Abstract
The incidence of hepatocellular carcinoma (HCC) is rapidly rising largely because of increased obesity leading to nonalcoholic steatohepatitis (NASH), a known HCC risk factor. There are no approved treatments to treat NASH. Here, we first used single-nucleus RNA sequencing to characterize a mouse model that mimics human NASH–driven HCC, the MUP-uPA mouse fed a high-fat diet. Activation of endoplasmic reticulum (ER) stress and inflammation was observed in a subset of hepatocytes that was enriched in mice that progress to HCC. We next treated MUP-uPA mice with the ER stress inhibitor BGP-15 and soluble gp130Fc, a drug that blocks inflammation by preventing interleukin-6 trans-signaling. Both drugs have progressed to phase 2/3 human clinical trials for other indications. We show that this combined therapy reversed NASH and reduced NASH-driven HCC. Our data suggest that these drugs could provide a potential therapy for NASH progression to HCC.
Clinically safe drugs that target cellular stress and inflammation could successfully treat NASH and liver cancer.
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common and fatal cancers worldwide (1). Over the past decade, global HCC incidence has increased by ~40%, rendering it the fastest-rising cause of cancer-related deaths (2). The rise in HCC coincides with patients presenting with obesity and symptoms associated with the metabolic syndrome, especially metabolic dysfunction–associated fatty liver disease (MAFLD) [formerly known as nonalcoholic fatty liver disease (NAFLD)] and the more aggressive nonalcoholic steatohepatitis (NASH), an established HCC risk factor (3). Globally, a staggering 25% of the population are estimated to suffer from MAFLD, with around 15% of those exhibiting signs of NASH (1). A subset of patients with NASH will develop HCC as a result of neoplastic transformations that occur following repetitive cycles of cell death and repair associated with lipotoxicity (4). The low survival rate of HCC is, in large part, due to lack of effective therapies and preventive measures (5, 6). The most widely used HCC drug, sorafenib, is a general tyrosine kinase inhibitor (TKI) that extends survival time by only 3 months (7). Current efforts in HCC therapeutic development is centered on development of more targeted systemic therapies and combining TKIs with other drugs, such as monoclonal antibodies and immune checkpoint inhibitors (ICIs) leading to several recent phase 2 and 3 clinical trials (6). Nevertheless, a dire need persists for successful strategies to prevent and combat NASH-induced HCC because, at present, alleviating NASH severity to prevent HCC relies entirely on lifestyle interventions and bariatric surgery aimed at reducing body weight (4).
NASH itself is one of the fastest-growing metabolic diseases and has become the leading cause of liver transplantation in the United States, constituting a major unmet clinical need. As expected, the NASH drug market is estimated to reach $40 billion by 2025 (5). Now, there are several clinical trials of new NASH drug candidates (5, 6, 8). Most attempts to treat NASH have focused on pathways that affect the balance between fatty acid uptake and export, de novo lipogenesis (DNL), and fatty acid oxidation (FAO) in the liver. While some drug candidates, such as incretins, are effective in treating MAFLD, there are few successful therapies that target liver lipids in patients with NASH, who present with advanced steatosis and secondary liver pathologies, such as inflammation and fibrosis. Drug candidates to treat NASH by reducing liver steatosis have shown promise but have failed because of unwanted side effects (9, 10). These side effects often include hyperlipidemia and/or hypercholesterolemia, which increase the risk of cardiovascular disease (11). Previous work from our group has demonstrated that NASH progression to HCC in high-fat diet (HFD)– or high-fructose diet–fed MUP-uPA mice, a preclinical model that faithfully mimics human NASH to HCC progression (5), is associated with inflammation and endoplasmic reticulum (ER) stress (12–14). To examine this association more deeply, we first profiled liver biopsies of HFD-fed MUP-uPA mice at 24 weeks, a time point where all mice have NASH but not HCC, by single-nucleus RNA sequencing (snRNA-seq). This analysis revealed an enrichment of recently identified disease-associated hepatocytes (daHeps) (15) and Trem2-positive macrophages (16–18) in livers of animals that manifested HCC at the experimental end point (40 weeks) compared with animals that remained HCC-free. In addition to Trem2-positive macrophages, daHep cells present a strong component of ER stress response in their transcriptome.
On the basis of the snRNA-seq data, we reasoned that the targeting inflammation and ER stress could be a viable approach to NASH, even without direct effects on lipid metabolism. Of note, ER stress inhibitors or chemical chaperones, such as tauroursodeoxycholic acid, prevent MAFLD/NASH in mice (12, 14), and we were involved in development of the hydroxylamine-based small-molecule BGP-15, which inhibits ER stress (19), fibrosis (20, 21), and oxidative stress (22), which are common features of NASH-driven HCC. BGP-15 has entered human clinical trials and has an excellent safety profile (23). Interleukin-6 (IL-6) signaling can either be detrimental (24, 25) or protective (14) in the etiology of NASH and/or HCC. The beneficial effects are likely due to activation of the membrane-bound IL-6 receptor (IL-6R)/gp130 signaling pathway that supports maintenance of gut barrier integrity (14, 26) and adenosine monophosphate (AMP)–activated protein kinase (AMPK)–mediated increases in FAO (27, 28), resulting in prevention of hepatic steatosis (28, 29). In contrast, IL-6 trans-signaling, a state where soluble IL-6 combines with a soluble IL-6R/gp130, transduces an indirect signal that accounts for the proinflammatory effects of IL-6 in both sepsis (30) and atherosclerosis (31). Of note, in obesity-driven insulin resistance, IL-6 trans-signaling blockade with soluble gp130Fc prevented macrophage accumulation in the adipose tissue and liver (32). The clinical form of sgp130Fc, termed Olamkicept, was found to be safe and effective in a phase 2 clinical trial for patients with active inflammatory bowel disease (33).
Given that ER stress and inflammation are common features of NASH-driven HCC and that BGP-15 and Olamkicept (sgp130Fc) have both entered phase 2/3 clinical trials and are safe, we tested whether these drugs as monotherapy, or in combination, would prevent NASH and/or NASH-driven HCC in HFD-fed MUP-uPA mice. Consistent with our hypothesis, we show that BGP-15/sgp130Fc treatment can regress NASH and markedly reduces NASH-driven HCC via inhibition of ER stress signaling and reduced inflammation.
RESULTS
snRNA-seq reveals cell states enriched in NASH conversion to HCC
To unveil cellular states associated with NASH progression to HCC, we first performed snRNA-seq on liver biopsies of HFD-fed MUP-uPA mice at a time point before malignant transformation (24 weeks) and then separated animals into tumor-bearing (TB, n = 6) and tumor-free (TF, n = 4) at end point (40 weeks) for downstream analyses (fig. S1A). Data were filtered, normalized, and scaled using Seurat 4.2 (34) and then batch-corrected using FastMNN (35) as a Seurat wrapper (see Materials and Methods). We obtained a total of 47,501 single-nucleus transcriptomes that clustered into 13 subsets following unsupervised clustering analysis. On the basis of cell-specific marker expression, we were able to annotate each cluster as one of the nine major liver cell types: Hnf4a (hepatocytes), F8 (endothelial cells), Adgre1 (myeloid cells), Pdgfrb (mesenchymal cells), Hnf1b (biliary epithelial cells), Skap1 [T and natural killer (NK) cells], Pax5 (B cells), Siglech [plasmacytoid dendritic cells (pDCs)], and Bnc2 (mesothelial cells) (fig. S1, B to D). Most cell types were represented by a single cluster, except by hepatocytes and endothelial cells that clustered into two subsets and myeloid cells that generated three clusters. Hierarchical clustering based on the top five markers of each cell type confirms appropriate annotation into nine major lineages (fig. S1E and table S1). Clusters representing the same major cell type were collapsed into one for downstream analyses. Cell type representation in each individual sample was calculated, revealing that immune cell types including myeloid, T and NK cells, and pDCs were enriched in animals that went on to develop HCC (fig. S1F). These observations are consistent with the notion that NASH-related inflammation is a major contributor to tumorigenesis. Reclustering of the myeloid lineage revealed eight transcriptionally distinct populations corresponding to well-characterized myeloid subsets (fig. S1G and table S2). The four most abundant represented macrophage subtypes include embryonic-derived Kupffer cells (Cd5l, Vsig4, and Clec4f); Cx3cr1+ macrophages recently shown to form a niche around portal tracts in both mouse and human liver (36, 37); Trem2+ macrophages (Gpnmb, Mmp12, and Lgals3), a subset frequently reported to arise and expand during liver disease (17, 18, 38); and undifferentiated monocytes, characterized by high Ccr2 expression. The remaining clusters corresponded to conventional DCs (cDC1 and cDC2), neutrophils, and “mature DCs enriched in immunoregulatory molecules” (Mreg_DCs) (39). Comparison of relative proportions of each subset between the TB and TF groups revealed that Trem2+ macrophages and Mreg_DCs were significantly increased, whereas Kupffer cells decreased in mice that went on to develop HCC, which may suggest exacerbated inflammation in the TB mice.
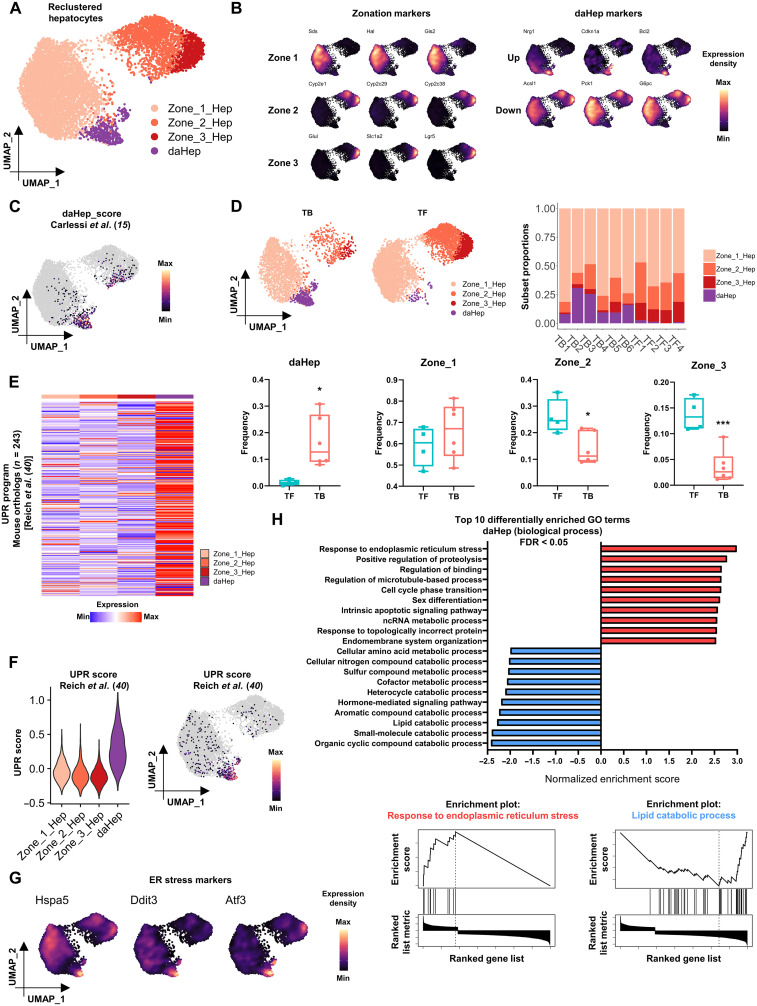
Since HCC is a hepatocyte malignancy, we explored this cluster in deeper detail. Hepatocyte nuclei in the main dataset were reclustered in isolation to identify further subpopulations and transitional states that could potentially distinguish TB from TF mice. This approach unveiled four subsets that could be annotated as three hepatocyte zonation phenotypes (zones 1, 2, and 3), and one cluster with a distinct expression signature similar to that of daHep, found in human liver (Fig. 1, A and B, and table S3) (15). High expression of Sds, Hal, and Gls2 distinguished zone 1 hepatocytes; Glul, Slac1a2, and Lgr5 characterized zone 3 cells; and a reducing gradient of the same markers expressed by zone 3 accompanied by high levels of a number of unique cytochrome P450 family genes, Cyp2e1, Cyp2c29, and Cyp2c38, was the hallmark of zone 2 hepatocytes. The daHep cluster was characterized by up-regulation of Nrg1, Cdkn1a, and Bcl2 and suppression of hepatocyte metabolic functional genes Acsl1, Pck1, and G6pc. To confirm the identity of the daHep cluster, we generated a gene module signature based on the top reported previously (15) using the AddModuleScore function in Seurat. This analysis showed that most nuclei that scored above the cutoff for the daHep module corresponded to nuclei annotated as daHep in our dataset (Fig. 1C). When frequencies of each hepatocyte subset were compared across groups, a remarkable increase was observed in daHep, which was paralleled by a decrease in zones 2 and 3 hepatocytes in TB mice (Fig. 1D). Higher numbers of daHep were previously shown to correlate with increased risk of HCC (15). Our data agree with this finding and further support the notion that, at least in NASH-driven HCC, daHep seem to replace zones 2 and 3 hepatocytes, even before HCC emergence.
