Abstract
Glutathione (GSH) is a critical component of the cellular redox system that combats oxidative stress. The glutamate-cystine antiporter, system xC−, is a key player in GSH synthesis that allows for the uptake of cystine, the rate-limiting building block of GSH. It is unclear whether GSH or GSH-dependent protein oxidation [protein S-glutathionylation (PSSG)] regulates the activity of system xC−. We demonstrate that an environment of enhanced PSSG promotes GSH increases via a system xC−–dependent mechanism. Absence of the deglutathionylase, glutaredoxin (GLRX), augmented SLC7A11 protein and led to significant increases of GSH content. S-glutathionylation of C23 or C204 of the deubiquitinase OTUB1 promoted interaction with the E2-conjugating enzyme UBCH5A, leading to diminished ubiquitination and proteasomal degradation of SLC7A11 and augmentation of GSH, effects that were reversed by GLRX. These findings demonstrate an intricate link between GLRX and GSH via S-glutathionylation of OTUB1 and system xC− and illuminate a previously unknown feed-forward regulatory mechanism whereby enhanced GSH protein oxidation augments cellular GSH.
Glutathione-dependent OTUB1 oxidation acts as a redox sensor to augment cellular GSH.
INTRODUCTION
Oxidants are key regulators of physiological processes (1, 2), and the highly abundant tripeptide molecule glutathione (GSH) is one of the major antioxidants that control cellular redox homeostasis. GSH plays critical roles in the function of regulatory T lymphocytes (3), activation of epithelial cells (4), and survival of cancer cells (5–7), among others. GSH consists of cysteine, glycine, and glutamic acid, and its synthesis occurs in a series of steps that involve the formation of gamma glutamyl-cysteine via glutamate cysteine ligase (GCL), followed by the addition of glycine via GSH synthetase (GS). Levels of GSH are tightly controlled via negative feedback regulation of the catalytic subunit of GCL and degradation of GSH via a series of enzymes [reviewed in (8)]. To maintain GSH redox homeostasis, cells adopt multiple strategies that include the transcriptional activation of GSH-synthesizing enzymes, the uptake of precursor amino acids, and changes in metabolism. Alterations in glycolysis, glutaminolysis, or serine/glycine metabolism in cancer cells promote survival as a result of increased levels of GSH, which allow tumor cells to withstand their enhanced oxidative environment (5, 9–11), and the pentose phosphate pathway, an offshoot of glycolysis, provides NADPH [reduced form of nicotinamide adenine dinucleotide phosphate (NADP+)]–reducing equivalents to maintain GSH homeostasis (12). A critical determinant of GSH levels is system xC−, the cystine-glutamate antiporter expressed on the cell surface that is composed of solute carrier family 7 member 11 (SLC7A11; xCT) linked via a disulfide bridge to SLC3A2 (4F2). SLC7A11 is the functional light chain that confers specificity for cystine uptake, which, intracellularly, is rapidly reduced to cysteine, the rate-limiting amino acid in GSH synthesis (8, 13). Lung adenocarcinomas (LUADs) displaying Kirsten rat sarcoma virus (KRAS) mutations have elevated levels of intracellular cystine and GSH in association with enhanced expression of SLC7A11 (13).
It is well known that GSH can exist in the thiol-reduced (GSH) and thiol-oxidized GSH disulfide forms [reviewed in (8)]. However, one facet of GSH biochemistry that remains less well recognized is its ability to control the oxidation state of proteins. Protein S-glutathionylation (PSSG), the covalent attachment of GSH to reactive cysteines within proteins via a cysteine-cysteine disulfide linkage, represents a key regulatory mechanism whereby GSH itself affects cellular processes. PSSG affects the structure and function of target proteins due to the changes in charge and/or size and also prevents the overoxidation of the cysteine moieties. The thiol transferase, glutaredoxin (GLRX), is the major deglutathionylase that reestablishes the reduced protein thiol group. In this manner, GLRX controls diverse (patho)physiological processes that include inflammation, energy metabolism, and fibrosis (14–17), and multiple S-glutathionylation targets that regulate these processes have been identified (18–21).
To date, it remains also unknown whether GLRX controls GSH levels. A recent S-glutathionylated proteome screen revealed that, in lung epithelial cells stimulated with interleukin-1β (IL-1β), the ovarian tumor (OTU) deubiquitinase, OTUB1, was a target for PSSG (20). OTUB1 was recently shown to interact with SLC7A11, thereby enhancing its stability and augmenting system xC− activity in cancer cells (22). IL-1β also increases glycolysis (23), which has been linked to enhanced system xC− activity (24). These collective findings prompted us to investigate the mechanism by which GSH is regulated in primary lung epithelial progenitor or LUAD cells and whether PSSG plays a functional role in this regulation. We used the cytokine IL-1β in our studies, as IL-1β is a major driver of glycolysis and proliferation in primary lung epithelial progenitor cells, induces oxidative stress (23, 25–27), and has been linked to lung cancer (28, 29). Here, we demonstrate that an environment of enhanced PSSG promotes increases in GSH via a system xC−–dependent mechanism. Notably, we demonstrate that S-glutathionylation of C23 or C204 of OTUB1 promotes binding of the E2-conjugating enzyme UBCH5A, leading to stabilization of SCL7A11 and augmentation of GSH in a GLRX-reversible manner. These findings describe an intricate link between GLRX and GSH status and point to a feed-forward regulatory mechanism wherein enhanced GSH-dependent OTUB1 oxidation acts as a redox sensor to augment cellular GSH.
RESULTS
GSH is increased in primary lung epithelial cells exposed to IL-1β in a system xC−–dependent manner and promotes secretion of proinflammatory cytokines
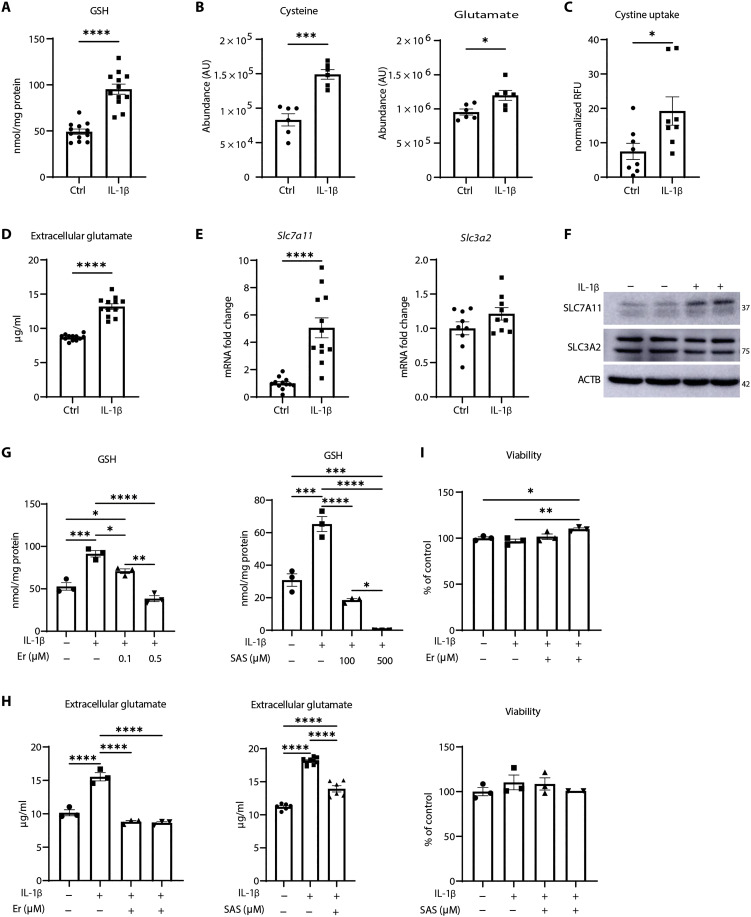
As stated above, IL-1β augments glycolysis in lung epithelial cells (23) and induces oxidative stress (25, 26). The pentose phosphate pathway, an offshoot of glycolysis, regulates the redox environment by providing NADPH-reducing equivalents necessary for GSH homeostasis (12). These collective observations prompted us to evaluate changes in GSH in response to IL-1β. Overall GSH levels significantly increased in response to 24 hours of IL-1β (Fig. 1A), and 13C-glucose tracing analysis demonstrated that carbons from glucose were used in synthesis of GSH (fig. S1A).
Fig. 1. IL-1β increases GSH and SLC7A11 levels in primary airway epithelial cells.
(A) GSH levels in control or IL-1β–stimulated mouse tracheal epithelial (MTE) cells following 24 hours of incubation with of IL-1β (10 ng/ml). (B) Levels of intracellular cysteine (left) and glutamate (right) in control and IL-1β–stimulated cells. AU, arbitrary units. (C) Fluorescein isothiocyanate–cystine uptake in control or IL-1β–treated cells. Results are expressed as relative fluorescence units (RFU) normalized to Hoechst staining in each well. (D) Levels of glutamate in conditioned medium from MTE cells in response to IL-1β. (E) Assessment of Slc7a11 and Slc3a2 mRNA. Ctrl, control. (F) Western blot analysis of SLC7A11 and SLC3A2 levels in response to IL-1β. ACTB, loading control. (G) GSH measurement in cells exposed to increasing doses of erastin (Er) or sulfasalazine (SAS). Cells were pretreated for 1 hour, followed by IL-1β treatment for 24 hours. (H) Extracellular glutamate levels in conditioned medium of cells pretreated with 0.5 μM Er or 100 μM SAS for 1 hour before IL-1β stimulation for 24 hours. (I) Assessment of cell viability in cells treated with 0.5 μM Er or 500 μM SAS and vehicle/IL-1β. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
GSH is a tripeptide composed of cysteine, glycine, and glutamic acid. Levels of cysteine and glutamate were increased in response to IL-1β, while glycine was not detected (Fig. 1B and fig. S1B). Tumor necrosis factor–α (TNF-α) also led to increases in GSH, while, conversely, interferon-γ (IFN-γ) decreased GSH levels (fig. S1C), pointing to divergent modulation by GSH by these proinflammatory stimuli.
We next sought to explore the mechanism(s) whereby IL-1β increased GSH. Enzymes involved in GSH biosynthesis comprise GCL (catalytic and modulatory subunits GCLC and GCLM) and GS [reviewed in (8)]. The transcription factor nuclear factor erythroid 2–related factor 2 (NRF2) controls expression GSH synthesis genes (30). Time course studies show that increases in NRF2 occur 2 hours after IL-1β treatment followed by decreases at later time points. No differences in GCLC or GCLM were observed at any times (fig. S2A). GSH started to increase 4 hours after IL-1β, peaked at 24 hours, and decreased below baseline levels 48 hours after IL-1β treatment (fig. S2B), a time when GS was also decreased (fig. S2A). To address the functional role of NRF2 in the IL-1β–mediated GSH increases, we exposed cells to the NRF2 inhibitor, ML385 (31). Although ML385 decreased the overall GSH levels, it did not dampen the IL-1β–induced increases in GSH (fig. S2C). Levels of cystathionase (CTH) and cystathionine β-synthase (CBS), members of the transsulfuration pathway as a potential source of cysteine (32), also remained unchanged or decreased in response to IL-1β (fig. S2D). Collectively, these observations suggest that the IL-1β–mediated increases in GSH occur independent from NRF2 or the transsulfuration pathways.
A key regulator of GSH homeostasis is the transporter system xC− that exports glutamate and concomitantly imports cystine in a 1:1 ratio (33). Intracellularly, cystine is reduced to cysteine, the rate-limiting amino acid required for GSH synthesis (34). In addition to increases in intracellular cysteine (Fig. 1B), uptake of fluorescein isothiocyanate–labeled cystine (Fig. 1C) and extracellular glutamate (Fig. 1D) were also significantly increased in response to IL-1β, results that point to increases in system xC− activity. mRNA expression and protein levels of the system xC− subunit, SLC7A11, were also increased in response to IL-1β, with no significant changes in SLC3A2 subunit (Fig. 1, E and F, and fig. S2E). To address whether IL-1β–induced increases in GSH are system xC−−dependent, we used two system xC− inhibitors: erastin (Er) and sulfasalazine (SAS). Both inhibitors significantly decreased IL-1β–induced increases in GSH and extracellular glutamate levels (Fig. 1, G and H). System xC− inhibition is well known to induce ferroptosis (33). However, we did not detect any changes in cell viability using Er or SAS (Fig. 1I). In contrast, secretion of various IL-1β–induced proinflammatory cytokines (20, 23, 35), notably of thymic stromal lymphopoetin (TSLP), chemokine (C-C motif) ligand 20 (CCL20), and chemokine (C-X-C motif) ligand 1 (CXCL1), was significantly attenuated by Er (fig. S3A). As expected, the inhibitor of GCLC, buthionine sulfoximine (BSO), depleted GSH and also led to significant decreases in TSLP and CCL20 (fig. S3B). Collectively, these data demonstrate that system xC− is critical in enhancing GSH levels in IL-1β–stimulated cells and that GSH, in turn, promotes proinflammatory signaling independently of cell death.
