Abstract
We have developed small-volume (50 or 250 μl)-format branched-DNA assays for human immunodeficiency virus type 1 (HIV-1) RNA for use with specimens in which the volume is limited and/or a high viral load is anticipated. These formats exhibited good correlation with the standard 1-ml format; high specificity, reproducibility, and linearity; and no significant difference in the quantification of HIV-1 subtypes.
Human immunodeficiency virus type 1 (HIV-1) RNA level has been recognized as one of the most important indicators of disease progression in HIV-1-infected adults (3, 5). Direct viral load measurements can be obtained by use of a number of commercially available quantitative methods (5). One of these, the branched-DNA (bDNA) assay (Quantiplex HIV-1 RNA; Chiron Corporation, Emeryville, Calif.), is easy to use and yields highly reproducible results (2, 4, 6). However, the relatively large 1-ml plasma volume requirement potentially limits its application in the testing of small-volume samples such as those from pediatric patients. We report the development of small-volume formats for the bDNA assay that require either 50 or 250 μl of plasma for each determination.
Banked acid-citrate-dextrose plasma samples (n = 134) were available from 37 children (ages, 0 to 162 months) born to HIV-1-infected mothers attending the Chicago Children’s Memorial, Cook County, and Wyler Children’s Hospitals. Of these, 5 (13.5%) were perinatally exposed but not infected (category E), 3 (8%) were asymptomatic (category N), 16 (43%) were mildly symptomatic (category A), 5 (13.5%) were moderately symptomatic (category B), and 8 (22%) were AIDS patients (category C) as defined by the 1994 HIV-1 classification system for children (1). Sixteen patients were receiving either zidovudine, dideoxyinosine, dideoxycytosine, or stavudine alone or combined antiretroviral therapies when samples were collected. All pediatric plasma samples were tested in duplicate with the 50-μl-format bDNA assay (described below). Samples with values below the detection limit were retested as singles with the 1-ml-format bDNA assay when a sufficient volume was available. In addition, 132 EDTA plasma samples from HIV-1-infected adults undergoing various antiviral therapies were obtained from one clinical site in the Bahamas and four sites in Toronto associated with the Hospital for Sick Children. Informed consent was obtained from patients or their parents or guardians, and human experimentation guidelines of each country and institution were followed in the conduct of clinical research.
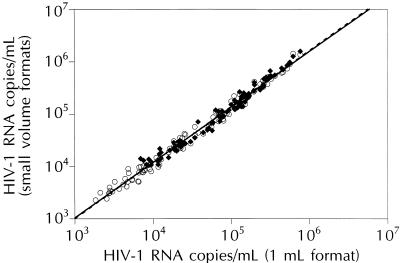
For HIV-1 RNA quantification, the Quantiplex HIV-1 RNA 2.0 assay was performed in accordance with manufacturer’s instructions, with the following exceptions. For the 250-μl format, 250 μl of plasma was centrifuged and the resulting viral pellet was tested. For the 50-μl format, 50 μl of plasma was tested directly without centrifugation. The relationships between HIV-1 RNA quantification values measured with the 1-ml versus the 250- and 50-μl formats were evaluated by testing specimens with viral levels that spanned the dynamic range of each format (500 to 800,000, 2,000 to 3,200,000, and 10,000 to 16,000,000 copies/ml for the 1-ml, 250-μl, and 50-μl formats, respectively). As shown in Fig. 1, the quantification values of the 1-ml format were highly correlated with those of the 250-μl (R2 = 0.988) and 50-μl formats (R2 = 0.976).
FIG. 1.
Correlation between HIV-1 RNA quantification values obtained with the 1-ml-versus the 250-μl (○; n = 118)- and 50-μl (⧫; n = 90)-format bDNA assays.
