In this review, Cai and de Lange discuss the role of the CST–Polα/Primase complex in telomere length regulation. Providing insight into its structure, evolution, and molecular interactions, they illustrate that CST–Polα/Primase not only effectuates fill-in strand synthesis but also controls telomerase to maintain telomere length, while genetic perturbations in its components result in telomeric disorders.
Keywords: CST, telomerase, telomere
Abstract
It has been known for decades that telomerase extends the 3′ end of linear eukaryotic chromosomes and dictates the telomeric repeat sequence based on the template in its RNA. However, telomerase does not mitigate sequence loss at the 5′ ends of chromosomes, which results from lagging strand DNA synthesis and nucleolytic processing. Therefore, a second enzyme is needed to keep telomeres intact: DNA polymerase α/Primase bound to Ctc1–Stn1–Ten1 (CST). CST–Polα/Primase maintains telomeres through a fill-in reaction that replenishes the lost sequences at the 5′ ends. CST not only serves to maintain telomeres but also determines their length by keeping telomerase from overelongating telomeres. Here we discuss recent data on the evolution, structure, function, and recruitment of mammalian CST–Polα/Primase, highlighting the role of this complex and telomere length control in human disease.
Telomeres and human health
Human telomeres contain an array of double-stranded (ds) TTAGGG repeats that end in a single-stranded (ss) 3′ overhang estimated to be 40–500 nt (Fig. 1). The binding of shelterin to this repeat array prevents activation of the DNA damage response (DDR) at the natural ends of chromosomes (for review, see de Lange 2018). Shelterin contains two ds telomeric DNA-binding proteins (TRF1 and TRF2) that, through their interactions with TIN2, bring the POT1/TPP1 heterodimer to the ss telomeric DNA (Fig. 1). Shelterin prevents the activation of ATM and ATR signaling at chromosome ends and blocks double-stranded break (DSB) repair pathways from inadvertently acting at telomere ends. Telomere protection by shelterin involves the remodeling of the telomeric DNA into the t-loop, in which the 3′ overhang invades the ds telomeric DNA (Fig. 1). T-loops effectively sequester the telomere end, thereby rendering telomeres impervious to signaling and repair pathways that act on DSBs. Shelterin also is required to recruit the two telomere maintenance machines: telomerase and CST–Polα/Primase (Fig. 1).
Figure 1.

Shelterin-mediated telomere maintenance and protection. Cartoon schematic of the three major complexes involved in telomere maintenance and protection. The six-subunit human shelterin complex associates with ds and ss telomeric DNA and forms t-loops to protect telomeres from DSB signaling and repair pathways. Note that two copies of TIN2, TPP1, and POT1 can be associated with the TRF1 and TRF2 homodimers (Zinder et al. 2022), but only one of each subunit is shown for simplicity. Shelterin recruits and regulates two telomere maintenance machines. Telomerase is recruited by TPP1 and uses the 3′ end as a primer for G-strand synthesis. CST–Polα/Primase is responsible for C-strand fill-in and is recruited by POT1/TPP1. CST also negatively regulates telomerase.
Because telomeres prevent DNA damage signaling at chromosome ends, their shortening limits the long-term survival and proliferation of human cells. There are several telomere disorders, including dyskeratosis congenita (DC) and Hoyeraal-Hreidarsson syndrome (for review, see Savage 2022), that result from mutations affecting the telomerase-mediated telomere maintenance pathway, including mutations in genes encoding the hTERT reverse transcriptase, the hTR telomerase RNA, telomerase biogenesis factors, TIN2, and the telomerase recruitment factor TPP1 (Fig. 1). These disorders are characterized by short telomeres and often present with anemia and bone marrow failure but can also affect other organs, such as the liver and lungs. The onset and severity of these diseases is variable, presumably due to the magnitude of the telomere length defect at birth and the residual telomerase level in stem cell compartments.
A different telomere syndrome, Coats plus syndrome (CP), is caused by mutations in Ctc1, Stn1, or POT1 that affect the function of the CST–Polα/Primase tract (Anderson et al. 2012; Keller et al. 2012; Polvi et al. 2012; Walne et al. 2013; Netravathi et al. 2015; Simon et al. 2016; Takai et al. 2016; Lin et al. 2017; Hidalgo-Sanz et al. 2019; Passi et al. 2020). This rare pediatric condition is characterized by the ophthalmological disorder Coats disease plus additional associated systemic disorders primarily affecting the brain, bones, and gastrointestinal tract. Coats plus is discussed in further detail below.
These diseases illustrate the importance of sufficient telomere reserve at birth. However, excessive telomere length at birth also carries a health risk, in this case due to cancer predisposition (for review, see Maciejowski and de Lange 2017). Two types of evidence support the idea that long telomeres at birth predispose to cancer. First, GWAS and WGS studies showed that a set of SNPs previously associated with long telomeres in adult blood predicted a greater risk of a variety of malignancies, including sarcoma, glioblastoma, neuroblastoma, and ovarian, lung, bladder, and skin cancers (Walsh et al. 2015; Haycock et al. 2017; Ballinger et al. 2023). Second, a number of cancer-prone families were found to carry POT1 and TIN2 mutations that induce excessively long telomeres at birth but no other telomere dysfunction (Schmutz et al. 2020; Kim et al. 2021). Patients with long telomeres due to TIN2 or POT1 impairment often develop multiple primary tumors, including thyroid, breast, lung, colon, and brain cancers. The high frequency and wide spectrum of the cancers in these patients is comparable with Li-Fraumeni syndrome, speaking to the importance of correct telomere length at birth to prevent cancer.
The increased cancer risk associated with long telomeres is due to a failure in the telomere tumor suppressor pathway (for review, see Maciejowski and de Lange 2017). It is thought that the long telomeres in the patients’ somatic cells delay the onset of the Hayflick limit, the point in clonal proliferation when critically short telomeres impede further cell divisions and thus kill developing malignant clones. The additional cell divisions afforded by the long telomeres likely allow early cancer clones to accumulate mutations, including mutations that facilitate the escape from this barrier. Thus, telomeres not only need to be maintained, but their length at birth must be regulated to prevent organ failure on the one hand and cancer on the other.
This review focuses on the CST–Polα/Primase complex that, together with telomerase, maintains human telomeres and regulates their length. We discuss recent work on the evolution, function, structure, and regulation of CST–Polα/Primase that has provided a clearer view on how this machine works and point out questions that merit attention.
Telomere end replication: two strands, two problems
The G-strand of vertebrate telomeres extends beyond the C-strand, forming an overhang that is needed for telomere extension by telomerase, t-loop formation, and telomere protection (Fig. 1). When leading strand DNA synthesis reaches the end of the chromosome, it creates a blunt end that is not protected because it lacks a 3′ overhang (Fig. 2). Initial resection of leading end telomeres by the TRF2-bound exonuclease Apollo and subsequent long-range resection by Exo1 can restore the overhang and thus telomere protection (Lenain et al. 2006; Lam et al. 2010; Wu et al. 2010, 2012). However, the G-strand of the leading end telomere will have been shortened by the length of the original 3′ overhang, representing the end replication problem that is solved by telomerase (Fig. 2, bottom). This problem was first identified on theoretical grounds (Lingner et al. 1995) and then directly observed at budding yeast telomeres (Soudet et al. 2014). Telomerase can extend the 3′ end and thereby counteract the loss of the G-strand sequences (for review, see Hockemeyer and Collins 2015). It is the absence of this elongation, together with the effect of resection, that underlies the shortening of telomeres by 50–100 bp/PD observed in human cells lacking telomerase.
Figure 2.
CST–Polα/Primase solves a second end replication problem. While telomerase can solve the leading end replication problem (loss of the G-strand overhang), CST–Polα/Primase-mediated fill-in is required to replenish the C-rich sequences at the lagging and leading end telomeres. (Inset) When the replisome reaches the telomere end, the last Okazaki fragment starts >40 nt from the ds–ss junction. This second end replication problem cannot be solved by telomerase and requires CST–Polα/Primase-mediated fill-in. (Pol δ) DNA polymerase δ, (PCNA) proliferating cell nuclear antigen, (Pol ε) DNA polymerase ε, (CMG) Cdc45–Mcm2-7–GINS complex. (Purple blocks) RNA primers.
It has recently become clear that there is a second end replication problem resulting in loss of C-strand sequences from telomeres replicated by lagging strand DNA synthesis (H Takai, V Aria, P Borges, JTP Yeeles, and T de Lange, in prep.) (Fig. 2). This C-strand loss is due to the replisome's inability to support Okazaki fragment synthesis using the G-strand overhang as a template. In vitro, the last Okazaki fragment is initiated by Polα/Primase >40 nt before the end of the duplex linear DNA, leaving the new 5′-ended strand >40 nt shorter than the original one (Fig. 2, inset). This sequence loss cannot be counteracted by telomerase, and its cumulative effect over successive rounds of replication will shorten telomeres. This second end replication problem is solved by replisome-independent fill-in synthesis by CST–Polα/Primase. In addition, fill-in synthesis by CST–Polα/Primase can mitigate the loss of C-strand sequences due to 5′ end resection and elongate the C-strand after telomerase has added an array of ss TTAGGG repeats (Fig. 2).
