO, and Keap1/NRF2 double KO H1299 cells and found that additional NRF2 KO did not reverse Keap1 KO-induced elevation of phospho-AKT (S473) levels (Fig. 3F, Supplementary Fig. 2C). Moreover, Keap1 overexpression in parental and NRF2 KO A549 or H1437 cells led to similar downregulation of mTOR (pS2481), AKT (pS473), and FOXO1 (pT24) levels (Fig. 3G, Supplementary Fig. 5B). We depleted Keap1 in EBC-1 cells and found that NRF2 protein levels remained unchanged. This is consistent with a previous study reporting that the NRF2D77V mutant impairs recognition by Keap1 [27]. Additionally, IKKβ, a proteolytic substrate for the CRL3Keap1 ubiquitin ligase complex [28], was stabilized, indicating that Keap1 is functional in this cell line. Finally, we observed that the mTORC2-AKT activity was elevated in EBC-1 cells depleted of Keap1, but not NRF2 (Fig. 3H).
Taken together, these results indicate that Keap1 negatively regulates the mTORC2-AKT pathway in an NRF2-independent manner.
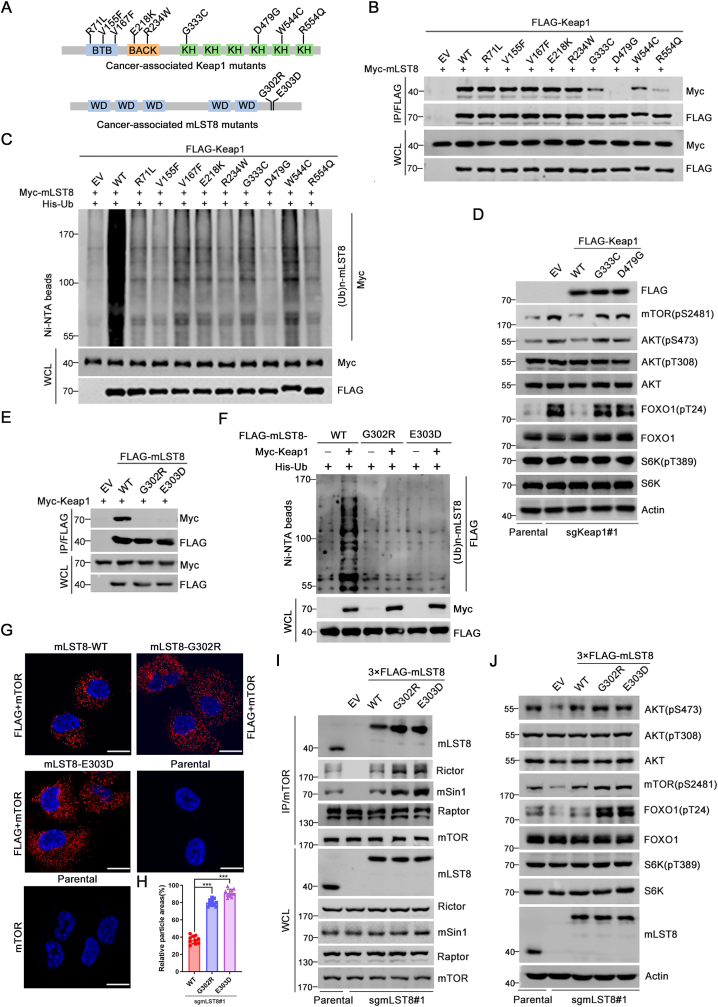
3.4. Keap1-mediated mTORC2 inhibition was disrupted by the cancer-derived mutants of Keap1 or mLST8
Loss-of-function mutations in Keap1 have been reported in various cancer types. Among these, Keap1 mutations are most frequently observed in NSCLC [12]. We investigated whether NSCLC-derived Keap1 mutants were defective in mediating mLST8 ubiquitination. We first examined the interaction between the NSCLC-derived Keap1 mutants and mLST8 (Fig. 4A). As shown in Fig. 4B, the mLST8-binding ability of the Keap1 mutants in the KELCH domains was severely impaired. In contrast, the Keap1 mutants in the BTB and BACK domains showed mLST8-binding abilities similar to those of Keap1-WT. This phenomenon was expected because the BTB and BACK domains are responsible for CUL3 binding and self-oligomerization, but not for substrate binding. Keap1-mediated mLST8 ubiquitination was markedly attenuated in all the mutants (Fig. 4C). Moreover, re-introduction of Keap1-WT, but not the cancer-derived Keap1 mutants, reversed Keap1 KO-induced mTORC2-AKT activation (Fig. 4D).
Fig. 4.
Keap1-mediated mTORC2 inhibition was disrupted by cancer-derived mutants of Keap1 or mLST8.
(A) Schematic representation of nine NSCLC-derived Keap1 mutants and two cancer-derived mLST8 mutants.
(B) 293T cells were transfected with the indicated plasmids. WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(C) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids.
(D) WB analysis of the indicated proteins in WCL from parental and Keap1 KO H1299 cells stably overexpressing EV, FLAG-Keap1-WT, -G333C or -D479G mutant.
(E) 293T cells were transfected with the indicated plasmids. WCL were prepared and subjected to co-IP with anti-FLAG antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(F) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids.
(G, H) In situ PLA analysis of the interaction between FLAG-mLST8 and endogenous mTOR in mLST8 KO H1299 cells stably overexpressing FLAG-mLST8-WT, -G302R, or -E303D mutant. The puncta area per cell were quantified and shown in (H). n = 10 cells. Scale bar, 20 μm. Single antibody (FLAG or mTOR) for PLA analysis were shown as negative controls. P values are calculated by the One-way ANOVA test. ***p < 0.001. (I) WCL from parental and mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant were prepared and subjected to co-IP with anti-mTOR antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(J) WB analysis of the indicated proteins in WCL from parental and mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant.