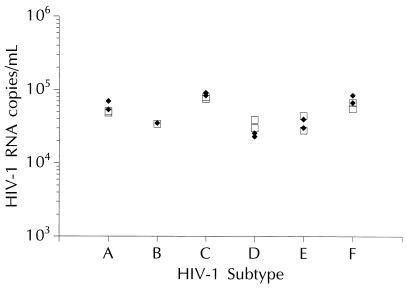
Performance characteristics, including specificity, reproducibility, linearity, and quantification of HIV-1 subtypes, were analyzed by using the 50-μl-format bDNA assay. Specificity was determined by analyzing plasma specimens from 116 HIV-1-seronegative blood donors. The number of relative luminescence units for all seronegative specimens fell well below the cutoff number of relative luminescence units for the assay (10,000 copies/ml), indicating that the 50-μl-format bDNA assay provides 100% specificity. Reproducibility was evaluated by testing replicates of plasma specimens from HIV-1-seropositive individuals. Three specimens were tested by using two lots of reagents in two assay runs performed by one operator for a total of 12 determinations per specimen. Mean quantification values were 10,500, 12,993, and 13,812 HIV-1 RNA copies/ml. The overall percent coefficient of variance was 23% or less, and 95% of the quantification values were within 1.59- to 1.99-fold of the geometric mean. Linearity was assessed as previously described (2). Measured HIV-1 RNA concentrations ranged from ∼5.4 × 102 to ∼3 × 106 copies/ml, and linear regression analysis yielded a high coefficient of determination (R2 = 0.998), indicating that the bDNA assay is linear over a >3-log10 range. The impact of HIV-1 genetic variability was evaluated by testing stock HIV-1 culture isolates from patients infected with divergent HIV-1 subtypes (A to F) diluted into HIV-1-seronegative plasma and normalized according to virion-associated p24 measurements as previously described (7). As shown in Fig. 2, HIV-1 RNA levels ranged from 25,000 to 86,000 copies/ml and quantification was virtually unaffected by HIV-1 genotypic variability. In other experiments, the 250-μl-format assay exhibited specificity (100%) and reproducibility (within-run coefficient of variance, <25%) comparable to those of the 1-ml- and 50-μl-format assays (data not shown).
FIG. 2.
Quantification of RNAs from HIV-1 subtypes A to F using the 1-ml (□)- and 50-μl (⧫)-format bDNA assays. For this analysis, two isolates of each HIV-1 subtype were tested, except for subtype B, for which only a single isolate was tested.
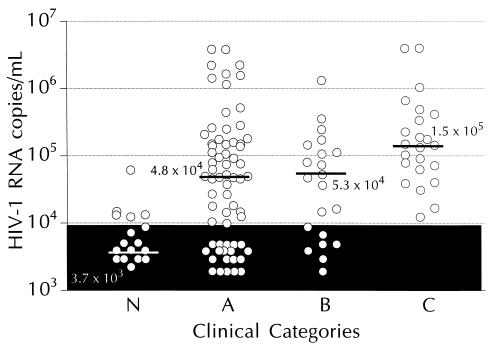
The clinical utility of the 50-μl-format bDNA assay was demonstrated by measuring HIV-1 RNA levels in 134 banked specimens from 37 pediatric patients. As shown in Fig. 3, the median HIV-1 RNA level varied with infection status. All 23 samples (100%) from AIDS patients (category C, n = 8) had detectable HIV-1 RNA, and the median HIV-1 RNA level in this group was significantly (P < 0.02) higher than that of the other groups. Of the samples from the moderately (category B, n = 5) and mildly (category A, n = 16) symptomatic groups, 67% (14 of 21) and 66% (44 of 67), respectively, had detectable HIV-1 RNA. No significant difference in median HIV-1 RNA levels was found between categories A and B (P = 0.55). Of the samples from asymptomatic patients (category N, n = 3), 28% (5 of 18) had detectable HIV-1 RNA, and the median level of HIV-1 RNA in this group was significantly (P < 0.02) less than that of the other three groups. No HIV-1 RNA was detected in samples from uninfected controls (category E, n = 5; data not shown). A gradual decrease in viral load with increasing age also was noted—mean HIV-1 RNA levels were 8.87 × 105, 2.67 × 105, and 6.23 × 104 copies/ml for groups aged 0 to 12, 13 to 24, and >24 months, respectively.