S-glutathionylation controls GSH and SLC7A11 levels
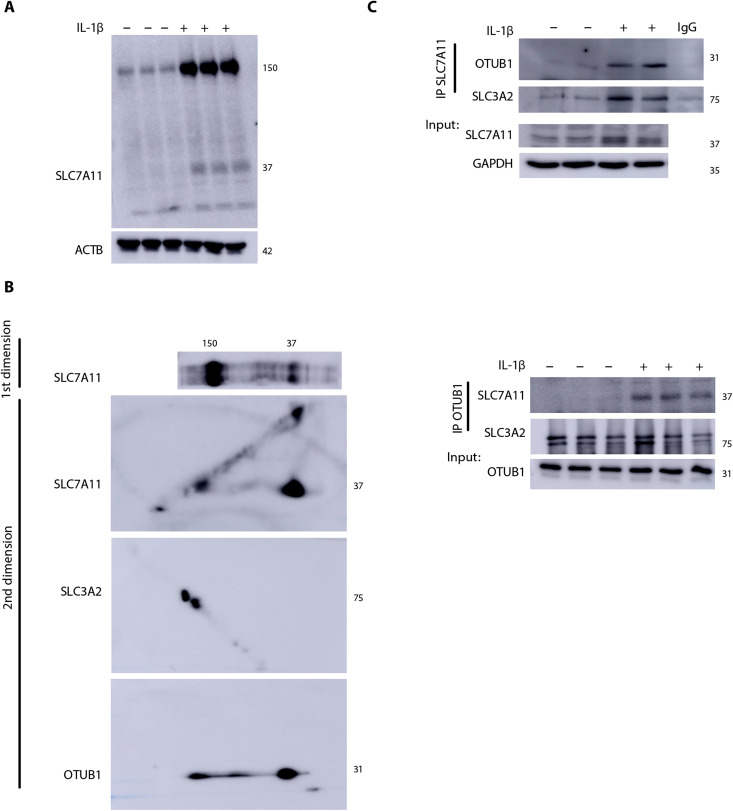
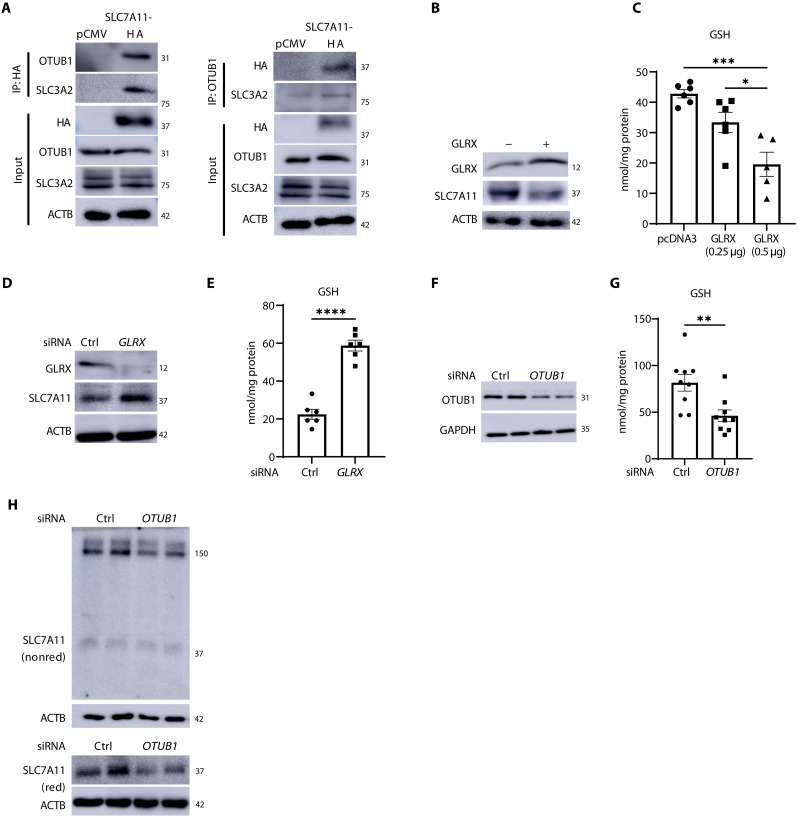
We have previously reported that OTUB1 is a target for PSSG in epithelial cells stimulated with IL-1β (20). OTUB1, a member of the OTU protease subfamily, directly interacts with and stabilizes SLC7A11 in cancer cells by preventing its ubiquitination (22). Therefore, we assessed whether system xC− is regulated in a redox-dependent manner. Nonreducing Western blots for SLC7A11 revealed the formation of a high–molecular weight (HMW) species appearing at ~150 kDa that increased in response to IL-1β (Fig. 2A and fig. S2E). Two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) showed that SLC7A11, OTUB1, and SLC3A2 were detected in association with the ~150-kDa complex (Fig. 2B), and co-immunoprecipitation analysis further confirmed the association between SLC7A11, SLC3A2, and OTUB1 after IL-1β stimulation (Fig. 2C). Although the transmembrane glycoprotein, CD44, is also known to bind and stabilize the SLC7A11 and OTUB1 complex (22), we were not able to detect CD44 in primary lung epithelial cells in the absence or presence of IL-1β stimulation.
Fig. 2. Formation of an oxidation-dependent HMW complex of SLC7A11, SLC3A2, and OTUB1 in response to IL-1β.
(A) Nonreducing Western blot of SLC7A11 showing a high–molecular weight (HMW) complex around ~150 kDa in MTE cells that is further increased upon treatment with IL-1β as described in Fig. 1. ACTB, loading control. (B) Two-dimensional (2D) gel analysis of the nonreduced gel strip containing SLC7A11 (top). SLC7A11, SLC3A2, and OTUB1 were evaluated via Western blotting in the second dimension. (C) Top: Immunoprecipitation of SLC7A11 in MTE cells and Western blot analysis of OTUB1 and SLC3A2. SLC7A11 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from whole-cell lysates were used as the input control. Bottom: Immunoprecipitation of OTUB1 protein followed by Western blotting analysis of SLC7A11 and SLC3A2. OTUB1 levels from whole-cell lysates were used as the input control. IgG, immunoglobulin G; IP, immunoprecipitation.
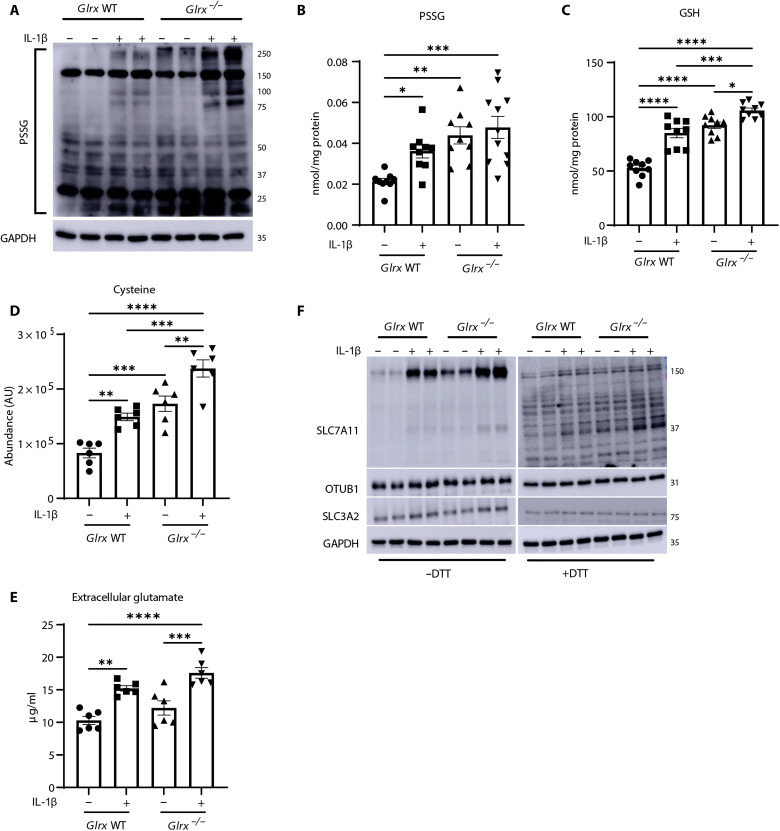
PSSG is a key regulator of biological responses induced by redox perturbations. PSSG occurs through addition of a GSH moiety to a reactive cysteine in a target protein leading to changes in protein’s structure and function (36–38). In line with our previous observations (20), overall PSSG was increased in response to IL-1β (Fig. 3, A and B). GLRX is the main enzyme responsible for deglutathionylating proteins, thereby reestablishing reduced protein thiol groups under physiological conditions. Accordingly, further increases in baseline or IL-1β–stimulated PSSG occurred in cells lacking Glrx (Fig. 3, A and B).
Fig. 3. S-glutathionylation regulates GSH in association with modulation of xC−.
(A) Assessment of PSSG in wild-type (WT) or Glrx−/− MTE cells treated with vehicle or IL-1β as described in Fig. 1. GAPDH, loading control. PSSG (B) or GSH (C) levels in WT or Glrx−/− epithelial cells stimulated with IL-1β for 24 hours. Intracellular cysteine (D) or glutamate content in conditioned medium (E) of WT or Glrx−/− epithelial cells stimulated with vehicle or IL-1β for 24 hours. (F) Nonreducing [−dithiothreitol (−DTT); left] and reducing (+DTT; right) Western blots of SLC7A11 (37 kDa), OTUB1, and SLC3A2 in WT and Glrx−/− cells. GAPDH, loading control. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
GLRX-mediated deglutathionylation per se is not expected to increase overall GSH pools due to the small fraction of GSH bound to proteins (<0.05%; see Fig. 3, B and C). Unexpectedly, total GSH levels were significantly elevated in Glrx−/− cells compared to wild-type (WT) cells, and treatment with IL-1β led to further increases in GSH in Glrx−/− cells compared to that in the respective WT group (Fig. 3C). Intracellular cysteine was also increased in Glrx−/− cells compared to that in WT cells, while extracellular glutamate was not significantly different between WT and Glrx−/− cells (Fig. 3, D and E). Given the increases in GSH and cysteine in Glrx−/− cells, we next assessed whether system xC− was affected by GLRX. Under nonreducing conditions [−dithiothreitol (−DTT)], Glrx−/− cells exhibited higher levels of HMW SLC7A11 compared to control cells, with further elevations occurring following IL-1β (Fig. 3F). Under reducing conditions (+DTT), SLC7A11 immunoreactivity was no longer observed as a prominent HMW complex, pointing to its oxidation-dependent nature. However, increases in the 37-kDa species of SLC7A11 were apparent in unstimulated Glrx−/− cells or in response to IL-1β, while levels of SLC3A2 and OTUB1 were not affected (Fig. 3F). These collective findings demonstrate that PSSG and GLRX control GSH levels through redox regulation of SLC7A11.