As argued previously (de Lange 2004, 2015; Lue 2018), the 3′-ended DNA strand of the first linear chromosomes could have been maintained—prior to the advent of telomerase—through a break-induced replication (BIR)-type mechanism involving strand invasion and extension of the 3′ end by canonical DNA polymerases. Because BIR only synthesizes the 3′-ended strand (for review, see Liu and Malkova 2022), the necessity for a mechanism to extend the 5′-ended strand may explain the emergence of the CST–Polα/Primase fill-in machinery (Lue 2018).
The structure and evolution of CST
CST was first identified in extracts of mouse cells as a Polα/Primase accessory factor (α accessory factor [AAF]) that stimulates the activities of both Polα and Primase and acts as a template affinity factor that promotes complete replication of long ss templates (Goulian and Heard 1990; Goulian et al. 1990). AAF was also shown to increase the processivity of Polα from synthesis of ∼20 nt without AAF to >100 nt with AAF (Goulian and Heard 1990). When two of the AAF subunits (Ctc1 and Stn1) were cloned (Casteel et al. 2009), it became clear that AAF was related to CST, a complex long known to function as the main protector of yeast telomeres (for review, see Bertuch and Lundblad 2006). Independently, mammalian CST also emerged from database searches for genes orthologous to yeast and plant subunits (Miyake et al. 2009; Surovtseva et al. 2009).
Mammalian CST is a trimeric ssDNA-binding complex related to replication protein A (RPA) (Fig. 3A; Casteel et al. 2009; Miyake et al. 2009; Lim et al. 2020). The 134-kDa Ctc1 subunit contains seven tandem oligosaccharide/oligonucleotide-binding (OB) folds with structural homologies with the four OB folds of RPA70. The N-terminal three OB folds of Ctc1 likely originated from a duplication and are separated by a three-helix bundle motif (Lim et al. 2020). Stn1 (44 kDa, comprising an OB fold and tandem winged helix–turn–helix [wHTH] domains) and Ten1 (14 kDa, a single OB fold) are structurally homologous to RPA32 and RPA14, respectively (Fig. 3A; Bryan et al. 2013; Lim et al. 2020).
Figure 3.
Evolution of CST–Polα/Primase. (A) Model for the independent evolution of CST from an archaeal RPA. Compared with a generalized archaeal RPA, CST retains an AROD-like (ARODL) three-helix bundle domain (inset; cf. PabRPA AROD) (Madru et al. 2023) in Ctc1. The last eukaryotic common ancestor (LECA) CST is drawn as predicted based on sequence alignments and predicted structures, suggesting that basal eukaryotic CSTs duplicated three OB folds in Ctc1 and the Stn1 wHTH domain, whereas eukaryotic RPAs contain neither the ARODL domain nor the duplications. It is also possible that eukaryotic RPA is derived from a separate archaeal precursor. CST itself has diverged in some eukaryotes through deletion of domains (Tetrahymena thermophila and Schizosaccharomyces pombe) or through a more complex independent evolution (Saccharomyces cerevisiae). (B) Comparison of telomere maintenance systems. Budding yeast, Tetrahymena, and humans differ in their telomeric proteins and mechanisms of recruiting telomerase and CST–Polα/Primase for G-strand and C-strand synthesis, respectively.
CST binds DNA with some sequence preference and binds to G-rich ssDNA with a KD in the 1–20 nM range when the DNA is at least 18 nt (Hom and Wuttke 2017). For instance, CST binds [TTAGGG]3 with a KD of ∼1 nM, forming a complex that is stable for hours (Chen et al. 2012). CST can also bind in a sequence-independent manner to longer (>30-nt) ssDNAs with a KD in the 100 nM range (Miyake et al. 2009) and can bind substrates with ss–ds junctions regardless of the sequence or the 5′/3′ orientation of the transition (Bhattacharjee et al. 2017). Interestingly, whereas Ctc1 and Stn1 are required for the interaction of CST with DNA, Ten1 is not, although its presence increases the stability of the protein–DNA complex (Feng et al. 2018). Finally, CST has some ability to unwind G-quadruplex structures (G4) that are readily formed by ss TTAGGG repeats and, when persistent, result in unreplicated gaps in telomeres formed by lagging strand DNA synthesis (Qureshi et al. 2012; Bhattacharjee et al. 2017; Zhang et al. 2019; Yang et al. 2020). RPA and CST share some similar properties, but RPA binds ssDNA at higher affinity without sequence preference and is more efficient at unwinding G4s (Olson et al. 2023).
New structural data suggest that CST evolved from an archaeal RPA rather than from eukaryotic RPA (Madru et al. 2023; for a review on eukaryotic evolution, see Eme et al. 2017). The largest subunit of RPA from Pyrococcus abyssi (PabRPA) contains a signature three-helix bundle at its N terminus called the acidic Rpa1 OB-binding domain (AROD) that is found in most archaeal RPAs but is absent from eukaryotic RPA. It functions to oligomerize PabRPA by bridging to an OB fold from a second PabRPA molecule. Strikingly, this OB fold-bridging three-helix bundle is present in human CST as the connector between the two sets of OB folds in Ctc1 (Fig. 3A; Lim et al. 2020; Madru et al. 2023). The presence of the AROD-like (ARODL) three-helix bundle suggests that eukaryotic CST is derived from an archaeal RPA that had the AROD motif and evolved separately from modern eukaryotic RPAs (Fig. 3A). Furthermore, the predicted structures of Ctc1 orthologs in some basal fungal phyla such as Zoopagomycota (e.g., Linderina pennispora; AF-A0A1Y1WN78-F1) and Mucoromuycota (e.g., Lichtheimia corymbifera; AF-A0A068RQH6-F1), as well as in slime molds (e.g., Dictyostelium discoideum; AF-Q54WQ3), align well with human Ctc1 (Varadi et al. 2022), suggesting that the first eukaryotes had Ctc1-like proteins (Fig. 3A).
The early emergence of CST is consistent with the C-strand-centric model of telomere evolution, in which the C-strand synthesis machinery evolved first to complement a primordial recombination-based telomere maintenance system operating prior to the advent of telomerase (de Lange 2015; Lue 2018). In this model, CST emerged in the eukaryotic lineage as a specialized ssDNA-binding protein to support telomere maintenance by Polα/Primase. Polα is also thought to have originated at the time when linear chromosomes evolved, in this case emerging from a viral origin (Forterre 2013; Lue 2018). The acquisition of CST–Polα/Primase prior to the emergence of eukaryotes from an Asgard archaeal ancestor would have facilitated the maintenance of linear chromosomes before telomerase created uniform telomeres that could function in conjunction with sequence-specific telomere-binding proteins.
As detailed below, the N-terminal domain of Ctc1, with its four OB folds and the ARODL motif, plays a critical role in the interactions with Polα/Primase and POT1/TPP1 and contains numerous Coats plus mutations (Cai et al. 2022, 2023). Although this part of Ctc1 is highly conserved, it is absent from Ctc1 orthologs in some unicellular organisms, particularly in several model systems for telomere biology (Fig. 3A). In contrast, the C-terminal half of Ctc1, which includes the C-terminal OB fold that is part of the trimerization core, is conserved in all eukaryotic Ctc1 orthologs (Fig. 3A). This part of Ctc1 interacts with Polα/Primase in a highly conserved manner, as discussed below.
Whereas budding yeast Stn1 and Ten1 clearly are orthologous to their mammalian counterparts, the Cdc13 subunit of budding yeast CST evolved independently of the Ctc1 lineage. Other than its trimerization core OB fold, Cdc13 has limited structural homology with mammalian Ctc1 or RPA1 (Fig. 3A; Yu et al. 2012; Rice and Skordalakes 2016; Ge et al. 2020; Lim et al. 2020). The function of budding yeast CST is also quite distinct from its mammalian counterpart, as it is the primary protector of the ss telomeric DNA (for review, see Bertuch and Lundblad 2006). Although there is a conserved interaction between budding yeast CST and Polα/Primase, Cdc13 is distinct from mammalian Ctc1 in that it also recruits telomerase. Nonetheless, C-strand fill-in and G-strand extension appear to be separate steps, since Stn1 and Ten1, which are involved in fill-in, prevent binding of Cdc13 to Est1, the component of yeast telomerase required for recruitment (Fig. 3B; Pennock et al. 2001; Chen et al. 2018; Ge et al. 2020).
In Tetrahymena, a CST-like complex that binds Polα/Primase has been identified as a constitutive part of the telomerase holoenzyme (Fig. 3; Jiang et al. 2015; He et al. 2022b). The abundant telomeres in the macronuclei of Tetrahymena carry a distinct set of telomere binding proteins whose functional interaction with telomerase and/or CST is unclear (Premkumar et al. 2014). Although more work is needed to investigate how the Tetrahymena telomere-binding proteins affect telomerase and CST, telomerase itself has been the subject of extensive structural studies (He and Feigon 2022). Notably, Tetrahymena could coordinate G-strand synthesis and C-strand fill-in within a single complex (discussed further below), a feature that may be specific to ciliates (Fig. 3B; He et al. 2022b). This review focuses on the mammalian telomere maintenance system, which can be generalized to most metazoans (Myler et al. 2021). Despite differences in regulation across species, the core functional interaction between CST and Polα/Primase remains constant (Fig. 3B). Given the presence of CST in metazoans, fungi, ciliates, and plants, it is reasonable to conclude that CST was already present in the first eukaryote, as predicted by Lue (2018).
