Abstract
Metastasis and chemoresistance in colorectal cancer are mediated by certain poorly differentiated cancer cells, known as cancer stem cells, that are maintained by Notch downstream signaling initiated upon Notch cleavage by the metalloprotease ADAM10. It has been shown that ADAM10 overexpression correlates with aberrant signaling from Notch, erbBs, and other receptors, as well as a more aggressive metastatic phenotype, in a range of cancers including colon, gastric, prostate, breast, ovarian, uterine, and leukemia. ADAM10 inhibition, therefore, stands out as an important and new approach to deter the progression of advanced CRC. For targeting the ADAM10 substrate-binding region, which is located outside of the catalytic domain of the protease, we generated a human anti-ADAM10 monoclonal antibody named 1H5. Structural and functional characterization of 1H5 reveals that it binds to the substrate-binding cysteine-rich domain and recognizes an activated ADAM10 conformation present on tumor cells. The mAb inhibits Notch cleavage and proliferation of colon cancer cell lines in vitro and in mouse models. Consistent with its binding to activated ADAM10, the mAb augments the catalytic activity of ADAM10 towards small peptide substrates in vitro. Most importantly, in a mouse model of colon cancer, when administered in combination with the therapeutic agent Irinotecan, 1H5 causes highly effective tumor growth inhibition without any discernible toxicity effects. Our singular approach to target the ADAM10 substrate-binding region with therapeutic antibodies could overcome the shortcomings of previous intervention strategies of targeting the protease active site with small molecule inhibitors that exhibit musculoskeletal toxicity.
Keywords: ADAM10, Monoclonal antibody, Notch, COLO205, Colorectal cancer, Chemotherapy, Xenograft
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the USA. A subgroup of cancer cells with stem cell-like properties, known as cancer stem cells (CSCs) initiate and sustain tumor growth and promote metastasis and chemoresistance in CRC [1]. A key determinant of the CSC phenotype is activation of Notch receptor signaling. Ligand-activated Notch signaling involves sequential cleavage of the extracellular and intracellular domains by ADAM10 metalloprotease (functioning as alpha-secretase) and γ-secretase activity, respectively, to modulate downstream transcription of target genes [2]. In addition to Notch receptor signaling, EGFR/erbB signaling that is essential for the development and metastasis of CRC, is also dependent on ADAM10 activity [3]. Notch signaling, and CSCs are also associated with drug resistance, and inhibition of Notch signaling is widely reported to increase sensitivity to both chemo- and targeted therapies [4-9]. However, targeting Notch using pan-specific γ-secretase inhibitors [10] is hampered by intestinal toxicity, reflecting the diversity of γ-secretase targets [11]. Nevertheless, Notch-specific inhibitory monoclonal antibodies (mAbs), avoiding this toxicity, have validated Notch inhibition as a promising anti-cancer therapeutic approach [12].
The ADAM (‘A Disintegrin And Metalloprotease’) proteases catalyze the release of cell surface proteins implicated in Notch, erbB, cytokine, chemokine and adhesion receptor signaling, thereby activating key oncogenic pathways, and are recognized targets for therapeutic intervention [2,3]. ADAMs are transmembrane proteins with an N-terminal pro-domain, followed by metalloprotease (M), disintegrin (D), cysteine-rich (C), transmembrane and cytoplasmic domains [13]. Their proteolytic specificity does not simply rely on a typical substrate cleavage signature, but also on non-catalytic interactions of substrates with the ADAM C domain to align the substrate and the protease domain for effective cleavage [14-16]. ADAM10 is principally involved in activation of Notch [17], and Eph [14,16] receptor signaling, while both ADAM10 and its close relative ADAM17 (TNFα-activating enzyme, or TACE) activate erbB/EGFR receptors via shedding of their ligands with differing specificities [3]. Previous attempts to deter oncogenic pathways by developing small molecules inhibitors targeting the protease active site of ADAM10 and ADAM17 failed due to musculoskeletal toxicity thought to be from inhibiting certain matrix metalloproteinases (MMPs) [18,19]. Studies developing more specific antibody-based inhibitors have been limited to ADAM17, confirming effects on EGFR/erbB signaling and cell proliferation in vitro [20,21] and in vivo [22], while prior to our work, no inhibitory/therapeutic ADAM10 mAb has been developed to the best of our knowledge. We previously identified the ligand-recognition region in ADAM10, (D+C domain region) and generated murine monoclonal antibodies (mAbs) against it [23]. One of the most potent anti-ADAM10 mAb, 8C7, inhibited Notch activity, as well as reduced the shedding of the erbB2 ectodomain and the expression of multiple cell-surface receptors, in mouse models of CRC, without any evidence of non-target toxicity. Remarkably, 8C7 selectively targeted CSCs by recognizing an active form of ADAM10, preferentially present in tumors as compared to normal tissue [24]. Consistent with its specificity, 8C7 was most effective in inhibiting tumor relapse when combined with chemotherapy [24]. Here we report the generation and characterization of a fully human anti-ADAM10 monoclonal antibody, 1H5, that binds to a similar ADAM10 epitope, preferentially recognizes ADAM10 on cancer cells, and inhibits Notch cleavage and proliferation of cancer cells in cell-based assays. Most importantly, in preclinical studies with the colon cancer cell line COLO205, 1H5 causes over 80 % tumor growth inhibition in combination with the chemotherapeutic agent, Irinotecan.