Considering that mLST8 binds Keap1 in an ETGE motif-dependent manner, we further explored whether the cancer-derived mutations that occur in the ETGE motif of mLST8 would attenuate the Keap1-mLST8 interaction and permit mLST8 to evade Keap1-mediated ubiquitination. Accordingly, we examined cancer sequencing data deposited in the Catalogue of Somatic Mutations in Cancer (COSMIC) database. A G302R mutation was observed in cervical cancer specimens and an E303D mutation was observed in thyroid cancer specimens (Fig. 4A). The Keap1-binding capacity of the cancer-derived mLST8 mutants was completely abolished (Fig. 4E). Keap1-mediated ubiquitination was completely abolished in mLST8 mutants (Fig. 4F). The mLST8 KO disrupted mTORC2 complex integrity, as previously reported [13]. The re-introduction of mLST8-WT into mLST8 KO cells restored mTORC2 complex integrity, whereas the re-introduction of cancer-derived mLST8-G302R or -E203D mutants further increased mTORC2 complex integrity, as demonstrated by the co-IP and PLA assays (Fig. 4G–I). The re-introduction of cancer-derived mLST8-G302R or -E203D mutant into mLST8 KO cells led to a remarkable increase in mTORC2-AKT activation compared to that in mLST8-WT-reintroduced cells (Fig. 4J). In contrast, mLST8-WT, -G302R, and -E303D mutants showed similar effects on mTORC2 complex integrity and mTORC2-AKT activation in Keap1-null A549 cells, implying that the differential effect on mTORC2-AKT activation by mLST8-WT and mutants is dependent on a functional Keap1 in cells (Supplementary Figs. 4C and D).
Taken together, these results indicate that the disruption of Keap1-mediated mLST8 ubiquitination by cancer-derived mutations allows mLST8 to evade Keap1-mediated ubiquitination, thereby enhancing mTORC2 complex integrity and mTORC2-AKT activation.
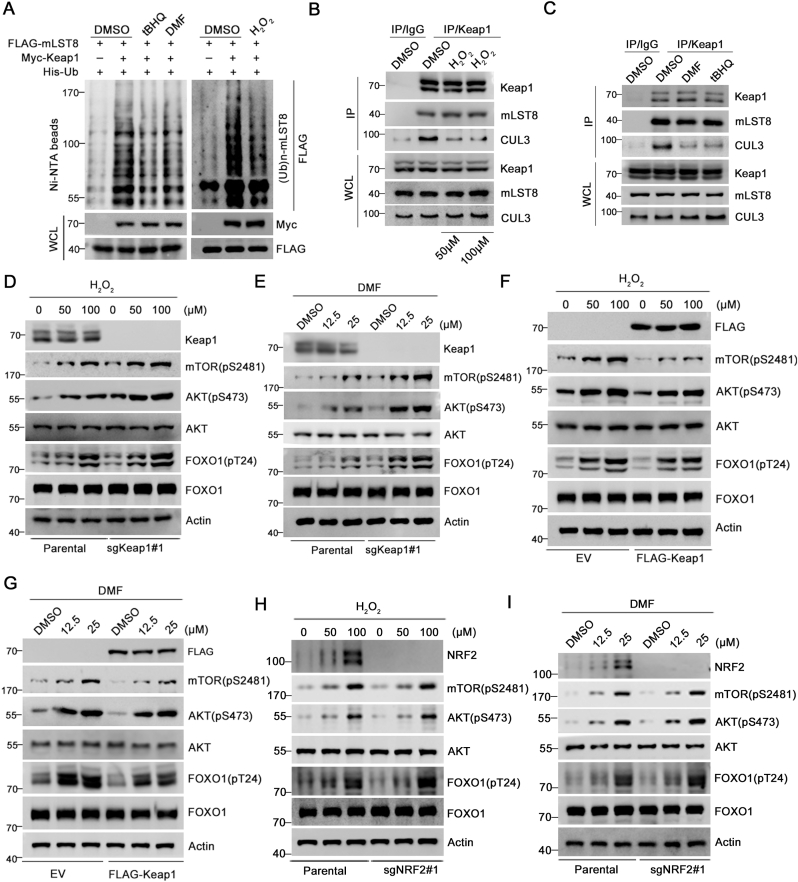
3.5. Oxidative/electrophilic stresses potentiate the mTORC2 pathway by inactivating Keap1
The redox balance is essential for mediating normal cellular physiology and signal transduction [29,30]. mTORC2 and AKT are critical downstream mediators of survival signaling in response to oxidative stress [31,32]. Keap1 contains multiple reactive cysteine residues that can be covalently modified by endogenous or exogenous electrophiles and oxidants. This reaction abrogates Keap1-mediated NRF2 degradation [13]. Thus, we investigated whether upstream signals affected Keap1-mediated mTORC2-AKT suppression. Exposure to hydrogen peroxide (H2O2) is widely used to elicit oxidative stress in cellular models. H2O2 treatment decreased Keap1-mediated mLST8 ubiquitination. Similarly, the treatment of cells with tert-butylhydroquinone (tBHQ) and dimethyl fumarate (DMF), which are electrophilic reagents for Keap1, also decreased Keap1-mediated mLST8 ubiquitination (Fig. 5A). A previous study showed that tBHQ treatment decreased Keap1-CUL3 interaction [10]. Similarly, we found that H2O2-, tBHQ-, or DMF-induced Keap1 inhibition was correlated with decreased Keap1-CUL3 interaction but not with Keap1-NRF2 interaction (Fig. 5B, C). Moreover, H2O2, tBHQ, or DMF did not affect the Keap1-mLST8 interaction (Fig. 5B, C). H2O2 or DMF treatment led to the activation of the mTORC2-AKT pathway both in Keap1 KO cells and parental cells, but at each time point, the phosphorylation levels of AKT and mTOR were higher than those in parental cells (Fig. 5D, E). Conversely, Keap1 overexpression suppressed H2O2- or DMF-induced activation of the mTORC2-AKT pathway in parental (Fig. 5F, G). Additionally, we showed that NRF2 KO did affect H2O2- or DMF -induced activation of the mTORC2-AKT pathway in H1299 cells (Fig. 5H, I). Taken together, these results suggest that oxidative/electrophilic stress may enhance the mTORC2 pathway, at least in part, by inactivating Keap1.