C-strand maintenance by CST–Polα/Primase
The role of CST in telomere fill-in was first observed in mouse cells. At mouse telomeres, CST is recruited by one of the two POT1 paralogs (POT1a and POT1b) that evolved through a duplication of the POT1 gene and gained distinct functions (Hockemeyer et al. 2006; Wu et al. 2006). POT1a is dedicated to the repression of ATR signaling at the ss telomeric DNA, whereas POT1b serves to recruit CST (Denchi and de Lange 2007; Wu et al. 2012). The first hint that POT1b is instrumental for telomere maintenance came from telomerase-negative mouse cells, where POT1b deletion results in exaggerated telomere shortening that ultimately impairs telomere function (Hockemeyer et al. 2008). This phenotype was not understood until it became clear that POT1b recruits CST to telomeres (Wu et al. 2012). When POT1b is mutated so that it no longer interacts with CST, the telomeric overhangs remain excessively long, as they do if Stn1 is depleted with an shRNA. In the presence of CST and wild-type POT1b, the fill-in synthesis at both daughter telomeres shortens the overhangs in late S/G2 (Wu et al. 2012). Similarly, in human cells, Stn1 depletion leads to abnormally long 3′ overhangs due to a defect in the late S/G2 fill-in synthesis (Wang et al. 2012).
Clever experiments using density gradient separation of BrdU-labeled DNA have shown that fill-in synthesis takes place after telomerase has extended the 3′ end (Zhao et al. 2009; Wang et al. 2012). Since telomerase requires a 3′ overhang (Rivera and Blackburn 2004; Lei et al. 2005), the elongation of the G-strand must be preceded by 5′ resection of the blunt leading end telomere. The order of events at telomeres therefore appears to be replication, resection, extension by telomerase, and last, CST–Polα/Primase fill-in (Fig. 2).
Human cells tolerate lack of telomere fill-in synthesis to some extent. Depending on the length of the telomeres, it can take many cell divisions before lagging strand synthesis and resection have shortened the C-rich strands enough to curb telomere protection. Analogously, the long telomeres of POT1b-deficient mouse embryonic fibroblasts (MEFs) sustain tens of population doublings in the absence of CST-mediated fill-in (Hockemeyer et al. 2008). On the other hand, human cancer cell lines with very short telomeres are highly sensitive to depletion of CST (Hu et al. 2021).
Interestingly, POT1b KO mice are viable only when telomerase is present, and the rate of telomere shortening in POT1b KO MEFs is diminished by telomerase expression (Hockemeyer et al. 2008). How could telomerase make a difference between life and death if it has no role in C-strand maintenance? The most reasonable explanation is that telomerase, freed from its inhibition by CST, generates very long overhangs in POT1b KO cells. These extended overhangs may overwhelm the ability of POT1a to block ATR kinase signaling, leading to ATR- and 53BP1-dependent recruitment of CST–Polα/Primase, similar to what happens at resected DSBs or telomeres lacking both POT1a and POT1b (Mirman et al. 2018, 2023; Mirman and de Lange 2020). This DDR-dependent loading and fill-in by CST–Polα/Primase likely mitigates C-strand loss (and dampens further ATR signaling) and thus promotes the viability of POT1b KO mice expressing telomerase.
A remaining question regarding the fill-in synthesis reaction concerns where fill-in starts and how the initiation is regulated. Since all telomeres carry a 3′ overhang even when telomerase is absent, fill-in is unlikely to start at the 3′ tip as seen at some DSBs (Schimmel et al. 2021). Given the preference of Primase for initiating RNA synthesis at two consecutive pyrimidines (Hay et al. 1984; Yamaguchi et al. 1985), the telomeric TTAGGG repeats represent a unique template with Primase start sites every 6 nt. Indeed, in vitro, CST–Polα/Primase initiates primer synthesis at sites spaced 6 nt apart along a telomeric template (He et al. 2022a; Zaug et al. 2022). On the shortest templates that yield a product (five or six repeats), synthesis starts ∼6 nt from the 3′ end. Similarly, the most terminal initiation event on a template of nine repeats appears to take place in the second repeat (Zaug et al. 2022). This would suggest that CST–Polα/Primase can create telomeres with an overhang of just 6 nt. This is puzzling because telomeric overhangs are generally >40 nt in length. Although the addition of purified POT1/TPP1 to the in vitro reactions did not alter the products, it may be that the recently described POT1/TPP1 complex bound to CST will have a different effect (Cai et al. 2023).
Another longstanding curiosity is that most human telomeres terminate with the sequence CCAATC-5′ (Sfeir et al. 2005). One possible explanation for this register specificity again invokes the biochemistry of Primase (Sheaff and Kuchta 1993), where Primase could synthesize one major 10-nt product (5′-AACCCUAACC-3′) on the telomeric template, and removal of this specific terminal RNA primer dictates the 5′ end sequence. In vitro, CST–Polα/Primase does initiate with a major RNA product, but it is 8 nt long (5′-AACCCUAA-3′), and removal of this primer would result in 5′ ends with the sequence AATCCC-5′ (Zaug et al. 2022). It remains to be determined whether CST–Polα/Primase synthesizes the same length RNA primer in vivo. Whether telomeric fill-in products are processed similarly to Okazaki fragments is another open question in this area.
Telomere length control by CST
It has long been known that TRF1, TIN2, and POT1 prevent excessive telomerase-mediated elongation at individual telomere ends (van Steensel and de Lange 1997; Kim et al. 1999; Loayza and De Lange 2003), enforcing the telomere length homeostasis critical to human health. The telomere length effects of these three shelterin subunits were primarily observed with dominant-negative alleles that, while affecting telomere length control, are not sufficiently penetrant to interfere with the essential protective role of shelterin. Indeed, TIN2 and POT1 are haploinsufficient for telomere length control, but loss of one allele of TIN2 or POT1 does not diminish telomere protection (Schmutz et al. 2020; Kim et al. 2021).
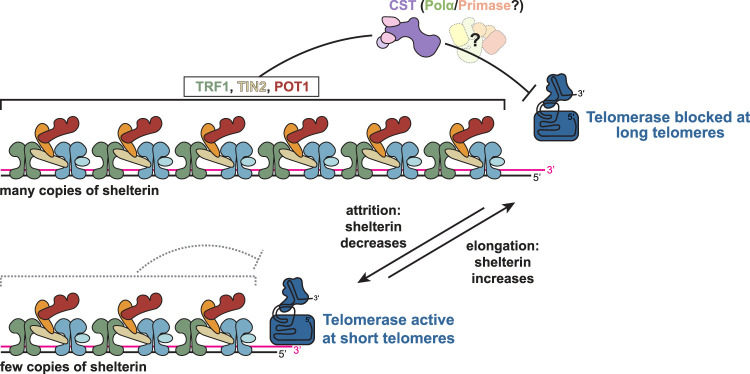
A common model for telomere length regulation (Fig. 4) poses that long telomeres contain more TRF1/TIN2/POT1, so that within each nucleus, telomerase is blocked at long telomeres but not at the short ones (for review, see Hockemeyer and Collins 2015). Indeed, mammalian telomerase extends the shortest telomeres preferentially (Zhu et al. 1998; Ouellette et al. 2000). Chromatin immunoprecipitation (ChIP) experiments showed that the density of these three proteins per kilobase of telomeric DNA is the same in cells with long and short telomeres (Loayza and de Lange 2003). Therefore, the higher total number of TRF1/TIN2/POT1 per telomere is predicted to enforce greater inhibition of telomerase at long telomeres. However, the critical question of whether the shortest telomeres in each nucleus indeed contain less shelterin than the longer telomeres has not been addressed.
Figure 4.
Protein-counting model for telomere length homeostasis. (Top) At long telomeres, more shelterin is present, and telomerase is prevented from acting (in cis) by TRF1, TIN2, and POT1. (Bottom) At short telomeres, which contain less shelterin, the inhibition of telomerase is alleviated, allowing telomere elongation. CST is thought to inhibit telomerase as a downstream effector of shelterin, but the mechanism by which it factors into the protein-counting model described remains an outstanding question. The role of Polα/Primase in telomerase inhibition is unknown.
Mounting evidence indicates that CST, possibly together with Polα/Primase, acts as the effector in the TRF1–TIN2–POT1 telomere length homeostasis pathway. Inhibition of either Ctc1 or Stn1 leads to dramatic telomere elongation mediated by telomerase (Chen et al. 2012; Feng et al. 2017), and a POT1 point mutation that leads to loss of fill-in synthesis by CST–Polα/Primase also abrogates telomere length control (Takai et al. 2016). Ten1, on the other hand, is not required for telomerase inhibition (Feng et al. 2018), though telomere length increase was observed in another study using a Ten1 shRNA (Chen et al. 2012).
Two nonmutually exclusive models for CST-dependent telomere length control can be considered. In the first model, the binding of CST to the 3′ overhang blocks telomerase from further extending its product (Chen et al. 2012; Chen and Lingner 2013; Zaug et al. 2021). Telomerase has been shown to act at all telomeres immediately after their replication, implying that this first extension step is unlikely to be regulated by telomere length (Zhao et al. 2009), but these experiments were done under conditions in which all telomeres were approximately equal in length. CST-mediated fill-in synthesis occurs hours later, and it may be at this point that CST prevents telomerase from executing a second round of G-strand extension at the shortest telomeres. In vitro, CST binding to ss TTAGGG repeats blocks telomerase from acting at the 3′ end, though CST is incapable of evicting telomerase that is already engaged on the DNA terminus (Chen et al. 2012; Zaug et al. 2021). Furthermore, CST competes for the ss telomeric DNA with POT1/TPP1 (Chen et al. 2012), which when bound to the telomerase primer can promote the processivity of telomerase repeat addition (Wang et al. 2007). These biochemical findings suggest that CST itself could limit telomerase through its DNA-binding activity.
