Abstract
The genetic relatedness among 29 enterotoxigenic Escherichia coli strains of serotype O6:H16 was investigated by randomly amplified polymorphic DNA (RAPD) analysis. The strains were isolated in different parts of the world, displayed CS1-CS3 or CS2-CS3 profiles, and expressed heat-labile toxin (LT) and heat-stable toxin; a single strain expressed only LT. Ten RAPD types were distinguished and showed significant similarity, having on average 82% of the amplified bands in common. These results indicated that, irrespective of the different geographical origin or virulence factors, these strains belonged to a widespread clonal group.
Enterotoxigenic Escherichia coli (ETEC) is one of the most common causes of childhood diarrhea in developing countries and also the major cause of diarrhea among travelers from developed countries that visit regions of poor hygiene (13). ETEC is identified by its ability to produce enterotoxins, either heat-labile toxin (LT), heat-stable toxin (ST), or both. These bacteria also express surface adhesins known as colonization factors (CFs) or coli surface (CS) antigens that mediate attachment to the small intestine (13).
In a first attempt to characterize this pathogenic group, a set of ETEC strains from various parts of the world were serotyped for O and H antigens (8). This study showed that most of the isolates belonged to a limited number of serotypes (8). Besides the association between enterotoxigenicity and serotype, the authors observed that members of the same serotype also shared the K antigen and biotype (9). These findings led to the concept of a clonal structure for ETEC populations (8, 9). As further studies reported the isolation of ETEC strains in many geographical areas, an increasing diversity of serotypes was recognized, but certain O:H combinations, such as O6:H16, were found to be prevalent in different parts of the world (3, 4, 12, 19, 20). It was also shown that the restriction of ETEC strains to certain serotypes applies particularly to those that produce both LT and ST (LT/ST) (3, 5, 12, 20). This toxigenic profile has been frequently associated with CF antigen II (CFA/II) expression, especially among strains of serotype O6:H16 (3, 4, 19, 20). These observations supported the notion that ETEC strains sharing many phenotypic traits (such as serotype, CF, and toxin profiles) owe their similarity to recent descent from a common ancestor, thus representing a clonal group (14). However, in the case of O6:H16 ETEC strains, experimental demonstration of the genetic similarity of this phenotypically uniform group has yet to be shown.
We have previously reported the application of random amplification of polymorphic DNA (RAPD) to investigate the genetic relationship among ETEC strains (11). We observed that isolates sharing the same serotype as well as other phenotypic traits had similar amplification patterns, indicating that RAPD profiles were useful in revealing clonal groups among these organisms. To further investigate the genetic diversity among ETEC strains, we have applied this kind of analysis to the worldwide-distributed O6:H16 ETEC serotype.
A total of 29 ETEC strains belonging to serotype O6:H16 were included in this work: 6 strains from Brazil (provided by B. E. Guth, Escola Paulista de Medicina, São Paulo, Brazil); 5 strains from Mexico (provided by A. Cravioto, Universidad Nacional Autonoma de Mexico, Mexico City, Mexico); 1 strain from Honduras and 4 from The Philippines (provided by P. Echeverria, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand); 4 strains from Bangladesh (provided by A. Svennerholm, Göteborg University, Göteborg, Sweden); and 9 strains from Argentina (selected from our own bacterial collection) (3). The toxin and CF types of most of the strains were determined by the laboratories that provided them. In addition, in the present work, all strains were subjected to colony hybridization assays using LT, STaI, and STaII probes, as previously described (15, 16), and a CF-specific enzyme-linked immunosorbent assay (ELISA) using monoclonal antibodies against CS1, CS2, and CS3 antigens (17). The geographical origin and the virulence-associated properties of the strains are listed in Table 1. The results shown are derived from our toxin and CF assays, except for those strains which gave negative results, in which case the original information about these traits was considered. All typed strains except one displayed the LT/ST profile and expressed either CS1-CS3 (12 strains) or CS2-CS3 (16 strains) antigens. The only exception was strain Ph 25, from the Philippines, which expressed only LT. The CF type of this strain could not be determined since no previous information was available, and it did not express either CS1, CS2, or CS3, as determined by ELISA.
