Abstract
Scarcity of labels for medical images is a significant barrier for training representation learning approaches based on deep neural networks. This limitation is also present when using imaging data collected during routine clinical care stored in picture archiving communication systems (PACS), as these data rarely have attached the high-quality labels required for medical image computing tasks. However, medical images extracted from PACS are commonly coupled with descriptive radiology reports that contain significant information and could be leveraged to pre-train imaging models, which could serve as starting points for further task-specific fine-tuning.
In this work, we perform a head-to-head comparison of three different self-supervised strategies to pre-train the same imaging model on 3D brain computed tomography angiogram (CTA) images, with large vessel occlusion (LVO) detection as the downstream task. These strategies evaluate two natural language processing (NLP) approaches, one to extract 100 explicit radiology concepts (Rad-SpatialNet) and the other to create general-purpose radiology reports embeddings (DistilBERT). In addition, we experiment with learning radiology concepts directly or by using a recent self-supervised learning approach (CLIP) that learns by ranking the distance between language and image vector embeddings. The LVO detection task was selected because it requires 3D imaging data, is clinically important, and requires the algorithm to learn outputs not explicitly stated in the radiology report.
Pre-training was performed on an unlabeled dataset containing 1,542 3D CTA - reports pairs. The downstream task was tested on a labeled dataset of 402 subjects for LVO. We find that the pre-training performed with CLIP-based strategies improve the performance of the imaging model to detect LVO compared to a model trained only on the labeled data. The best performance was achieved by pre-training using the explicit radiology concepts and CLIP strategy.
Index Terms: Large vessel occlusion, ischemic stroke, CLIP, BERT
1. INTRODUCTION
Scarcity of high quality labeled medical imaging data is a major hurdle for the development of machine learning models, and especially for deep neural networks. This is because data annotation is a manual and time-consuming process that can be expensive and necessitates medical expertise. Medical imaging data is typically stored in a picture archiving and communications system (PACS) server, to facilitate storage and sharing. In addition to images, PACS also store radiology reports that contain significant information that could be leveraged to pre-train imaging models with large datasets and no manual labeling. Several model pre-training and self-supervision approaches have been proposed to reduce model dependency on larger annotated datasets [1, 2, 3]. Recent studies [4, 5] have shown that the network can be trained without any explicit data annotation by joint modeling of radiology reports and medical images. These approaches have been using pre-trained imaging models and 2D X-ray images and, with a downstream task closely resembling what has been explicitly described in the radiology reports. It is not clear how self-supervised strategies trained on radiology reports would work on more complex downstream tasks and with 3D image data.
Here, we use 3D Brain CT angiography (CTA) data with a large unlabeled dataset containing radiology reports, and a smaller labeled dataset with Large vessel occlusion (LVO). LVO is the obstruction of large, proximal cerebral arteries which accounts for 24–46% of acute ischemic stroke (AIS), when including both A2 and P2 segments of the anterior and posterior cerebral arteries [6]. Significant brain regions are frequently damaged by the involvement of proximal vasculature, leading to significant neurological impairments. LVO is never explicitly mentioned in the reports and its detection requires the use of 3D-based feature representation as vessel occlusions can happen in any direction and cannot be visible from 2D slices evaluated independently. We experiment with three different strategies using the same imaging model backbone and compare them to a model trained only on labeled data. In the first strategy, we use a natural language processing (NLP) model, Rad-SpatialNet [7], to extract 100 explicit concepts from the reports and use them as a target for pre-training the imaging network with a cross-entropy loss; in the second strategy, we use the DistilBERT NLP model [8] to generate a radiology report vector embedding and then pre-train the imaging model by minimizing the image model vector embeddings to the matching report vector embeddings using the CLIP strategy [9], which is a recently developed type of contrasting learning for language–image training; in the third strategy, we pre-train the imaging model using the explicit concepts from reports, as in the first experiment, but this time by using the CLIP strategy to minimize the distance between the image model vector embeddings to the vector embeddings generated by the explicit concepts from reports. The performance of these different strategies demonstrated that explicit labels are not required to perform well on interpretation tasks when corresponding radiology reports/concepts are used for pre-training. This could be because the reports are naturally labeled and can provide a natural source of supervision. The LVO detection task was selected because it requires 3D imaging data, it is clinically relevant, and because radiology reports will not verbatim state the presence of an LVO. To the best of our knowledge, our approach is the first to compare and contrast pre-training strategies using radiology reports and 3D imaging data.