In the second model, CST would prevent the recruitment of telomerase via protein–protein interactions. Telomerase is recruited to telomeres through an interaction between hTERT and the TEL patch of TPP1 (Abreu et al. 2010; Nandakumar et al. 2012; Zhong et al. 2012; Liu et al. 2022; Sekne et al. 2022). Live-cell imaging suggests that this interaction is labile and distinct from the more stable interaction of telomerase with the 3′ overhang (Schmidt et al. 2016; Laprade et al. 2020). Since telomere elongation is abrogated when telomeres lack the TPP1 TEL patch, it appears that the TPP1-dependent initial recruitment of telomerase is a required first step in shepherding the enzyme to the 3′ end. Like telomerase, CST binds to TPP1 (as well as POT1), and it is conceivable that its presence on the POT1/TPP1 heterodimer interferes with the loading of telomerase. However, recent cryo-EM structures of CST–POT1/TPP1, discussed further below, argue against inhibition by direct protein–protein competition, as CST does not interact with the TPP1 TEL patch (Cai et al. 2023). Instead, CST interacts with the TPP1 TIN2-interacting domain, and loss of this interaction results in extensive telomere elongation (Cai et al. 2023; Wang et al. 2023). This interaction is competitive with TPP1 binding to TIN2, which is required to keep POT1/TPP1 associated with the rest of shelterin (Takai et al. 2011; Frescas and de Lange 2014; Cai et al. 2023). The mechanism by which the CST–TPP1 interaction inhibits telomerase is unclear.
The recruitment of telomerase by TPP1 represents a confounding aspect of telomere length regulation. A priori, one could expect that longer telomeres, which contain more TPP1, will recruit more telomerase and become preferentially elongated. This is clearly not the case. One possibility is that telomerase recruitment by TPP1, though dependent on the number of TPP1 molecules per telomere, does not affect how telomerase acts at the telomere terminus. Indeed, most TPP1-dependent interactions of telomerase with telomeres are transient and most likely will not result in telomere extension (Schmidt et al. 2016). A second possibility is that the initial recruitment of telomerase by TPP1 relates only to the first round of extension by telomerase, when all telomeres are elongated regardless of their length. After this initial extension, telomerase could remain associated with the 3′ end, and telomere length homeostasis could act at the second step, preventing telomerase from further extension at the longest telomeres. In agreement, the POT1ΔOB allele that leads to excessive telomere elongation (Loayza et al. 2004) does not affect the recruitment of telomerase by TPP1 but increases the stable association of telomerase with the ss telomeric DNA (Laprade et al. 2020). If telomere length control—and thus CST—indeed plays out at this second step, the model posing that CST binding to the ss telomeric DNA blocks telomerase is more likely to be correct.
A second issue that remains to be addressed is the role of Polα/Primase in telomere length control. In vitro, Polα/Primase does not affect CST's ability to prevent telomerase initiation by DNA binding (Zaug et al. 2021). However, in both yeast and mammals, temperature-sensitive mutations in Polα lead to telomere elongation (Carson and Hartwell 1985; Nakamura et al. 2005), suggesting that CST may work with Polα/Primase to control telomere length in vivo. Furthermore, disruption of the interaction between CST and Polα/Primase increases the recruitment of telomerase to telomeres (Gu et al. 2018). If Polα/Primase contributes to telomere length control, it will be important to understand in what state it acts (recruitment complex-like or preinitiation complex-like) (discussed below).
Regardless of these mechanistic considerations, several questions deserve immediate attention. First, it needs to be established that longer telomeres in any given nucleus contain more TRF1, TIN2, POT1, and perhaps CST than the shorter ones. Because of the low abundance of both POT1 and CST, antibodies to the endogenous proteins are not likely to be of use, and new tools will have to be developed for quantitative analysis. Second, it is predicted that the dominant-negative alleles of TRF1, TIN2, and POT1 will reduce the loading of CST at telomeres. If this is not the case, it will be hard to argue that CST acts downstream from these shelterin subunits in a protein-counting system, and alternative models will have to be entertained. Finally, it will be important to determine the positional distribution of TRF1, TIN2, POT1, and CST throughout the telomeric chromatin, as previously suggested (Lim and Cech 2021).
Cryo-EM structures of CST–Polα/Primase: recruitment and preinitiation complexes
Recent cryo-EM structures of CST–Polα/Primase have illuminated some of the molecular cogs and levers of this enigmatic complex, providing insights into regulation and activation of the fill-in machinery (Fig. 5; Cai et al. 2022; He et al. 2022a,b). These structures show CST is indeed a specialized accessory factor for the regulation of Polα/Primase, participating in multiple modes of interaction with the enzyme.
Figure 5.
Cryo-EM structures of CST–Polα/Primase and CST–POT1/TPP1. (A,B) Structures of CST–Polα/Primase in the recruitment (Cai et al. 2022) and preinitiation (He et al. 2022a) complex conformations, shown in cartoon and surface representations. The cartoon representation of CST is colored by domain as in Figure 3A, and the surface representation is colored as in Figure 1 ([purple] Ctc1, [light pink] Stn1, [lavender] Ten1). ssDNA is shown as a magenta surface, and residues mutated in CP are shown as spheres and colored as in D. (C) Structure of human CST bound to POT1/TPP1 in the presence of telomeric ssDNA (Cai et al. 2023). Colors are the same as in D and E. (D) Domain schematics of CST and Polα/Primase. Hypomorphic CP mutations in CST are indicated and colored as described here. (Red) Mutations described to disrupt Polα/Primase association, (lime green) mutations affecting POT1 binding, (blue) mutations affecting DNA binding, (orange) mutations disrupting CST trimerization and complex formation, (black) mutations with unknown mechanism. Null mutations in CST are not shown. CST domains are colored as in Figure 3A. (NTD) N-terminal domain, (EXO) inactive exonuclease, (CTD) C-terminal domain, (OB) OB fold, (PDE) phosphodiesterase-like domain, (wH) winged helix–turn–helix. Dashed lines indicate regions not modeled in cryo-EM structures. (Bottom left) POLA1–Ctc1 interaction in the RC. The CRL is highlighted in cyan. (Bottom right) Comparison of POLA1CTD AlphaFold2 (Jumper et al. 2021) models from invertebrates and Tetrahymena showing that the CRL is not conserved in organisms that do not have a corresponding Ctc1 N-terminal domain. (E) Domain schematics and cartoon representation of POT1/TPP1. (HJRL) Holliday junction resolvase-like domain, (RD) POT1 recruitment domain, (TID) TIN2-interacting domain. Dashed lines indicate regions not modeled in cryo-EM structures.
The first cryo-EM structure of human CST–Polα/Primase showed the complex in what is proposed to be the recruitment state (Fig. 5A; Cai et al. 2022). In this recruitment complex (RC), CST binds Polα/Primase in an autoinhibited state stabilized by chemical cross-linking (Baranovskiy et al. 2016; Kilkenny et al. 2022). The RC may represent the structure of CST–Polα/Primase as it is recruited to the telomere, prior to activation of the complex for fill-in synthesis. Although it is not visible in the cryo-EM map, ss telomeric DNA is present in the cross-linked sample, and its position is inferred by the loss of Stn1C binding at the DNA anchor site on Ctc1, which has been previously observed in ssDNA-bound CST (Lim et al. 2020; Cai et al. 2022).
The binding of CST to Polα/Primase in the RC is mediated by the POLA1 C-terminal domain (POLA1CTD) and the Ctc1 N-terminal domain (Fig. 5D). Half of the interacting cleft in Ctc1 is formed by OB-D, a specialized OB fold that adopts a stretched architecture that is divergent from any other described OB fold in RPA or other CST complexes (Lim et al. 2020). The other half of the Ctc1 cleft interacts with a conserved loop in POLA1CTD, termed the Ctc1 recognition loop (CRL) (Fig. 5D). The two relevant motifs for RC formation, the CRL and the Ctc1 N-terminal domain, are conserved in metazoans, whereas CST–Polα/Primase in many unicellular eukaryotes have lost these features (Fig. 5D; Cai et al. 2022). Importantly, the RC and RC-like conformations could potentially explain why CP mutations in the N-terminal domain have been observed to disrupt CST–Polα/Primase complex formation (Chen et al. 2013; Gu et al. 2018). Specifically, the V665G mutation resides on a β-strand in OB-D, which constitutes a major part of the RC interface. A destabilizing mutation in this OB fold would affect the interaction. The A227V and V265M mutations are found in OB-B, and many more mutations have since been identified in this OB fold after the initial characterization of A227V and V265M (Fig. 5D). It is likely that the other mutations in OB-B may affect CST–Polα/Primase complex formation in a similar manner. Interestingly, OB-B is not involved in the interface of the 4.6-Å RC structure, but low-resolution cryo-EM data suggest that OB-B interacts with POLA2 in an RC-like conformation of CST–Polα/Primase (Cai et al. 2022). Indeed, the POLA2 N-terminal domain (POLA2NTD) is predicted to interact with the N-terminal domain of Ctc1 (Cai et al. 2023).