GLRX decreases GSH levels in association with diminished S-glutathionylation of OTUB1 and decreases in SLC7A11
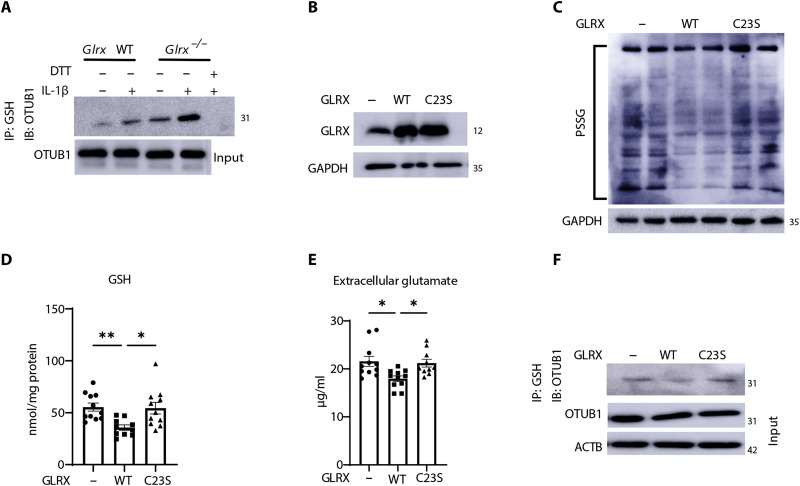
As stated earlier, OTUB1 was one of the PSSG targets detected in IL-1β–stimulated epithelial cells (20), raising the possibility that S-glutathionylation of OTUB1 affects the stability of SLC7A11. Immunoprecipitation of PSSG followed by Western blotting of OTUB1 showed increases in OTUB1 S-glutathionylation (OTUB1-SSG) in response to IL-1β, with further increases occurring in cells lacking Glrx (Fig. 4A), consistent with the role of GLRX as a deglutathionylating enzyme. Direct administration of recombinant WT GLRX protein to cells (Fig. 4B) decreased the overall PSSG and GSH levels (Fig. 4, C and D) and decreased extracellular glutamate (Fig. 4E), while catalytically inactive C23S GLRX (17) did not elicit these effects. WT GLRX but not C23S GLRX decreased OTUB1-SSG (Fig. 4F), pointing to a putative role of OTUB1-SSG in the regulation of GSH levels.
Fig. 4. OTUB1 is a target for S-glutathionylation and is regulated by GLRX.
(A) Detection of OTUB1-SSG in WT or Glrx−/− MTE cells stimulated with vehicle or IL-1β for 24 hours. PSSG was immunoprecipitated with anti-GSH followed by Western blotting of OTUB1. DTT was incubated with cell lysates before immunoprecipitation as a negative control. OTUB1 levels from whole-cell lysates were used as the input control. (B) Evaluation of GLRX in lysates prepared from cells 5 days after treatment with WT-GLRX or C23S-mutant GLRX recombinant protein. GAPDH, loading control. (C) Assessment of total PSSG in WT MTE cells treated with WT GLRX or C23S-GLRX recombinant protein. Shown are nonreducing Western blots probed with anti-GSH antibody. GAPDH, loading control. (D) GSH levels in MTE cells treated with recombinant WT-GLRX or C23S-GLRX. (E) Glutamate in cell culture supernatant of MTE cells treated with WT-GLRX or C23S-GLRX. (F) Detection of OTUB1-SSG in cells treated with recombinant WT-GLRX or C23S-GLRX protein following immunoprecipitation of PSSG using GSH antibody, followed by Western blotting analysis of OTUB1. Total OTUB1 levels from whole-cell lysates were used as the input control. *P < 0.05 and **P < 0.01. IB, immunoblot.
System xC− has been extensively studied in lung cancer where it controls sensitivity to ferroptosis (33, 39). We next sought to better unravel the functional importance of OTUB1-SSG in the regulation of SLC7A11 and GSH using the H522 LUAD cell line. H522 cells express system xC− and are sensitive to GSH depletion (40). Immunoprecipitation of hemagglutinin (HA)–tagged SLC7A11 or OTUB1 shows a constitutive interaction between SLC7A11, SLC3A2, and OTUB1 (Fig. 5A). Overexpression of GLRX in H522 cells (Fig. 5B) decreased GSH (Fig. 5C) and SLC7A11 levels (Fig. 5B). Conversely, knockdown of GLRX increased GSH and SLC7A11 levels (Fig. 5, D and E), consistent with our observations in primary epithelial cells. In addition, knockdown of OTUB1 diminished the HMW SLC7A11 complex formation and the overall levels of SLC7A11 and GSH in H522 cells (Fig. 5, F to H). Knockdown of OTUB1 also decreased GSH in primary lung epithelial cells (fig. S4, A and B). These findings collectively demonstrate functional importance of OTUB1 in the regulation of system xC− and GSH levels.
Fig. 5. GLRX and OTUB1 control SLC7A11 and GSH levels in H522 LUAD cells.
(A) Hemagglutinin (HA)–tagged SLC7A11 overexpressed in H522 cells was immunoprecipitated followed by Western blotting for OTUB1 and SLC3A2 (left). OTUB1 was immunoprecipitated followed by Western blotting for HA-SLC7A11 and SLC3A2 (right). Western blots of HA-SLC7A11, OTUB1, SLC3A2, and ACTB from total lysates were used as input. (B) GLRX and SLC7A11 Western blots of H522 cells overexpressing GLRX or pcDNA3 empty plasmid. ACTB, loading control. (C) Assessment of GSH levels in cells overexpressing GLRX. (D) Western blots of GLRX and SLC7A11 in H522 cells following small interfering RNA (siRNA)–mediated knockdown of GLRX. ACTB, loading control. (E) GSH levels in H522 cells following knockdown of GLRX. (F) Confirmation of OTUB1 via Western blot analysis in H522 cells following siRNA-mediated knockdown. GAPDH, loading control. (G) GSH levels in H522 cells following knockdown of OTUB1. (H) Nonreducing (nonred) (top) and reducing (bottom) Western blots of SLC7A11 in H522 cells with siRNA-mediated OTUB1 knockdown. ACTB, loading controls. *P < 0.05, **P < 0.01, and ***P < 0.001, ****P < 0.0001.
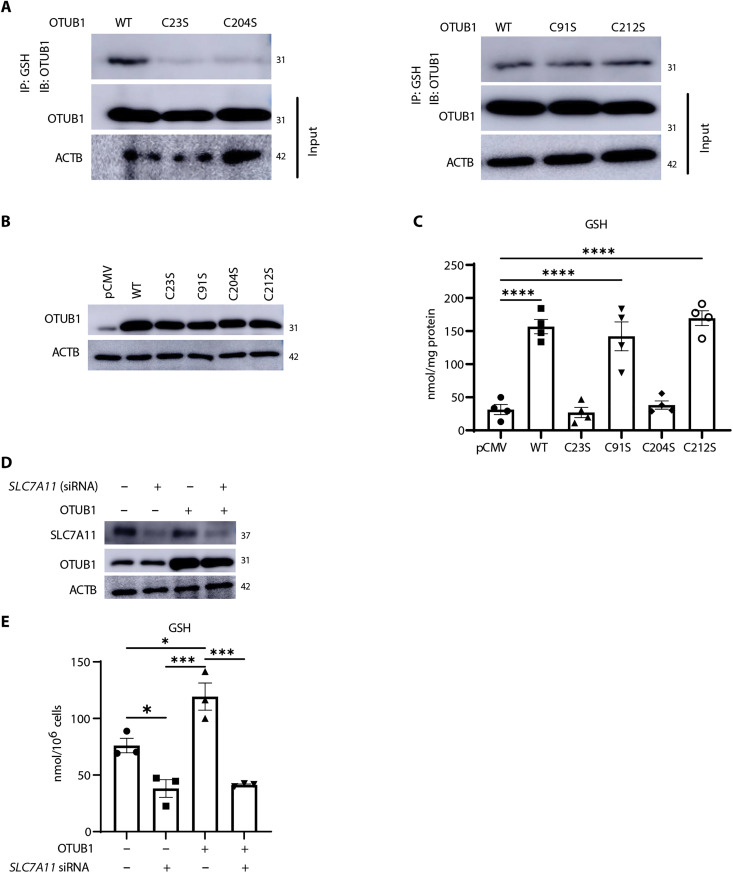
S-glutathionylation of OTUB1 at C23 or C204 increases levels of SLC7A11 and GSH
OTUB1 contains four cysteines (C23, C91, C204, and C212), with C91 required for OTUB1’s canonical deubiquitinase activity (41, 42). To identify the cysteines in OTUB1 that are targets for S-glutathionylation, we mutated each of the four cysteines to serines and expressed these constructs in H522 cells. OTUB1-WT, OTUB1-C91S, and OTUB1-C212S were constitutively S-glutathionylated. In contrast, OTUB1-C23S and OTUB1-C204S mutants were refractory to S-glutathionylation (Fig. 6A). While expression of WT, C91S, and C212S mutants of OTUB1 increased GSH, comparable expression of C23S- or C204S-OTUB1 mutants did not induce any significant increases in GSH (Fig. 6, B and C, and fig. S4, C and D). Similar results were obtained in H522 cells in which the OTUB1 gene was ablated via CRISPR-Cas9 wherein reexpression of WT OTUB1, C91S, or C212S, but not the S-glutathionylation resistant mutants C23S- or C204S-OTUB1, elevated GSH (fig. S4, E to G). Consistent with its ability to increase GSH, expression of WT OTUB1 increased SLC7A11 levels and its HMW complex observed under nonreducing conditions (fig. S4H) and interaction between SLC7A11 and SLC3A2 assessed via proximity ligation assay (fig. S4I). WT OTUB1 failed to induce increases in GSH when SLC7A11 was knocked down (Fig. 6D), demonstrating the requirement of SLC7A11 in the OTUB1-mediated increases in GSH. Overall, these results demonstrate that S-glutathionylation of OTUB1 at C23 or C204 augments GSH levels, in association with stabilization of SLC7A11.
Fig. 6. OTUB1 C23 and C204 are S-glutathionylation targets that increase GSH in a system XC−–dependent manner.
(A) OTUB1-WT or OTUB1-C23S or OTUB1-C204S (left) and OTUB1-WT, OTUB1-C91S, or OTUB1-C212S (right) were expressed in H522 cells. PSSG was immunoprecipitated using GSH antibody followed by detection of OTUB1-SSG via Western blot analysis. OTUB1 and ACTB from total cell lysates were used as input. (B) Confirmation of equal expression of OTUB1 plasmids used in (C). ACTB, loading control. (C) Assessment of GSH in H522 cells overexpressing 125 ng of OTUB1-WT, OTUB1-C23S, OTUB1-C91S, OTUB1-C204S, and OTUB1-C212S plasmids or pCMV vector control. (D) Western blot analysis of SLC7A11 and OTUB1 in H522 cells transfected with control or SLC7A11 siRNA and WT OTUB1 plasmid or pCMV empty vector control. ACTB, loading control. (E) GSH in H522 cells following knockdown of SLC7A11 and overexpression of WT OTUB1. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
S-glutathionylation of OTUB1 enhances interaction with the E2 Ub-conjugating enzyme, UBCH5A
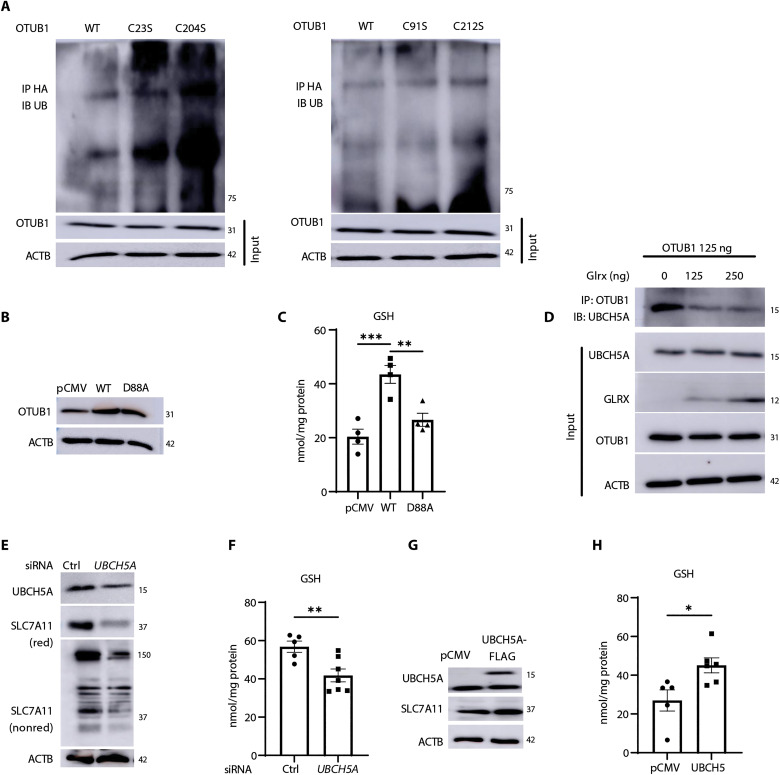
OTUB1 was previously shown to interfere with ubiquitination and subsequent proteasomal degradation of SLC7A11 (22). As expected, overexpression of OTUB1 decreased ubiquitinated SLC7A11 (fig. S4J). However, cells expressing OTUB1-C23S or OTUB1-C204S showed increased ubiquitination of SLC7A11 compared to cells expressing OTUB1-WT, OTUB1-C91S, or OTUB1-C212S mutants (Fig. 7A). The OTUB1-mediated attenuation of ubiquitination can occur through multiple mechanisms. OTUB1 can act as a canonical deubiquitinase that cleaves K48 chains. Alternatively, OTUB1 also can inhibit the function of E2-conjugating enzymes, including UBCH5 (43–45), and OTUB1 has been found in a complex with E2 enzymes. In addition, binding of E2 enzymes to OTUB1 can also promote OTUB1 deubiquitinase activity dependent on the ratio of ubiquitin (Ub)–charged E2 (E2-Ub) to uncharged E2 enzyme and levels of free Ub (46). OTUB1 D88 binds E2 enzymes and suppresses Ub-conjugating activity (47). Stabilization of SLC7A11 by OTUB1 has been speculated to involve direct binding between OTUB1 and SLC7A11 as well as the inhibition of E2 enzymes (22). Consistent with this scenario, OTUB1 D88A failed to induce increases in GSH levels in H522 cells (Fig. 7, B and C). These collective observations, along with observed increases in GSH following expression of OTUB1-C91S (Fig. 6C), support a noncanonical role of OTUB1 in augmenting GSH. Separate domains within OTUB1 are required for noncanonical inhibition via binding of E2-Ub enzyme and free Ub, in a configuration that mimics the product of OTUB1-mediated deubiquitination (48). These domains in OTUB1 are in close proximity to C23 and C204, raising the possibility that S-glutathionylation at either site regulates binding or orientation of either free Ub or E2-Ub enzyme. To address this possible scenario, we determined whether OTUB1-SSG affected the interaction between OTUB1 and UBCH5A that requires the N-terminal region containing C23 (43). Our previous data show that GLRX decreases OTUB1-SSG (Fig. 4F). GLRX also decreases the interaction between OTUB1 and UBCH5A (Fig. 7D). Knockdown of UBCH5A attenuated SLC7A11 levels and diminished GSH levels, while, conversely, overexpression of UBCH5A augmented SLC7A11 and GSH (Fig. 7, E to H).