TABLE 1.
Characteristics of the O6:H16 ETEC strains studied
| Country of origin and strain | Virulence factors
|
RAPD profile
|
RAPD type | ||
|---|---|---|---|---|---|
| Toxin(s)a | CFs | Primer 1254 | Primer 1290 | ||
| Brazil | |||||
| Br 15 | LT/ST | CS1-CS3 | A1 | B1 | 1 |
| Br 16 | LT/ST | CS2-CS3 | A1 | B1 | 1 |
| Br 23 | LT/ST | CS1-CS3 | A2 | B2 | 3 |
| Br 26 | LT/ST | CS2-CS3 | A2 | B5 | 5 |
| Br 64 | LT/ST | CS2-CS3 | A2 | B5 | 5 |
| Br 45 | LT/ST | CS1-CS3 | A2 | B2 | 3 |
| Mexico | |||||
| Mx 6 | LT/ST | CS2-CS3 | A2 | B1 | 2 |
| Mx 7 | LT/ST | CS2-CS3 | A2 | B1 | 2 |
| Mx 8 | LT/ST | CS2-CS3 | A2 | B1 | 2 |
| Mx 9 | LT/ST | CS2-CS3 | A2 | B8 | 8 |
| Mx 10 | LT/ST | CS2-CS3 | A2 | B2 | 3 |
| Honduras | |||||
| Hd 35 | LT/ST | CS1-CS3 | A2 | B2 | 3 |
| Philippines | |||||
| Ph 25 | LT | Negative | A3 | B4 | 10 |
| Ph 28 | LT/ST | CS1-CS3 | A2 | B2 | 3 |
| Ph 35 | LT/ST | CS1-CS3 | A2 | B1 | 2 |
| Ph 55 | LT/ST | CS1-CS3 | A2 | B1 | 2 |
| Argentina | |||||
| Ag 7 | LT/ST | CS1-CS3 | A2 | B6 | 6 |
| Ag 8 | LTST | CS2-CS3 | A2 | B3 | 4 |
| Ag 9 | LT/ST | CS1-CS3 | A2 | B2 | 3 |
| Ag 18 | LT/ST | CS2-CS3 | A2 | B3 | 4 |
| Ag 19 | LT/ST | CS2-CS3 | A2 | B3 | 4 |
| Ag 20 | LT/ST | CS1-CS3 | A2 | B6 | 6 |
| Ag 21 | LT/ST | CS2-CS3 | A2 | B1 | 2 |
| Ag 22 | LT/ST | CS2-CS3 | A2 | B9 | 9 |
| Ag 23 | LT/ST | CS1-CS3 | A2 | B2 | 3 |
| Bangladesh | |||||
| Bg 1 | LT/ST | CS2-CS3 | A2 | B8 | 8 |
| Bg 2 | LT/ST | CS2-CS3 | A2 | B8 | 8 |
| Bg 3 | LT/ST | CS2-CS3 | A2 | B7 | 7 |
| Bg 4 | LT/ST | CS1-CS3 | A2 | B1 | 2 |
LT and ST refer to the subtypes LTI and STaII, respectively.
A single nonenterotoxigenic E. coli strain expressing serotype O6:H31, isolated from a case of urinary tract infection in Brazil, was included for comparative purposes. This strain represents a common RAPD type associated with the O6:H31 serotype and was considered as an out-group sample to estimate the degree of similarity among O6:H16 strains, based on RAPD profiles (10).
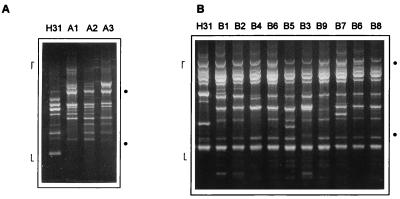
RAPD typing consisted of bacterial growth, template preparation, amplification reactions, and electrophoretic analysis of products, performed as previously described (10, 11). Each strain was tested with two primers, 1254 (5′-CCGCAGCCAA-3′) and 1290 (5′-GTGGATGCGA-3′). The whole procedure, from bacterial growth to RAPD profile recording, was repeated at least three times for each sample to confirm the results. Those bands that were consistently detected in different amplification reactions were used to define RAPD profiles. Representative RAPD profiles and the bands selected for recording are shown in Fig. 1. Bands with extremely low or high molecular weight were not considered because of their weak appearance on the gels and poor reproducibility. Different RAPD profiles were designated by different letters; minor variations (at least one polymorphic band) were indicated by different numbers.