Fig. 5.
Oxidative/electrophilic stresses potentiate the mTORC2-AKT pathway through inactivating Keap1.
(A) WB analysis of the indicated proteins in Ni-NTA pull-downs products and WCL from 293T cells transfected with the indicated plasmids and treated with DMSO, tBHQ (25 μM) or DMF (25 μM) for 24 h or H2O2 (100 μM) for 3 h
(B) H1299 cells were treated with DMSO or H2O2 (50,100 μM) for 3 h, then WCL were prepared and subjected to co-IP with anti-Keap1 antibody. DMSO-treated H1299 cells were prepared and subjected to co-IP with control IgG. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(C) H1299 cells were treated with DMSO, DMF (25 μM), or tBHQ (25 μM) for 3 h, then WCL were prepared and subjected to co-IP with anti-Keap1 antibody. DMSO-treated H1299 cells were prepared and subjected to co-IP with control IgG. The immunoprecipitates were analyzed by WB with the indicated antibodies.
(D) WB analysis of the indicated proteins in WCL from parental and Keap1 KO H1299 cells treated with H2O2 (0,50,100 μM) for 3 h
(E) WB analysis of the indicated proteins in WCL from parental and Keap1 KO H1299 cells treated with DMSO, or DMF (12.5,25 μM) for 3 h
(F) WB analysis of the indicated proteins in WCL from H1299 cells stably overexpressing EV or FLAG-Keap1 and treated with DMSO or H2O2 (50,100 μM) for 3 h
(G) WB analysis of the indicated proteins in WCL from H1299 cells stably overexpressing EV or FLAG-Keap1 and treated with DMSO or DMF (12.5,25 μM) for 3 h
(H) WB analysis of the indicated proteins in WCL from parental and NRF2 KO H1299 cells treated with H2O2 (0,50,100 μM) for 3 h
(I) WB analysis of the indicated proteins in WCL from parental and NRF2 KO H1299 cells treated with DMSO, or DMF (12.5,25 μM) for 3 h.
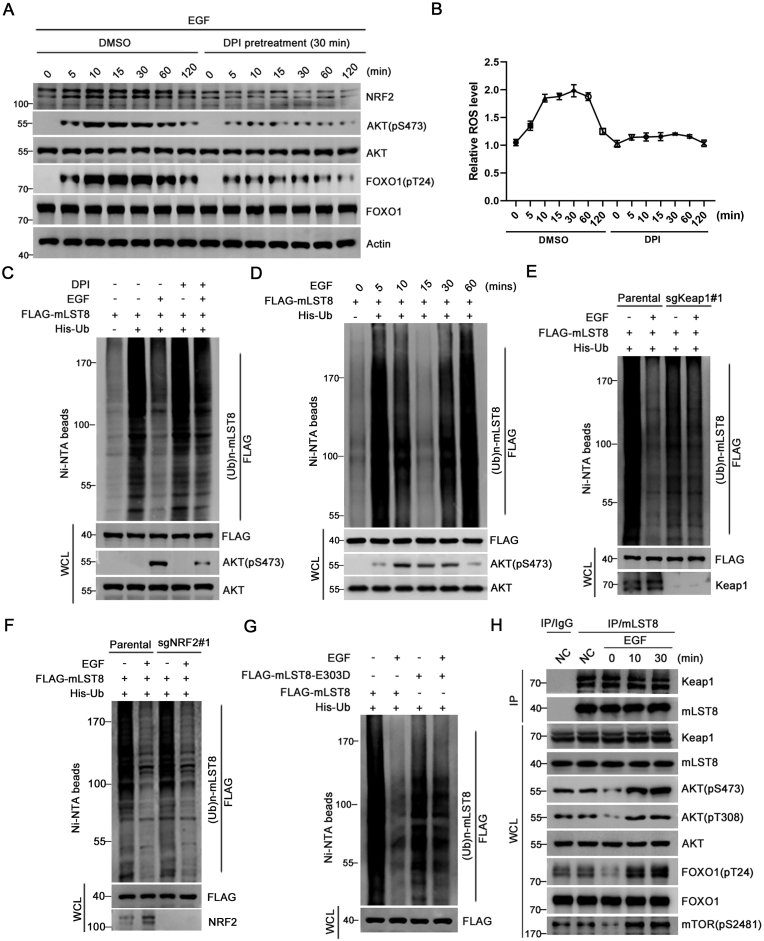
3.6. EGF-induced intracellular ROS burst mitigates Keap1-mediated mLST8 ubiquitination
Endogenous ROS—H2O2 is not only a source of oxidative stress but also plays an important role as a secondary messenger in the signal transduction networks of cells [33,34]. Treatment with growth factors ((EGF, Insulin, etc.) results in the assembly and activation of NADPH oxidase (NOX) complexes, which generate endogenous H2O2 [35,36]. EGF-stimulated generation of H2O2 and AKT activation were attenuated by NOX inhibitors, such as apocynin and Diphenyleneiodonium chloride (DPI) [37,38]. On the other hand, mLST8 ubiquitination was markedly reduced upon stimulation with growth factors, which is critical for mTORC2-AKT activation [19]. These facts prompted us to examine the impact of EGF-triggered intracellular ROS burst on mST8 ubiquitination and mTORC2-AKT activation.