2. MATERIALS AND METHODS
Materials:
The data for LVO detection includes unlabeled data (brain CTA images with corresponding radiologist reports) and labeled data (brain CTA images without any reports). This study was performed in accordance with the guidelines from the Helsinki Declaration and under IRB HSC-MS-19–0630 approved by the University of Texas Health Science Center at Houston (UTHealth Houston) IRB and Memorial Hermann Hospital. The unlabeled dataset contains 1542 subjects. The labeled dataset contains 402 ischemic stroke subjects, 170 without LVO and 232 with LVO. The data was acquired at Memorial Hermann hospital system, Houston, Texas, USA. LVO labels were manually extracted from the stroke center’s radiologist reports where LVO is defined as an occlusion in the ICA, M1, M2, A1 brain vasculature, high-grade stenosis, or near-complete occlusions that are not considered LVOs.
Method:
Figure 1 depicts the network architecture with four distinct experiment pipelines. Let and represent the CTA image and label from the labeled dataset, and and represent the CTA image and text report from the unlabeled dataset. and as the image and text encoder in stage 1 respectively to learn image and text feature representations and is the image encoder used in stage 2, to learn image feature representations . In all experiments, the same image encoder backbone, a standard 3D ResNet18, is used.
Fig. 1.
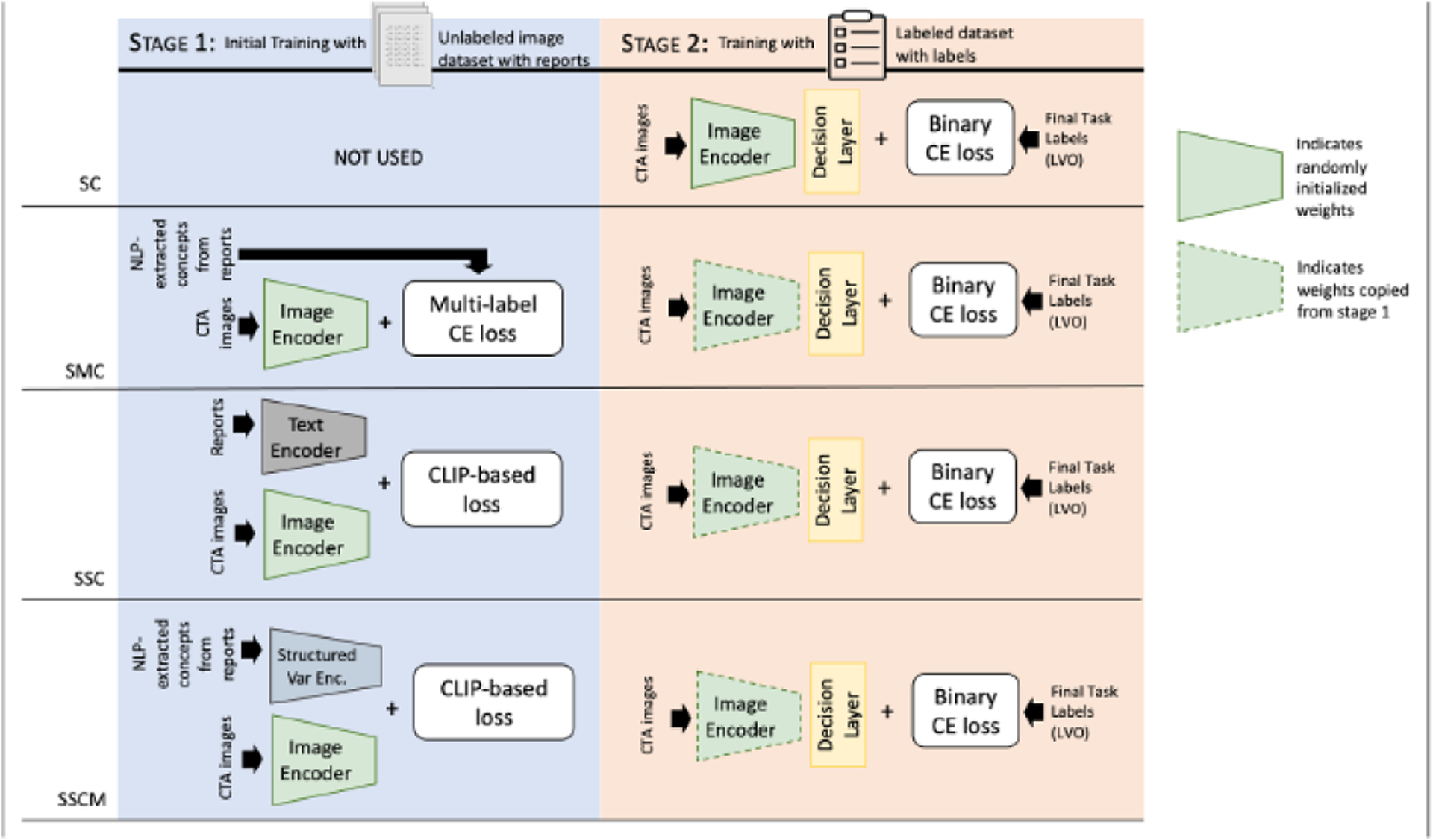
Four training strategies compared. Stage 1 is the training pipeline with the unlabeled dataset. The model learns features from raw radiologist reports, which serve as a natural source of supervision. Stage 2 is the training pipeline with the labeled dataset. SC: supervised classifier, SMC: supervised multi-label classifier, SSC: self-supervised classifier, and SSCM: self-supervised classifier using multi-labeled data.