FIG. 1.
RAPD analysis using primers 1254 (A) and 1290 (B) of ETEC strains belonging to serotype O6:H16 from different countries. RAPD profiles, designated as shown in Table 1, are indicated at the tops of the lanes. Dots indicate the positions of 600 and 2,000 bp; markers limit the range of bands considered for analysis.
Amplifications performed with primer 1254 resulted in the identification of three RAPD profiles among the 29 O6:H16 strains (Table 1 and Fig. 1A). These variant RAPD profiles were distinguished by the presence of 4 polymorphic bands in a total of 14 amplified bands. In contrast, the representative O6:H31 strain was clearly different from the O6:H16 strains (Fig. 1A). Amplifications using primer 1290 revealed nine RAPD profiles (Table 1 and Fig. 1B) and a higher proportion of polymorphic bands among O6:H16 strains, 12 in a total of 18 amplified bands. When these profiles were compared with that of the O6:H31 strain, the difference was not so prominent as that observed with primer 1254 (Fig. 1B); nevertheless, the presence of 3 bands unique to the O6:H31 profile distinguished it as a less related strain. By combining the data obtained with the two primers, a total of 32 bands (16 polymorphic) were amplified and 10 different RAPD types were defined among the O6:H16 strains (Fig. 1 and Table 1).
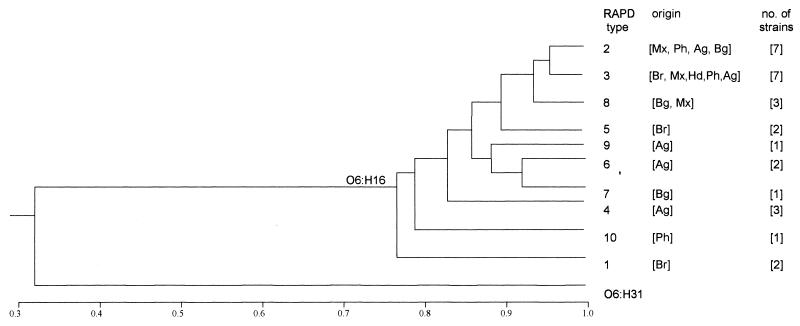
For each tested strain, the amplification profiles obtained with the two primers were considered for comparison. The relatedness among RAPD profiles was estimated by the proportion of bands in common, applying the coefficient defined by Jaccard (6). Data recording and calculations were performed by using RAPDistance programs, version 1.03 (2). The resulting matrix of pairwise distances was used to generate a phenogram based on the unweighted pair-group method with arithmetic mean (UPGMA) method, included in the MEGA software (7). Despite the presence of polymorphic bands, a significant similarity was observed among the O6:H16 strains, which had on average 82% of the amplified bands in common (minimum value, 70%; maximum, 96%). In contrast, the unrelated O6:H31 strain subjected to the same procedure had only 32% of the amplified bands (maximum, 36%) in common with the O6:H16 group, thus standing in a distantly related branch in the phenogram (Fig. 2). The high degree of similarity observed, based on the proportion of amplified bands in common, indicates that all the O6:H16 strains tested can be considered components of a clonal group.
FIG. 2.
Phenogram representing the relatedness of O6:H16 ETEC strains. Amplification profiles generated by two primers were considered, and the comparison of samples was based on the proportion of bands in common (indicated on the scale). Groups of similarity were established by using the UPGMA method. RAPD types and their respective geographical origins and numbers of strains are indicated on the right (designations as shown in Table 1). An RAPD profile of a non-ETEC O6:H31 strain obtained with the same primers was included for comparative purposes.