Fig. 1. daHeps are enriched in NASH before HCC and characterized by the exacerbated ER stress–related gene expression program.
(A) Uniform manifold approximation and projection (UMAP) visualization of reclustered hepatocytes. Four subsets identified representing hepatocyte zonation phenotypes (Zone_1_Hep, Zone_2_Hep, and Zone_3_Hep) and daHep. (B) Expression distribution of (left) zonation marker genes and (right) up-regulated and down-regulated daHep markers in the UMAP space. (C) Mapping of daHep scores generated from Carlessi et al. (15) onto the reclustered hepatocytes UMAP. (D) (Top left) Hepatocytes UMAP visualization split by HCC outcome at 40 weeks (TB, tumor-bearing; TF, tumor-free). (Top right) Relative frequencies of hepatocyte subsets in each sample. (Bottom) Box and whisker plots showing frequencies of each hepatocyte subset according to HCC outcome. *P < 0.05, ***P < 0.01 by unpaired t test, TF, n = 4 and TB, n = 6. (E) Heatmap showing average expression of 243 mouse orthologs of human UPR program from Reich et al. (40) across hepatocyte subsets. (F) Mapping of UPR scores generated from Reich et al. (40) across hepatocyte subsets: (left) violin plot and (right) UMAP visualization. (G) Expression distribution of ER stress markers in the UMAP space, showing high levels where daHep cells localize. (H) (Top) GSEA of log2FC ranked daHep DEGs showing the top 10 over- and underrepresented gene ontology (biological process) terms. (Bottom) Enrichment plots for “response to ER stress” and “lipid catabolic process” gene sets.
To gain insight into gene regulatory programs associated with conversion to HCC, nuclei from each major cell type were combined into a single expression matrix for pseudobulk differential expression analysis comparing the TB and TF groups. This analysis revealed differentially expressed genes (DEGs) associated with progression to HCC across all hepatic lineages (table S4). ER stress genes such as Ddit3, Hspa5, and Atf3 were up-regulated in TB versus TF hepatocytes, supporting the previously proposed role for ER stress in NASH to HCC progression (12). To determine how the ER stress program is expressed among hepatocyte subsets, we next examined average expression of n = 243 mouse orthologs previously demonstrated to represent the complete unfolded protein response in human cells (table S5) (40). This approach revealed that the vast majority of unfolded protein response (UPR) genes were up-regulated in daHep (Fig. 1E). We generated a signature module representing the top 25 DEGs in this list and then mapped the signature score across hepatocyte subsets. This confirmed that the UPR program, inclusive of known ER stress markers, is disproportionally overrepresented in daHep as compared with healthy zonated hepatocytes (Fig. 1, F and G). Last, we conducted gene set enrichment analysis (GSEA) on the log2FC-ranked list of DEGs in daHep (table S6). This analysis compared the daHep cluster to the combined zonated hepatocyte clusters. GSEA highlighted “response to ER stress” as the top enriched biological process in daHep and confirmed the underrepresentation of several metabolic pathways commonly associated with normal hepatocyte identity in these cells (Fig. 1H). It must be acknowledged that the snRNA-seq experiments were conducted in a single, albeit appropriate, mouse model. This may question the clinical significance of the results. Accordingly, we investigated the expression levels of the same genes that were observed to be increased in daHeps from our MUP-uPA model (Ddit3, Hspa5, and Atf3), in a publicly available bulk RNA-seq human dataset (GSE126848), where subjects were grouped according to stage in the NAFLD spectrum (normal, obese, NAFL, and NASH) (41). All genes were highly up-regulated in patients with NAFL and, even more prominently, NASH compared with healthy individuals (fig. S2, A to C). Together, the data herein provide evidence that targeting ER stress in human NASH may hold clinical significance, suggesting that a dual approach targeting both inflammation and ER stress might represent an alternative strategy to treat NASH and prevent its progression to HCC.
sgp130Fc or BGP-15 monotherapies have mild, positive effects on NASH and HCC
Given the inflammatory and ER stress signatures described above, we crossed MUP-uPA mice (12) with mice that overexpress soluble gp130Fc (fig. S3A) (32, 42). At 6 weeks of age, we placed MUP-uPA, MUP-uPA/sgp130Fc, or littermate control [wild-type (WT)] mice on normal chow or HFD. Mice in each group were given BGP-15 in drinking water (100 mg/kg) or water alone. The dose of BGP-15 was based on a pilot bioavailability study (fig. S3A). Mice underwent liver biopsies and blood withdrawal at 24 weeks to assess the effect of drug treatment on NASH progression and were euthanized after 40 weeks to evaluate the effect on NASH-driven HCC (protocol summarized in fig. S3B). All animals survived the liver biopsy technique. In the three groups of mice, HFD increased body weight as expected, but the effect of BGP-15 administration was genotype dependent as it decreased (WT), increased (MUP-uPA/sgp130Fc), or had no effect (MUP-uPA) on body weight (fig. S3, C and D).
Initially, data from all groups were evaluated for markers of NASH including steatosis score and collagen deposition at 24 weeks and markers of HCC at 40 weeks. Treatment of MUP-uPA mice with BGP-15 alone had no effect on markers of NASH markers at 24 weeks, while untreated MUP-uPA/sgp130Fc showed some improvement in steatosis at 24 weeks (fig. S4, A to C). Monotherapy with either drug improved tumor incidence at 40 weeks but did not reduce either tumor number or tumor volume compared with untreated MUP-uPA mice placed on an HFD (fig. S5, A to C). These results were in marked contrast with combined therapy with BGP-15 and sgp130Fc, and, given these results, we subsequently focused our studies on MUP-uPA mice fed an HFD compared with MUP-uPA/sgp130Fc treated with BGP-15 (combined therapy).
BGP treatment of MUP-uPA/sgp130Fc mice ameliorates NASH
To gain insight into the effect of sgp130Fc and BGP-15 combined treatment on NASH progression, we first measured alanine aminotransferase (ALT), a marker of liver damage in blood at 24 weeks, and found it elevated in MUP-uPA relative to WT mice on an HFD, as previously reported (12). There was a tendency (not significant) for ALT to be reduced in MUP-uPA/sgp130Fc mice treated with BGP-15 (fig. S6A). To determine the effect of combined treatment on liver gene expression, we performed bulk RNA-seq on liver samples obtained at biopsy from MUP-uPA mice that ultimately developed tumors and MUP-uPA/sgp130Fc mice treated with BGP-15 that did not develop liver tumors (n = 4 in each group). Principal components analysis (PCA) and volcano plots demonstrated that the liver transcriptome was markedly different between the two datasets, with most significantly DEGs being down-regulated in the MUP-uPA/sgp130Fc BGP-15–treated group (fig. S7, A and B). The top-ranked DEG (fig. S7C) and pathways, as measured by GSEA (fig. S7D), were related to collagen deposition, and immunoregulatory interactions were down-regulated in the MUP-uPA/sgp130Fc BGP-15–treated group (fig. S7, E and F). Inflammatory (43), collagen biosynthetic (44), and extracellular matrix (45) pathways are all known to increase during the progression from simple steatosis to NASH.
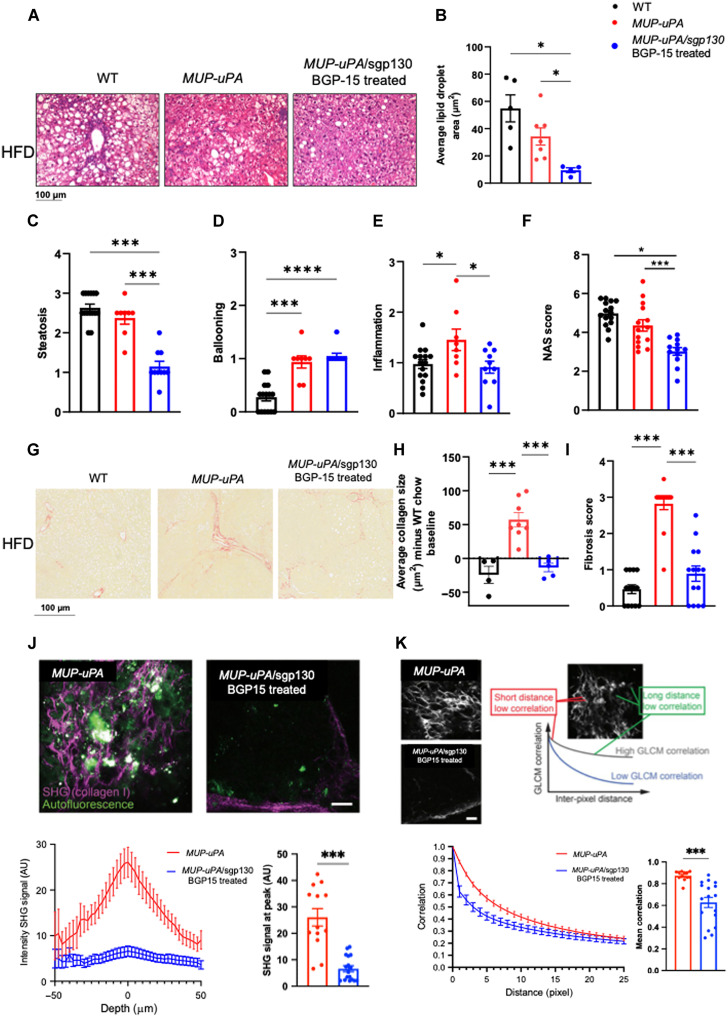
We next quantified hepatosteatosis, lipid droplets, hepatocyte ballooning, and inflammation in liver samples obtained at biopsy. MUP-uPA/sgp130Fc mice treated with BGP15 displayed markedly lower average lipid droplet area and steatosis score (Fig. 2, A to C), indicating that inhibition of ER stress and inflammation can ameliorate hepatosteatosis, even at this early disease stage. However, hepatocyte ballooning (Fig. 2D) was not affected, whereas lobular inflammation (Fig. 2E) and overall NAFLD activity score (NAS) (Fig. 2F) were reduced in MUP-uPA/sgp130Fc treated with BGP15 (Fig. 2, C to F). Liver fibrosis is defined as the excessive accumulation of extracellular matrix proteins including collagen that occurs in most types of chronic liver diseases. Accordingly, we quantified collagen fibers in biopsy samples using picrosirius red (PSR) staining. BGP treatment of MUP-uPA/sgp130Fc mice decreased average collagen size and fibrosis score compared with untreated MUP-uPA mice fed an HFD (Fig. 2, G to I). To verify PSR staining results, we used two independent imaging analyses and compared the MUP-uPA mice with the MUP-uPA/sgp130Fc treated with BGP-15 fed the HFD. Second-harmonic generation (SHG) microscopy is a powerful tool for imaging fibrillar collagen because it is highly sensitive to the collagen fiber structure and to changes that occur in fibrotic tumors and connective tissue disorders (46, 47). SHG imaging demonstrated that treatment of MUP-uPA/sgp130Fc mice with BGP-15 markedly reduced liver collagen deposition (Fig. 2J). Gray-level co-occurrence matrix (GLCM) allows for classification of different tissues based on the evaluation of geometrical arrangement of collagen fibrillar bundles (48). A lower mean GLCM correlation, indicative of lower collagen deposition in MUP-uPA/sgp130Fc mice treated with BGP-15 fed an HFD, compared with MUP-uPA mice fed an HFD was observed (Fig. 2K). Together, these data indicate that sgp130Fc overexpression, coupled with BGP-15 treatment, in our mouse model of NASH, ameliorates steatosis, inflammation, and collagen deposition, common features of fibrosis and NASH progression.
Fig. 2. Treatment of MUP-uPA/sgp130Fc mice with BGP-15 ameliorates markers of NASH.
Liver biopsy samples were obtained at 24 weeks. Representative H&E staining in WT, MUP-uPA that developed tumors and MUP-uPA/sgp130Fc BGP-15–treated mice that did not develop tumors (A). Quantification of average lipid droplet area (B), steatosis (C), hepatocyte ballooning (D), lobular inflammation (E), and NAS (F). The NAS is a measure of grade and is the sum of numerical scores applied to steatosis, hepatocellular ballooning, and lobular inflammation. Collagen deposition in WT, MUP-uPA, and MUP-uPA/sgp130Fc BGP-15–treated mice measured by PSR. Representative images (G), average collagen size (H), and fibrosis score (I). Fibrillar collagen measured by SHG microscopy (J). Representative images (top), intensity SHG signal versus depth of image (bottom left), and SHG signal at peak (bottom right). Geometrical arrangement of collagen fibrillar bundles measured by GLCM. (K) Representative images (top), correlation versus distance (bottom left), and mean correlation (bottom right). For (B), (C), (E), (F), (H), and (I): One-way analysis of variance (ANOVA) was performed. (J and K) Unpaired Welch’s t tests compared with MUP-uPA. The following numbers of biological replicates were used (independent mice) per group in each experiment: (B) 4 to 7; (C to F) 10 to 15; (H) 3 to 8; (I) 13 to 14; (J and K) 13 to 17. Data are expressed as means ± SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001.