Fig. 7. The E2-conjugating enzyme, UBCH5A, interacts with OTUB1 in a GLRX-sensitive manner to increase SLC7A11 and GSH.
(A) Levels of ubiquitinated SLC7A11 in H522 cells transfected with OTUB1-WT, OTUB1-C23S, or OTUB1-C204S (left) and OTUB1-WT, OTUB1-C91S, or OTUB1-C212S (right) and treated with 40 μg of MG-132 for 4 hours. OTUB1 and ACTB from total cell lysates were used as input controls. (B) Confirmation of expression of OTUB1-WT, OTUB1-D88A, or pCMV vector plasmids via Western blot analysis. ACTB, loading control (C) Assessment of GSH levels in H522 cells overexpressing OTUB1-WT or OTUB1-D88A plasmids or pCMV vector control. (D) Effect of overexpression of GLRX for the interaction between UBCH5A and OTUB1. OTUB1 was immunoprecipitated followed by detection of UBCH5A via Western blotting. Input: GLRX, UBCH5A, and OTUB1 levels in whole-cell lysates used for immunoprecipitation. ACTB, loading control (E) Impact of SiRNA-mediated knockdown of the E2 Ub-conjugating enzyme, UBCH5A, on SLC7A11 levels in H522 cells. UBCH5A knockdown and SLC7A11 were assessed via Western blot analysis under reducing or nonreducing conditions (for SLC7A11). (F) GSH levels in H522 cells subjected to UBCH5A knockdown. Effect of overexpression of UBCH5B on SLC7A11 levels (G) or GSH content (H). *P < 0.05, **P < 0.01, and ***P < 0.001.
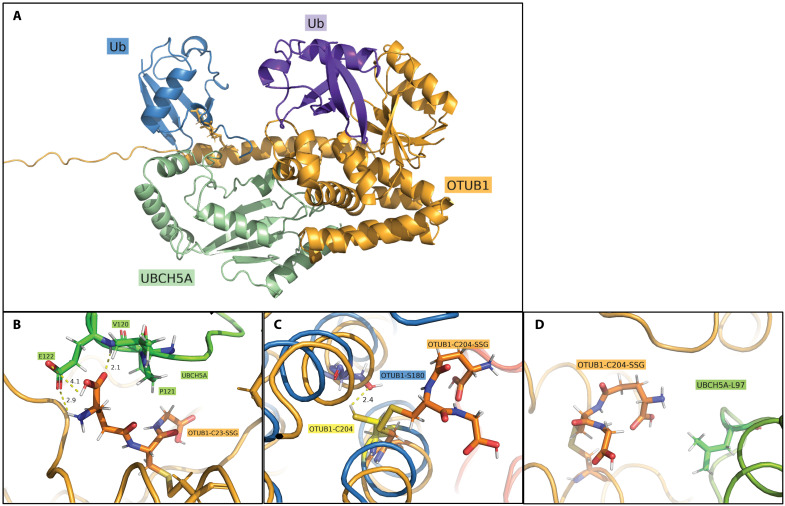
These findings suggest that UBCH5A augmented OTUB1’s noncanonical activity through a mechanism that involves S-glutathionylation of OTUB1 at either position C23 or C204. To understand how S-glutathionylation affects the OTUB1-UBCH5A complex, we created three different in silico models of OTUB1 in conjunction with Ub-charged UBCH5A and free Ub. In the first model, OTUB1 contains reduced cysteines, while the other models consist of OTUB1 S-glutathionylated at C23 (OTUB1-C23-SSG) (Fig. 8, A and B) or OTUB1 S-glutathionylated C204 (OTUB1-C204-SSG) (Fig. 8, C and D). The overall root mean square deviation of UBCH5A was shown to remain locked into place regardless of the S-glutathionylation status of OTUB1. OTUB1-C23-SSG rapidly formed a stable contact between the terminal glutamic acid moiety of GSH and a protein cleft at UBCH5A consisting of V120, P121, and E122 (Fig. 8B). Specifically, both the carbonyl and free amine of GSH were shown to bind to the backbone amine and side-chain carbonyl of E122 with contacts maintained under 3 Å (Fig. 8B). These contacts may serve to stabilize the protein-protein interaction of OTUB1-C23-SSG and UBCH5A when compared to reduced OTUB1. In contrast, OTUB1-C204-SSG was predicted to induce conformational changes in OTUB1 and to restrict the movement of C204 that is otherwise shown to oscillate toward Ser180 and form an internalized polar contact (Fig. 8C). Despite this change, no major stabilizing interactions were calculated between GSH and UBCH5A aside from the nonpolar contact with L97 (Fig. 8D), and the site remained relatively isolated throughout simulation.
Fig. 8. Molecular modeling visualizing the impact of OTUB1-SSG on the binding between OTUB1 and UBCH5A.
(A) Protein complex of OTUB1 (orange), UBCH5A with Ub bound (green/blue), and the distal Ub (purple). Models were constructed with GSH at either C23 (OTUB1-C23-SSG, as shown) or C204. (B) In silico binding site between GSH-C23 (OTUB1-C23-SSG) (orange) and V120, P121, and E122 of UBCH5A (green). Contacts were shown to optimize over simulation length, with convergence of two sub–3-Å contacts between the glutamic acid of GSH and E122 of UBCH5A. (C) Visualization of OTUB1-C204-SSG disrupting minor polar contact between C204 and Ser180 within OTUB1. (D) Final frame of C204-SSG simulation showcasing minor nonpolar contact observed between GSH and L97 at UBCH5A interface.
Decreases in GLRX and increases in overall PSSG and OTUB-SSG occur in LUAD
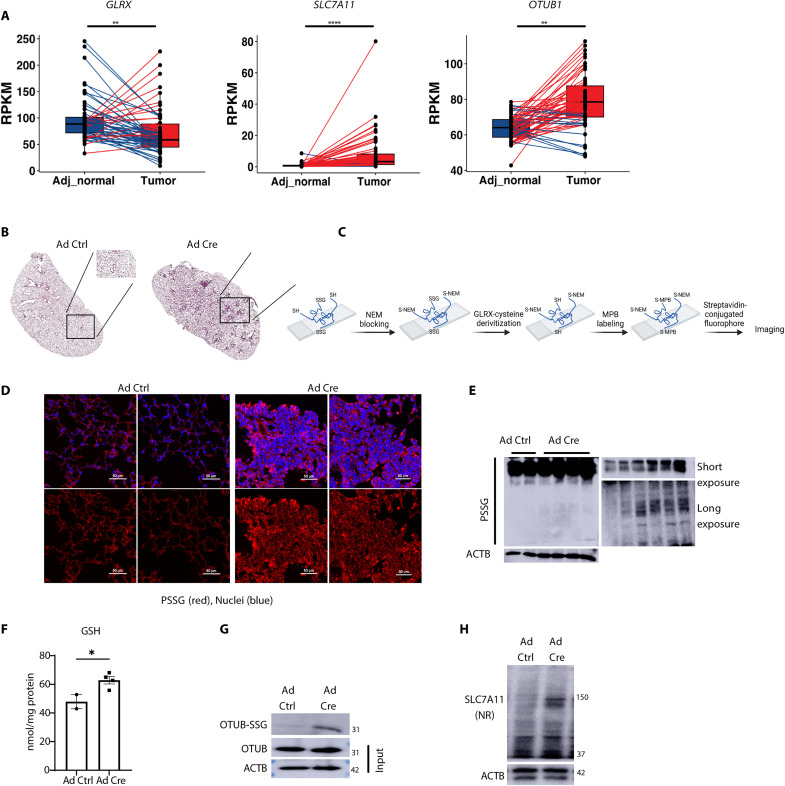
System xC− and OTUB1 have been extensively studied in cancer including LUAD (13, 49–51). To address the potential relevance of GLRX, SLC7A11, and OTUB1 in LUAD, we examined GLRX, SLC7A11, and OTUB1 expression in The Cancer Genome Atlas (TCGA) database. Evaluation of 58 LUADs with matched adjacent normal lung tissue showed a decreased expression of GLRX in LUAD (Fig. 9A), while, in contrast, SLC7A11 and OTUB1 expression was increased relative to the adjacent normal tissue (Fig. 9A).
Fig. 9. Altered expression of GLRX, SLC7A11, and OTUB1 in LUAD.
(A) Assessment of GLRX, SLC7A11, and OTUB1 in 58 LUADs (tumor) compared with matched adjacent normal lung tissue from the TCGA database. Lines connect matched samples from individual patients (adjacent normal to tumor). Blue lines indicate decreased expression, while red represents increased expression in LUAD compared to adjacent normal tissue. RPKM, reads per kilobase of transcript per million mapped reads. (B) Hematoxylin and eosin staining of tissue sections derived from lungs of mice expressing KrasLoxP-Stop-LoxP(LSL)-G12D/WT 15 weeks after the administration of adenovirus expressing Cre recombinase (Ad Cre; right) or control virus (Ad Ctrl; left). (C) Schematic of GLRX-based cysteine derivatization used in (D). MPB, 3-(N-maleimido-propionyl) biocytin; NEM, N-ethylmaleimide. (D) PSSG staining (in red) in lung sections from mice treated with Ad Ctrl (left) or Ad Cre (right). Ad Cre images represent tumor areas selected on the basis of morphology. The figure was created using Biorender.com. (E) Total PSSG in lung lysates detected by Western blotting. The top right blot shows the PSSG bands of the left blot under short exposure. The bottom right blot shows the PSSG bands of lower region of the left blot after long exposure. (F) Total GSH levels in lung lysates of mice treated with Ad ctrl or Ad cre. (G) Immunoprecipitation of glutathionylated proteins via GSH antibody followed by Western blot analysis of OTUB1 to detect OTUB-SSG (top). OTUB1 and ACTB from total cell lysates were used as input controls. (H) Nonreducing Western blotting for SLC7A11. ACTB, loading control. *P < 0.05,**P < 0.01, and ****P < 0.0001.