Supervised classifier (SC):
The network for SC consists of an image encoder that has been trained on the labeled dataset. Given brain CTA images from the labeled dataset and corresponding labels (n is the number of CTA images), the goal is to learn mapping that correctly classifies a brain CTA image as having LVO or no LVO . The weights for the image encoder were randomly initialized. The image representations generated by the image encoder are then passed to the decision layer which is a fully connected layer. The binary cross entropy (BCE) was used as the loss function for training the classifiers and the image encoder on the labeled data.
Supervised multi-label classifier (SMC):
First, NLP concepts were extracted using a two-stage relation extraction model [10], where the first stage extracts concepts (e.g., findings, devices, anatomical locations, spatial trigger terms), and the second stage connects the concepts through relations as well as identifying other secondary concepts (e.g., distance, laterality, negation). The underlying machine learning model used for each stage is a BERT [11] question-answering-style information extraction (QA-as-IE) approach. See [7] for more information on the underlying Rad-SpatialNet schema. The radiology reports described both brain and neck CTA acquisitions, since the images used were pre-processed to include the brain only, we removed any radiology concept referring to non-brain areas.
The SMC network consists of an image encoder that is trained as a multi-label classification problem on the labels extracted from NLP-extracted concepts. Given brain CTA images from the unlabeled dataset and corresponding multi-labels (n is the number of CTA images) and i = 1,2,…100, the goal is to learn mapping that correctly classifies a brain CTA image as having one or more of the multi-label which signifies the presence of LVO. Multi-label cross entropy (CE) was used as the loss function for training the image encoder:
| (1) |
where is the predicted class probability. Here, the unknown class labels were ignored while calculating CE loss. Then, using labeled data, the downstream task (stage 2) was trained and tested in the same manner as the SC method, with the only difference being that the weights are not initiated randomly, but rather copied from stage 1.
Self-supervised classifier (SSC):
The SSC network consists of an image encoder and a text encoder that were trained on images and reports from unlabeled datasets using CLIP. Given brain CTA images from the unlabeled dataset and corresponding text report , the goal is to learn a mapping that correctly classifies a brain CTA image as having LVO or no LVO. The image features and the text features are represented as 2048 and 768-dimensional vectors, respectively. A separate projection module was used to make the encodings have a similar shape (256 in our case). Then CLIP-based loss was then used as the loss function for jointly training the image and text encoders. The logits and target are calculated as and . Where sim(·, ·) is the similarity measurement defined as where is the transpose function, and is the temperature parameter, which is . Then, using labeled data downstream task (stage 2) was trained and tested in the same manner as the SC method, with the only difference being that the weights are not initiated randomly, but rather copied from stage 1.
Self-supervised classifier using multi-labeled data (SSCM):
The SSCM network consists of an image encoder trained on images and a structured variational encoder whose input is multi-label data used as features. The network is trained on the unlabeled dataset using CLIP. Given brain CTA images from the unlabeled dataset and corresponding NLP-extracted concepts , the goal is to learn a mapping that correctly classifies a brain CTA image as having LVO or not. The image features and the structured variational encoder features are represented as 2048 and 16-dimensional vectors, respectively. A separate projection module was used to make the encodings have a similar shape (256 in our case). Then a CLIP-based loss function was then used as the loss function for joint training. The logits and target are calculated as and . Where . Then, using labeled data, the downstream task (stage 2) was trained and tested using labeled data in the same manner as the SC method, with the only difference being that the weights are not initiated randomly, but rather copied from stage 1.
Pre-processing:
The skull stripping in each CTA volume was performed using the FSL-based pipeline described in [12] and then linearly registered to a common template with 1mm isometric voxels and image resolution of 182×218×182. All image registrations were manually checked for errors, when registrations errors were found, the images were not included in the dataset.