BGP-15 treatment of MUP-uPA/sgp130Fc mice reduces NASH-driven HCC
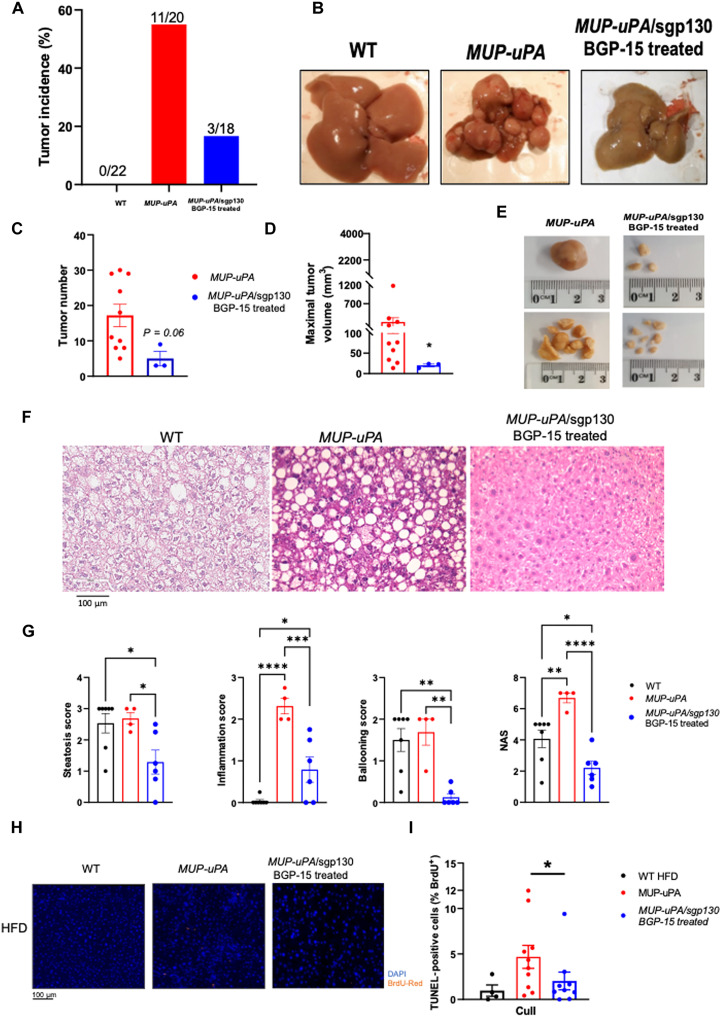
To determine whether attenuated NASH progression in BGP-15–treated MUP-uPA/sgp130 mice resulted in protection from HCC, livers were examined at 40 weeks. As expected, WT mice fed an HFD did not develop tumors, whereas ~55% of MUP-uPA mice fed an HFD developed tumors at 40 weeks (Fig. 3A). BGP-15 decreased HCC incidence to 16% (Fig. 3, A and B). Moreover, the three mice in this group that developed HCC tended to have fewer tumors (Fig. 3, C and E) with markedly lower tumor mass compared with MUP-uPA mice (Fig. 3, D and E). ALT levels were reduced in MUP-uPA/sgp130 mice treated with BGP15, relative to untreated MUP-uPA mice (fig. S6B). In addition, consistent with our data collected at biopsy, markers of NASH assessed by pathological analyses of the liver histology via hematoxylin and eosin (H&E) (Fig. 3, F and G) and PSR (fig. S8, A and B) were markedly attenuated when comparing BGP-15–treated MUP-uPA/sgp130 mice with untreated MUP-uPA mice. As previously presented (12), terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining showed that both apoptotic (nuclear fragmentation) and necrotic (diffuse cytoplasmic staining) cell deaths were increased in HFD-fed MUP-uPA livers, but this was reduced in the livers from the MUP-uPA/sgp130 mice treated with BGP-15 (Fig. 3, H and I).
Fig. 3. Treatment of MUP-uPA/sgp130Fc mice with BGP-15 ameliorates NASHdriven HCC.
- Liver samples were obtained at 40 weeks. Tumor incidence in WT mice, MUP-uPA mice, and MUP-uPA/sgp130Fc mice treated with BGP-15 (A). Representative liver images (B). Tumor number (C), maximal tumor volume (D), and representative images (E). Representative H&E staining in WT, MUP-uPA, and MUP-uPA/sgp130 BGP-15–treated mice (F). Quantification of average lipid steatosis, lobular inflammation, hepatocyte ballooning, and NAS (G). The NAS is a measure of grade and is the sum of numerical scores applied to steatosis, hepatocellular ballooning, and lobular inflammation. Representative images (H) and quantification (I) for TUNEL staining. (C) and (D) were analyzed by unpaired t tests. (G) to (I) were analyzed by one-way ANOVA. The following numbers of biological replicates were used (independent mice) per group in each experiment: (A) 18 to 22; (C and D) 3 to 10; (F and G) 4 to 10. Data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
BGP-15 treatment of MUP-uPA/sgp130Fc mice reduces ER stress and inflammation
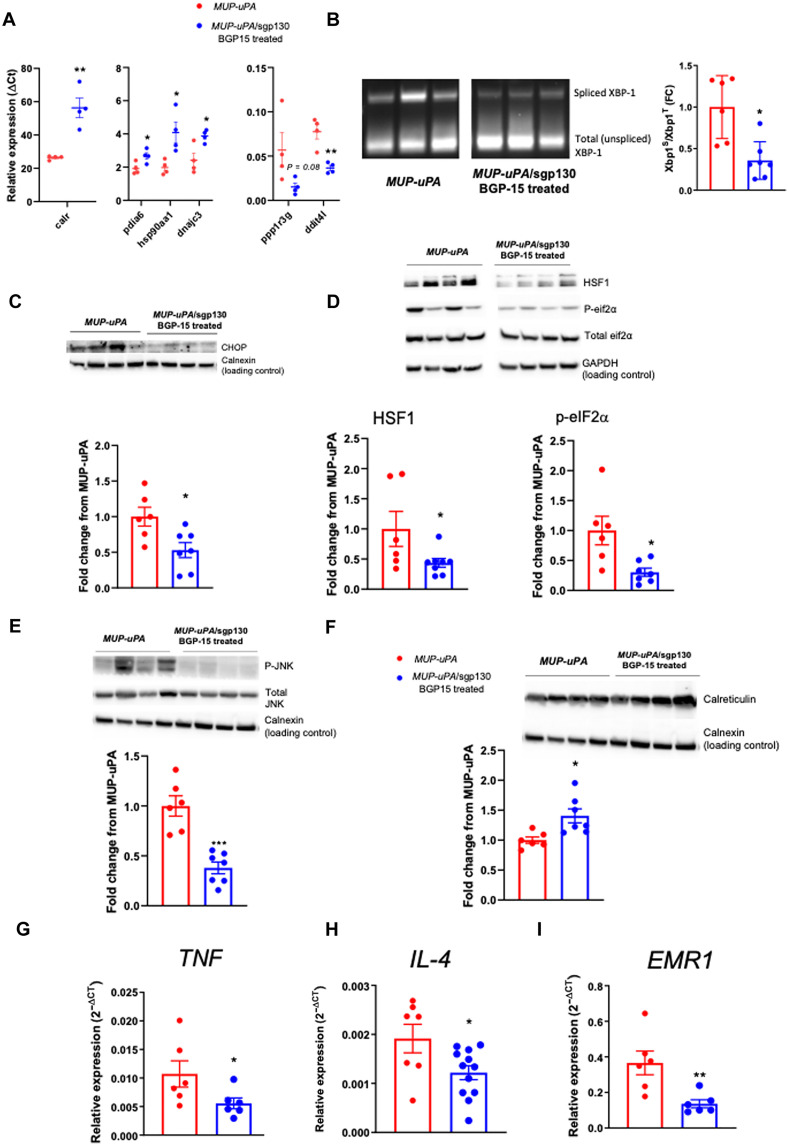
We next performed bulk RNA-seq on liver samples obtained from HFD-fed MUP-uPA mice and MUP-uPA/sgp130Fc mice treated with BGP-15 at 40 weeks. The PCA and volcano plots demonstrated that the liver transcriptome was markedly altered when comparing the two (fig. S9, A and B). The top-ranked DEG (fig. S9C) and pathways, as measured by GSEA (fig. S9D), revealed that some markers, such as immunoregulatory and cell surface interactions, were down-regulated, whereas other pathways such as those involved in activating chaperone genes and the UPR were up-regulated in the treated group (fig. S9, E and F). To validate these transcriptomic changes, we performed reverse transcription polymerase chain reaction (RT-PCR) analysis. Calr, a gene encoding calreticulin, which binds misfolded proteins and prevents them from being exported from the ER; Pdia6, a gene that inhibits aggregation of misfolded proteins; hsp90aa1, a gene that encodes the HSP90 protein, which aids in the proper folding of specific target proteins; and dnajc3, a gene that encodes a protein that inhibits double-stranded RNA (dsRNA)–activated protein kinase (PKR), were all elevated in livers of MUP-uPA/sgp130 mice treated with BGP-15 relative to livers of MUP-uPA mice (Fig. 4A). In contrast, ddit4l, a gene encoding DNA damage–inducible transcript 4–like protein, which inhibits cell growth by regulating the target of rapamycin (TOR) signaling pathway, was down-regulated in BGP-15–treated MUP-uPA/sgp130 livers relative to MUP-uPA livers (Fig. 4A). X-box binding protein 1 (XBP1) mRNA splicing is an event downstream of IRE1α activation, which is thought to be a critical UPR mediator and a classical marker of ER stress (49). XBP1 mRNA splicing was down-regulated in BGP-15–treated MUP-uPA/sgp130 livers relative to livers from MUP-uPA mice (Fig. 4B). Likewise, other markers of ER stress including C/EBP homologous protein (CHOP), heat shock factor 1 (HSF1), phosphorylated eIF2α (p-eIF2α), and phosphorylated c-Jun terminal kinase (p-JNK) were all reduced in BGP-15–treated MUP-uPA/sgp130 livers relative to those livers from the MUP-uPA mice (Fig. 4, C to E). In contrast, calreticulin was up-regulated in BGP-15–treated MUP-uPA/sgp130 mice (Fig. 4F). Together, these data demonstrate that BGP-15 treatment of MUP-uPA/sgp130 mice decreases ER stress and up-regulates the machinery that prevents export of misfolded proteins. We next examined markers of inflammation and initially quantified a panel of inflammatory genes by RT-PCR. Tumor necrosis factor (Tnf) mRNA was markedly reduced in MUP-uPA/sgp130Fc mice treated with BGP-15 compared with MUP-uPA mice (Fig. 4G). This pattern was also seen for Il4 (Fig. 4H) and Erm1, the gene that encodes F4/80 (Fig. 4I). We observed no differences in the mRNA expression of IL-1β, IL-6, or TGF-β (fig. S10, A to C).
Fig. 4. Treatment of MUP-uPA/sgp130Fc mice with BGP-15 reduces inflammation and ER stress.