Human CST–Polα/Primase was also captured in a distinct conformation that is dependent on ssDNA and proposed to represent a preinitiation complex (PIC) (Fig. 5B; He et al. 2022a). A major conformational change needs to take place to remodel the RC into the PIC. In the PIC, Polα/Primase is held in its extended state by Stn1, Ten1, and the Ctc1 C terminus. CST forms a platform for the ss telomeric DNA template and scaffolds the primase and polymerase catalytic centers in a single plane, providing a structural basis for CST-mediated stimulation of the enzymatic activity. Strikingly, both CST and Polα/Primase use a completely different set of interfaces in the RC and PIC. Neither the Ctc1 N-terminal domain nor the POLA1 CRL is involved in the PIC, and the residues mutated in CP that affect the association of CST with Polα/Primase (V665G, A227V, and V265M) (Chen et al. 2013; Gu et al. 2018) are not involved in PIC formation (Fig. 5B). It is interesting to note that CP mutations are not found at PIC-specific interfaces. Such mutations may still surface, or perhaps the predominance of mutations affecting RC formation indicates that it is the limiting step in the fill-in reaction. The other CP mutations in CST appear to affect CST recruitment (discussed below), DNA binding, or formation of the CST heterotrimer or are yet uncharacterized mutations in regions that do not map to protein–protein interfaces (Fig. 5D).
Stn1, Ten1, and the Ctc1 C terminus form the trimerization core of CST that is the most conserved half of the complex (Figs. 3A, 5D). Another recent structure determined of Tetrahymena thermophila (Tt) telomerase–CST–POLA1 bound to ss telomeric DNA supports a conserved mode of interaction between the trimerization core and Polα/Primase (He et al. 2022b). TtCST is different from metazoan CST in two major aspects: TtCST is a constitutive component of the Tetrahymena telomerase holoenzyme, and TtCtc1 only has three OB folds that are orthologous to the metazoan Ctc1 C-terminal OB folds E, F, and G (Fig. 3A). Although they are orthologous and the trimerization core is conserved, the Tetrahymena CST components differ in structure from metazoan CST, and there are altered CST–Polα interaction interfaces. Despite these differences, the overall architecture of TtCST bound to TtPOLA1 resembles the human PIC, where POLA1 sits on an interface formed by the Ctc1 C terminus, Stn1C, and ss telomeric DNA. The presence of this complex suggests that the synthesis of the telomeric G-rich and C-rich strands are executed by a single complex in Tetrahymena, but it remains to be tested whether and how this allows a direct handoff of the telomerase product to CST–Polα/Primase (He et al. 2022b).
Because metazoan CST–Polα/Primase is not embedded in telomerase and does not interact with the enzyme, it can be recruited to telomeres independently (Fig. 3B). Such independent recruitment allows the two telomere maintenance enzymes to be independently regulated and permits fill-in synthesis in cells lacking telomerase, a unique condition in humans and in some other metazoans.
POT1 recruits and regulates CST–Polα/Primase at telomeres
The DNA-binding activity of CST is not sufficient to ensure its presence at telomeres. CST–Polα/Primase is brought to telomeres through the interaction between CST and POT1/TPP1, the heterodimer in shelterin that interacts with ss telomeric DNA (Fig. 1). Human CST has been shown to interact with TPP1 in coimmunoprecipitation (co-IP) studies (Wan et al. 2009) and with POT1 in yeast two-hybrid experiments (Chen et al. 2012), and a CP mutation in POT1 (S322L) interferes with CST function at telomeres (Takai et al. 2016). Although CST is recruited to mouse telomeres by POT1b (Wu et al. 2012), human POT1 does not form a complex with CST detectable in co-IP experiments. It was therefore assumed that human CST, unlike mouse CST, is primarily recruited by TPP1. However, a mutation in Ctc1 that abolishes its interaction with TPP1 has no effect on telomere fill-in synthesis (Wang et al. 2023).
Recent cryo-EM structures of human CST bound to POT1/TPP1 reveal that POT1 is the main interactor that likely brings CST to telomeres (Fig. 5C; Cai et al. 2023). POT1 uses interfaces that are conserved with mouse POT1b to recruit CST. Importantly, the interaction between human POT1 and CST requires phosphorylation of POT1, explaining the negative co-IP data and providing a mechanism for cell cycle-dependent regulation of CST at telomeres.
POT1, which is known to be highly flexible (Smith et al. 2022; Zinder et al. 2022), is stabilized in a single conformation by Ctc1 binding at multiple interfaces (Fig. 5C,E; Cai et al. 2023). The primary interaction is formed between the POT1 C terminus (the split OB-3/HJRL) and the Ctc1 ARODL and OB-D. The POT1 CP mutation (S322L) likely prevents the formation of a critical intramolecular salt bridge that locks the Ctc1-interacting region of POT1 in place. The Ctc1 CP mutation H484P maps directly to the interface between Ctc1 and POT1, and Ctc1 G503R is predicted to destabilize the hydrophobic core of the ARODL, which is important in the interaction (Fig. 5C–E).
At a separate interface, the OB folds of the ssDNA-binding domain in the POT1 N terminus interact with Ctc1 and Stn1C (Fig. 5C). This region of CST is involved in several protein–protein interactions that depend on ssDNA, including PIC formation (Fig. 5B) and CST oligomerization (Lim et al. 2020; He et al. 2022a). In the CST–POT1/TPP1 complex, POT1 is bound to ssDNA and blocks the Ctc1 ssDNA anchor site. Comparison of the CST–POT1/TPP1 structure with the CST–Polα/Primase structures indicates that POT1/TPP1 binding to CST precludes PIC formation but is compatible with the RC (Fig. 5A–C; Cai et al. 2023).
The observation that POT1 binds to CST only when it is phosphorylated suggests a model for CST–Polα/Primase recruitment and regulation (Fig. 6; Cai et al. 2023). Following DNA replication, CST–Polα/Primase is recruited to the telomere in the RC state by a phosphorylated POT1. Sequestration of CST–Polα/Primase in the inactive state by POT1 can account for the temporal delay between replication and fill-in, creating a window for telomerase to act (Zhao et al. 2009; Wang et al. 2012). According to this model, a switch occurs upon dephosphorylation of POT1 that releases CST–Polα/Primase to form the PIC and initiate fill-in (Fig. 6; He et al. 2022a; Cai et al. 2023). It is predicted that the kinases involved are cell cycle-regulated, but their identity remains an open question.
Figure 6.

Model for fill-in regulation by shelterin. Cartoon schematic depicting the model for shelterin-mediated regulation of fill-in by CST–Polα/Primase. Following DNA replication, CST–Polα/Primase is recruited to the telomere in the autoinhibited RC state by phosphorylated POT1. 5′ end resection by Apollo and Exo1 creates a 3′ overhang, which can then be extended by telomerase. During the 5′ resection and 3′ extension steps, CST–Polα/Primase is held inactive by POT1, accounting for the known temporal delay between replication and fill-in. Upon dephosphorylation of POT1 by a hypothetical cell cycle-dependent switch, CST–Polα/Primase is released from shelterin to the ss telomeric DNA, where it can readily form the PIC and initiate fill-in synthesis to produce mature G1 telomeres.
The effects of Coats plus mutations on telomere maintenance
CP patients generally have compound heterozygous mutations in CTC1 or STN1, with one allele being nonfunctional (e.g., a frameshift mutation) and the other showing a partial loss of function (shown in Fig. 5D). This compound genotype explains why CP is rare, and the presence of one hypomorphic allele, rather than LOF mutations in both alleles, is consistent with CST being required for long-term cell viability. The parents of CP patients have no notable symptoms, indicating that the hypomorphic alleles do not lead to a phenotype unless the other allele is not functional (Anderson et al. 2012; Keller et al. 2012; Polvi et al. 2012; Walne et al. 2013; Netravathi et al. 2015; Simon et al. 2016; Takai et al. 2016; Lin et al. 2017; Hidalgo-Sanz et al. 2019; Passi et al. 2020). No CP mutations in the TEN1 gene have been reported but they may be identified in the future. There is also a possibility that mutations in Polα/Primase could be found to cause CP, though such mutations would have to be specific to the interaction with CST without affecting the canonical replication function of the enzyme. In the two (related) CP cases ascribed to a POT1 defect, both POT1 alleles have the same sequence alteration that results in a S322L change (Fig. 5E). Cells expressing only POT1S322L show defects in the CST-related functions of POT1 but retain the ability to protect their telomeres from ATR signaling, which is the essential function of POT1 (Takai et al. 2016).
CP mutations are expected to manifest milder versions of the telomeric phenotypes observed in cells lacking CST, which include C-strand shortening and extension of the G-strand (when telomerase is present), as well as stochastic telomere loss resulting from excessive and unmitigated C-strand resection (Stewart et al. 2012; Wang et al. 2012; Takai et al. 2016; Feng et al. 2017, 2018; Gu et al. 2018; Zhang et al. 2019). Additionally, metaphase chromosomes in cells lacking CST can show disrupted telomeric FISH signals (referred to as fragile telomeres) (Sfeir et al. 2009) that report on problems in the replication of the double-stranded telomeric DNA (Price et al. 2010; Wang et al. 2012).