We confirmed that EGF treatment led to mTORC2-AKT activation, intracellular ROS burst, and NRF2 protein stabilization, which were markedly mitigated by treatment with Diphenyleneiodonium chloride (DPI), a NOX inhibitor (Fig. 6A, B). Consistent with a previous study [19], EGF treatment markedly reduced mLST8 ubiquitination. However, EGF-induced reduction of mLST8 ubiquitination was compromised by DPI treatment (Fig. 6C). We found that reduced mLST8 ubiquitination by acute EGF (5–15 min) stimulation at early time points correlated with elevated AKT (473) phosphorylation. At a later time point (60 min), mLST8 ubiquitination had been restored to levels that were comparable to those in basal conditions, indicating that Keap1-mediated mLST8 is dynamically-regulated (Fig. 6D). Moreover, EGF-induced reduction in mLST8 ubiquitination was also compromised in Keap1 KO cells, but not in NRF2 KO cells (Fig. 6E, F). The mLST8-E303D mutant, which is incapable of binding to Keap1, was largely resistant to EGF-induced reduction of mLST8 ubiquitination (Fig. 6G). Although EGF treatment altered Keap1-mediated mLST8 ubiquitination, mLST8-Keap1 binding affinity was unchanged (Fig. 6H).
Fig. 6.
EGF-induced intracellular ROS burst mitigates Keap1-mediated mLST8 ubiquitination.
(A, B) WB analysis of the indicated proteins in WCL of serum-starved H1299 cells pretreated with DPI (10 μM) for 30 min prior to EGF (100 ng/ml) treatment for the indicated times. At each time point, the relative cellular ROS levels for each condition were measured and shown in (B), the P values are calculated by the Two-way ANOVA test.
(C) H1299 cells were transfected with the indicated plasmids and serum-starved for 24 h, then cells were pretreated with DPI (10 μM) for 30 min prior to EGF (100 ng/ml) treatment for 20 min. Ni-NTA pull-downs products and WCL were prepared for WB analysis.
(D) H1299 cells were transfected with the indicated plasmids and serum-starved for 24 h, then cells were treated with EGF (100 ng/ml) for the indicated times. Ni-NTA pull-downs products and WCL were prepared for WB analysis.
(E) Parental or Keap1 KO H1299 cells were transfected with the indicated plasmids and were serum-starved for 24 h, then cells were treated with EGF (100 ng/ml) for 20 min. Ni-NTA pull-downs products and WCL were prepared for WB analysis.
(F) Parental or NRF2 KO H1299 cells were transfected with the indicated plasmids and were serum-starved for 24 h, then cells were treated with EGF (100 ng/ml) for 15 min. Ni-NTA pull-downs products and WCL were prepared for WB analysis.
(G) H1299 cells were transfected with the indicated plasmids and serum-starved for 24 h, then cells were treated with EGF (100 ng/ml) for 20 min. Ni-NTA pull-downs products and WCL were prepared for WB analysis.
(H) WCL from serum-starved H1299 cells were stimulated with EGF (100 ng/ml) for indicated times, prepared and subjected to co-IP with IgG or anti-mLST8 antibody. The immunoprecipitates were analyzed by WB with the indicated antibodies.
Taken together, these results suggest that EGF-induced intracellular ROS burst mitigates Keap1-mediated mLST8 ubiquitination, thereby promoting mTORC2-AKT activation.
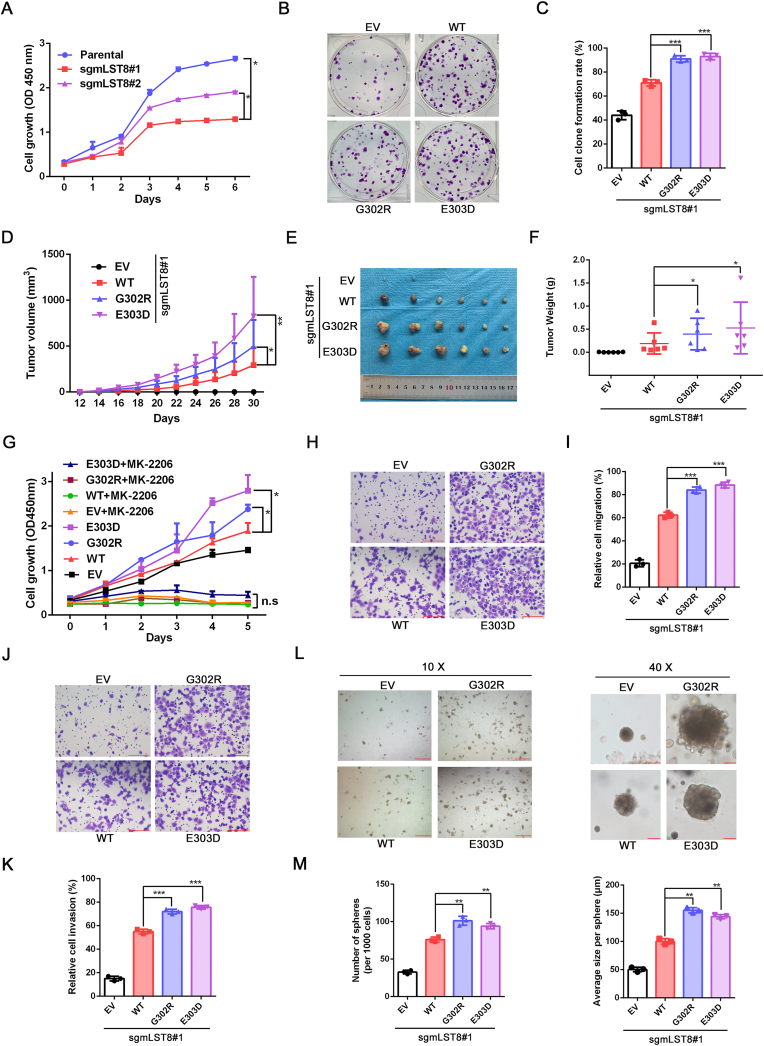
3.7. Loss of Keap1-mediated mLST8 ubiquitination enhances neoplastic phenotypes of cancer cells