Liver samples were obtained at 40 weeks. RT-PCR analyses of Calr, a gene that encodes calreticulin; Pdia6, a gene that inhibits aggregation of misfolded proteins; hsp90aa1, a gene that encodes HSP90; dnajc3, a gene that encodes a protein that inhibits PKR; ppp1r3g, a gene that encodes protein phosphatase 1 regulatory subunit 3G; and ddit4l, a gene encoding DNA damage-inducible transcript 4–like protein, which inhibits cell growth by regulating the TOR signaling pathway (A). Representative image and quantification of XBP-1 mRNA splicing (B). Representative Western blot and quantification of CHOP (C), HSF1, p-eIF2α (D), p-JNK (E), and calreticulin (F). RT-PCR analyses of TNF (G), IL-4 (H), and EMR1, the gene that encodes the F4/80 protein (I). Unpaired t tests were used to analyze all data. The following numbers of biological replicates were used (independent mice) per group in each experiment: (A) 4; (B to F) 6 to 7; (G and I) 6; (H) 7 to 12. Data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
BGP-15 treatment of MUP-uPA/sgp130Fc mice does not affect liver or plasma lipids
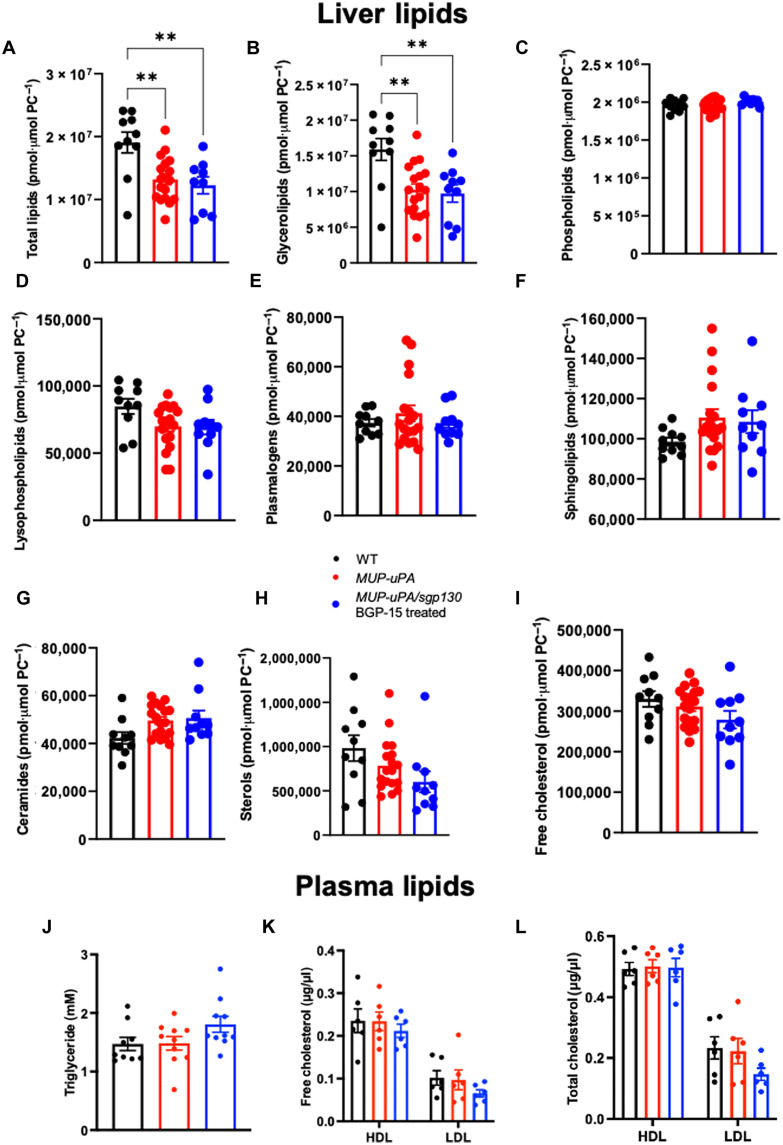
As discussed above, many therapeutic strategies to treat NASH have targeted pathways that affect the balance between fatty acid uptake and export, DNL and FAO. As we have recently discussed (11), these strategies have led to some promising drug candidates, but many have failed in late phase clinical trials due to adverse side effects such as hyperlipidemia and/or hypercholesterolemia. Accordingly, we performed a comprehensive lipidomic screen on liver and plasma samples from littermate control mice, MUP-uPA mice, and MUP-uPA/sgp130Fc mice treated with BGP-15 on samples collected at 40 weeks. We detected 359 liver lipid species in total (fig. S11, A to C). As shown in the volcano plot (fig. S11A), 23% of all lipid species were significantly altered when comparing WT with MUP-uPA mice, with most of these being reduced, consistent with the histological data we observed at the biopsy time point (see Fig. 2B). Accordingly, there was a small yet significant decrease in total lipids when comparing liver samples from MUP-uPA with WT mice (Fig. 5A). Given our histological data on steatosis (see Fig. 2, A to C), we were somewhat surprised that only 9% of the lipid species were significantly affected by the treatment (fig. S11C), and total lipids were similar when comparing the livers of MUP-uPA mice with those from MUP-uPA/sgp130Fc mice treated with BGP-15 (Fig. 5A). Histological steatosis scores measure lipid droplet area and size, but neither the concentration of lipid nor the lipid species within the droplet. We next examined major lipid classes that are dysregulated in NAFLD including glycerolipids, phospholipids, plasmalogens, sphingolipids, and sterols. Glycerolipids make up majority of the lipid pool (~70 to 80%) and total glycerolipids and their major subclasses, triacylglyerol and diacylglycerol, known to be elevated in fatty liver disease, followed the same pattern as total lipids, in that they were slightly reduced in MUP-uPA mice relative to WT, with no differences observed when comparing MUP-uPA with MUP-uPA/sgp130Fc mice treated with BGP-15 (Fig. 5B and fig. S12, A and B). Other lipid classes/subclass species, such as phospholipids (Fig. 5C and fig. S12C), lysophospholipids (Fig. 5D), plasmalogens (Fig. 5E), sphingolipids (Fig. 5, F and G, and fig. S12, E to K), and sterols (Fig. 5, H and I), were not different when comparing the three groups of mice. Consistent with the results observed in the liver lipidome, plasma triglycerides (Fig. 5J), free (Fig. 5K) and total (Fig. 5L) high-density lipoprotein and low-density lipoprotein cholesterol, and cholesterol ester (fig. S12L) were not different when comparing groups. Together, these data indicate that drugs that can effectively treat NASH and NASH-driven HCC can do so independent of liver lipid modification. The treatment intervention did not cause plasma hyperlipidemia and/or hypercholesterolemia, which is sometimes the case with other NASH treatment interventions.
Fig. 5. MUP-uPA/sgp130Fc mice treated with BGP-15 do not show reduced liver lipids.
Liver and blood samples were obtained at 40 weeks. Total lipids (A), glycerolipids (B), phospholipids (C), lysophospholipids (D), plamalogens (E), sphingolipids (F), ceramides (G), sterols (H), and free cholesterol (I) in liver samples and triglycerides (J), free (K) and total (L) cholesterol in high-density lipoprotein (HDL) and low-density lipoprotein (LDL) in plasma samples in WT mice, MUP-uPA mice, and MUP-uPA/sgp130 mice treated with BGP-15. One-way ANOVA with Tukey’s multiple comparisons where indicated. The following numbers of biological replicates were used (independent mice) per group in each experiment: (A to I) 10 to 18; (J to L) 6 to 10. Data are expressed as means ± SEM. **P < 0.01.
Treatment with BGP-15 and sgp130Fc after NASH onset results in disease regression
Our data to this point clearly demonstrated that MUP-uPA/sgp130Fc mice treated with BGP-15 displayed decreased NASH progression at 24 weeks and NASH-driven HCC at 40 weeks compared with MUP-uPA mice fed an HFD. These data, while important, do not provide evidence that this combination drug therapy can halt disease progression after onset because sgp130Fc was overexpressed from embryogenesis, and BGP-15 treatment was administered immediately after weaning. Accordingly, we next performed a drug intervention trial in WT and MUP-uPA mice (late-onset study). We commenced the HFD after 6 weeks of age and took biopsies at 12 weeks before commencing double treatment (DT), which included supplying BGP-15 in drinking water (as described in the previous experiment) and injecting sgp130Fc protein (0.5 mg/kg) into the intraperitoneal space twice weekly for a further 11 weeks, as previously described (32). Control mice received no BGP-15 in the drinking water and were injected with an equal volume of saline at the same intervals into the intraperitoneal space (CON) (protocol summarized in fig. S13A). This protocol was chosen because in preliminary experiments, we observed signs of NASH onset between 8 and 12 weeks in HFD-fed MUP-uPA mice. While MUP-uPA mice were slightly lighter than littermate controls, DT had no effect on body weight gain over the course of the experiment (fig. S13, B and C). Consistent with these data, neither food intake (fig. S13D) nor total energy expenditure (fig. S13E) was different when comparing groups. However, DT improved glucose tolerance as measured by an oral glucose tolerance test (OGTT) (fig. S14A) and decreased insulin levels for a given glucose concentration in animals in the fed state (fig. S14B), indicating that the DT improved metabolic homeostasis. Of note, consistent with the initial study, we also observed no difference in plasma total lipids (fig. S14C) or plasma free fatty acids (fig. S14D) when comparing MUP-uPA with DT.
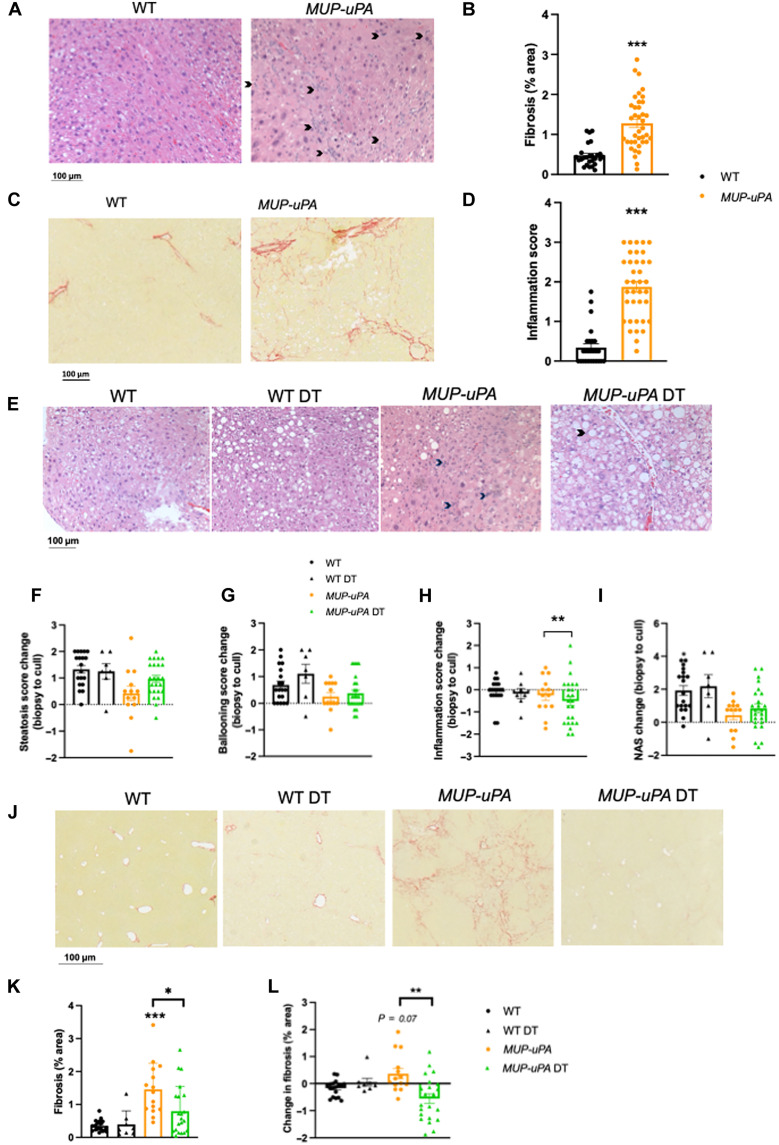
Consistent with our pilot data, before the intervention (12 weeks), both inflammation (Fig. 6, A and B) and fibrosis (Fig. 6, C and D) were higher in MUP-uPA compared with WT mice, akin to patients presenting with early-onset NASH. We next examined markers of NASH using H&E and PSR staining. Steatosis and hepatocyte ballooning increased over the 11-week intervention period, and neither was improved by DT (Fig. 6, E to G). Of interest, however, 13 of 24 mice in the DT group had lower inflammation score (Fig. 6, E to H). However, the DT group did not show an improved NAS compared with the MUP-UPA group (Fig. 6, E to I). As expected, WT mice showed no signs of fibrosis progression regardless of treatment (Fig. 6, J and K). In contrast, MUP-uPA mice administered CON showed significant increases in fibrosis progression, but this was markedly attenuated by DT. Not only did DT halt fibrosis progression, it also ameliorated the disease in ~75% (16 of 21) of the animals (Fig. 6, J to L). These data demonstrate that the BGP-15 and sgp130Fc treatment can regress inflammation and fibrosis, without affecting net steatosis, after onset in a mouse model that mimics human NASH.
Fig. 6. Treatment with BGP-15 and sgp130Fc after NASH onset results in disease regression.
WT and MUP-uPA mice were untreated or treated with BGP-15 [in drinking water and sgp130Fc (twice weekly injections of 0.5 mg/kg)] for 11 weeks (DT). Representative images of H&E (A) and Picosirus Red (PSR) (C) and quantification of fibrosis (B) and lobular inflammation (D) in WT and MUP-uPA mice at 12 weeks before DT or placebo treatment. Representative images of H&E (E) and quantification steatosis (F), ballooning hepatocytes (G), lobular inflammation (H), and NAS (I) in WT and MUP-uPA mice either untreated or DT. The NAS is a measure of grade and is the sum of numerical scores applied to steatosis, hepatocellular ballooning, and lobular inflammation. Data in (F) to (I) represent the change in score from biopsy (pre-treatment) to cull (post-treatment). Representative PSR images (J) and quantification of % fibrosis (K) and change in fibrosis area (pre-post treatment) (L) in WT, MUP-uPA, and MUP-uPA/sgp130 BGP-15–treated mice. (B) and (D), unpaired t tests; (F) to (I), (K), and (L), one-way ANOVA with Tukey’s multiple comparisons where indicated. The following numbers of biological replicates were used (independent mice) per group in each experiment: (B and D) 26 to 38; (F to I) 7 to 24; (K and L) 8 to 22. Data are expressed as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. In (L), P = 0.07 relative to WT DT.