The characterization of E. coli strains using pulse-field gel electrophoresis or RAPD has shown that these methods can readily distinguish genetically distant strains and can also disclose variation among phenotypically related strains (1, 11, 18). In the present study, the presence of polymorphic bands allowed us to detect 10 RAPD types among the 29 O6:H16 strains tested. No particular correlation was observed between the various RAPD types and the virulence factor profiles of the strains. The only apparent exception was a single strain originated from the Philippines (strain Ph 25), which displayed a toxigenic phenotype (LT only) different from that of the other strains and showed unique RAPD profiles with both primers (Table 1). In regard to the geographic distribution, 7 of the 10 RAPD types detected were restricted to single countries (Fig. 2). On the other hand, the RAPD types with the highest degree of similarity (RAPD types 2 and 3) included strains isolated from all countries covered by this study (Fig. 2). It could also be noted that strains from a certain location (e.g., Argentina or Mexico) were present in different branches of the O6:H16 similarity group in the phenogram (Fig. 2). Taking into account the dynamic state of bacterial genomes and the discriminatory power of PCR-based methods, the different O6:H16 RAPD types revealed in this study can be considered variant genotypes of a major clonal group, irrespective of variations in their geographical origin or toxin and CF phenotypes, representing the result of recent evolutionary divergence (1). In this sense, we believe that the definition of a similarity group based on having approximately 80% of the amplified bands in common as corresponding to a clonal group (used in this study) is a more comprehensive approach than the adoption of more strict limits such as the complete identity of band profiles.
Different aspects of the variability revealed by RAPD are apparent from the results. Typing with primer 1254 may be useful to group-related bacterial strains. With this primer, the O6:H16 strains showed on average 80% identity in their band profiles. In contrast, the representative O6:H31 strain showed on average only 20% of the bands in common with the O6:H16 group. This difference was not as apparent when primer 1290 was used. In that case, the O6:H31 strain and the O6:H16 group had on average 43% of the bands in common while the O6:H16 strains had 71% in common. On the other hand, amplifications using primer 1290 disclosed significant polymorphism among the O6:H16 strains, reflected in the high proportion of polymorphic bands and in the number of different RAPD profiles identified (Fig. 1). The different discriminatory properties of these two primers had been observed in other studies in which we investigated the relatedness of E. coli strains, leading us to suggest the use of primer 1254 to define major groups of related organisms and the use of primer 1290 to disclose intraclonal variations (10, 11). In addition, the overall results obtained with each primer are equivalent, which adds to the consistency of the established groups.
The use of RAPD to investigate the genetic diversity of human ETEC isolates from areas of endemicity in Brazil has shown that strains of different serotypes were genetically distinct while those with the same serotype were closely related (11). In this work, we further demonstrate the similarity of ETEC strains having the same O:H type but varying in geographical origin and virulence factors. Although the reduced number of tested strains limits generalizations, the results suggest that O:H serotypes in fact represent good indicators of the genetic relatedness among ETEC strains, as originally proposed by the Ørskov group (8, 9).
Acknowledgments
This work was supported by grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Centro Brasileiro Argentino de Biotecnologia (CBABIO), and Financiadora de Estudos e Projetos (FINEP).
We thank A. Cravioto, P. Echeverria, A. Svennerholm, and B. Guth for providing the bacterial strains and M. K. Wolf for providing unpublished data. We are greatly indebted to A. Svennerholm and H. Sommerfelt for help in testing the virulence factors of ETEC strains. We also thank Celso Pereira and Eduardo Camacho for technical assistance.