Ideally, the effects of CP mutations would be studied in cell culture systems that reflect the patient genotypes. However, preliminary studies using co-IP and overexpression systems have already revealed the mechanism by which a subset of CP mutations in Ctc1 affects CST–Polα/Primase function (Chen et al. 2013; Gu and Chang 2013). As mentioned above, three mutations (highlighted in red in Fig. 5), including one near the Ctc1–POLA1 CRL interface (Fig. 5A,D), diminish the interaction with Polα/Primase, resulting in little nuclear CST and defective telomeric accumulation of the complex (Chen et al. 2013; Gu et al. 2018). These data are consistent with the partial dependence of CST on Polα/Primase for nuclear import (Kelich et al. 2021).
Two mutations in the OB-F part of the Ctc1 DNA-binding domain (R975G and C985D) (colored blue in Fig. 5) strongly diminish the interaction of CST with DNA without affecting its association with Polα/Primase or Stn1/Ten1. Importantly, these versions of Ctc1 accumulate at telomeres, demonstrating that DNA binding by CST is not required for its recruitment to the telomeric chromatin. A third mutation in OB-F (R987W) similarly affects the DNA-binding activity without diminishing the interaction of Ctc1 with its binding partners. Interestingly, this mutant has a greater defect in nuclear localization even though its interaction with Polα/Primase in co-IP appears normal. However, the portion of nuclear CST in this mutant does still associate with telomeres based on ChIP (Chen et al. 2013).
The G503R in the ARODL domain appears to behave like wild-type Ctc1 in terms of DNA binding and interaction with Stn1/Ten1 and Polα/Primase (Chen et al. 2013). Nevertheless, this protein, while nuclear, does not accumulate at telomeres, suggesting a recruitment defect. Indeed, G503R is predicted to destabilize the ARODL domain, which is critical for POT1 binding (Cai et al. 2023). Remarkably, the expression of the G503R mutant interferes with the regulation of telomere length by the endogenous wild-type CST, indicating that the minute amount of G503R protein at telomeres exerts a dominant-negative effect (Chen et al. 2013).
Finally, the R840W and V871M mutants in OB-E are mysterious in that they appear to behave like wild-type Ctc1 in all assays (Chen et al. 2013). Perhaps these alleles and a handful of newly described patient mutations (colored black in Fig. 5) will reveal their defects when they are tested in a setting that recapitulates the genotype of the patient.
Why is the presentation of Coats plus so different from dyskeratosis congenita? One proffered explanation (Zhang et al. 2019) is that, unlike the mutations in the telomerase pathway, the defects in CST could also affect nontelomeric sites in the genome. Indeed, CST (possibly alongside Polα/Primase) functions at genome-wide sites of replication stress, where it promotes the unfolding of G4 structures and replication fork restart and additionally affects the firing of dormant origins. The reported interactions of CST with Rad51, AND-1, and the MCM proteins in the replicative helicase (for review, see Stewart et al. 2018) are likely relevant to these extratelomeric roles of CST. However, one argument against a role for loss of nontelomeric CST functions in the pathogenesis of CP is the finding of a POT1 mutation in CP patients (Takai et al. 2016). Since POT1 is unlikely to contribute to the genome-wide DNA replication function of CST, these CP cases show that the full spectrum of CP symptoms can arise from lack of the telomere-specific function of CST. At present, therefore, it remains a mystery why mutations in the two telomere maintenance machines lead to different disease manifestations. To make matters more complex, the Ctc1 C985D mutation found in CP patients is also the cause of at least three cases of DC or a DC-like presentation (Walne et al. 2013; Shen et al. 2019; Han et al. 2020).
Perspective
From its humble beginnings as yet another “accessory factor” to the replication machinery, CST has emerged as a new master regulator of telomere length that influences both C-strand and G-strand synthesis through CST–Polα/Primase fill-in and telomerase inhibition, respectively. Decades of genetic studies combined with the recent power of cryo-EM have provided insights into the structure, function, and evolution of this new (but in the evolutionary scale, ancient) telomere maintenance machine. It will be exciting to see further advances in our understanding of the interplay between CST–Polα/Primase, telomerase, and shelterin and in the characterization of molecular missteps in this dance that cause human disease.
Acknowledgments
We thank Neal Lue and members of the de Lange laboratory for comments on the manuscript. We thank Ludovic Sauguet for kindly sharing coordinates for the archaeal PabRPA AROD prior to publication. Tom Cech and one anonymous reviewer are thanked for their insightful comments. S.W.C. is supported by the David Rockefeller Graduate Program and a National Science Foundation Graduate Research Fellowship (1946429). Work on telomeres and CST in the de Lange laboratory is supported by grants from the National Institutes of Health (R35 CA210036) and the Breast Cancer Research Foundation (BCRF-22-036) to T.d.L.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350479.123.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
T.d.L. is a member of the Scientific Advisory Board of Calico Life Sciences LLC.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982. 10.1128/MCB.00240-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BH, Kasher PR, Mayer J, Szynkiewicz M, Jenkinson EM, Bhaskar SS, Urquhart JE, Daly SB, Dickerson JE, O'Sullivan J, et al. 2012. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat Genet 44: 338–342. 10.1038/ng.1084 [DOI] [PubMed] [Google Scholar]
- Ballinger ML, Pattnaik S, Mundra PA, Zaheed M, Rath E, Priestley P, Baber J, Ray-Coquard I, Isambert N, Causeret S, et al. 2023. Heritable defects in telomere and mitotic function selectively predispose to sarcomas. Science 379: 253–260. 10.1126/science.abj4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranovskiy AG, Babayeva ND, Zhang Y, Gu J, Suwa Y, Pavlov YI, Tahirov TH. 2016. Mechanism of concerted RNA–DNA primer synthesis by the human primosome. J Biol Chem 291: 10006–10020. 10.1074/jbc.M116.717405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V. 2006. The maintenance and masking of chromosome termini. Curr Opin Cell Biol 18: 247–253. 10.1016/j.ceb.2006.04.005 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Wang Y, Diao J, Price CM. 2017. Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res 45: 12311–12324. 10.1093/nar/gkx878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan C, Rice C, Harkisheimer M, Schultz DC, Skordalakes E. 2013. Structure of the human telomeric Stn1–Ten1 capping complex. PLoS One 8: e66756. 10.1371/journal.pone.0066756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SW, Zinder JC, Svetlov V, Bush MW, Nudler E, Walz T, de Lange T. 2022. Cryo-EM structure of the human CST–Polα/primase complex in a recruitment state. Nat Struct Mol Biol 29: 813–819. 10.1038/s41594-022-00766-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai SW, Takai H, Walz T, de Lange T. 2023. Structural basis of CST–Polα/primase recruitment and regulation by POT1 at telomeres. bioRxiv 10.1101/2023.05.08.539880 [DOI] [Google Scholar]
- Carson MJ, Hartwell L. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42: 249–257. 10.1016/s0092-8674(85)80120-3 [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. 2009. A DNA polymerase-α·primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818. 10.1074/jbc.M807593200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Lingner J. 2013. CST for the grand finale of telomere replication. Nucleus 4: 277–282. 10.4161/nucl.25701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Redon S, Lingner J. 2012. The human CST complex is a terminator of telomerase activity. Nature 488: 540–544. 10.1038/nature11269 [DOI] [PubMed] [Google Scholar]
- Chen LY, Majerská J, Lingner J. 2013. Molecular basis of telomere syndrome caused by CTC1 mutations. Genes Dev 27: 2099–2108. 10.1101/gad.222893.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue J, Churikov D, Hass EP, Shi S, Lemon LD, Luciano P, Bertuch AA, Zappulla DC, Géli V, et al. 2018. Structural insights into yeast telomerase recruitment to telomeres. Cell 172: 331–343.e13. 10.1016/j.cell.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2004. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol 5: 323–329. 10.1038/nrm1359 [DOI] [PubMed] [Google Scholar]
- de Lange T. 2015. A loopy view of telomere evolution. Front Genet 6: 321. 10.3389/fgene.2015.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2018. Shelterin-mediated telomere protection. Annu Rev Genet 52: 223–247. 10.1146/annurev-genet-032918-021921 [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071. 10.1038/nature06065 [DOI] [PubMed] [Google Scholar]
- Eme L, Spang A, Lombard J, Stairs CW, Ettema TJG. 2017. Archaea and the origin of eukaryotes. Nat Rev Microbiol 15: 711–723. 10.1038/nrmicro.2017.133 [DOI] [PubMed] [Google Scholar]
- Feng X, Hsu SJ, Kasbek C, Chaiken M, Price CM. 2017. CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res 45: 4281–4293. 10.1093/nar/gkx125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Hsu SJ, Bhattacharjee A, Wang Y, Diao J, Price CM. 2018. CTC1–STN1 terminates telomerase while STN1–TEN1 enables C-strand synthesis during telomere replication in colon cancer cells. Nat Commun 9: 2827. 10.1038/s41467-018-05154-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P. 2013. Why are there so many diverse replication machineries?. J Mol Biol 425: 4714–4726. 10.1016/j.jmb.2013.09.032 [DOI] [PubMed] [Google Scholar]