For the radiologist reports, we extracted the impressions section, since it contains a concise summary of the entire report. Text from reports was tokenized using the DistilBert Tokenizer. Two special tokens, CLS and SEP, were added to the actual input tokens. These special tokens indicate the beginning and end of a sentence. To capture the entirety of a sentence, we used the CLS token’s final representations, and this representation captures the overall meaning of the sentence.
Implementation Details:
The network architectures include a 3D ResNet18 [13], for the image encoder and a DistilBERT [8] for the text encoder. The 3D ResNet18 weights were not initialized using any type of external datasets, such as ImageNet. We used DistilBERT pre-trained on the Book-Corpus dataset in a self-supervised setting. This model is uncased (e.g., does not differentiate between “disease” and “Disease”). The image encoder has a base size of 33 million parameters and 70 layers of architecture, with 18 deep layers. The text encoder employs text embeddings with a maximum token length of 200. We used the Adam to minimize the loss with a learning rate of 0.0001 and 0.00001 for stage 1 and stage 2 respectively. The model has a batch size of 14 and is trained for 100 epochs. All of the images from the unlabeled dataset are used for training stage 1 and the labeled dataset is divided into train (60%), validation (20%), and test set (20%). This same data split is used for stage 2 of all four experiments. The final model performance is evaluated on the test set. In both stages, the validation mean area under the ROC (AUC) after each epoch is calculated, and the best model checkpoint is saved if the model outperforms the previous best model during training.
3. RESULTS
We used the AUC for our model evaluation. Table 1 gives the AUC score for SC, SMC, SSC, and SSCM networks. The AUC obtained for the self-supervised network using reports (0.80) and multi-label data as features derived from NLP-extracted concepts from reports (0.75) is better than the supervised network on binary data (0.73) and multi-label data (0.64). In self-supervised models, the SSC model recognizes the presence of LVO better than the SSCM model, which signifies the NLP-extracted concepts from reports might not capture as much information as the radiologist reports themselves.
Table 1.
AUC score for SC, SMC, SSC, and SSCM networks.
| Approach | SC | SMC | SSC | SSCM |
|---|---|---|---|---|
| AUC | 0.73 | 0.64 | 0.80 | 0.75 |
4. DISCUSSION AND CONCLUSION
Self-supervised strategies to pre-train medical image computing models using radiology reports have the potential of enabling researchers to leverage large amounts of unlabeled data, reducing the need for labeled datasets and the inefficiencies caused by extensive labeling efforts. In this work, we performed a head-to-head comparison of three different self-supervised strategies to pre-train the same imaging model on 3D brain CTA images, with LVO detection as the downstream task. The first strategy (SMC) evaluated an NLP approach with extracted radiology concepts as target labels. The second (SSC) and third (SSCM) strategies experimented with reports embeddings and NLP-extracted radiology concepts by using a self-supervised learning approach (CLIP) that learns by ranking the distance between language and image vector embeddings. Our results indicated that both pre-trained models using CLIP achieved higher performance compared to a model trained only on the labeled data. Specifically, the SSC and SSCM methods achieved an AUC +0.07 and +0.02 AUC points above that of the fully supervised model (SC). Surprisingly, the SMC model achieved lower performance than SC, this is likely due to a local minimum in the pre-training phase which cannot be overcome by the smaller learning rate used during the fine-tuning. Increasing the learning rate would lead to similar performance as SC, however, this would not be a head-to-head comparison with the other strategies.
The image encoder used is a standard 3D ResNet not specialized for LVO detection, other architectures [14, 15, 16] can lead to higher absolute performance, however, we decided not to use them to better evaluate any gains on a general 3D convolutional neural network that has not been specifically optimized for the downstream tasks, whose specialized design could bias the findings.
In addition, we did not evaluate the imaging model performance as a zero-shot learner by inputting text prompts to the text encoder as described in [4]. This is because it would have only be possible in the SSC approach, not allowing a head-to-head comparison, and it would have been severely limited by the fact that LVO is rarely explicitly stated in the radiology report making the design of the text prompt a challenge.
In future work, we will train Rad-SpatialNet on CTA reports for improving the text encoder, evaluate other strategies to initialize the model weights, and we will investigate the potential of SSC and SSCM with general-purpose image encoders based on vision transformers and other downstream tasks with alternative 3D imaging modalities such as MRI.
ACKNOWLEDGMENTS
This work is supported by NIH grants R01NS121154 and R21EB029575. Finally, we would like to thank the Memorial Hermann Health System for enabling the data collection effort.