DISCUSSION
NASH and NASH-driven HCC are major diseases of the new millennium and urgently require new therapies. Since the most successful, current therapies are bariatric surgery and lifestyle intervention, including diet and exercise, most pipeline therapeutics to treat NASH echo those used for obesity, type 2 diabetes, and MAFLD. The drug pipeline to target NASH and NASH-driven HCC has largely focused on drugs that target lipid metabolism in the liver including those that target acetyl–coenzyme A carboxylase (ACC), AMPK, farnesoid X receptor (FXR), and glucagon-like peptide–1 (GLP-1) (50). Recently, data from phase 3 SURPASS trial on the efficacy of Tirzepatide, a novel first-in-class dual gastric inhibitory polypeptide/GLP-1 agonist demonstrated improvement in overall glycemic control and weight loss (51). The emergence of the dual incretin agonists as drugs to treat obesity and MAFLD has led to growing interest and hope that such a therapy may revolutionize drug therapy for metabolic disease such as NASH since these drugs appear to target many tissues and organs dysregulated in obesity including the liver (52). Many patients present to the clinic with liver disease that has advanced from MAFLD. In addition, and of importance to our therapeutic strategy, advanced liver fibrosis in NASH is often accompanied by a reduction in hepatic fat to the point of complete fat loss, so-called “burnt-out NASH” (53). In these patients, with significant NASH, many drugs that target lipid metabolism have failed because of either unwanted side effects or a lack of efficacy as patients no longer have significant steatosis. For example, MK-4074, a liver-specific ACC1/2 inhibitor, or the FXR ligand obeticholic acid is quite effective in ameliorating liver fat but unfortunately markedly increases either plasma triglycerides or cholesterol (9, 10). Enanta Pharmaceuticals also recently discontinued development of EDP-305 and EDP-297, FXR agonists with mechanisms of actions similar to obeticholic acid, due to lack of efficacy in clinical trials. Likewise, the Gilead Sciences ATLAS study, which combined firsocostat, an ACC inhibitor, with cilofexor and FXR agonist, failed in phase 2 clinical trials. There are even fewer therapeutic options for the treatment of HCC, and therapeutic development has centered on combination therapies of TKIs with ICIs leading to several recent phase 2 and 3 clinical trials (6). However, these trials involve patients that present with HCC rather than patients with advanced NASH at risk for HCC.
Our data, albeit collected in a preclinical model, are important because we were able to track disease progression by performing liver biopsies on HFD-fed MUP-uPA mice at a time point where all mice displayed NASH, but none had HCC. snRNA-seq revealed an enrichment of recently identified daHep (54) and Trem2-positive macrophages (16–18) in livers of mice that later progressed to HCC, compared with animals that remained TF. NASH-afflicted livers presented a strong component of ER stress in their transcriptome, supporting the notion that the transition from NASH to HCC depends on ER stress and inflammation. Accordingly, our therapeutic strategy focused on reducing ER stress and inflammation. We were indeed successful in arresting NASH, and NASH progression to HCC, without affecting body weight or fat mass, food intake, liver lipids, or plasma lipids. Despite these metabolic parameters being unaltered, not only was NASH and NASH-driven HCC markedly improved, so too were markers of glucose intolerance and insulin resistance (fig. S13, A and B). Whether these positive outcomes will translate into human trials is unclear, but the available data give cause for encouragement. In a small phase 2 study of 42 patients, BGP-15 improved insulin sensitivity without affecting body weight or fasting lipids, and the drug was deemed very safe and well tolerated (23). Data from the Olamkicept clinical trial did not mention these parameters, but like BGP-15, the drug was well tolerated and safe (33). Recent reviews of therapeutic pipeline drugs to treat NASH have concluded that combination therapies addressing multiple aspects of NASH pathogenesis are expected to provide benefit for patients (50). Our data support the notion that combination therapies are superior to monotherapies. While BGP-15 or sgp130Fc therapy alone produced some moderate benefits, in terms of arresting NASH and/or reducing tumor burden, the combination of the two was either additive for tumor burden or indeed synergistic in terms of fibrosis. It is also possible that this dual therapy strategy could be used in combination with other existing drugs to successfully target NASH and NASH-driven HCC.
In conclusion, we demonstrate that it is possible to successfully treat and block NASH progression and NASH-driven HCC in a mouse model that faithfully mimic human NASH progression using a therapeutic strategy that targets ER stress and inflammation without affecting liver lipid content. Moreover, BGP-15 and Olamkicept (sgp130Fc) are well tolerated and safe in human clinical trials for metabolic and/or inflammatory diseases. Our data suggest, therefore, that this drug combination therapy is a realistic therapeutic strategy for treating human NASH and progression to HCC, diseases that are rapidly growing with a clinical unmet need.
MATERIALS AND METHODS
Experimental procedures
Mouse models
MUP-uPA mice (generated by E. Sandgren at School of Veterinary Medicine, University of Wisconsin-Madison) originating from the Karin laboratory (12, 26) were cross-bred with sgp130Fc transgenic mice from the Rose-John laboratory, which overexpress human sgp130Fc, as previously described (32). Heterozygous animals were bred together, and litters were genotyped (Transnetyx, TN, USA). Animals negative for the transgene were the designated WT, and animals expressing both uPA and the sgp130Fc gene (MUP/+sgp/+) were designated MUP-uPA/sgp130Fc. Male littermate WT and MUP-uPA/sgp130Fc mice were used for the study. MUP-uPA/sgp130Fc transgenic, MUP-uPA, and C57BL/6 WT mice were bred and housed at the Biological Testing Facility (Garvan Institute of Medical Research, Sydney, Australia) and the Murine Disease Modeling Facility (Monash University, Parkville, Australia) in a pathogen-free facility under controlled environmental conditions and exposed to a 12-hour light/12-hour dark cycle. Animal experiments were approved by the Garvan/St. Vincent’s Animal Ethics Committee (AEC) and the Monash Institute of Pharmaceutical Sciences Ethics Committee (MIPS AEC) and conducted in accordance with the National Health and Medical Research Council (NHMRC) of Australia Guidelines for Animal Experimentation. All experiments commenced when mice were 6 weeks of age. Mice were fed either a chow diet (5% of total energy from fat) or an HFD (36% of total energy from fat; SF03-002, Speciality Feeds, WA, Australia) for 17 to 34 weeks depending on the experiment. Animals were given their prescribed diet and autoclaved water ad libitum and group housed (two to four mice per cage) in individually ventilated cages, with weekly bedding changes. Mice were treated with or without BGP-15 (100 mg/kg) in their drinking water from 6 weeks of age (40-week study). A liver biopsy was performed after 18 weeks of HFD (40-week study) or after 17 weeks (late-onset study). For mice in the late-onset study, treatment with or without BGP-15 (100 mg/kg; N-Gene) in their drinking water as well as weekly intraperitoneal injections of sgp130 Fc (500 mg/kg; S.R.-J.) or a saline control commenced following the liver biopsy at 12 weeks.
Liver biopsy
Mice underwent a liver biopsy at 24 weeks of age (40-week study) or 12 weeks of age (late-onset study). Following aesthetic induction (4% isoflurane) and analgesic administration presurgery (buprenorphine, 0.1 mg/kg), a transverse incision (~1 cm) through skin and muscle layers was performed to expose the liver. A sterile dry cotton tip and blunt forceps were used to maneuver the lowest liver lobe outside the abdominal cavity to then excise a small wedge of liver and replace it with gel-foam to stem bleeding. This liver wedge was halved and then either fixed [10% neutral-buffered formalin (NBF)] for paraffin embedding and snap-frozen for RNA extraction in liquid nitrogen. The liver was returned into the abdominal cavity and then muscle layer was closed with a continuous suture (absorbable 5′0 suture). The skin was stapled closed with suture clips and then post-surgery local analgesia was applied to the suture site (bupivacaine, 0.1 mg/kg). Skin clips were removed 5 to 6 days after surgery after wound healing.
Nucleus isolation
Nuclei were isolated from flash-frozen liver biopsies as previously described (15). Briefly, biopsies were homogenized with a Kimble Dounce tissue grinder (Sigma-Aldrich, D8938) in 2 ml of ice-cold nuclei lysis buffer (10 mM tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL, CA-630); then, another 2 ml of nuclei lysis buffer was added to each sample, and lysis proceeded on ice for 10 min followed by straining the lysates through 40-μm cell strainers (Falcon, Corning). Lysates were centrifuged at 500g for 5 min at 4°C and resuspended in 4 ml of nuclei wash buffer [phosphate-buffered saline (PBS) supplemented with 1% bovine serum albumin (BSA) and RNasin Plus ribonuclease inhibitor (0.2 U/μl; Promega, N2615)]. Following another round of centrifugation, nuclei were resuspended in 700 μl of nuclei wash buffer and stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) at 0.1 μg/ml and propidium iodide (PI) at 2.5 μg/ml. Nuclei preparations were strained through 70-μm cell strainers before fluorescence-activated cell sorting (FACS) with a two-laser configuration (488 nm, 80 mW and 640 nm, 50 mW) BD FACSJazz stream-in-air cell sorter (BD Biosciences), equipped with a 100-μm nozzle operating at a sheath pressure of 27 psi. Machine calibration was performed by flow cytometry facility staff before each sort using Sphero 8-peak rainbow calibration beads (BioLegend) to achieve optimal stream alignment, laser alignment, and target mean fluorescence intensities for each detector. The drop delay was determined by setting a value that resulted in total side-stream deflection of Accudrop Beads (BD Biosciences) sorted through a 640-nm, 5-mW laser that bisected center and side streams. Nuclei were identified through an initial forward scatter height/side scatter height (FSC-H/SSC-H) gate, followed by the discrimination of single events exhibiting proportional FSC-W/FSC-H profiles. Within single events, nuclei were identified as PI-positive events. Two peaks of PI-positive events were visualized corresponding to 2n and 4n nuclei; both peaks were pooled together for sorting and downstream 10x Genomics analysis to maintain hepatic cell representation unaltered. A total of 50,000 PI-positive events were sorted per sample and then post-sort nuclei concentration and quality were checked using a fluorescence microscope and a hemocytometer.
Single-nucleus RNA library preparation and sequencing
For the construction of snRNA-seq libraries, Chromium Next GEM Single Cell 3′ v3.1 - Dual Index reagent kits were used according to the manufacturer’s instructions. A total of 10,000 freshly sorted nuclei were loaded onto a 10x Genomics Chromium Single Cell 3′ v3.1 chip G and processed immediately in a 10x Chromium controller. Specifically, we used 19 PCR cycles for cDNA amplification. Sequencing of libraries was performed as in (55). Briefly, libraries were quantified with quantitative PCR (qPCR) using the NEBnext Library Quant Kit for Illumina and fragment size assessed with TapeStation D1000 kit (Agilent). Libraries were pooled in equimolar concentration and sequenced using an Illumina NovaSeq 6000 and S2 flow cells (100-cycle kit) with a read one length of 28 cycles and a read two length of 94 cycles.
snRNA-seq data processing and downstream analysis
Binary Base Call (BCL) files were demultiplexed and converted into FASTQ using bcl2fastq utility of Illumina BaseSpace Sequence Hub. FASTQ files were processed using Cell Ranger 7.0.0. using the mm10-2020-A reference and include introns option set to True. Raw gene-barcode matrices from Cell Ranger outputs were read into Seurat R package v4.2 (34) using the functions Read10x and then CreateSeuratObject. The dataset was initially filtered on the basis of a minimum number of 200 unique features (genes) and less than 3% of mitochondrial genes per barcode. Further filtering excluded barcodes with less than 500 and greater than 15,000 unique molecular identifiers. Data from each original sample were individually normalized and scaled, and the top 2000 variable features were identified by applying the SplitObject function by “orig.ident” and then passing the functions NormalizeData, ScaleData, and FindVariableFeatures, respectively, to each orig.ident split object. Next, we implemented Batchelor (35) using the FastMNN Seurat wrapper for sample integration and batch correction and then passed the top 25 components of the MNN output to the RunUMAP and FindNeighbors functions and calculated Louvain clusters using the FindClusters function. Next, we implemented a manual supervised approach to remove low-quality and doublet barcodes. The approach was based on successive rounds of clustering, identification, and removal of clusters corresponding to low-quality and doublet nuclei. Low-quality clusters likely corresponded to empty droplets that were contaminated with ambient RNA, and these were characterized by presenting a low-average number of features and expression of highly expressed cell type–specific genes from multiple cell types. Doublets were identified and removed on the basis of high expression of canonical cell type–specific genes from two cell types; these clusters also presented an average number of features above the mean of other clusters in the dataset. This approach was implemented until no low-quality or doublet clusters could be identified after passing the FindClusters function with a resolution of 0.25. The final filtered and curated dataset resulted in clusters driven by cell type, and a total of 47,501 nuclei from n = 10 mice (TF, n = 4 and TB, n = 6) and 25,308 genes were detected. Differential expression analysis was conducted using the default Wilcoxon rank sum test with the FindAllMarkers function retaining only those genes expressed in at least 25% of the cells in a given cluster and a log-fold change of at least 0.25 compared to all remaining cells. Clusters corresponding to hepatocytes and myeloid lineages were subset in separate objects for reclustering. Each of these subsets were reanalyzed in isolation similarly to above. For pseudobulk differential expression analysis in cell types, we first applied the function AverageExpression for each cell type cluster and then ran FindMarkers with default parameters comparing TB versus TF samples. Specifically, for the daHep cluster of hepatocytes, we ran the FindMarkers function versus the combined zonated clusters with a less stringent filter, retaining genes expressed in at least 10% of the cluster cells to capture a larger gene set for downstream GSEA analysis. GSEA was conducted on WebGestalt (56) by uploading the log2FC ranked gene list to the web server. Functional database was set to geneontology (Biological Process) and advanced parameters set to default. Enriched categories were first ranked on the basis of false discovery rate (FDR) and then the top 10 significant categories were selected for plotting.