- Frescas D, de Lange T. 2014. Binding of TPP1 protein to TIN2 protein is required for POT1a,b protein-mediated telomere protection. J Biol Chem 289: 24180–24187. 10.1074/jbc.M114.592592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Wu Z, Chen H, Zhong Q, Shi S, Li G, Wu J, Lei M. 2020. Structural insights into telomere protection and homeostasis regulation by yeast CST complex. Nat Struct Mol Biol 27: 752–762. 10.1038/s41594-020-0459-8 [DOI] [PubMed] [Google Scholar]
- Goulian M, Heard CJ. 1990. The mechanism of action of an accessory protein for DNA polymerase α/primase. J Biol Chem 265: 13231–13239. 10.1016/S0021-9258(19)38289-4 [DOI] [PubMed] [Google Scholar]
- Goulian M, Heard CJ, Grimm SL. 1990. Purification and properties of an accessory protein for DNA polymerase α/primase. J Biol Chem 265: 13221–13230. 10.1016/S0021-9258(19)38288-2 [DOI] [PubMed] [Google Scholar]
- Gu P, Chang S. 2013. Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell 12: 1100–1109. 10.1111/acel.12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Jia S, Takasugi T, Smith E, Nandakumar J, Hendrickson E, Chang S. 2018. CTC1-STN1 coordinates G- and C-strand synthesis to regulate telomere length. Aging Cell 17: e12783. 10.1111/acel.12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han E, Patel NA, Yannuzzi NA, Laura DM, Fan KC, Negron CI, Prakhunhungsit S, Thorson WL, Berrocal AM. 2020. A unique case of coats plus syndrome and dyskeratosis congenita in a patient with CTC1 mutations. Ophthalmic Genet 41: 363–367. 10.1080/13816810.2020.1772315 [DOI] [PubMed] [Google Scholar]
- Hay RT, Hendrickson EA, DePamphilis ML. 1984. Sequence specificity for the initiation of RNA-primed simian virus 40 DNA synthesis in vivo. J Mol Biol 175: 131–157. 10.1016/0022-2836(84)90471-6 [DOI] [PubMed] [Google Scholar]
- Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J, Wade KH, Timpson NJ, Evans DM, Willeit P, et al. 2017. Association between telomere length and risk of cancer and non-neoplastic diseases: a Mendelian randomization study. JAMA Oncol 3: 636–651. 10.1001/jamaoncol.2016.5945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Feigon J. 2022. Telomerase structural biology comes of age. Curr Opin Struct Biol 76: 102446. 10.1016/j.sbi.2022.102446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Lin X, Chavez BL, Agrawal S, Lusk BL, Lim CJ. 2022a. Structures of the human CST–Polα–primase complex bound to telomere templates. Nature 608: 826–832. 10.1038/s41586-022-05040-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Song H, Chan H, Liu B, Wang Y, Sušac L, Zhou ZH, Feigon J. 2022b. Structure of Tetrahymena telomerase-bound CST with polymerase α–primase. Nature 608: 813–818. 10.1038/s41586-022-04931-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo-Sanz J, Pérez-Delgado R, García-Jiménez I, López-Pisón J, Castillo-Castejón O, Poch ML, Izquierdo-Álvarez S. 2019. Lactante con calcificaciones intracraneales y retinopatía. Rev Neurol 69: 289–292. 10.33588/rn.6907.2019166 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Collins K. 2015. Control of telomerase action at human telomeres. Nat Struct Mol Biol 22: 848–852. 10.1038/nsmb.3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. 2006. Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77. 10.1016/j.cell.2006.04.044 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Palm W, Wang RC, Couto SS, de Lange T. 2008. Engineered telomere degradation models dyskeratosis congenita. Genes Dev 22: 1773–1785. 10.1101/gad.1679208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom RA, Wuttke DS. 2017. Human CST prefers G-rich but not necessarily telomeric sequences. Biochemistry 56: 4210–4218. 10.1021/acs.biochem.7b00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K, Ghandi M, Huang FW. 2021. Integrated evaluation of telomerase activation and telomere maintenance across cancer cell lines. Elife 10: e66198. 10.7554/eLife.66198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O'Brien Johnson R, Collins K, Loo JA, et al. 2015. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350: aab4070. 10.1126/science.aab4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Applying and improving AlphaFold at CASP14. Proteins 89: 1711–1721. 10.1002/prot.26257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelich JM, Papaioannou H, Skordalakes E. 2021. Pol α–primase dependent nuclear localization of the mammalian CST complex. Commun Biol 4: 349. 10.1038/s42003-021-01845-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller RB, Gagne KE, Usmani GN, Asdourian GK, Williams DA, Hofmann I, Agarwal S. 2012. CTC1 mutations in a patient with dyskeratosis congenita. Pediatr Blood Cancer 59: 311–314. 10.1002/pbc.24193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny ML, Veale CE, Guppy A, Hardwick SW, Chirgadze DY, Rzechorzek NJ, Maman JD, Pellegrini L. 2022. Structural basis for the interaction of SARS-CoV-2 virulence factor nsp1 with DNA polymerase α–primase. Protein Sci 31: 333–344. 10.1002/pro.4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J. 1999. TIN2, a new regulator of telomere length in human cells. Nat Genet 23: 405–412. 10.1038/70508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WT, Hennick K, Johnson J, Finnerty B, Choo S, Short SB, Drubin C, Forster R, McMaster ML, Hockemeyer D. 2021. Cancer-associated POT1 mutations lead to telomere elongation without induction of a DNA damage response. EMBO J 40: e107346. 10.15252/embj.2020107346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S. 2010. SNMIB/apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J 29: 2230–2241. 10.1038/emboj.2010.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade H, Querido E, Smith MJ, Guérit D, Crimmins H, Conomos D, Pourret E, Chartrand P, Sfeir A. 2020. Single-molecule imaging of telomerase RNA reveals a recruitment-retention model for telomere elongation. Mol Cell 79: 115–126.e6. 10.1016/j.molcel.2020.05.005 [DOI] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR. 2005. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem 280: 20449–20456. 10.1074/jbc.M502212200 [DOI] [PubMed] [Google Scholar]
- Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. 2006. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol 16: 1303–1310. 10.1016/j.cub.2006.05.021 [DOI] [PubMed] [Google Scholar]
- Lim CJ, Cech TR. 2021. Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat Rev Mol Cell Biol 22: 283–298. 10.1038/s41580-021-00328-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Barbour AT, Zaug AJ, Goodrich KJ, McKay AE, Wuttke DS, Cech TR. 2020. The structure of human CST reveals a decameric assembly bound to telomeric DNA. Science 368: 1081–1085. 10.1126/science.aaz9649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Gong L, Zhan S, Wang Y, Liu A. 2017. Novel biallelic missense mutations in CTC1 gene identified in a Chinese family with Coats plus syndrome. J Neurol Sci 382: 142–145. 10.1016/j.jns.2017.09.041 [DOI] [PubMed] [Google Scholar]
- Lingner J, Cooper JP, Cech TR. 1995. Telomerase and DNA end replication: no longer a lagging strand problem?. Science 269: 1533–1534. 10.1126/science.7545310 [DOI] [PubMed] [Google Scholar]
- Liu L, Malkova A. 2022. Break-induced replication: unraveling each step. Trends Genet 38: 752–765. 10.1016/j.tig.2022.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, He Y, Wang Y, Song H, Zhou ZH, Feigon J. 2022. Structure of active human telomerase with telomere shelterin protein TPP1. Nature 604: 578–583. 10.1038/s41586-022-04582-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, de Lange T. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018. 10.1038/nature01688 [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. 2004. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem 279: 13241–13248. 10.1074/jbc.M312309200 [DOI] [PubMed] [Google Scholar]
- Lue NF. 2018. Evolving linear chromosomes and telomeres: a C-strand-centric view. Trends Biochem Sci 43: 314–326. 10.1016/j.tibs.2018.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, de Lange T. 2017. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18: 175–186. 10.1038/nrm.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madru C, Martínez-Carranza M, Laurent S, Alberti AC, Chevreuil M, Raynal B, Haouz A, Le Meur RA, Delarue M, Henneke G, et al. 2023. DNA-binding mechanism and evolution of replication protein A. Nat Commun 14: 2326. 10.1038/s41467-023-38048-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z, de Lange T. 2020. 53BP1: a DSB escort. Genes Dev 34: 7–23. 10.1101/gad.333237.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z, Lottersberger F, Takai H, Kibe T, Gong Y, Takai K, Bianchi A, Zimmermann M, Durocher D, de Lange T. 2018. 53BP1–RIF1–shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 560: 112–116. 10.1038/s41586-018-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z, Cai S, de Lange T. 2023. CST/polα/primase-mediated fill-in synthesis at DSBs. Cell Cycle 22: 379–389. 10.1080/15384101.2022.2123886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. 2009. RPA-like mammalian Ctc1–Stn1–Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206. 10.1016/j.molcel.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Myler LR, Kinzig CG, Sasi NK, Zakusilo G, Cai SW, de Lange T. 2021. The evolution of metazoan shelterin. Genes Dev 35: 1625–1641. 10.1101/gad.348835.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Nabetani A, Mizuno T, Hanaoka F, Ishikawa F. 2005. Alterations of DNA and chromatin structures at telomeres and genetic instability in mouse cells defective in DNA polymerase α. Mol Cell Biol 25: 11073–11088. 10.1128/MCB.25.24.11073-11088.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289. 10.1038/nature11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netravathi M, Kumari R, Kapoor S, Dakle P, Dwivedi MK, Roy SD, Pandey P, Saini J, Ramakrishna A, Navalli D, et al. 2015. Whole exome sequencing in an Indian family links Coats plus syndrome and dextrocardia with a homozygous novel CTC1 and a rare HES7 variation. BMC Med Genet 16: 5. 10.1186/s12881-015-0151-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CL, Barbour AT, Wieser TA, Wuttke DS. 2023. RPA engages telomeric G-quadruplexes more effectively than CST. Nucleic Acids Res 51: 5073–5086. 10.1093/nar/gkad315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette MM, Liao M, Herbert BS, Johnson M, Holt SE, Liss HS, Shay JW, Wright WE. 2000. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J Biol Chem 275: 10072–10076. 10.1074/jbc.275.14.10072 [DOI] [PubMed] [Google Scholar]