FIG. 3.
HIV-1 RNA levels in plasma specimens from pediatric patients classified by infection stage. Median levels for the groups are indicated by bars. The shaded area represents values below the detection limit of the 50-μl-format bDNA assay.
Detection rates were evaluated by comparing the number of plasma specimens quantified by the 1-ml, 250-μl, and 50-μl assay formats. For this analysis, samples were categorized based on HIV-1 RNA levels—category 1, <10,000 copies/ml (n = 44); category 2, 10,000 to 100,000 copies/ml (n = 44); category 3, >100,000 copies/ml (n = 44). All samples with HIV-1 RNA levels above 10,000 copies/ml (categories 2 and 3) were detected by all three formats. Of the 44 category 1 samples, 42 (95.4%) were detected by the 1-ml format and 33 (75%) were detected by the 250-μl format.
In summary, we found the small-volume-format bDNA assays to be an accurate and reproducible means for quantifying HIV-1 RNA in plasma. Given the low sample volume requirement, these assay formats may be particularly well suited for testing of viral load in children, particularly HIV-infected neonates, who often have very high HIV-1 RNA levels and very limited sample volume. Advantages of the bDNA assay over other HIV-1 RNA quantification methods include its ease of use, sample volume flexibility, and precision of quantification. Together with the standard 1-ml-format bDNA assay, the 50- and 250-μl formats provide greater versatility for testing of small-volume samples. The 50-μl format requires no sample preparation and offers a detection limit of 10,000 copies/ml. The 250-μl format requires a 1-h concentration step but offers a lower detection limit of 2,000 copies/ml. As reliable methods for viral load testing, these small-volume-format bDNA assays provide valuable tools for assessing the efficacy of antiviral therapy and predicting disease progression.
Acknowledgments
We thank Laurie Senn, Nancy Chorne, and Aftab Keval for technical assistance; Peter Dailey for critical review of the manuscript; Kristina Whitfield for graphics; and Patricia Laurenson and Linda Wuestehube for writing and editorial assistance.
This work was supported in part by grants from the Chicago Pediatric Faculty Foundation and the National Institutes of Health (1R29-AI40891-01 [Y.Z.]).
REFERENCES
- 1.Centers for Disease Control. Revised classification system for human immunodeficiency virus infection in children less than 13 years of age. Morbid Mortal Weekly Rep. 1994;43:1–10. [Google Scholar]
- 2.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 4.Pachl C, Todd J A, Kern D G, Sheridan P J, Fong S-F, Stempien M, Hoo B, Besemer D, Yeghiazarian T, Irvine B, Kolberg J, Kokka R, Neuwald P, Urdea M S. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA (bDNA) signal amplification assay. J Acquired Immune Defic Syndr Hum Retrovirol. 1995;8:446–454. doi: 10.1097/00042560-199504120-00003. [DOI] [PubMed] [Google Scholar]
- 5.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV-1 viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 6.Todd, J., C. Pachl, R. White, T. Yeghiazarian, P. Johnson, B. Taylor, M. Holodniy, D. Kern, S. Hamren, D. Chernoff, and M. Urdea. 1995. Performance characteristics for the quantitation of plasma HIV-1 RNA using branched DNA signal amplification technology. J. Acquired Immune Defic. Syndr. Hum. Retroviral. 10:(Suppl. 2):S35–S44. [PubMed]
- 7.Todd J, Yeghiazarian T, Hoo B, Detmer J, Kolberg J, White R, Wilber J, Urdea M. Quantitation of human immunodeficiency virus plasma RNA by branched DNA and reverse transcription coupled polymerase chain reaction assay methods: a critical evaluation of accuracy and reproducibility. Serodiagn Immunother Infect Dis. 1994;6:1–7. [Google Scholar]