Metabolic measurements
EchoMRI
Mouse body composition [fat mass and lean body mass (LBM)] was measured weekly with a 4-in-1 EchoMRI body composition analyzer (EchoMRI, USA) and standard laboratory scales.
Oral glucose tolerance test
OGTTs (2 g/kg LBM) were performed in 5-hour–fasted mice, as previously described (29). Blood was sampled from a lateral tail vein nick at 0, 15, 30, 45, 60, 90, and 120 min after glucose bolus delivery, and glucose was measured using test strip and a glucometer (Accu-Chek Performa, 22001BO, McFarlane Medical and Scientific, VIC, Australia).
Plasma measurements
Insulin concentrations were measured using a Mouse Ultrasensitive Insulin ELISA kit (ALPCO) according to the manufacturer’s instructions.
Indirect calorimetry
Mice were placed into Promethion (Sable Systems International) metabolic cages to adapt to their surroundings for 48 hours before study. Data from two light-dark cycles were used in the analysis. Rates of O2 consumption (VO2; ml kg–1 hour–1) and CO2 production (VCO2) were continuously acquired as was food intake. These data were used to calculate the total energy expenditure, which is the kilocalories used for all bodily activity, corrected for lean mass (LM) over a 24-hour period.
Histology, NAS, and imaging
Freshly collected liver tissues from MUP-uPA, MUP-uPA/sgp130Fc transgenic and C57BL/6 mice were fixed in 10% NBF or 4% paraformaldehyde, embedded in paraffin [formalin-fixed, paraffin-embedded (FFPE)], sectioned, and stained with H&E and PSR. For frozen-block preparation, tissues were embedded in Tissue-Tek optimum cutting temperature compound and frozen in a 2-methylbutane/dry-ice slurry and stored at −80°C until sectioning for SHG microscopy of collagen. Representative images were captured on an upright light (H&E and PSR), fluorescence (TUNEL), or confocal (SHG) microscopes (Zeiss) equipped with an AxioCam camera. The H&E-stained liver sections were scored by a vet pathologist and three independent assessors for signs of NASH as per the NASH Clinical Research Network histological scoring system (57). Each individual point represents a mean score from the assessors per section.
The resulting NAS is a sum of steatosis, ballooning, and inflammation scores. Fibrosis was scored as per NASH CRN guidelines and reported separately. Fibrosis was quantified in PSR-stained liver sections with image quantification (FIJI software) and reported as average collagen size in square micrometers minus WT samples (normal collagen staining around vessels and liver capsule).
ALT assay
Plasma ALT levels were measured from plasma samples taken at 24 weeks (at biopsy) and 40 weeks (end of study) as a measure of liver damage as per the manufacturer’s instructions (ALT Reagent, Thermo Fisher Scientific, TR71121).
Tumor biology
At the end of the 40-week study, the remaining whole livers were fixed (10% NBF), and tumors were microdissected out of the liver, counted, and sized. Tumor volume was calculated using the modified ellipsoid method [0.5 (length × width2)] after sizing with a digital caliper. Tumor markers alpha-fetoprotein (ab46799, Abcam, 1:100 dilution) and p62 (23214, Cell Signaling Technology, 1:250 dilution) were detected within dewaxed, rehydrated liver FFPE sections and stained overnight at 4°C in blocking reagent (Ultracruz, Santa Cruz Biotechnology) following heat-based antigen retrieval (15 min, 95°C in 10 mM sodium citrate buffer with buffer refresh at 10 min). Tumor markers were visualized with chromogen DAB/0.5% hydrogen peroxide [0.1 M tris-HCl (pH 7.6)] following horseradish peroxidase (HRP)–conjugated secondary antibody incubation (90 min, GTX213110-01, GeneTex, 1:250) and three PBS washes. Sections were counterstained with hematoxylin, washed in distilled water, dehydrated through ethanol and xylene, and then mounted in limonene organic mounting media (Santa Cruz Biotechnology) before visualization via light microscopy. Area of brown DAB staining was quantified (FIJI software).
TUNEL staining
TUNEL staining was performed on liver sections using a TUNEL assay kit (Abcam, ab66110) according to the manufacturer’s instructions and counterstained with DAPI. Positive areas of BrDU incorporation into apoptotic cells were counted (FIJI software) and reported as percentage of all cells (DAPI+) in the field of view.
GLCM analysis
To assess stromal collagen fiber organization and cross-linking, GLCM analysis was used to characterize the texture of the organotypic matrix samples by determining the correlation of the SHG signal intensity within the matrix, as a function of distance, where a slower decay shows a more organized and correlated network of collagen fibers (58). SHG images were acquired with the laser power adjusted to give an approximately uniform intensity between images. For each matrix within a triplicate, five representative regions were taken with a field-of-view of 512 by 512 μm, a line averaging of 32, and a scan speed of 400 Hz. The average GLCM texture parameters were calculated between pixel offsets in 1-pixel increments, up to 100 pixels, at 0°, 90°, 180°, and 270° orientations. The image correlation, as a function of distance, was plotted, and the mean correlation distance D = ∑idic(di)/∑ic(di), where di is the offset of the ith pixel and c(d) is the GLCM correlation, as a function of distance.
RNA-seq analysis
RNA-seq analysis from frozen liver samples was performed as previously described (14). RNA was extracted from snap-frozen liver using the NucleoSpin RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. Library preparation for RNA-seq was performed using TruSeq Stranded mRNA Library Prep Kit (Illumina), following the manufacturer’s guidelines and best practices. For quality control of the libraries, the samples were inspected with a Bioanalyzer chip (DNA 1000, Agilent). Sequencing was performed for paired-end samples on a HiSeq 2500 v4.0 system. The resulting fastq files were quality controlled using FastQC, and adapters were trimmed using TrimGalore! v0.4.0 (both developed at the Babraham Institute) for paired-end sequenced samples. The trimmed fastq files were aligned to the mouse reference genome (Mus_musculus.GRCm38.83) using STAR aligner (v2.5.1) (59). RSEM (v1.3.0) (60) was used to estimate gene expression levels. Downstream analysis for DEGs from the raw count table was performed using edgeR (61) in R. Genes were considered differentially expressed at Benjamini-Hochberg’s FDR < 0.05. Heatmaps were drawn using the R package gplots – heatmap.2; R package version 3.1.1; https://CRAN.R-project.org/package=gplots. Clustering columns and rows and the values for each row were centered and scaled. Mapping of gene expression levels to biological pathways was performed using GSEA in Reactome (https://reactome.org) following mouse-to-human conversion of the DEGs list. Gene sets were obtained from the Molecular Signatures Database (Human MsigDB, version 5.0, collections).
qPCR
RNA samples were prepared from frozen liver tissue using Isolate II Mini kit (Bioline) as per the manufacturer’s instructions. RNA was reverse-transcribed using a Tetro cDNA synthesis kit (Bioline), and qPCR was performed using Taqman probes (Thermo Fisher Scientific) on a Bio-Rad CFX96 machine. Relative mRNA expression was calculated from the comparative threshold cycle (Ct) values relative to hypoxanthine guanine phosphoribosyl transferase housekeeping mRNA. PCR primers were designed using online using the Taqman assay search tool for best coverage against the below gene transcripts: tnf (Mm00443258_m1), il4 (Mm00445259_m1), emr1 (F4/80) (Mm00802529_m1), il6 (Mm00446190_m1), il1beta (Mm00434228_m1), and tgfbeta (Mm01178820_m1).
XBP-1 splicing assay
RNA transcripts were probed to assay the level of XBP-1 splicing as described previously (62). Briefly, total isolated RNA from livers was reverse-transcribed to cDNA (see qPCR) and primers designed specifically to amplify XBP-1 mRNA across the splice junction (5′ oligo: AAACAGAGTAGCAGCGCAGACTGC, 3′ oligo: GGATCTCTAAAACTAGAGGCTTGGTG) were used in a hot start PCR under the following cycling conditions: 94°C for 30 s (initial denaturation), 94°/51°/68°C for 30/30/60 s, 30 cycles (denaturation/annealing/extension) and 68°C for 5 min (final extension) using 5 μl of cDNA (Hot start Taq DNA Polymerase, NEB, M0495S). Incubation of the amplicon at 4°C occurred for 5 min before the addition of 25 μl of the Pst I restriction enzyme in restriction buffer with nuclease-free water (Pst I–HF, NEB, and R0410S) and incubation at 37°C for 15 min. The restriction reaction was terminated by the addition of 10 μl of 6× loading dye at room temperature, and the entire 50 μl of digested cDNA was run on a 1.5% agarose gel in 1× tris-acetate-EDTA buffer against a 100–base pair DNA ladder (N3231S, NEB). Total XBP-1 (unspliced) contains a Pst I restriction site, whereas spliced (active) XBP-1 does not; therefore, the Pst I–digested total XBP-1 doublet runs faster (the lower doublet band) on the gel. XBP-1 activity was quantified as the ratio of spliced over total XBP-1 (expressed as fold control).
Immunoblotting and antibodies
Immunoblotting analysis was performed on liver tissue lysates that were separated by SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. Blots were incubated with 5% (w/v) non-fat dry milk in tris-buffered saline (TBS) with 0.1% (w/v) Tween 20 (MilliporeSigma) (TBST) at room temperature for 1 hour to block nonspecific binding, overnight at 4°C with primary antibodies in 2.5% BSA (w/v) in TBST and finally with HRP-conjugated secondary antibody in 2.5% BSA (w/v) in TBST. Blots were developed using the Clarity Western ECL Substrate detection kit (Bio-Rad). Immunoblotting was performed with antibodies against p-JNK (9251, Cell Signaling Technology), total JNK (9252, Cell Signaling Technology), CHOP/ddit3 (ab11419, Abcam), calnexin (2679, Cell Signaling Technology), heat HSF1 (4356, Cell Signaling Technology), phospho-eif2α (3579, Cell Signaling Technology), total eif2α (9722, Cell Signaling Technology), and calreticulin (2891S, Cell Signaling Technology) or glyceraldehyde-3-phosphate dehydrogenase (2118S, Cell Signaling Technology) loading controls. All primary antibodies were diluted 1:1000. Secondary antibodies were diluted 1:10,000.
Lipidomics
Lipid species from frozen liver samples were prepared and assayed as previously described (14). Liver tissue was homogenized in one volume of PBS (pH 7.4) and sonicated. Next, a bicinchoninic acid assay protein assay was performed, and 25 μg of each liver sample was aliquoted and used for lipid extraction. An internal standard mixture and CHCl3-methanol (2:1) mixture was added to each sample before it was vortexed, mixed, sonicated, and centrifuged. Lipid-containing supernatants were removed and dried before being resuspended in H2O-saturated butanol and sonicated again. Methanol with 10 mM ammonium was added, the resuspended samples were centrifuged, and the supernatants were transferred to individual glass vials. Lipidomic determination was performed by liquid chromatography–electrospray ionization tandem mass spectrometry, combined with a triple quadrupole mass spectrometer. Data were analyzed using Multiquant software v1.2, and all lipid species were normalized to the total phosphatidylcholine levels of each sample.
Statistical analysis
All data are expressed as means ± SEM. Statistical analyses was performed in Prism and involved either one or two-way analyses of variance (ANOVAs) with multiple comparisons or t tests between groups indicated, following confirmation that the data were normally distributed via Kolmogorov-Smironov test and others (Shapiro-Wilk, D’Agostino-Pearson, and Anderson-Darling). If the data were not normally distributed, equivalent nonparametric tests were instead performed, such as Freidmen’s or Mann-Whitney tests.