Consistent with the substantial role of mTORC2 in cellular homeostasis, mLST8 KO markedly reduced cell growth rate (Fig. 7A). Colony formation and xenograft tumor assays showed that re-introduction of mLST8-WT restored cell growth in vitro and tumor growth in vivo. Notably, mLST8 KO cells re-introduced with mLST8-G302R or -E303D mutant showed a higher growth rate than mLST8 KO cells reconstituted with mLST8-WT (Fig. 7B-F). The mLST8-G302R and mLST8-E303D mutant-mediated acceleration of cell growth was largely blocked by treatment with a specific AKT inhibitor MK-2206 (Fig. 7G). Transwell assay results showed that mLST8-G302R or -E303D mutant expression increased cell migration and invasion compared to mLST8-WT (Fig. 7H-K). Furthermore, 3D sphere-formation assay results showed that the number and size of spheres in mLST8-KO cells reintroduced with mLST8-G302R or -E303D mutants were markedly increased, indicating increased anchorage-independent cell growth by cancer-derived mLST8 mutants (Fig. 7L-M). In contrast, the mLST8-WT, -G302R, and -E303D mutants showed comparable effects in terms of cell growth, migration, and invasion in A549 cells (Supplementary Figs. 6A–G).
Fig. 7.
Deficiency in Keap1-mediated mLST8 ubiquitination confers oncogenicity. (A) CCK-8 assays in parental and mLST8 KO H1299 cells for the indicated days. Data are shown as mean ± SD (n = 3).
(B, C) Colony formation assays in mLST8 KO H1299 cells stably overexpressing FLAG-mLST8-WT, -G302R, or -E303D mutant. Quantitative data are shown in (C). Data are shown as mean ± SD (n = 3).
(D-F) mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant were injected subcutaneously into the right flank of nude mice. Tumor volume was measured every other day since day 12 (D). Tumors in each group at day 30 were harvested and photographed (E) and tumor weight was documented (F). Data are shown as means ± SD (n = 6).
(G) CCK-8 assays in mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant treated with DMSO or MK-2206 (1 μM) for the indicated days. Data are shown as means ± SD (n = 3).
(H, I) Transwell migration assays in mLST8 KO H129 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant. Quantitative data are shown in (I). Data are shown as the means ± SD (n = 3).
(J, K) Transwell invasion assays in mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant. Quantitative data are shown in (K). Data are shown as the means ± SD (n = 3). Scale bar, 300 μm
(L, M) 3D sphere formation assays in mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant. Quantitative data are shown in (M). Data are shown as the mean ± SD (n = 3). Scale bars, 10x image: 600 μm; 40x image: 50 μm
P values were calculated by the one-way ANOVA test in (C, F, I, K, and M), and the two-way ANOVA test in (A, D, and G). *p < 0.05, **p < 0.01, ***p <0.001.
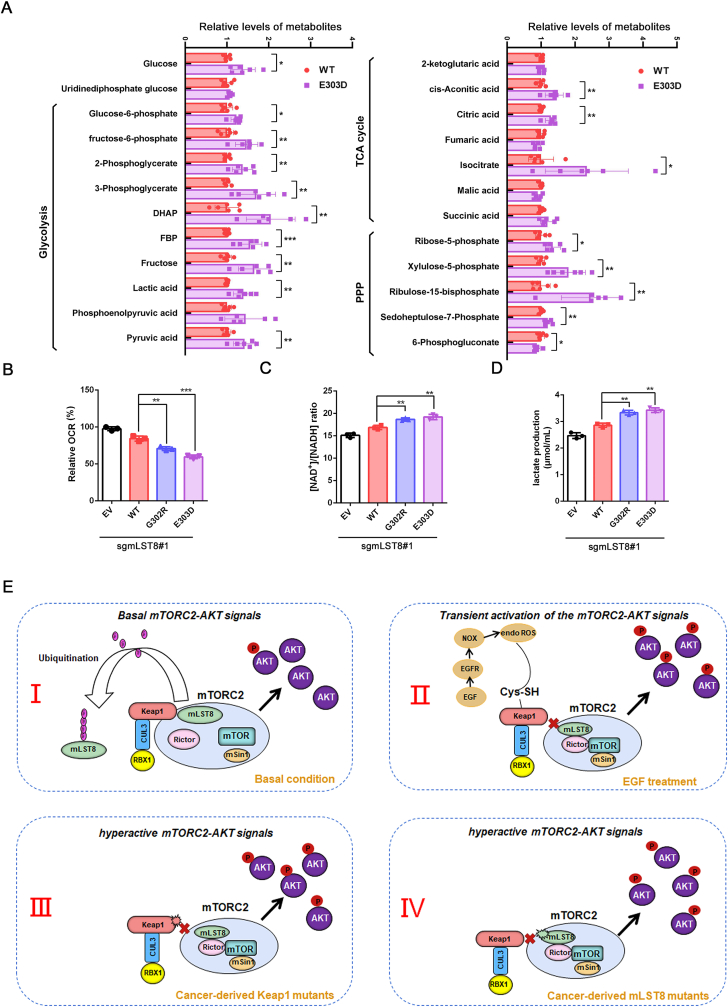
Aerobic glycolysis (the Warburg effect) is a core hallmark of cancer [39]. mTORC2 plays a pivotal role in controlling glucose metabolism [2]. Notably, using a high-throughput metabolomics approach, we found that multiple intermediates of glycolysis, TCA cycle, and the pentose phosphate (PPP) pathway were increased in mLST8 mutant expressing cells (Fig. 8A). We also found that mLST8 mutant expression led to a decreased oxygen consumption rate (Fig. 8B), an increased NAD+/NADPH ratio and lactate production (Fig. 8C-D), indicating an increased glycolysis. Taken together, these results indicate that loss of Keap1-mediated mLST8 ubiquitination promotes cell malignancy and remodels cell energy metabolism.