Visualizing GLRX expression and genomic alterations using Oncoprint revealed no clear association between GLRX and common LUAD oncogenic drivers (fig. S5). We next used an adenovirus-expressing Cre-recombinase (AdCre) system to induce early tumors in KrasG12D mice (Fig. 9B). Using GLRX-mediated cysteine derivatization to illuminate regions of PSSG, we showed increases in PSSG in early tumor regions compared to the control mice (Fig. 9, C and D). Last, overall PSSG, GSH, and OTUB-SSG were significantly increased in lungs with activated KrasG12D-induced tumors, accompanied by increases in SLC7A11 protein levels (Fig. 9, E to H). We did not detect changes in GLRX levels in the homogenized whole lung tissue, possibly due to the small representation of tumor area. Despite these different observations, the decreases in GLRX in LUAD and the increases in PSSG and OTUB1-SSG in KrasG12D-driven tumors in mice point to the relevance of findings here for LUAD. These overall findings illuminate a previously unknown mechanism of regulation of system xC− through GSH-dependent protein oxidation that is under OTUB1-dependent GLRX-directed redox control (Fig. 10) and highlight its putative relevance in lung cancer.
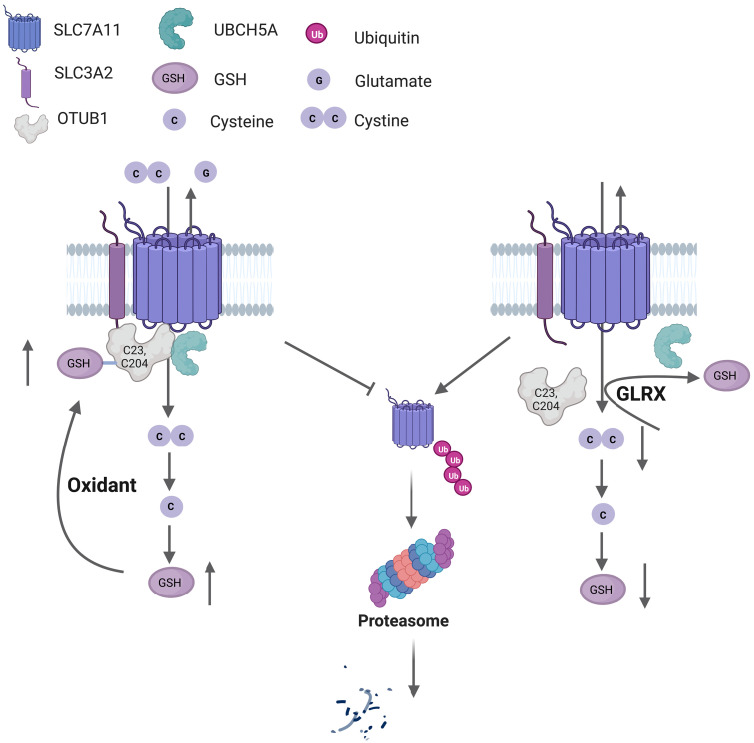
Fig. 10. Graphical abstract depicting system xC− regulation in a redox-dependent manner through OTUB1-SSG.
S-glutathionylation of the deubiquitinase OTUB1 promotes stabilization of SLC7A11, the active subunit of system xC− transporter. An enhanced oxidative environment leads to increases in S-glutathionylation of OTUB1 (OTUB1-SSG), at C23 and C204, which increases interaction with the E2 Ub-conjugating enzyme, UBCH5A, to prevent ubiquitination and proteasomal degradation of SLC7A11. The resultant increase in system xC− transporter activity allows enhanced cystine uptake that will be incorporated in GSH synthesis following its reduction to cysteine intracellularly (left). The deglutathionylase, GLRX, removes GSH from OTUB1, leading to ubiquitination of SLC7A11 and its subsequent proteasomal degradation, resulting in lower GSH content (right). This pathway represents a feed-forward regulatory mechanism whereby enhanced GSH-dependent protein oxidation augments cellular GSH. The figure was created using BioRender.com.
DISCUSSION
The GLRX–S-glutathionylation redox axis has emerged as a key regulator of cellular processes as it controls protein structure and function and prevents overoxidation of reactive cysteines within proteins. Numerous S-glutathionylation targets in diverse pathways have been described, and, with advances in redox proteomics, the list of glutathionylated protein targets continues to grow (20, 36, 37, 52–54). To date, the interplay between GLRX and GSH levels has largely remained unknown. In the present study, we illuminate a new dimension of GLRX’s action by demonstrating that GLRX controls GSH levels via the regulation of system xC−. Notably, we demonstrate that S-glutathionylation of OTUB1 at C23 or C204 regulates SLC7A11 stability through noncanonical regulation of the E2-conjugating enzyme UBCH5A and subsequent stabilization of the system xC− complex, in turn, leading to cystine uptake and increases in GSH.
OTUB1 was previously shown to stabilize SLC7A11 via a CD44-dependent mechanism. CD44 increased the interaction between OTUB1 and SCL7A11, and the CD44-mediated enhancement of SLC7A11 stability depended on OTUB1 (22). As stated above, in the present study, we did not detect CD44 interaction with SLC7A11 in primary lung epithelial cells and obtained inconclusive results in H522 LUAD cells. The findings here showing that OTUB1-C91S increases SLC7A11 and GSH similar to OTUB1-WT, while OTUB1-D88A disrupts GSH, and that OTUB1-SSG promotes binding of the E2-conjugating enzyme, UBCH5A, collectively confirm a noncanonical mechanism of action of OTUB1 in the stabilization of SLC7A11. The precise mechanism whereby a GSH moiety at C23 or C204 of OTUB1 regulates interaction with UBCH5A or free Ub with OTUB1 and the implications for the OTUB1-dependent regulation of SLC7A11 will require further study. The function of UBCH5A as a Ub-conjugating enzyme depends on its Ub charge and the levels of free Ub. E2-conjugating enzymes have intrinsic reactivity toward small-molecule nucleophiles (55). One possible scenario is that a GSH-mixed disulfide at cysteine 23 of OTUB1 also attracts and stabilizes UBCH5A, consistent with our in silico findings (Fig. 8B). Alternatively, disulfide bond disruption or trans-glutathionylation between OTUB1 and UBCH5A has the potential to inhibit UBCH5A activity, and the latter possibility is supported by a study demonstrating that E2 Ub-conjugating enzymes are inhibited via S-glutathionylation (56). In addition, OTUB1-C23-SSG or C204-SSG could realign the OTUB1-bound UBCH5 and free Ub, into a configuration that increases the potency of OTUB1-mediated inhibition of ubiquitination. Last, GLRX or PSSG may also regulate free Ub or Ub charge of UBCH5A. Elucidation of these various scenarios awaits additional analyses.
In addition to SLC7A11, OTUB1 also has other targets including multiple oncoproteins such as forkhead box protein M1, tumor protein P53, cellular inhibitor of apoptosis protein, and murine double minute X, which it stabilizes through a nonconventional mechanism (57–59). Further analysis is necessary to address whether S-glutathionylation of OTUB1 also regulates those targets. Besides OTUB1, another deubiquitinase also regulates SLC7A11. The tumor suppressor breast cancer type 1 (BRCA1)-associated protein 1 (BAP1) is a nuclear deubiquitinase that reduces chromatin histone 2A ubiquitination. BAP1 was shown to repress SLC7A11 expression in a deubiquitination-dependent manner, thereby promoting ferroptosis (60). BAP1 C91 is required for deubiquitinase activity and repression of SLC7A11. Future studies aimed at elucidating whether C91 is a target for S-glutathionylation will be important, as interference with BAP1’s deubiquitinase activity could serve as a potential additional mechanism whereby S-glutathionylation chemistry augments SLC7A11. Furthermore, under conditions of GSH depletion, cancer cells rely on deubiquitinases for survival. In those settings, combined depletion of GSH and inhibition of deubiquitinases caused an accumulation of ubiquitinated proteins, proteotoxic stress, endoplasmic reticulum stress, and tumor cell death (40). In light of the latter findings, future studies addressing whether conditions of diminished GSH levels or increased GSH oxidation affect deubiquitinase S-glutathionylation and implications for their canonical or alternative functions, therefore, appears well warranted.
The OTUB1-mediated stabilization of SLC7A11 following its S-glutathionylation depicts a regulatory feed forward mechanism whereby GSH, in a form of a protein-mixed disulfide with OTUB1, regulates its own synthesis in an SLC7A11-dependent manner. Besides the system xC−-mediated import of cystine, the trans-sulfuration pathway also provides a source of cysteine. As described above, CBS catalyzes the first and rate-limiting step in the trans-sulfuration pathway to convert homocysteine to cysteine. S-glutathionylation of C346 was reported to augment the activity of CBS, leading to increases in cysteine (61). Last, the Kelch-like ECH-associated protein 1 (KEAP1)-NRF2 pathway plays a key role in the transcriptional up-regulation of SLC7A11 (62). S-glutathionylation of KEAP1 has been described as a mechanism that promotes the activation of NRF2 (63), providing an additional pathway through which enhanced S-glutathionylation can up-regulate SLC7A11. Here, we detected transient increases in NRF2 and no changes in CBS or CTH content in response to IL-1β. However, future studies will be needed to address the relative importance of GLRX-mediated deglutathionylation of these diverse targets for the control of system xC− and GSH levels.
In the present study, we also illuminate the importance of system xC− and GSH levels in IL-1β–induced proinflammatory signaling in primary airway epithelial cells, independently of ferroptosis. Our results showing diminished secretion of proinflammatory cytokines, notably TSLP and CCL20 in IL-1β–stimulated epithelial cells following GSH depletion by Er or BSO, point to the importance of GSH and system xC− in regulating proinflammatory cascades. Our previous studies showed that the absence of Glrx enhanced glycolysis and promoted IL-1β–stimulated TSLP (20). IL-1β–stimulated glycolysis in epithelial cells primes epithelial cells for subsequent stimulation with allergens and contributes to allergic airway disease (23). Thus, analogous to the glucose dependence of SLC7A11-expressing tumors (64), primary airway epithelial cells stimulated with IL-1β also rely on glucose and SLC7A11 expression to induce proinflammatory cytokines. Further studies are required to understand the mechanistic details whereby cystine and GSH regulate the transcriptional activation and release of these and other cytokines and whether this involves S-glutathionylation of OTUB1 or other redox sensitive targets. The role of system xC− in regulating responses to IL-1β also merits future study given the pleiotropic role of this cytokine and its emerging role in lung cancer (65). Intriguingly, a recent study reported that IL-1 receptor accessory protein can bind to system xC− to increase uptake of cystine and augment GSH levels (66), pointing to intriguing interplay between IL1 signaling and system xC− activity.
Our findings here demonstrating that OTUB1-SSG promotes OTUB1 and that UBCH5A binding stabilizes SLC7A11, enhances system xC− activity, and subsequently augments GSH have important implications for tumor biology. Enhanced expression of OTUB1 and SLC7A11 occurs in numerous cancers. Increased OTUB1 expression is associated with aggressive disease, poor prognosis, and worse patients’ survival. SLC7A11 expression is positively correlated with chemotherapeutic resistance and worsening survival in cancer patients (49, 67). Cystine uptake requires reduction to cysteine, a process that consumes NADPH. SLC7A11-expressing tumors were found to be reliant on glucose metabolism and activation of the pentose phosphate pathway to maintain redox control. Limitation of the glucose supply or inhibition of glucose transporters resulted in selective killing of SLC7A11-high cancer cells and suppressed tumor growth (64). Our current findings suggest a role for PSSG and, specifically, for OTUB-SSG in Kras G12D adenocarcinomas. Additional investigations are required to confirm the exact role played by GLRX, SLC7A11, and OTUB1-SSG in the development and/or progression of SLC7A11-expressing tumors. Last, more studies are needed to test whether targeting OTUB-SSG by small molecules is sufficient to improve outcomes in mouse models of lung cancer.
In summary, results from the present study provide new insights into the regulation of system xC− by illuminating its control by GLRX and S-glutathionylation of OTUB1. These findings demonstrate a positive regulatory mechanism whereby S-glutathionylation augments GSH synthesis and has potential impact for the management of diseases that are accompanied by increases in activity of system xC−, including cancer.