Supplementary Material
Acknowledgments
We acknowledge the work of the staff at the animal research facilities at both the Garvan Institute of Medical Research and Monash Institute of Pharmaceutical Sciences, Monash University. We also acknowledge the contribution of J. Singh and J. Lu for contribution to this dataset.
Funding: M.A.F. is a senior principal research fellow of the NHMRC (APP1116936) and is also supported by an NHMRC Investigator Grant (APP1194141). Research in his laboratory was supported by project grants from the NHMRC (APP1042465, APP1041760, and APP1156511 to M.A.F. and APP1122227 to M.A.F. and M.K.). M.K. is an American Cancer Research Society professor and holds the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases. His research was supported by grants from the NIH (P42ES010337, R01DK120714, R01CA198103, R37AI043477, R01CA211794, and R01CA234128). R.C. is the recipient of a Cancer Council WA Postdoctoral Research Fellowship. The single nucleus work was enabled by a collaborative cancer research grant provided by the Cancer Research Trust “Enabling advanced single-cell cancer genomics in Western Australia” and an enabling grant from the Cancer Council of Western Australia to J.E.E.T.-P. and R.C.
Author contributions: Conceptualization: M.A.F. and M.K. Methodology: R.C., S.R., C.N., N.H., M.J., P.T., J.E.E.T.-P., P.J.M., M.K., and M.A.F. Analysis: E.B., R.C., S.R., B.S., C.L.E., E.M., M.N., C.N., N.H., D.C.H., and N.A.M. Bioinformatics: R.C., S.R., S.T., A.M., and E.D. Resources: M.A.F., S.R.-J., M.K., and A.R.R.F. Writing—original draft: E.B., B.S., R.C., and M.A.F. Writing—review and editing: all authors. Visualization: E.B., R.C., C.N., M.N., and N.H. Supervision: P.T., J.E.E.T.-P., P.J.M., and M.A.F. Project administration: E.B. Funding acquisition: M.A.F., M.K., J.E.E.T.-P., and R.C.
Competing interests: M.K. holds U.S. Patent no. 10034462 B2 on the use of MUP-uPA mice for the study of NASH and NASH-driven HCC. M.A.F. holds U.S. Patent no. 20200179363 on the methods for treating NASH and for preventing NASH-induced HCC. M.A.F. is a shareholder and scientific advisor for N-Gene Pharmaceuticals. M.A.F. is the founder and shareholder of Celesta Therapeutics. S.R.-J. has acted as a consultant and speaker for AbbVie, Chugai, Genentech Roche, Pfizer, and Sanofi. He also declares that he is an inventor on patents owned by CONARIS Research Institute, which develops the sgp130Fc protein Olamkicept together with the companies Ferring and I-Mab. S.R.-J. has stock ownership in CONARIS. The authors declare that they have no other competing interests.
Data and materials availability: GLCM analysis was performed using a custom Matlab script, available at https://github.com/timpsonlab/shg-quantification-tools. The analysis was performed and the code accessed on 15 or 16 December 2019 and version 1.5.0-d468570 (last updated 10 September 2019) was used. For snRNA-seq data processing and downstream analysis, visualizations were made using Seurat R package v4.2, scCustomize v1.0.2 (https://doi.org/10.5281/zenodo.5706430), or ggplot2 v3.3.6 R package. Code and data related to snRNA-seq and bulk RNA-seq analyses are available at https://zenodo.org/record/8214157. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, M.A.F., mark.febbraio@monash.edu. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Fitzmaurice C., Akinyemiju T. F., Al Lami F. H., Alam T., Alizadeh-Navaei R., Allen C., Alsharif U., Alvis-Guzman N., Amini E., Anderson B. O., Aremu O., Artaman A., Asgedom S. W., Assadi R., Atey T. M., Avila-Burgos L., Awasthi A., Saleem H. O. B., Barac A., Bennett J. R., Bensenor I. M., Bhakta N., Brenner H., Cahuana-Hurtado L., Castaneda-Orjuela C. A., Catala-Lopez F., Choi J. J., Christopher D. J., Chung S. C., Curado M. P., Dandona L., Dandona R., das Neves J., Dey S., Dharmaratne S. D., Doku D. T., Driscoll T. R., Dubey M., Ebrahimi H., Edessa D., El-Khatib Z., Endries A. Y., Fischer F., Force L. M., Foreman K. J., Gebrehiwot S. W., Gopalani S. V., Grosso G., Gupta R., Gyawali B., Hamadeh R. R., Hamidi S., Harvey J., Hassen H. Y., Hay R. J., Hay S. I., Heibati B., Hiluf M. K., Horita N., Hosgood H. D., Ilesanmi O. S., Innos K., Islami F., Jakovljevic M. B., Johnson S. C., Jonas J. B., Kasaeian A., Kassa T. D., Khader Y. S., Khan E. A., Khan G., Khang Y. H., Khosravi M. H., Khubchandani J., Kopec J. A., Kumar G. A., Kutz M., Lad D. P., Lafranconi A., Lan Q., Legesse Y., Leigh J., Linn S., Lunevicius R., Majeed A., Malekzadeh R., Malta D. C., Mantovani L. G., McMahon B. J., Meier T., Melaku Y. A., Melku M., Memiah P., Mendoza W., Meretoja T. J., Mezgebe H. B., Miller T. R., Mohammed S., Mokdad A. H., Moosazadeh M., Moraga P., Mousavi S. M., Nangia V., Nguyen C. T., Nong V. M., Ogbo F. A., Olagunju A. T., Pa M., Park E. K., Patel T., Pereira D. M., Pishgar F., Postma M. J., Pourmalek F., Qorbani M., Rafay A., Rawaf S., Rawaf D. L., Roshandel G., Safiri S., Salimzadeh H., Sanabria J. R., Milicevic M. M. S., Sartorius B., Satpathy M., Sepanlou S. G., Shackelford K. A., Shaikh M. A., Sharif-Alhoseini M., She J., Shin M. J., Shiue I., Shrime M. G., Sinke A. H., Sisay M., Sligar A., Sufiyan M. B., Sykes B. L., Tabares-Seisdedos R., Tessema G. A., Topor-Madry R., Tran T. T., Tran B. X., Ukwaja K. N., Vlassov V. V., Vollset S. E., Weiderpass E., Williams H. C., Yimer N. B., Yonemoto N., Younis M. Z., Murray C. J. L., Naghavi M., Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: A systematic analysis for the global burden of disease study. JAMA Oncol. 4, 1553–1568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z., Suo C., Mao X., Jiang Y., Jin L., Zhang T., Chen X., Global incidence trends in primary liver cancer by age at diagnosis, sex, region, and etiology, 1990-2017. Cancer 126, 2267–2278 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Cohen J. C., Horton J. D., Hobbs H. H., Human fatty liver disease: Old questions and new insights. Science 332, 1519–1523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diehl A. M., Day C., Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N. Engl. J. Med. 377, 2063–2072 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Febbraio M. A., Reibe S., Shalapour S., Ooi G. J., Watt M. J., Karin M., Preclinical models for studying NASH-driven HCC: How useful are they? Cell Metab. 29, 18–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smeuninx B., Boslem E., Febbraio M. A., Current and future treatments in the fight against non-alcoholic fatty liver disease. Cancers (Basel) 12, 1714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llovet J. M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J. F., de Oliveira A. C., Santoro A., Raoul J. L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T. F., Galle P. R., Seitz J. F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J., Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Lazaridis N., Tsochatzis E., Current and future treatment options in non-alcoholic steatohepatitis (NASH). Expert Rev. Gastroenterol. Hepatol. 11, 357–369 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Kim C. W., Addy C., Kusunoki J., Anderson N. N., Deja S., Fu X., Burgess S. C., Li C., Ruddy M., Chakravarthy M., Previs S., Milstein S., Fitzgerald K., Kelley D. E., Horton J. D., Acetyl CoA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: A bedside to bench investigation. Cell Metab. 26, 576 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Chapman R. W., Lynch K. D., Obeticholic acid—A new therapy in PBC and NASH. Br. Med. Bull. 133, 95–104 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Febbraio M. A., Karin M., “Sweet death”: Fructose as a metabolic toxin that targets the gut-liver axis. Cell Metab. 33, 2316–2328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa H., Umemura A., Taniguchi K., Font-Burgada J., Dhar D., Ogata H., Zhong Z., Valasek M. A., Seki E., Hidalgo J., Koike K., Kaufman R. J., Karin M., ER stress cooperates with hypernutrition to trigger TNF-dependent spontaneous HCC development. Cancer Cell 26, 331–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shalapour S., Lin X. J., Bastian I. N., Brain J., Burt A. D., Aksenov A. A., Vrbanac A. F., Li W., Perkins A., Matsutani T., Zhong Z., Dhar D., Navas-Molina J. A., Xu J., Loomba R., Downes M., Yu R. T., Evans R. M., Dorrestein P. C., Knight R., Benner C., Anstee Q. M., Karin M., Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 551, 340–345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todoric J., Di Caro G., Reibe S., Henstridge D. C., Green C. R., Vrbanac A., Ceteci F., Conche C., McNulty R., Shalapour S., Taniguchi K., Meikle P. J., Watrous J. D., Moranchel R., Najhawan M., Jain M., Liu X., Kisseleva T., Diaz-Meco M. T., Moscat J., Knight R., Greten F. R., Lau L. F., Metallo C. M., Febbraio M. A., Karin M., Fructose stimulated de novo lipogenesis is promoted by inflammation. Nat. Metab. 2, 1034–1045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlessi R., Denisenko E., Boslem E., Köhn-Gaone J., Main N., Abu Bakar N. D. B., Shirolkar G. D., Jones M., Beasley A. B., Poppe D., Dwyer B. J., Jackaman C., Tjiam M. C., Lister R., Karin M., Fallowfield J. A., Kendall T. J., Forbes S. J., Gray E. S., Olynyk J. K., Yeoh G., Forrest A. R. R., Ramm G. A., Febbraio M. A., Tirnitz-Parker J. E. E., Single-nucleus RNA sequencing of pre-malignant liver reveals disease-associated hepatocyte state with HCC prognostic potential. Cell Genomics 3, 100301 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou J., Zhang J., Cui P., Zhou Y., Liu C., Wu X., Ji Y., Wang S., Cheng B., Ye H., Shu L., Zhang K., Wang D., Xu J., Shu Q., Colonna M., Fang X., TREM2 sustains macrophage-hepatocyte metabolic coordination in nonalcoholic fatty liver disease and sepsis. J. Clin. Invest. 131, e135197 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramachandran P., Dobie R., Wilson-Kanamori J. R., Dora E. F., Henderson B. E. P., Luu N. T., Portman J. R., Matchett K. P., Brice M., Marwick J. A., Taylor R. S., Efremova M., Vento-Tormo R., Carragher N. O., Kendall T. J., Fallowfield J. A., Harrison E. M., Mole D. J., Wigmore S. J., Newsome P. N., Weston C. J., Iredale J. P., Tacke F., Pollard J. W., Ponting C. P., Marioni J. C., Teichmann S. A., Henderson N. C., Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong X., Kuang H., Ansari S., Liu T., Gong J., Wang S., Zhao X. Y., Ji Y., Li C., Guo L., Zhou L., Chen Z., Leon-Mimila P., Chung M. T., Kurabayashi K., Opp J., Campos-Perez F., Villamil-Ramirez H., Canizales-Quinteros S., Lyons R., Lumeng C. N., Zhou B., Qi L., Huertas-Vazquez A., Lusis A. J., Xu X. Z. S., Li S., Yu Y., Li J. Z., Lin J. D., Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L. L., Russell D. L., Wong S. L., Chen M., Tsai T. S., St John J. C., Norman R. J., Febbraio M. A., Carroll J., Robker R. L., Mitochondrial dysfunction in oocytes of obese mothers: Transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development 142, 681–691 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Gehrig S. M., van der Poel C., Sayer T. A., Schertzer J. D., Henstridge D. C., Church J. E., Lamon S., Russell A. P., Davies K. E., Febbraio M. A., Lynch G. S., Hsp72 preserves muscle function and slows progression of severe muscular dystrophy. Nature 484, 394–398 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Sapra G., Tham Y. K., Cemerlang N., Matsumoto A., Kiriazis H., Bernardo B. C., Henstridge D. C., Ooi J. Y., Pretorius L., Boey E. J., Lim L., Sadoshima J., Meikle P. J., Mellet N. A., Woodcock E. A., Marasco S., Ueyama T., Du X. J., Febbraio M. A., McMullen J. R., The small-molecule BGP-15 protects against heart failure and atrial fibrillation in mice. Nat. Commun. 