REFERENCES
- [1].Tan Chuanqi, Sun Fuchun, Kong Tao, Zhang Wenchang, Yang Chao, and Liu Chunfang, “A survey on deep transfer learning,” in International conference on artificial neural networks. Springer, 2018, pp. 270–279. [Google Scholar]
- [2].Chen Ting, Kornblith Simon, Norouzi Mohammad, and Hinton Geoffrey, “A simple framework for contrastive learning of visual representations,” in International conference on machine learning. PMLR, 2020, pp. 1597–1607. [Google Scholar]
- [3].He Kaiming, Fan Haoqi, Wu Yuxin, Xie Saining, and Girshick Ross, “Momentum contrast for unsupervised visual representation learning,” in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2020, pp. 9729–9738. [Google Scholar]
- [4].Tiu Ekin, Talius Ellie, Patel Pujan, Langlotz Curtis P, Ng Andrew Y, and Rajpurkar Pranav, “Expert-level detection of pathologies from unannotated chest x-ray images via self-supervised learning,” Nature Biomedical Engineering, pp. 1–8, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chauhan Geeticka, Liao Ruizhi, Wells William, Andreas Jacob, Wang Xin, Berkowitz Seth, Horng Steven, Szolovits Peter, and Golland Polina, “Joint modeling of chest radiographs and radiology reports for pulmonary edema assessment,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2020, pp. 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rennert Robert C, Wali Arvin R, Steinberg Jeffrey A, Santiago-Dieppa David R, Olson Scott E, Pannell J Scott, and Khalessi Alexander A, “Epidemiology, natural history, and clinical presentation of large vessel ischemic stroke,” Neurosurgery, vol. 85, no. suppl 1, pp. S4–S8, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Datta Surabhi, Ulinski Morgan, Godfrey-Stovall Jordan, Khanpara Shekhar, Riascos-Castaneda Roy F, and Roberts Kirk, “Rad-spatialnet: a frame-based resource for fine-grained spatial relations in radiology reports,” in International Conference on Language Resources and Evaluation, 2020, vol. 2020, p. 2251. [PMC free article] [PubMed] [Google Scholar]
- [8].Sanh Victor, Debut Lysandre, Chaumond Julien, and Wolf Thomas, “Distilbert, a distilled version of bert: smaller, faster, cheaper and lighter,” arXiv preprint arXiv:1910.01108, 2019. [Google Scholar]
- [9].Radford Alec, Kim Jong Wook, Hallacy Chris, Ramesh Aditya, Goh Gabriel, Agarwal Sandhini, Sastry Girish, Askell Amanda, Mishkin Pamela, Clark Jack, et al. , “Learning transferable visual models from natural language supervision,” in International Conference on Machine Learning. PMLR, 2021, pp. 8748–8763. [Google Scholar]
- [10].Datta Surabhi and Roberts Kirk, “Fine-grained spatial information extraction in radiology as two-turn question answering,” International journal of medical informatics, vol. 158, pp. 104628, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina, “Bert: Pre-training of deep bidirectional transformers for language understanding,” arXiv preprint arXiv:1810.04805, 2018. [Google Scholar]
- [12].Muschelli John, Ullman Natalie L, Mould W Andrew, Vespa Paul, Hanley Daniel F, and Crainiceanu Ciprian M, “Validated automatic brain extraction of head ct images,” Neuroimage, vol. 114, pp. 379–385, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].He Kaiming, Zhang Xiangyu, Ren Shaoqing, and Sun Jian, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778. [Google Scholar]
- [14].Czap Alexandra L., Bahr-Hosseini Mersedeh, Singh Noopur, Yamal Jose-Miguel, and et al. , “Machine Learning Automated Detection of Large Vessel Occlusion From Mobile Stroke Unit Computed Tomography Angiography,” Stroke, vol. 53, no. 5, pp. 1651–1656, May 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rodrigues Gabriel, Barreira Clara M., Bouslama Mehdi, Haussen Diogo C., Al-Bayati Alhamza, Pisani Leonardo, and et al. , “Automated Large Artery Occlusion Detection in Stroke: A Single-Center Validation Study of an Artificial Intelligence Algorithm,” Cerebrovascular Diseases, pp. 1–6, Oct. 2021. [DOI] [PubMed] [Google Scholar]
- [16].Barman Arko, Inam Mehmet E., Lee Songmi, Savitz Sean, Sheth Sunil, and Giancardo Luca, “Determining Ischemic Stroke From CT-Angiography Imaging Using Symmetry-Sensitive Convolutional Networks,” in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Apr. 2019, pp. 1873–1877. [Google Scholar]