Limitation of the study
Additional studies involving testing the OTUB1-SSG/GLRX/xC− paradigm in mouse models and clinical specimens will be required to elucidate the potential impact of our work for tumorigenesis and proinflammatory signaling in vivo. Our study was also limited to the use of cells that express the xC− transporter. Future studies will be required to address whether the GLRX/glutathionylation redox node regulates GSH in cells that do not express the transporter by the aforementioned regulation of other redox-dependent pathways. Studies here were also limited to testing only one of the E2-conjugating enzymes (UBCH5A). Additional analysis will also be required to elucidate the impact of S-glutathionylation of OTUB1 for the inhibition of other E2-conjugating enzymes.
MATERIALS AND METHODS
Recombinant mouse IL-1β/ IL-1F2, TNF-α, and IFN-γ proteins were purchased from R&D Systems. 13−C glucose, SAS, and BSO were obtained from Sigma-Aldrich. Er was purchased from Selleckchem. OTUB1, GCLC, and GS antibodies were obtained from Abcam, while beta-actin (ACTB), glyceraldehyde-3-phosphate dehydrogenase, HA, and Ub antibodies were obtained from Cell Signaling Technology. SLC7A11 antibody was a gift from M. Conrad (Helmholtz Zentrum München, Germany), and GSH antibody was from Virogen.
Cell culture
Primary mouse tracheal epithelial (MTE) cells were isolated from C57BL/6NJ WT or C57BL6/NJ mice lacking the Glrx gene and cultured as previously described (68, 69). Cells were grown on collagen-coated plastic plates or transwell inserts with medium change every 2 days until confluency. For IL-1β treatment, cells were incubated in plain Dulbecco’s modified Eagle’s medium (DMEM):F12 medium containing 6 mM glucose and 2 mM glutamine overnight followed by stimulation with IL-1β (10 ng/ml) or 0.1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) (vehicle control). Cells were treated with IL-1β for 24 hours throughout the whole study unless otherwise stated. Treatment with Er (0.5 μM; Selleckchem), SAS (100 μM; Sigma-Aldrich), BSO (1 mM; Sigma-Aldrich), or ML385 (10 μM; Sigma-Aldrich) occurred 1 hour before IL-1β stimulation. Medium and cells were collected for further analysis. H22 LUAD cells were obtained from American Type Culture Collection (CRL-5810). H522 or OTUB1-CRISPR-H522 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Plasmid overexpression and OTUB1 knockdown experiments were done when cells were 60 to 70% confluent.
Animal studies
KrasG12Dlox-stop-lox(LSL) (B6.129S4-krastm4Tyi/J, 08179) mice were obtained from the Jackson Laboratory (70) and further bred in the animal facility at the University of Vermont. KrasG12D in the lungs of age-matched 7- to 10-week-old mice was activated by the oropharyngeal administration of cytomegalovirus (CMV) AdCre or Ad5 control virus (Ad5; 5 × 107 plaque-forming units, Iowa Vector Core) as a control. Mice were euthanized 15 weeks after the adenovirus installation, and lungs were flash-frozen in liquid nitrogen for biochemical analysis. Animals were maintained in pathogen-free housing with a 12-hour light/dark cycle and ad libitum access to food and water. The University of Vermont Institutional Animal Care and Use Committee (IACUC) approved all animal work (IACUC no. 202000035).
13C-glucose labeling and metabolomics analysis
MTE cells were grown to confluency in six-well transwells as described above. After reaching confluency, cells were incubated overnight in plain DMEM:F12 medium containing 6 mM glucose and 2 mM glutamine. The next day, cells were washed thoroughly in glucose-free medium before stimulating with IL-1β or 0.1% BSA in PBS in medium containing 6 mM of 13C-glucose (Sigma-Aldrich) and 2 mM glutamine. Twenty-four hours after, cells were pelleted, and mass spectrometry–based metabolomics was performed at Metabolomics Core, School of Medicine, University of Colorado, as described here (20, 71–73).
Western blotting and 2D gel analysis
A Bio-Rad detergent compatible (DC) protein estimation kit was used to determine protein concentration in cell lysates. Equal amounts of proteins were resolved using SDS-PAGE and transferred to polyvinylidene difluoride membranes, blocked in 5% BSA, and incubated with primary antibodies overnight. Membranes were subsequently incubated with peroxidase-conjugated secondary antibodies for 1 hour and then visualized using chemiluminescence.
For 2D gels: The first-dimension lysates were run in the same manner as described above with omitting DTT from the lysis buffer. Gel strips were cut and incubated in NuPAGE sample buffer containing 50 mM DTT for 15 min at room temperature. After decanting the solution, gel strips were placed in an alkylating solution containing 50 mM dimethylamine for 15 min. Last, the strips were quenched in a solution containing 5 mM DTT and 20% ethanol before running in the second dimensional SDS-PAGE.
For immunoprecipitation assays: 150 to 250 μg of proteins were used in the pull-down assays. One hour of incubation with 50 mM DTT was used as a negative control for S-glutathionylated immunoprecipitated protein.
Total GSH and PSSG measurement
Cell lysates were prepared in 100 mM potassium phosphate buffer containing 0.6% sulfosalicylic acid, 1 mM EDTA, and 0.01% Triton X-100. To measure total GSH, equal amounts of proteins were incubated with 3 μM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) reagent and GSH reductase (GR; 2.25 μg/ml) followed by the addition of 240 μM NADP+. Kinetic absorbance was measured every minute at 412 nm for 20 min (74). For PSSG measurement, 400 μg of protein was acetone precipitated and washed thoroughly to remove any free GSH. Pellets were reconstituted in 100 mM potassium phosphate buffer with 1 mM EDTA. Samples were then incubated with 1 mM sodium borohydride (NaBH4) or water (negative control) for 1 hour at room temperature. Metaphosphoric acid (10%) was added to all the samples, including controls, and left on ice for 10 min. Samples were centrifuged at 1000g for 15 min, and 20 μl was loaded in a 96-well plate to measure PSSG using DTNB recycling enzyme method as stated above (74).
Glutamate measurement
Glutamate in medium was measured using a BioVision kit following the manufacturer’s protocol.
Calcein AM viability
An Invitrogen Live/Dead viability/cytotoxicity kit was used to measure cell viability according to manufacturer’s instruction (Invitrogen). Data were expressed as percentage survival compared to control untreated cells.
Real-time quantitative polymerase chain reaction
RNeasy mini columns were used to extract total RNA according to the manufacturer’s instruction. First-strand cDNA was synthesized using 0.5 μg of RNA and used for reverse-transcribed gene analysis using SYBR Green. cDNA was further amplified by real-time quantitative polymerase chain reaction Slc7a11 primers. Data were normalized to Actb.
Enzyme-linked immunosorbent assay
TSLP, granulocyte-macrophage colony-stimulating factor, CCL20, and CXCL1 levels in cell culture supernatants were detected using enzyme-linked immunosorbent assay kits according to the manufacturer’s instructions (R&D Systems).
Molecular modeling
Our all-atom models were created from the alignment of two crystal structures of OTUB1, with one containing the charged Ub and UBCH5A [Protein Data Bank ID (PDBID): 2ESK] and another providing the second Ub in the distal site (PDBID: 4I6L). The thioester bond between the Ub and UBCH5A was then created by hand in each model, and GSH was added to the corresponding cysteines via Schrödinger’s Maestro (75). Each model was then minimized and solvated with transferable intermolecular potential 3P (TIP3P) water and neutralized with salt using Schrödinger’s System Builder software (75). After equilibration, each model was run through an initial Desmond (76) relaxation followed by simulation in an NPT ensemble at 300 K for 100 ns with a recording interval of 4.8 ps. Nose-Hoover thermostat and MartynaTobiasKlein (MTK) barostat were combined with a 9-Å cutoff for Coulombic interactions. All minimization, relaxation, and production simulation were done with the OPLS4 force field (77). Visual Molecular Dynamics (VMD) and PyMOL were used for visualization and the generation of all figures (78, 79).
CRISPR-Cas9 generation of OTUB1 knockout cells
CRISPR-Cas9–mediated knockout of OTUB1 in H522 cells was generated as previously described (80, 81). Briefly, three 20–base pair targeting OTUB1 sequences were generated [single-guide RNA 1 (sgRNA1): (forward) CAC CGG GAT GTA CGA GTA CTT TTT G and (reverse) AAA CCA AAA AGT ACT CGT ACA TCC C; sgRNA2: (forward) CAC CGA TCC GCA AGA CCA GGC CTG A and (reverse) AAA CTC AGG CCT GGT CTT GCG GAT C; and sgRNA3: (forward) CAC CGA GGC CAG ACA GTT AAC ACC T and (reverse) AAA CAG GTG TTA ACT GTC TGG CCT C] and transfected in H522 cells followed by flow sorting for positively transfected cells. Only pools of cells that successfully exhibited OTUB1 knockout were propagated and used in experiments.
Plasmids and mutagenesis
Human untagged clone of OTUB1 was purchased from OriGene (catalog no. SC108722), and site-directed mutagenesis was performed with an Agilent QuickChange system using the following primers: for C23S, 5′-catcataggccagactgttaacaccttcggagtcg-3′ and 5′-cgactccgaaggtgttaacagtctggcctatgatg-3′; for C91S, 5′-agcccgatagaaactgttgccgtcaggcc-3′ and 5′-ggcctgacggcaacagtttctatcgggct-3′; for C204S, 5′-cacctcctgctggctgaactccttgacag-3′ and 5′-ctgtcaaggagttcagccagcaggaggtg-3′; and for C212S, 5′-tcgctctccttgctcatgggctccacc-3′ and 5′-ggtggagcccatgagcaaggagagcga-3′. Following site-directed mutagenesis, OTUB1 plasmid mutations were sequence verified, and large-scale plasmid preparations were made using Qiagen Endofree Plasmid Maxi Kits.
Proximity ligation assay
OTUB1 CRISPR H522 cells were platted in glass chamber slides and transfected with SLC7A11-HA in combination with OTUB1-WT or pCMV vector control. A Duolink proximity ligation assay (PLA) kit was used to label the HA tag (for SLC7A11) and SLC3A2 according to the manufacturer’s protocol. Images were obtained using the confocal microscopy with 4′,6-diamidino-2-phenylindole nuclei as a counter stain.
In situ PSSG staining
PSSG staining was performed as described previously (82). Briefly, tissue sections were dewaxed, and free thiols were blocked using N-ethylmaleimide (Thermo Fisher Scientific). GLRX switch–based reaction mix containing GLRX, GR, GSH, NADPH, EDTA, and tris-HCl was then incubated with the tissue for an hour followed by labeling the newly reduced cysteines with 1 mM 3-(N-maleimido-propionyl) biocytin (Sigma-Aldrich). Tissue sections were incubated with a streptavidin-conjugated fluorophore before imaging using the confocal microscopy.
Bioinformatics/TCGA database
All TCGA data including LUAD genotype and RNA sequencing datasets were downloaded from the National Cancer Institute GDC Data Portal (Portal.gdc.cancer.gov). Before and after plots were generated with custom R scripts [R version 4.2.2 (2022-10-31)], while OncoPrint plots were created using the ComplexHeatmap package v.2.14.0.
Statistical analysis
Experiments presented here were repeated at least three times with n = 3 per group (unless otherwise stated). Results are expressed as means ± SEM. Analysis and statistical differences were determined using GraphPad Prism software (Graphpad 8.2.1) using t test (for groups of two) or one-way analysis of variance (ANOVA) with Tukey post hoc correction test for multiple comparisons. TCGA gene expression values in tumor versus adjacent normal are expressed as reads per kilobase of transcript per million mapped reads. Welch two-sample t test analysis was performed using base R [R version 4.2.2 (2022-10-31)]. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We thank A. D’Alessandro and J. A. Reisz for help with the metabolomics study and R. J. Hondal for helpful discussions. We would like to thank M. Conrad for sharing SLC7A11 antibody. The results published here are, in part, based on data generated by the TCGA Research Network: www.cancer.gov/tcga.
Funding: This work was supported by the National Institute of Health (grants R35HL135828 and R01CA273238 to Y.M.W.J.H. and R01CA219156 to J.v.d.V.) and the American Lung Association (no. 909299) (Y.M.W.J.H.).