REFERENCES
- 1.Arbeit R D, Arthur M, Dunn R, Kim C, Selander R K, Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulse field electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J, Gibbs A, Peakall R, Weiller G. RAPDistance programs, version 1.03, for the analysis of patterns of RAPD fragments. Canberra: Australian National University; 1994. [Google Scholar]
- 3.Binsztein N, Jouve M J, Viboud G I, Moral L L, Rivas M, Ørskov I, Ahren C, Svennerholm A. Colonization factors of enterotoxigenic Escherichia coli isolated from children with diarrhea in Argentina. J Clin Microbiol. 1991;29:1893–1898. doi: 10.1128/jcm.29.9.1893-1898.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cravioto A, Scotland S M, Rowe B. Hemagglutination activity and colonization factor antigens I and II in enterotoxigenic and nonenterotoxigenic strains of Escherichia coli isolated from humans. Infect Immun. 1982;36:189–197. doi: 10.1128/iai.36.1.189-197.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Echeverria P, Ørskov F, Ørskov I, Plianbangchang D. Serotypes of enterotoxigenic Escherichia coli in Thailand and The Philippines. Infect Immun. 1982;36:851–856. doi: 10.1128/iai.36.3.851-856.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaccard P. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull Soc Vaud Sci Nat. 1901;37:547–579. [Google Scholar]
- 7.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.02. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 8.Ørskov F, Ørskov I, Evans D J, Sack R B, Sack D A, Wadström T. Special serotypes among enterotoxigenic strains from diarrhoea in adults and children. Med Microbiol Immunol (Berlin) 1976;162:73–80. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- 9.Ørskov I, Ørskov F. Special O:K:H serotypes among enterotoxigenic E. coli strains from diarrhea in adults and children. Med Microbiol Immunol. 1977;163:99–110. doi: 10.1007/BF02121825. [DOI] [PubMed] [Google Scholar]
- 10.Pacheco A B F, Guth B E C, Soares K C C, Almeida D F, Ferreira L C S. Clonal relationships among Escherichia coli serogroup O6 isolates based on RAPD. FEMS Microbiol Lett. 1997;148:255–260. doi: 10.1111/j.1574-6968.1997.tb10297.x. [DOI] [PubMed] [Google Scholar]
- 11.Pacheco A B F, Guth B E C, Soares K C C, Almeida D F, Ferreira L C S. Random amplification of polymorphic DNA reveals serotype-specific genetic clusters among enterotoxigenic Escherichia coli (ETEC) isolated from humans. J Clin Microbiol. 1997;35:1521–1525. doi: 10.1128/jcm.35.6.1521-1525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reis M H L, Matos D P, Pestana de Castro A F, Toledo M R F, Trabulsi L R. Relation among enterotoxigenic phenotypes, serotypes, and sources of strains in enterotoxigenic Escherichia coli. Infect Immun. 1980;28:24–27. doi: 10.1128/iai.28.1.24-27.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowe B, Scotland S M, Gross R J. Enterotoxigenic Escherichia coli causing infantile enteritis in Britain. Lancet. 1977;i:90–91. doi: 10.1016/s0140-6736(77)91099-6. [DOI] [PubMed] [Google Scholar]
- 14.Selander R K, Caugant D A, Whittam T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 15.Sommerfelt H, Grewal H M, Bhan M K. Simplified and accurate nonradioactive polynucleotide gene probe assay for identification of enterotoxigenic Escherichia coli. J Clin Microbiol. 1990;28:49–54. doi: 10.1128/jcm.28.1.49-54.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommerfelt H, Steinsland H, Grewal H M S, Viboud G I, Bhandari N, Gaastra W, Svennerholm A, Bhan M. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J Infect Dis. 1996;174:768–776. doi: 10.1093/infdis/174.4.768. [DOI] [PubMed] [Google Scholar]
- 17.Viboud G I, Binsztein N, Svennerholm A. Characterization of monoclonal antibodies against putative colonization factors of enterotoxigenic Escherichia coli and their use in an epidemiological study. J Clin Microbiol. 1993;31:558–564. doi: 10.1128/jcm.31.3.558-564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf M K, Taylor D N, Boedeker E C, Hyams K C, Maneval D R, Levine M M, Tamura K, Wilson R A, Echeverria P. Characterization of enterotoxigenic Escherichia coli isolated from U.S. troops deployed to the Middle East. J Clin Microbiol. 1993;31:851–856. doi: 10.1128/jcm.31.4.851-856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf M K. Occurrence, distribution, and associations of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin Microbiol Rev. 1997;10:569–584. doi: 10.1128/cmr.10.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]