Fig. 8.
Deficiency in Keap1-mediated mLST8 ubiquitination remodels energy metabolism.
(A) The levels of representative metabolites of the glycolysis, TCA cycle and PPP pathway extracted from mLST8 KO H1299 cells stably overexpressing FLAG-mLST8-WT and E303D mutant were measured by UHPLC-MS/MS and normalized against the amount of total cellular proteins. Data are shown as means ± SD (n = 3).
(B) The OCR of mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant were measured by OCR assay Kit. Data are shown as means ± SD (n = 3).
(C) The intracellular NAD+/NADH ratio of mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant were measured by NAD+/NADH assay Kit. Data are shown as means ± SD (n = 3).
(D) The intracellular lactate levels of mLST8 KO H1299 cells stably overexpressing EV, FLAG-mLST8-WT, -G302R, or -E303D mutant were measured by Lactate Assay Kit. (E) A model summarizing the findings of the present study.
P values are calculated by the One-way ANOVA test in (A) and the Two-way ANOVA test in (B, C, D). *p < 0.05, **p < 0.01, ***p <0.001.
4. Discussion
Cancer cells usually utilize abnormal mTOR signaling to drive neoplastic growth and progression [40]. Oncogenic signal transduction through mTOR is common in cancer and leads to metabolic remodeling, enhanced proliferation, and increased metastatic potential. Different therapeutic strategies have been developed to target this pathway, resulting in the emergence of unique pharmacological compounds that have already been approved for use in clinical oncology or are currently under preclinical or clinical development [41]. Aberrant mTOR signaling in cancer is commonly achieved by loss-of-function mutations in tumor suppressors or gain-of-function mutations in oncogenes that feed into the mTOR pathway. Although the oncogenic effect of Keap1 mutations is often interpreted as increased NRF2 activity, accumulating evidence suggests that Keap1 plays pleiotropic roles in cellular hemostasis by regulating non-NRF2 substrates such as IKKβ and p62 [13]. The results of the present study showed that Keap1 acts as an upstream repressor of mTORC2 by promoting the non-degradative ubiquitination of mLST8 under physiological and pathological conditions (Fig. 8E). Although Keap1-mediated mLST8 ubiquitination decreases mTORC2 complex integrity, the biochemical details underlying this process remain unknown. However, the polyubiquitin chains on mLST8 may directly hinder complex assembly or indirectly recruit specific ubiquitin-binding proteins. Considering the high prevalence of Keap1 mutations in some cancer types, the biological significance of mTORC2-AKT hyperactivation in Keap1 mutation-induced tumorigenesis should be evaluated further in mouse models and clinical tumor samples. While our findings suggested that keap1 loss-induced mTORC2-AKT activation is mostly independent of NRF2, another group had reported that cancer-derived NRF2 mutants activates the mTORC1 pathway by increasing RagD expression [42]. This suggests that the functional interplay between Keap1, NRF2, and mTOR may be very intricate and dependent on the specific contexts. It would be necessary to experimentally validate their relationship in diverse cancer models by other research groups in the future.
Furthermore, we found that the cancer patient-derived mLST8 mutant (G302R and E303D), which locate in the ETGE motif recognized by Keap1, were unable to bind with Keap1. As a result, these two mLST8 mutants escaped CRL3Keap1-mediated ubiquitination and became more oncogenic than mLST8-WT, favoring mTORC-AKT activation and tumorigenesis. In addition, we observed that the dynamic regulation of mLST8 ubiquitination in response to growth factor was also lost due to mLST8 mutations. However, it is important to note that the mutation frequency of mLST8-ETGE motif is relatively low in human cancers compared to the high mutation frequency of NRF2-ETGE motif.
Excessive ROS have been shown to be toxic and are involved in the etiology and progression of various human diseases such as cancer, diabetes, and neurodegenerative disorders [43]. Over the past decades, however, a new paradigm has emerged that ROS function as fundamental intracellular second messenger with regulatory roles in a plethora of physiological processes [33,44]. ROS modulate protein structure and function through reverse protein oxidation, providing a mechanism by which changes in cellular redox states can be sensed and exploited [45]. The essential roles of endogenous ROS generation in the activation of growth factor signaling have been clearly established [35,36]. Paulsen et al. showed that EGF-mediated signaling results in H2O2 production and oxidation of the EGFR active site (Cys797) to enhance kinase activity and downstream AKT and ERK activation [38]. Su et al. identified two cysteines (Cys60 and C77) in the PH domain of AKT that can be oxidized, which promotes AKT recruitment to the plasma membrane and subsequent activation [37,46]. Our study provides an alternative possible explanation that exogenous or endogenous ROS promotes mTORC2-AKT activation by disrupting the Keap1-mLST8 axis. These findings may not be mutually exclusive. Considering the profound effects of ROS on cellular processes, multiple regulatory mechanisms underlying ROS-mediated AKT activation may simultaneously function in cells. In addition to AKT, mTORC2 has multiple downstream kinases such as SGK1, PKC, and PKN1 [47]. It will be interesting to investigate whether these mTORC2 effectors are regulated by Keap1 and exogenous/endogenous ROS.