Author contributions: R.A., J.v.d.V., Z.P., C.E., S.W., and M.M. conducted experiments and designed constructs and cell lines. E.F.M.W., N.L.R., and Y.M.W.J.-H. provided critical scientific suggestions and aided with experimental design. D.S. conducted bioinformatics analysis. N.H. and J.L. conducted molecular modeling studies. R.A. and Y.M.W.J.-H. wrote the manuscript and all authors made edits.
Competing interests: Y.M.W.J.-H. and N.L.R. hold patents: U.S. patent no. 8,679,811, “Treatments involving glutaredoxins and similar agents”; and U.S. patent no. 8,877,447, “Detection of glutathionylated proteins”. Y.M.W.J.-H. also holds patents: U.S. patent nos. 9,907,828 and 10,688,150, “Treatments of oxidative stress conditions,” and has received consulting fees from Celdara Medical LLC for contributions to the proposed commercialization of GLRX for the treatment of pulmonary fibrosis. Y.M.W.J.-H., R.A., J.v.d.V., and J.L. are inventors on the U.S. provisional patent application “Ovarian tumor deubiquitinase oxidation and uses thereof,” reference no. 63/388,203, filed on 11 July 2022. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Code used to generate Oncoprint image (fig. S5) is available at DOI: 10.5281/zenodo.8147215 and at https://github.com/bebojs/Aboushousha-et-al-2023/tree/mainhttps://nam02.safelinks.protection.outlook.com/?url=https%3A%2F%2Fgithub.com%2Fbebojs%2FAboushousha-et-al-2023%2Ftree%2Fmain&data=05%7C01%7Cyvonne.janssen%40med.uvm.edu%7C00971e91835a45be1d3308db77dd1971%7Ced03ff7aba9f420480a6b226316c919d%7C0%7C0%7C638235563717581200%7CUnknown%7CTWFpbGZsb3d8eyJWIjoiMC4wLjAwMDAiLCJQIjoiV2luMzIiLCJBTiI6Ik1haWwiLCJXVCI6Mn0%3D%7C3000%7C%7C%7C&sdata=mmU%2B%2F1xDE9YaV38qtuUweJjCY2ZlBIGk8JJWf7tknDo%3D&reserved=0
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.Janssen-Heininger Y., Reynaert N. L., van der Vliet A., Anathy V., Endoplasmic reticulum stress and glutathione therapeutics in chronic lung diseases. Redox Biol. 33, 101516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssen-Heininger Y. M. W., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A., Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurniawan H., Franchina D. G., Guerra L., Bonetti L., Baguet L. S., Grusdat M., Schlicker L., Hunewald O., Dostert C., Merz M. P., Binsfeld C., Duncan G. S., Farinelle S., Nonnenmacher Y., Haight J., das Gupta D., Ewen A., Taskesen R., Halder R., Chen Y., Jäger C., Ollert M., Wilmes P., Vasiliou V., Harris I. S., Knobbe-Thomsen C. B., Turner J. D., Mak T. W., Lohoff M., Meiser J., Hiller K., Brenner D., Glutathione restricts serine metabolism to preserve regulatory T cell function. Cell Metab. 31, 920–936.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reynaert N. L., van der Vliet A., Guala A. S., McGovern T., Hristova M., Pantano C., Heintz N. H., Heim J., Ho Y. S., Matthews D. E., Wouters E. F. M., Janssen-Heininger Y. M. W., Dynamic redox control of NF-κB through glutaredoxin-regulated S-glutathionylation of inhibitory κB kinase β. Proc. Natl. Acad. Sci. U.S.A. 103, 13086–13091 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anastasiou D., Poulogiannis G., Asara J. M., Boxer M. B., Jiang J. K., Shen M., Bellinger G., Sasaki A. T., Locasale J. W., Auld D. S., Thomas C. J., Vander Heiden M. G., Cantley L. C., Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lien E. C., Lyssiotis C. A., Juvekar A., Hu H., Asara J. M., Cantley L. C., Toker A., Glutathione biosynthesis is a metabolic vulnerability in PI(3)K/Akt-driven breast cancer. Nat. Cell Biol. 18, 572–578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogiwara H., Takahashi K., Sasaki M., Kuroda T., Yoshida H., Watanabe R., Maruyama A., Makinoshima H., Chiwaki F., Sasaki H., Kato T., Okamoto A., Kohno T., Targeting the vulnerability of glutathione metabolism in ARID1A-deficient cancers. Cancer Cell 35, 177–190.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Chia S. B., Elko E. A., Aboushousha R., Manuel A. M., van de Wetering C., Druso J. E., van der Velden J., Seward D. J., Anathy V., Irvin C. G., Lam Y. W., van der Vliet A., Janssen-Heininger Y. M. W., Dysregulation of the glutaredoxin/S-glutathionylation redox axis in lung diseases. Am. J. Physiol. Cell Physiol. 318, C304–C327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeNicola G. M., Chen P.-H., Mullarky E., Sudderth J. A., Hu Z., Wu D., Tang H., Xie Y., Asara J. M., Huffman K. E., Wistuba I. I., Minna J. D., DeBerardinis R. J., Cantley L. C., Erratum: NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet. 48, 473 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Jin H., Wang S., Zaal E. A., Wang C., Wu H., Bosma A., Jochems F., Isima N., Jin G., Lieftink C., Beijersbergen R., Berkers C. R., Qin W., Bernards R., A powerful drug combination strategy targeting glutamine addiction for the treatment of human liver cancer. eLife 9, e56749 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukha A., Kahya U., Linge A., Chen O., Löck S., Lukiyanchuk V., Richter S., Alves T. C., Peitzsch M., Telychko V., Skvortsov S., Negro G., Aschenbrenner B., Skvortsova I.-I., Mirtschink P., Lohaus F., Hölscher T., Neubauer H., Rivandi M., Labitzky V., Lange T., Franken A., Behrens B., Stoecklein N. H., Toma M., Sommer U., Zschaeck S., Rehm M., Eisenhofer G., Schwager C., Abdollahi A., Groeben C., Kunz-Schughart L. A., Baretton G. B., Baumann M., Krause M., Peitzsch C., Dubrovska A., GLS-driven glutamine catabolism contributes to prostate cancer radiosensitivity by regulating the redox state, stemness and ATG5-mediated autophagy. Theranostics 11, 7844–7868 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patra K. C., Hay N., The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu K., Li K., Lv J., Feng J., Chen J., Wu H., Cheng F., Jiang W., Wang J., Pei H., Chiao P. J., Cai Z., Chen Y., Liu M., Pang X., Suppression of the SLC7A11/glutathione axis causes synthetic lethality in KRAS-mutant lung adenocarcinoma. J. Clin. Invest. 130, 1752–1766 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller O. G., Behring J. B., Siedlak S. L., Jiang S., Matsui R., Bachschmid M. M., Zhu X., Mieyal J. J., Upregulation of glutaredoxin-1 activates microglia and promotes neurodegeneration: Implications for parkinson's disease. Antioxid. Redox Signal. 25, 967–982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weinberg E. O., Ferran B., Tsukahara Y., Hatch M. M. S., Han J., Murdoch C. E., Matsui R., IL-33 induction and signaling are controlled by glutaredoxin-1 in mouse macrophages. PLOS ONE 14, e0210827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao D., Han J., Hou X., Fry J., Behring J. B., Seta F., Long M. T., Roy H. K., Cohen R. A., Matsui R., Bachschmid M. M., Glutaredoxin-1 deficiency causes fatty liver and dyslipidemia by inhibiting sirtuin-1. Antioxid. Redox Signal. 27, 313–327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anathy V., Lahue K. G., Chapman D. G., Chia S. B., Casey D. T., Aboushousha R., van der Velden J. L. J., Elko E., Hoffman S. M., McMillan D. H., Jones J. T., Nolin J. D., Abdalla S., Schneider R., Seward D. J., Roberson E. C., Liptak M. D., Cousins M. E., Butnor K. J., Taatjes D. J., Budd R. C., Irvin C. G., Ho Y. S., Hakem R., Brown K. K., Matsui R., Bachschmid M. M., Gomez J. L., Kaminski N., van der Vliet A., Janssen-Heininger Y. M. W., Reducing protein oxidation reverses lung fibrosis. Nat. Med. 24, 1128–1135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelton M. D., Mieyal J. J., Regulation by reversible S-glutathionylation: Molecular targets implicated in inflammatory diseases. Mol. Cells 25, 332–346 (2008). [PMC free article] [PubMed] [Google Scholar]
- 19.Salzano S., Checconi P., Hanschmann E.-M., Lillig C. H., Bowler L. D., Chan P., Vaudry D., Mengozzi M., Coppo L., Sacre S., Atkuri K. R., Sahaf B., Herzenberg L. A., Herzenberg L. A., Mullen L., Ghezzi P., Linkage of inflammation and oxidative stress via release of glutathionylated peroxiredoxin-2, which acts as a danger signal. Proc. Natl. Acad. Sci. U.S.A. 111, 12157–12162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aboushousha R., Elko E., Chia S. B., Manuel A. M., van de Wetering C., van der Velden J., MacPherson M., Erickson C., Reisz J. A., D’Alessandro A., Wouters E. F. M., Reynaert N. L., Lam Y.-W., Anathy V., van der Vliet A., Seward D. J., Janssen-Heininger Y. M. W., Glutathionylation chemistry promotes interleukin-1 beta-mediated glycolytic reprogramming and pro-inflammatory signaling in lung epithelial cells. FASEB J. 35, e21525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maillouxm R. J., Protein S-glutathionylation reactions as a global inhibitor of cell metabolism for the desensitization of hydrogen peroxide signals. Redox Biol. 32, 101472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T., Jiang L., Tavana O., Gu W., The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 79, 1913–1924 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian X., Aboushousha R., van de Wetering C., Chia S. B., Amiel E., Schneider R. W., van der Velden J. L. J., Lahue K. G., Hoagland D. A., Casey D. T., Daphtary N., Ather J. L., Randall M. J., Aliyeva M., Black K. E., Chapman D. G., Lundblad L. K. A., McMillan D. H., Dixon A. E., Anathy V., Irvin C. G., Poynter M. E., Wouters E. F. M., Vacek P. M., Henket M., Schleich F., Louis R., van der Vliet A., Janssen-Heininger Y. M. W., IL-1/inhibitory κB kinase ε-induced glycolysis augment epithelial effector function and promote allergic airways disease. J. Allergy Clin. Immunol. 142, 435–450.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koppula P., Zhang Y., Shi J., Li W., Gan B., The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 292, 14240–14249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q., Harraz M. M., Zhou W., Zhang L. N., Ding W., Zhang Y., Eggleston T., Yeaman C., Banfi B., Engelhardt J. F., Nox2 and Rac1 regulate H2O2-dependent recruitment of TRAF6 to endosomal interleukin-1 receptor complexes. Mol. Cell. Biol. 26, 140–154 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakley F. D., Smith R. L., Engelhardt J. F., Lipid rafts and caveolin-1 coordinate interleukin-1beta (IL-1beta)-dependent activation of NFkappaB by controlling endocytosis of Nox2 and IL-1beta receptor 1 from the plasma membrane. J. Biol. Chem. 284, 33255–33264 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi J., Park J. E., Tsagkogeorga G., Yanagita M., Koo B. K., Han N., Lee J. H., Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill W., Lim E. L., Weeden C. E., Lee C., Augustine M., Chen K., Kuan F.-C., Marongiu F., Evans E. J. Jr., Moore D. A., Rodrigues F. S., Pich O., Bakker B., Cha H., Myers R., van Maldegem F., Boumelha J., Veeriah S., Rowan A., Naceur-Lombardelli C., Karasaki T., Sivakumar M., De S., Caswell D. R., Nagano A., Black J. R. M., Martínez-Ruiz C., Ryu M. H., Huff R. D., Li S., Favé M.-J., Magness A., Suárez-Bonnet A., Priestnall S. L., Lüchtenborg M., Lavelle K., Pethick J., Hardy S., McRonald F. E., Lin M.-H., Troccoli C. I., Ghosh M., Miller Y. E., Merrick D. T., Keith R. L., Al Bakir M., Bailey C., Hill M. S., Saal L. H., Chen Y., George A. M., Abbosh C., Kanu N., Lee S.-H., McGranahan N., Berg C. D., Sasieni P., Houlston R., Turnbull C., Lam S., Awadalla P., Grönroos E., Downward J., Jacks T., Carlsten C., Malanchi I., Hackshaw A., Litchfield K.; TRACERx Consortium, DeGregori J., Jamal-Hanjani M., Swanton C., Lung adenocarcinoma promotion by air pollutants. Nature 616, 159–167 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan B., Clowers M. J., Velasco W. V., Peng S., Peng Q., Shi Y., Ramos-Castaneda M., Zarghooni M., Yang S., Babcock R. L., Chang S. H., Heymach J. V., Zhang J., Ostrin E. J., Watowich S. S., Kadara H., Moghaddam S. J., Targeting IL-1β as an immunopreventive and therapeutic modality for K-ras-mutant lung cancer. JCI Insight 7, e157788 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giustarini D., Milzani A., Dalle-Donne I., Rossi R., How to increase cellular glutathione. Antioxidants (Basel) 12, 1094 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh A., Venkannagari S., Oh K. H., Zhang Y.