5, 5705 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Chung J., Nguyen A. K., Henstridge D. C., Holmes A. G., Chan M. H., Mesa J. L., Lancaster G. I., Southgate R. J., Bruce C. R., Duffy S. J., Horvath I., Mestril R., Watt M. J., Hooper P. L., Kingwell B. A., Vigh L., Hevener A., Febbraio M. A., HSP72 protects against obesity-induced insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 105, 1739–1744 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Literati-Nagy B., Kulcsar E., Literati-Nagy Z., Buday B., Peterfai E., Horvath T., Tory K., Kolonics A., Fleming A., Mandl J., Koranyi L., Improvement of insulin sensitivity by a novel drug, BGP-15, in insulin-resistant patients: A proof of concept randomized double-blind clinical trial. Horm. Metab. Res. 41, 374–380 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Naugler W. E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A. M., Karin M., Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317, 121–124 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Park E. J., Lee J. H., Yu G. Y., He G., Ali S. R., Holzer R. G., Osterreicher C. H., Takahashi H., Karin M., Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 140, 197–208 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniguchi K., Wu L. W., Grivennikov S. I., de Jong P. R., Lian I., Yu F. X., Wang K., Ho S. B., Boland B. S., Chang J. T., Sandborn W. J., Hardiman G., Raz E., Maehara Y., Yoshimura A., Zucman-Rossi J., Guan K. L., Karin M., A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature 519, 57–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carey A. L., Steinberg G. R., Macaulay S. L., Thomas W. G., Holmes A. G., Ramm G., Prelovsek O., Hohnen-Behrens C., Watt M. J., James D. E., Kemp B. E., Pedersen B. K., Febbraio M. A., Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes 55, 2688–2697 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Findeisen M., Allen T. L., Henstridge D. C., Kammoun H., Brandon A. E., Baggio L. L., Watt K. I., Pal M., Cron L., Estevez E., Yang C., Kowalski G. M., O'Reilly L., Egan C., Sun E., Thai L. M., Krippner G., Adams T. E., Lee R. S., Grotzinger J., Garbers C., Risis S., Kraakman M. J., Mellet N. A., Sligar J., Kimber E. T., Young R. L., Cowley M. A., Bruce C. R., Meikle P. J., Baldock P. A., Gregorevic P., Biden T. J., Cooney G. J., Keating D. J., Drucker D. J., Rose-John S., Febbraio M. A., Treatment of type 2 diabetes with the designer cytokine IC7Fc. Nature 574, 63–68 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Matthews V. B., Allen T. L., Risis S., Chan M. H., Henstridge D. C., Watson N., Zaffino L. A., Babb J. R., Boon J., Meikle P. J., Jowett J. B., Watt M. J., Jansson J. O., Bruce C. R., Febbraio M. A., Interleukin-6-deficient mice develop hepatic inflammation and systemic insulin resistance. Diabetologia 53, 2431–2441 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Barkhausen T., Tschernig T., Rosenstiel P., van Griensven M., Vonberg R. P., Dorsch M., Mueller-Heine A., Chalaris A., Scheller J., Rose-John S., Seegert D., Krettek C., Waetzig G. H., Selective blockade of interleukin-6 trans-signaling improves survival in a murine polymicrobial sepsis model. Crit. Care Med. 39, 1407–1413 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Schuett H., Oestreich R., Waetzig G. H., Annema W., Luchtefeld M., Hillmer A., Bavendiek U., von Felden J., Divchev D., Kempf T., Wollert K. C., Seegert D., Rose-John S., Tietge U. J., Schieffer B., Grote K., Transsignaling of interleukin-6 crucially contributes to atherosclerosis in mice. Arterioscler. Thromb. Vasc. Biol. 32, 281–290 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Kraakman M. J., Kammoun H. L., Allen T. L., Deswaerte V., Henstridge D. C., Estevez E., Matthews V. B., Neill B., White D. A., Murphy A. J., Peijs L., Yang C., Risis S., Bruce C. R., Du X. J., Bobik A., Lee-Young R. S., Kingwell B. A., Vasanthakumar A., Shi W., Kallies A., Lancaster G. I., Rose-John S., Febbraio M. A., Blocking IL-6 trans-signaling prevents high-fat diet-induced adipose tissue macrophage recruitment but does not improve insulin resistance. Cell Metab. 21, 403–416 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Schreiber S., Aden K., Bernardes J. P., Conrad C., Tran F., Hoper H., Volk V., Mishra N., Blase J. I., Nikolaus S., Bethge J., Kuhbacher T., Rocken C., Chen M., Cottingham I., Petri N., Rasmussen B. B., Lokau J., Lenk L., Garbers C., Feuerhake F., Rose-John S., Waetzig G. H., Rosenstiel P., Therapeutic interleukin 6 trans-signaling inhibition by olamkicept (sgp130Fc) in patients with active inflammatory bowel disease. Gastroenterology 160, 2354–2366.e11 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. 3rd, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haghverdi L., Lun A. T. L., Morgan M. D., Marioni J. C., Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.English K., Tan S. Y., Kwan R., Holz L. E., Sierro F., McGuffog C., Kaisho T., Heath W. R., MacDonald K. P., McCaughan G. W., Bowen D. G., Bertolino P., The liver contains distinct interconnected networks of CX3CR1+ macrophages, XCR1+ type 1 and CD301a+ type 2 conventional dendritic cells embedded within portal tracts. Immunol. Cell Biol. 100, 394–408 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guilliams M., Bonnardel J., Haest B., Vanderborght B., Wagner C., Remmerie A., Bujko A., Martens L., Thone T., Browaeys R., De Ponti F. F., Vanneste B., Zwicker C., Svedberg F. R., Vanhalewyn T., Goncalves A., Lippens S., Devriendt B., Cox E., Ferrero G., Wittamer V., Willaert A., Kaptein S. J. F., Neyts J., Dallmeier K., Geldhof P., Casaert S., Deplancke B., Ten Dijke P., Hoorens A., Vanlander A., Berrevoet F., Van Nieuwenhove Y., Saeys Y., Saelens W., Van Vlierberghe H., Devisscher L., Scott C. L., Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrikx T., Porsch F., Kiss M. G., Rajcic D., Papac-Milicevic N., Hoebinger C., Goederle L., Hladik A., Shaw L. E., Horstmann H., Knapp S., Derdak S., Bilban M., Heintz L., Krawczyk M., Paternostro R., Trauner M., Farlik M., Wolf D., Binder C. J., Soluble TREM2 levels reflect the recruitment and expansion of TREM2+ macrophages that localize to fibrotic areas and limit NASH. J. Hepatol. 77, 1373–1385 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Maier B., Leader A. M., Chen S. T., Tung N., Chang C., LeBerichel J., Chudnovskiy A., Maskey S., Walker L., Finnigan J. P., Kirkling M. E., Reizis B., Ghosh S., D'Amore N. R., Bhardwaj N., Rothlin C. V., Wolf A., Flores R., Marron T., Rahman A. H., Kenigsberg E., Brown B. D., Merad M., A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich S., Nguyen C. D. L., Has C., Steltgens S., Soni H., Coman C., Freyberg M., Bichler A., Seifert N., Conrad D., Knobbe-Thomsen C. B., Tews B., Toedt G., Ahrends R., Medenbach J., A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites. Nat. Commun. 11, 2936 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suppli M. P., Rigbolt K. T. G., Veidal S. S., Heeboll S., Eriksen P. L., Demant M., Bagger J. I., Nielsen J. C., Oro D., Thrane S. W., Lund A., Strandberg C., Konig M. J., Vilsboll T., Vrang N., Thomsen K. L., Gronbaek H., Jelsing J., Hansen H. H., Knop F. K., Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am. J. Physiol. Gastrointest. Liver Physiol. 316, G462–G472 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Rabe B., Chalaris A., May U., Waetzig G. H., Seegert D., Williams A. S., Jones S. A., Rose-John S., Scheller J., Transgenic blockade of interleukin 6 transsignaling abrogates inflammation. Blood 111, 1021–1028 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Font-Burgada J., Sun B., Karin M., Obesity and cancer: The oil that feeds the flame. Cell Metab. 23, 48–62 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Karsdal M. A., Daniels S. J., Holm Nielsen S., Bager C., Rasmussen D. G. K., Loomba R., Surabattula R., Villesen I. F., Luo Y., Shevell D., Gudmann N. S., Nielsen M. J., George J., Christian R., Leeming D. J., Schuppan D., Collagen biology and non-invasive biomarkers of liver fibrosis. Liver Int. 40, 736–750 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Zheng X., Liu W., Xiang J., Liu P., Ke M., Wang B., Wu R., Lv Y., Collagen I promotes hepatocellular carcinoma cell proliferation by regulating integrin β1/FAK signaling pathway in nonalcoholic fatty liver. Oncotarget 8, 95586–95595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X., Nadiarynkh O., Plotnikov S., Campagnola P. J., Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 7, 654–669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vennin C., Chin V. T., Warren S. C., Lucas M. C., Herrmann D., Magenau A., Melenec P., Walters S. N., Del Monte-Nieto G., Conway J. R., Nobis M., Allam A. H., McCloy R. A., Currey N., Pinese M., Boulghourjian A., Zaratzian A., Adam A. A., Heu C., Nagrial A. M., Chou A., Steinmann A., Drury A., Froio D., Giry-Laterriere M., Harris N. L., Phan T., Jain R., Weninger W., McGhee E. J., Whan R., Johns A. L., Samra J. S., Chantrill L., Gill A. J., Kohonen-Corish M., Harvey R. P., Biankin A. V.; I. Australian Pancreatic Cancer Genome, Evans T. R., Anderson K. I., Grey S. T., Ormandy C. J., Gallego-Ortega D., Wang Y., Samuel M. S., Sansom O. J., Burgess A., Cox T. R., Morton J. P., Pajic M., Timpson P., Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl. Med. 9, eaai8504 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cicchi R., Kapsokalyvas D., De Giorgi V., Maio V., Van Wiechen A., Massi D., Lotti T., Pavone F. S., Scoring of collagen organization in healthy and diseased human dermis by multiphoton microscopy. J. Biophotonics 3, 34–43 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Romero F. A., Jones C. T., Xu Y., Fenaux M., Halcomb R. L., The race to bash NASH: Emerging targets and drug development in a complex liver disease. J. Med. Chem. 63, 5031–5073 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Inagaki N., Takeuchi M., Oura T., Imaoka T., Seino Y., Efficacy and safety of tirzepatide monotherapy compared with dulaglutide in Japanese patients with type 2 diabetes (SURPASS J-mono): A double-blind, multicentre, randomised, phase 3 trial. Lancet Diabetes Endocrinol. 10, 623–633 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Hammoud R., Drucker D. J., Beyond the pancreas: Contrasting cardiometabolic actions of GIP and GLP1. Nat. Rev. Endocrinol. 19, 201–216 (2023). [DOI] [PubMed] [Google Scholar]
- 53.van der Poorten D., Samer C. F., Ramezani-Moghadam M., Coulter S., Kacevska M., Schrijnders D., Wu L. E., McLeod D., Bugianesi E., Komuta M., Roskams T., Liddle C., Hebbard L., George J., Hepatic fat loss in advanced nonalcoholic steatohepatitis: Are alterations in serum adiponectin the cause? Hepatology 57, 2180–2188 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Carlessi R., Denisenko E., Boslem E., Koehn-Gaone J., Main N., Abu Bakar N. D. B., Shirolkar G. D., Jones M., Poppe D., Dwyer B. J., Jackaman C., Tjiam M. C., Lister R., Karin M., Fallowfield J. A., Kendall T. J., Forbes S. J., Olynyk J. K., Yeoh G., Forrest A. R. R., Ramm G. A., Febbraio M. A., Tirnitz-Parker J. E. E., Single nucleus rna sequencing of pre-malignant liver reveals disease-associated hepatocyte state with HCC prognostic potential. Cell Genome 3, 100301 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denisenko E., Guo B. B., Jones M., Hou R., de Kock L., Lassmann T., Poppe D., Clement O., Simmons R. K., Lister R., Forrest A. R. R., Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21, 130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y., Wang J., Jaehnig E. J., Shi Z., Zhang B., WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burt A. D., Lackner C., Tiniakos D. G., Diagnosis and assessment of NAFLD: Definitions and histopathological classification. Semin. Liver Dis. 35, 207–220 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Conway J. R. W., Vennin C., Cazet A. S., Herrmann D., Murphy K. J., Warren S. C., Wullkopf L., Boulghourjian A., Zaratzian A., Da Silva A. M., Pajic M., Morton J. P., Cox T. R., Timpson P., Three-dimensional organotypic matrices from alternative collagen sources as pre-clinical models for cell biology. Sci. Rep. 7, 16887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan J. Y., Luzuriaga J., Maxwell E. L., West P. K., Bensellam M., Laybutt D. R., The balance between adaptive and apoptotic unfolded protein responses regulates β-cell death under ER stress conditions through XBP1, CHOP and JNK. Mol. Cell Endocrinol. 413, 189–201 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Legends for tables S1 to S6
Tables S1 to S6