Covalent modification of one or more critical cysteine residues in Keap1 represents a potential chemico-biological sensor for the activation of the NRF2 pathway. In addition to ROS, the disruption of glucose, lipid, and amino acid metabolism may lead to the accumulation of electrophilic intermediate metabolites that directly modify Keap1 [13]. In hereditary type 2 papillary renal cell carcinoma, fumarate accumulation, which results from fumarate hydratase mutations, leads to Keap1 inactivation and NRF2 activation [48,49]. Methylglyoxal and itaconate, which are endogenous metabolites derived from glycolysis and the TCA cycle, respectively, covalently modify Keap1 and promote NRF2 activation [[50], [51], [52]]. Given the coordinated interconnection between energy metabolism and the mTORC2 pathway, the effect of these metabolites on mTORC2 activity via modulation of Keap1-mediated mLST8 ubiquitination is worth investigating.
The Keap1-NRF2 interaction is susceptible to competition or interference from other proteins containing the ET(S)GE motif. The landscape of the Keap1 interactome has been exhaustively mapped in human cells and dozens of proteins have been identified [17,53]. Some of these Keap1 interactors, such as p62, DPP3, and PLAB2, reduce the Keap1 pool available for binding to NRF2, thus effectively protecting NRF2 from degradation and promoting cytoprotective gene expression [17,18,54]. NRF2 stability was not altered by mLST8 KO or overexpression. However, we cannot exclude the possibility that mLST8 still exerts an mTORC2-independent function by regulating NRF2 stability in specific tissues or under certain internal/external stimuli. Furthermore, although mLST8 and Keap1 genes are present in Drosophila, the Drosophila mLST8 homolog does not harbor the ETGE motif, implying that mLST8 is unlikely to be regulated by Keap1 in Drosophila. The ETGE motif of mLST8 has emerged relatively later in eukaryote evolution, which may reflect the demand for fine-tuning mTORC2 activity in higher organisms [55].
In conclusion, our study revealed an unrecognized role for Keap1 in the regulation of the mTORC2 pathway. A thorough understanding of the effect of the Keap1-mTORC2-AKT axis would guide the use of mTOR and AKT inhibitors targeting Keap1 mutation-driven cancers.
Ethics statement
Our study did not include human participants, data or tissue samples. Animal studies were approved by the Ethics Review Committee for Animal Experimentation of Fudan University.
Author contribution statement
CJ.W. conceived the study. YJ.C., DY.J., HY.H., HR.S., and YJ.L. performed experiments and data analyses. YJ.C., DY.J., HY.H., HR.S., YJ.L., R.M., K.G., Y.L., PZ.Z., and CJ. W. analyzed and interpreted the data. CJ.W. wrote and revised the manuscript.
Funding statement
This work was supported in part by the National Natural Science Foundation of China (No. 32370726, 81972396, 91957125, and 81672558 to CJ.W.; No. 82272992, 91954106, and 81872109 to K.G.; No. 81760463 to R.M., and No. 82172938 to PZ.Z.), State Key Development Programs of China (No. 2022YFA1104200 to C.W.), Natural Science Foundation of Shanghai (No. 22ZR1449200 to K.G; 22ZR1406600 to CJ.W.), Open Research Fund of the State Key Laboratory of Genetic Engineering, Fudan University (No. SKLGE-2111 to K.G.), Science and Technology Research Program of Shanghai (No. 9DZ2282100), and Central Guidance on Local Science and Technology Development Foundation (No. 2021ZY0037 to R.M.).
Declaration of competing interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102872.
Contributor Information
Ren Mo, Email: moren325@163.com.
Kun Gao, Email: kungao@tongji.edu.cn.
Chenji Wang, Email: chenjiwang@fudan.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Kim J., Guan K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 2.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., et al. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 5.Szwed A., Kim E., Jacinto E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol. Rev. 2021;101:1371–1426. doi: 10.1152/physrev.00026.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu W., Hall M.N. Regulation of mTORC2 signaling. Genes. 2020;11 doi: 10.3390/genes11091045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genschik P., Sumara I., Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J. 2013;32:2307–2320. doi: 10.1038/emboj.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerins M.J., Ooi A. A catalogue of somatic NRF2 gain-of-function mutations in cancer. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-31281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloer E.W., Goldfarb D., Schrank T.P., Weissman B.E., Major M.B. NRF2 activation in cancer: from DNA to protein. Cancer Res. 2019;79:889–898. doi: 10.1158/0008-5472.CAN-18-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W.L., Papagiannakopoulos T. The pleiotropic role of the KEAP1/NRF2 pathway in cancer. Annu. Rev. Cell Biol. 2020;4:413–435. [Google Scholar]
- 14.Hwang Y., Kim L.C., Song W., Edwards D.N., Cook R.S., Chen J. Disruption of the scaffolding function of mLST8 selectively inhibits mTORC2 assembly and function and suppresses mTORC2-dependent tumor growth in vivo. Cancer Res. 2019;79:3178–3184. doi: 10.1158/0008-5472.CAN-18-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao K., Zhang Y., Shi Q., Zhang J., Zhang L., Sun H., et al. iASPP-PP1 complex is required for cytokinetic abscission by controlling CEP55 dephosphorylation. Cell Death Dis. 2018;9:528. doi: 10.1038/s41419-018-0561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huttlin E., Ting L., Bruckner R., et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu K., Alcivar A.L., Ma J., Foo T.K., Zywea S., Mahdi A., et al. NRF2 induction supporting breast cancer cell survival is enabled by oxidative stress-induced DPP3-KEAP1 interaction. Cancer Res. 2017;77:2881–2892. doi: 10.1158/0008-5472.CAN-16-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Martin P., Komatsu M. p62/SQSTM1 - steering the cell through health and disease. J. Cell Sci. 2018:131. doi: 10.1242/jcs.222836. [DOI] [PubMed] [Google Scholar]