-Q., Rohde J. M., Liu L., Nimmagadda S., Sudini K., Brimacombe K. R., Gajghate S., Ma J., Wang A., Xu X., Shahane S. A., Xia M., Woo J., Mensah G. A., Wang Z., Ferrer M., Gabrielson E., Li Z., Rastinejad F., Shen M., Boxer M. B., Biswal S., Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem. Biol. 11, 3214–3225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Combs J. A., DeNicola G. M., The non-essential amino acid cysteine becomes essential for tumor proliferation and survival. Cancers (Basel) 11, 678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. III, Stockwell B. R., Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewerenz J., Hewett S. J., Huang Y., Lambros M., Gout P. W., Kalivas P. W., Massie A., Smolders I., Methner A., Pergande M., Smith S. B., Ganapathy V., Maher P., The cystine/glutamate antiporter system x(c)(−) in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 18, 522–555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van de Wetering C., Manuel A. M., Sharafi M., Aboushousha R., Qian X., Erickson C., MacPherson M., Chan G., Adcock I. M., ZounematKermani N., Schleich F., Louis R., Bohrnsen E., D'Alessandro A., Wouters E. F., Reynaert N. L., Li J., Wolf C. R., Henderson C. J., Lundblad L. K. A., Poynter M. E., Dixon A. E., Irvin C. G., van der Vliet A., van der Velden J. L., Janssen-Heininger Y. M., Glutathione-S-transferase P promotes glycolysis in asthma in association with oxidation of pyruvate kinase M2. Redox Biol. 47, 102160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohr S., Hallak H., de Boitte A., Lapetina E. G., Brüne B., Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 274, 9427–9430 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Velu C. S., Niture S. K., Doneanu C. E., Pattabiraman N., Srivenugopal K. S., Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry 46, 7765–7780 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cha S. J., Kim H., Choi H.-J., Lee S., Kim K., Protein glutathionylation in the pathogenesis of neurodegenerative diseases. Oxid. Med. Cell. Longev. 2017, 2017, 2818565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon S. J., Patel D. N., Welsch M., Skouta R., Lee E. D., Hayano M., Thomas A. G., Gleason C. E., Tatonetti N. P., Slusher B. S., Stockwell B. R., Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 3, e02523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris I. S., Endress J. E., Coloff J. L., Selfors L. M., McBrayer S. K., Rosenbluth J. M., Takahashi N., Dhakal S., Koduri V., Oser M. G., Schauer N. J., Doherty L. M., Hong A. L., Kang Y. P., Younger S. T., Doench J. G., Hahn W. C., Buhrlage S. J., DeNicola G. M., Kaelin W. G. Jr., Brugge J. S., Deubiquitinases maintain protein homeostasis and survival of cancer cells upon glutathione depletion. Cell Metab. 29, 1166–81.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edelmann M. J., Iphöfer A., Akutsu M., Altun M., di Gleria K., Kramer H. B., Fiebiger E., Dhe-Paganon S., Kessler B. M., Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem. J. 418, 379–390 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Balakirev M. Y., Tcherniuk S. O., Jaquinod M., Chroboczek J., Otubains: A new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4, 517–522 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S.-I., Juang Y.-C., O’Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A.-C., Natsume T., Suda T., Durocher D., Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Markson G., Kiel C., Hyde R., Brown S., Charalabous P., Bremm A., Semple J., Woodsmith J., Duley S., Salehi-Ashtiani K., Vidal M., Komander D., Serrano L., Lehner P., Sanderson C. M., Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res. 19, 1905–1911 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiener R., DiBello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., Wolberger C., E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun X. X., Challagundla K. B., Dai M. S., Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 31, 576–592 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juang Y. C., Landry M. C., Sanches M., Vittal V., Leung C. C. Y., Ceccarelli D. F., Mateo A. R. F., Pruneda J. N., Mao D. Y. L., Szilard R. K., Orlicky S., Munro M., Brzovic P. S., Klevit R. E., Sicheri F., Durocher D., OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim J. K. M., Delaidelli A., Minaker S. W., Zhang H. F., Colovic M., Yang H., Negri G. L., von Karstedt S., Lockwood W. W., Schaffer P., Leprivier G., Sorensen P. H., Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc. Natl. Acad. Sci. U.S.A. 116, 9433–9442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badgley M. A., Kremer D. M., Maurer H. C., DelGiorno K. E., Lee H.-J., Purohit V., Sagalovskiy I. R., Ma A., Kapilian J., Firl C. E. M., Decker A. R., Sastra S. A., Palermo C. F., Andrade L. R., Sajjakulnukit P., Zhang L., Tolstyka Z. P., Hirschhorn T., Lamb C., Liu T., Gu W., Seeley E. S., Stone E., Georgiou G., Manor U., Iuga A., Wahl G. M., Stockwell B. R., Lyssiotis C. A., Olive K. P., Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science 368, 85–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baietti M. F., Simicek M., Abbasi Asbagh L., Radaelli E., Lievens S., Crowther J., Steklov M., Aushev V. N., Martínez García D., Tavernier J., Sablina A. A., OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol. Med. 8, 288–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nulton-Persson A. C., Starke D. W., Mieyal J. J., Szweda L. I., Reversible inactivation of alpha-ketoglutarate dehydrogenase in response to alterations in the mitochondrial glutathione status. Biochemistry 42, 4235–4242 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Anathy V., Aesif S. W., Guala A. S., Havermans M., Reynaert N. L., Ho Y. S., Budd R. C., Janssen-Heininger Y. M. W., Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J. Cell Biol. 184, 241–252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sánchez G., Pedrozo Z., Domenech R. J., Hidalgo C., Donoso P., Tachycardia increases NADPH oxidase activity and RyR2 S-glutathionylation in ventricular muscle. J. Mol. Cell. Cardiol. 39, 982–991 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Stewart M. D., Ritterhoff T., Klevit R. E., Brzovic P. S., E2 enzymes: More than just middle men. Cell Res. 26, 423–440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jahngen-Hodge J., Obin M. S., Gong X., Shang F., Nowell T. R. Jr., Gong J., Abasi H., Blumberg J., Taylor A., Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 272, 28218–28226 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Karunarathna U., Kongsema M., Zona S., Gong C., Cabrera E., Gomes A. R., Man E. P. S., Khongkow P., Tsang J. W. H., Khoo U. S., Medema R. H., Freire R., Lam E. W. F., OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene 35, 1433–1444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun X. X., Dai M. S., Deubiquitinating enzyme regulation of the p53 pathway: A lesson from Otub1. World J. Biol. Chem. 5, 75–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y., Wang Y. G., Li Y., Sun X. X., Dai M. S., Otub1 stabilizes MDMX and promotes its proapoptotic function at the mitochondria. Oncotarget 8, 11053–11062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y., Shi J., Liu X., Feng L., Gong Z., Koppula P., Sirohi K., Li X., Wei Y., Lee H., Zhuang L., Chen G., Xiao Z. D., Hung M. C., Chen J., Huang P., Li W., Gan B., BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat. Cell Biol. 20, 1181–1192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niu W. N., Yadav P. K., Adamec J., Banerjee R., S-glutathionylation enhances human cystathionine β-synthase activity under oxidative stress conditions. Antioxid. Redox Signal. 22, 350–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu D. S., Duong C. P., Haupt S., Montgomery K. G., House C. M., Azar W. J., Pearson H. B., Fisher O. M., Read M., Guerra G. R., Haupt Y., Cullinane C., Wiman K. G., Abrahmsen L., Phillips W. A., Clemons N. J., Inhibiting the system xC−/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat. Commun. 8, 14844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carvalho A. N., Marques C., Guedes R. C., Castro-Caldas M., Rodrigues E., van Horssen J., Gama M. J., S-Glutathionylation of Keap1: A new role for glutathione S-transferase pi in neuronal protection. FEBS Lett. 590, 1455–1466 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Liu X., Olszewski K., Zhang Y., Lim E. W., Shi J., Zhang X., Zhang J., Lee H., Koppula P., Lei G., Zhuang L., You M. J., Fang B., Li W., Metallo C. M., Poyurovsky M. V., Gan B., Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 22, 476–486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ridker P. M., MacFadyen J. G., Thuren T., Everett B. M., Libby P., Glynn R. J.; CANTOS Trial Group , Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: Exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 390, 1833–1842 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Zhang H.-F., Hughes C. S., Li W., He J.-Z., Surdez D., El-Naggar A. M., Cheng H., Prudova A., Delaidelli A., Negri G. L., Li X., Ørum-Madsen M. S., Lizardo M. M., Oo H. Z., Colborne S., Shyp T., Scopim-Ribeiro R., Hammond C. A., Dhez A.-C., Langman S., Lim J. K. M., Kung S. H. Y., Li A., Steino A., Daugaard M., Parker S. J., Geltink R. I. K., Orentas R. J., Xu L.-Y., Morin G. B., Delattre O., Dimitrov D. S., Sorensen P. H., Proteomic screens for suppressors of anoikis identify IL1RAP as a promising surface target in Ewing sarcoma. Cancer Discov. 11, 2884–2903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyoshi S., Tsugawa H., Matsuzaki J., Hirata K., Mori H., Saya H., Kanai T., Suzuki H., Inhibiting xCT improves 5-fluorouracil resistance of gastric cancer induced by CD44 variant 9 expression. Anticancer Res. 38, 6163–6170 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Wu R., Smith D., Continuous multiplication of rabbit tracheal epithelial cells in a defined, hormone-supplemented medium. In Vitro 18, 800–812 (1982). [DOI] [PubMed] [Google Scholar]
- 69.Alcorn J. F., Guala A. S., van der Velden J., McElhinney B., Irvin C. G., Davis R. J., Janssen-Heininger Y. M. W., Jun N-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-beta1. J. Cell Sci. 121, 1036–1045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jackson E. L., Willis N., Mercer K., Bronson R. T., Crowley D., Montoya R., Jacks T., Tuveson D. A., Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nemkov T., Reisz J. A., Gehrke S., Hansen K. C., D’Alessandro A., High-throughput metabolomics: Isocratic and gradient mass spectrometry-based methods. Methods Mol. Biol. 1978, 13–26 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Nemkov T., D’Alessandro A., Hansen K. C., Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids 47, 2345–2357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nemkov T., Hansen K. C., D'Alessandro A., A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun. Mass Spectrom. 31, 663–673 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rahman I., Kode A., Biswas S. K., Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Schrödinger Release 2023-1: Maestro S, LLC, New York, NY, 2021.
- 76.Schrödinger Release 2023-1: Desmond Molecular Dynamics System DESR, New York, NY, 2021. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY, 2021.
- 77.Lu C., Wu C., Ghoreishi D., Chen W., Wang L., Damm W., Ross G. A., Dahlgren M. K., Russell E., Von Bargen C. D., Abel R., Friesner R. A., Harder E. D., OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 17, 4291–4300 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 79.The PyMOL Molecular Graphics System VS, LLC.
- 80.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giuliano C. J., Lin A., Girish V., Sheltzer J. M., Generating single cell-derived knockout clones in mammalian cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol. 128, e100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aesif S. W., Anathy V., Havermans M., Guala A. S., Ckless K., Taatjes D. J., Janssen-Heininger Y. M. W., In situ analysis of protein S-glutathionylation in lung tissue using glutaredoxin-1-catalyzed cysteine derivatization. Am. J. Pathol. 175, 36–45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5