- Passi GR, Shamim U, Rathore S, Joshi A, Mathur A, Parveen S, Sharma P, Crow YJ, Faruq M. 2020. An Indian child with Coats plus syndrome due to mutations in STN1. Am J Med Genet A 182: 2139–2144. 10.1002/ajmg.a.61737 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396. 10.1016/s0092-8674(01)00226-4 [DOI] [PubMed] [Google Scholar]
- Polvi A, Linnankivi T, Kivelä T, Herva R, Keating JP, Mäkitie O, Pareyson D, Vainionpää L, Lahtinen J, Hovatta I, et al. 2012. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am J Hum Genet 90: 540–549. 10.1016/j.ajhg.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar VL, Cranert S, Linger BR, Morin GB, Minium S, Price C. 2014. The 3′ overhangs at Tetrahymena thermophila telomeres are packaged by four proteins, Pot1a, Tpt1, Pat1, and Pat2. Eukaryot Cell 13: 240–245. 10.1128/EC.00275-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. 2010. Evolution of CST function in telomere maintenance. Cell Cycle 9: 3157–3165. 10.4161/cc.9.16.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi MH, Ray S, Sewell AL, Basu S, Balci H. 2012. Replication protein A unfolds G-quadruplex structures with varying degrees of efficiency. J Phys Chem B 116: 5588–5594. 10.1021/jp300546u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C, Skordalakes E. 2016. Structure and function of the telomeric CST complex. Comput Struct Biotechnol J 14: 161–167. 10.1016/j.csbj.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MA, Blackburn EH. 2004. Processive utilization of the human telomerase template: lack of a requirement for template switching. J Biol Chem 279: 53770–53781. 10.1074/jbc.M407768200 [DOI] [PubMed] [Google Scholar]
- Savage SA. 2022. Dyskeratosis congenita and telomere biology disorders. Hematology Am Soc Hematol Educ Program 2022: 637–648. 10.1182/hematology.2022000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J, Muñoz-Subirana N, Kool H, van Schendel R, Tijsterman M. 2021. Small tandem DNA duplications result from CST-guided Pol α–Primase action at DNA break termini. Nat Commun 12: 4843. 10.1038/s41467-021-25154-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Zaug AJ, Cech TR. 2016. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166: 1188–1197.e9. 10.1016/j.cell.2016.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz I, Mensenkamp AR, Takai KK, Haadsma M, Spruijt L, de Voer RM, Choo SS, Lorbeer FK, van Grinsven EJ, Hockemeyer D, et al. 2020. TINF2 is a haploinsufficient tumor suppressor that limits telomere length. Elife 9: e61235. 10.7554/eLife.61235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekne Z, Ghanim GE, van Roon AM, Nguyen THD. 2022. Structural basis of human telomerase recruitment by TPP1-POT1. Science 375: 1173–1176. 10.1126/science.abn6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE. 2005. Telomere-end processing the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138. 10.1016/j.molcel.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103. 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaff RJ, Kuchta RD. 1993. Mechanism of calf thymus DNA primase: slow initiation, rapid polymerization, and intelligent termination. Biochemistry 32: 3027–3037. 10.1021/bi00063a014 [DOI] [PubMed] [Google Scholar]
- Shen W, Kerr CM, Przychozen B, Mahfouz RZ, LaFramboise T, Nagata Y, Hanna R, Radivoyevitch T, Nazha A, Sekeres MA, et al. 2019. Impact of germline CTC1 alterations on telomere length in acquired bone marrow failure. Br J Haematol 185: 935–939. 10.1111/bjh.15862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AJ, Lev A, Zhang Y, Weiss B, Rylova A, Eyal E, Kol N, Barel O, Cesarkas K, Soudack M, et al. 2016. Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects. J Exp Med 213: 1429–1440. 10.1084/jem.20151618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EW, Lattmann S, Liu ZB, Ahsan B, Rhodes D. 2022. Insights into POT1 structural dynamics revealed by cryo-EM. PLoS One 17: e0264073. 10.1371/journal.pone.0264073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Jolivet P, Teixeira MT. 2014. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol Cell 53: 954–964. 10.1016/j.molcel.2014.02.030 [DOI] [PubMed] [Google Scholar]
- Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD, Wright WE, Price CM. 2012. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 31: 3537–3549. 10.1038/emboj.2012.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Wang Y, Ackerson SM, Schuck PL. 2018. Emerging roles of CST in maintaining genome stability and human disease. Front Biosci (Landmark Ed) 23: 1564–1586. 10.2741/4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. 2009. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218. 10.1016/j.molcel.2009.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. 2011. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell 44: 647–659. 10.1016/j.molcel.2011.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Jenkinson E, Kabir S, Babul-Hirji R, Najm-Tehrani N, Chitayat DA, Crow YJ, de Lange T. 2016. A POT1 mutation implicates defective telomere end fill-in and telomere truncations in Coats plus. Genes Dev 30: 812–826. 10.1101/gad.276873.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743. 10.1038/385740a0 [DOI] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. 2022. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50: D439–D444. 10.1093/nar/gkab1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Bhagat T, Kirwan M, Gitiaux C, Desguerre I, Leonard N, Nogales E, Vulliamy T, Dokal IS. 2013. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica 98: 334–338. 10.3324/haematol.2012.071068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KM, Codd V, Rice T, Nelson CP, Smirnov IV, McCoy LS, Hansen HM, Elhauge E, Ojha J, Francis SS, et al. 2015. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget 6: 42468–42477. 10.18632/oncotarget.6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D. 2009. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284: 26725–26731. 10.1074/jbc.M109.021105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510. 10.1038/nature05454 [DOI] [PubMed] [Google Scholar]
- Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM. 2012. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep 2: 1096–1103. 10.1016/j.celrep.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ma T, Zhang X, Chen W, Lan Y, Kuang G, Hsu SJ, He Z, Chen Y, Stewart J, et al. 2023. CTC1 OB–B interaction with TPP1 terminates telomerase and prevents telomere overextension. Nucleic Acids Res 51: 4914–4928. 10.1093/nar/gkad237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. 2006. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62. 10.1016/j.cell.2006.05.037 [DOI] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T. 2010. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell 39: 606–617. 10.1016/j.molcel.2010.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T. 2012. Telomeric 3′ overhangs derive from resection by Exo1 and apollo and fill-in by POT1b-associated CST. Cell 150: 39–52. 10.1016/j.cell.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Hendrickson EA, DePamphilis ML. 1985. DNA primase–DNA polymerase α from simian cells: sequence specificity of initiation sites on simian virus 40 DNA. Mol Cell Biol 5: 1170–1183. 10.1128/mcb.5.5.1170-1183.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Takai KK, Lovejoy CA, de Lange T. 2020. Break-induced replication promotes fragile telomere formation. Genes Dev 34: 1392–1405. 10.1101/gad.328575.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Sun J, Lei M, Lue NF. 2012. Analyses of Candida Cdc13 orthologues revealed a novel OB fold dimer arrangement, dimerization-assisted DNA binding, and substantial structural differences between Cdc13 and RPA70. Mol Cell Biol 32: 186–198. 10.1128/MCB.05875-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Lim CJ, Olson CL, Carilli MT, Goodrich KJ, Wuttke DS, Cech TR. 2021. CST does not evict elongating telomerase but prevents initiation by ssDNA binding. Nucleic Acids Res 49: 11653–11665. 10.1093/nar/gkab942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Goodrich KJ, Song JJ, Sullivan AE, Cech TR. 2022. Reconstitution of a telomeric replicon organized by CST. Nature 608: 819–825. 10.1038/s41586-022-04930-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang B, Li T, Liu R, Xiao Y, Geng X, Li G, Liu Q, Price CM, Liu Y, et al. 2019. Mammalian CST averts replication failure by preventing G-quadruplex accumulation. Nucleic Acids Res 47: 5243–5259. 10.1093/nar/gkz264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475. 10.1016/j.cell.2009.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494. 10.1016/j.cell.2012.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Hathcock KS, Hande P, Lansdorp PM, Seldin MF, Hodes RJ. 1998. Telomere length regulation in mice is linked to a novel chromosome locus. Proc Natl Acad Sci 95: 8648–8653. 10.1073/pnas.95.15.8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder JC, Olinares PDB, Svetlov V, Bush MW, Nudler E, Chait BT, Walz T, de Lange T. 2022. Shelterin is a dimeric complex with extensive structural heterogeneity. Proc Natl Acad Sci 119: e2201662119. 10.1073/pnas.2201662119 [DOI] [PMC free article] [PubMed] [Google Scholar]