- 19.Wang B., Jie Z., Joo D., Ordureau A., Liu P., Gan W., et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545:365–369. doi: 10.1038/nature22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichard E.L., Chirico G.G., Dewey W.J., Nassif N.D., Bard K.E., Millas N.E., et al. Substrate ubiquitination controls the unfolding ability of the proteasome. J. Biol. Chem. 2016;291:18547–18561. doi: 10.1074/jbc.M116.720151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., et al. Defining a cancer dependency map. Cell. 2017;170:564–576 e516. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price C., Gill S., Ho Z.V., Davidson S.M., Merkel E., McFarland J.M., et al. Genome-wide interrogation of human cancers identifies EGLN1 dependency in clear cell ovarian cancers. Cancer Res. 2019;79:2564–2579. doi: 10.1158/0008-5472.CAN-18-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayraktar E.C., La K., Karpman K., Unlu G., Ozerdem C., Ritter D.J., et al. Metabolic coessentiality mapping identifies C12orf49 as a regulator of SREBP processing and cholesterol metabolism. Nat. Metab. 2020;2:487–498. doi: 10.1038/s42255-020-0206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A., Misra V., Thimmulappa R.K., Lee H., Ames S., Hoque M.O., et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3(10) doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R., An J., Ji F., Jiao H., Sun H., Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008;373(1):151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein L.D., Lee J., Gnad F., Klijn C., Schaub A., Reeder J., et al. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep. 2016;16(10):2605–2617. doi: 10.1016/j.celrep.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. U. S. A. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee D.F., Kuo H.P., Liu M., Chou C.K., Xia W., Du Y., et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol. Cell. 2009;36(1):131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trachootham D., Lu W., Ogasawara M.A., et al. Redox regulation of cell survival. Antioxidants Redox Signal. 2008;10:1343–1374. doi: 10.1089/ars.2007.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mittler R. ROS are good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Cai W., Andres D.A. mTORC2 associates with Rit and is required for Rit-mediated Akt phosphorylation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koundouros N., Poulogiannis G. Phosphoinositide 3-kinase/akt signaling and redox metabolism in cancer. Front. Oncol. 2018;8:160. doi: 10.3389/fonc.2018.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulsen C.E., Carroll K.S. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem. Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finkel T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller E.W., Tulyathan O., Isacoff E.Y., Chang C.J. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat. Chem. Biol. 2007;3:263–267. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 36.Woo H.A., Yim S.H., Shin D.H., Kang D., Yu D.Y., Rhee S.G. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Su Z., Burchfield J.G., Yang P., Humphrey S.J., Yang G., Francis D., et al. Global redox proteome and phosphoproteome analysis reveals redox switch in Akt. Nat. Commun. 2019;10:5486. doi: 10.1038/s41467-019-13114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen C.E., Truong T.H., Garcia F.J., Homann A., Gupta V., Leonard S.E., Carroll K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat. Chem. Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Kim L.C., Cook R.S., Chen J. mTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene. 2017;36:2191–2201. doi: 10.1038/onc.2016.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wander S.A., Hennessy B.T., Slingerland J.M. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J. Clin. Invest. 2011;121:1231–1241. doi: 10.1172/JCI44145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata T., Saito S., Kokubu A., Suzuki T., Yamamoto M., Hirohashi S. Global downstream pathway analysis reveals a dependence of oncogenic NF-E2-Related factor 2 mutation on the mTOR growth signaling pathway. Cancer Res. 2010;70(22):9095–9105. doi: 10.1158/0008-5472.CAN-10-0384. [DOI] [PubMed] [Google Scholar]
- 43.Houstis N., Rosen E.D., Lander E.S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 44.D'Autréaux B., Toledano M.B. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 45.Veal E.A., Day A.M., Morgan B.A. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 46.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic. Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Battaglioni S., Benjamin D., Wälchli M., Maier T., Hall M.N. mTOR substrate phosphorylation in growth control. Cell. 2022;185:1814–1836. doi: 10.1016/j.cell.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Kinch L., Grishin N.V., Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell. 2011;20:418–420. doi: 10.1016/j.ccr.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adam J., Hatipoglu E., O'Flaherty L., Ternette N., Sahgal N., Lockstone H., et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bambouskova M., Gorvel L., Lampropoulou V., Sergushichev A., Loginicheva E., Johnson K., et al. Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature. 2018;556:501–504. doi: 10.1038/s41586-018-0052-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bollong M.J., Lee G., Coukos J.S., Yun H., Zambaldo C., Chang J.W., et al. A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling. Nature. 2018;562:600–604. doi: 10.1038/s41586-018-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hast B.E., Goldfarb D., Mulvaney K.M., Hast M.A., Siesser P.F., Yan F., et al. Proteomic analysis of ubiquitin ligase KEAP1 reveals associated proteins that inhibit NRF2 ubiquitination. Cancer Res. 2013;73:2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma J., Cai H., Wu T., Sobhian B., Huo Y., Alcivar A., et al. PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol. Cell Biol. 2012;32:1506–1517. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beauchamp E.M., Platanias L.C. The evolution of the TOR pathway and its role in cancer. Oncogene. 2013;32:3923–3932. doi: 10.1038/onc.2012.567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.