2. Materials and methods
Cell lines:
The triple-negative breast cancer (TNBC) cell line MDA-MB-231 was cultured in Dulbecco’s Modified Eagle Medium (DMEM), 10 % Fetal Bovine Serum (FBS), 1 % Penicillin/Streptomycin (P/S) and 2 mM L-Glutamine whereas SKBR-3 was grown in McCoy’s 5a, 10 % FBS and 1 % P/S. The ovarian carcinoma cell line OVCAR-3 was cultured in RPMI-1640, 10 % FBS, 1 % P/S, 10 mM HEPES and 0.2 units/ml Insulin while SKOV-3 was grown in DMEM, 10 % FBS and 1 % P/S. The lung adenocarcinoma cell line HCC-827 and the COLO205 cell line (made up of epithelial cells from ascitic fluid derived from a 70-year-old, Caucasian man with colon cancer), were cultured in RPMI-1640, 10 % FBS and 1 % P/S. The glioma cell line U-87 MG was propagated in Eagle’s Minimum Essential Medium, 10 % FBS and 1 % P/S All these cell lines described above were purchased from American Type Culture Collection (ATCC). The LIM1215 colon cancer cell line [25], was purchased from Cell Bank Australia and cultured in RPMI1640 with 2 mM L-Glutamine, 25 mM HEPES, 10 % FBS, 0.6 μg/ml Insulin, 1 μg/ml Hydrocortisone and 10 μM Thiolglycerol. The cell lines were regularly tested for mycoplasma contamination and subcultured according to the instruction manual.
Expression and purification of the ADAM constructs:
The human ADAM10 cDNA and the human ADAM19 cDNA were purchased from GenScript. The bovine ADAM10 cDNA and the human ADAM17 cDNA were gifts from Dr. Carl Blobel, Hospital for Special Surgery, New York. The bovine ADAM10 extracellular domain, ECD, (20–646) as well as the D+C domains (455–646) were cloned, expressed, and purified from HEK293 cells using a custom made pcDNA™ 3.1+ vector [14]. The constructs were fused to a removable Fc-tag at the C-terminus. The C-terminal Fc-tag was used to facilitate protein-A affinity chromatography and removed by thrombin cleavage afterwards. Likewise, we purified the human ADAM19 D+C construct (438–646) using this mammalian expression system. The human ADAM10 ECD (20–650), human ADAM10 D+C (455–650), human ADAM17 ECD (20–655) and the human ADAM17 D+C (482–648) were cloned into a custom-made pMA152a baculovirus vector [26]. pMA152a is based on the pAcGP67B vector (BD Biosciences) with an incorporated removable Fc-tag (human). Secreted recombinant proteins were produced by baculovirus-infected Hi5 insect cell following the protocol provided by BD Biosciences. The proteins were purified from the culture supernatants of the insect cells using protein-A. Size-exclusion chromatography was performed as the final step to obtain all the purified recombinant proteins described here.
Generation of 1H5:
The bovine ADAM10 D+C was used as an antigen to pan a large naïve human Fab library. For panning, two ICAT5 and ICAT5–1 phage libraries were pre-blocked with 3 % skim-milk in PBS (w/v) for 1 h at 25 °C. Blocked phages were incubated with 100 nM biotinylated human ADAM10 D+C for 1 h at 25 °C. Bound phages were separated by streptavidin coated magnetic beads (Invitrogen, 11–205-D) and washed 10 times with 1 ml of PBS pH 7.4 containing 0.1 % Tween-20 (w/v). Elution of bound phages was conducted by adding 1 μM of non-biotinylated antigen or 8C7 for O/N at 4°C. For 2nd and 3rd rounds of panning, reduced concentration (50 nM and 5 nM respectively) of biotinylated ADAM10 D+C was applied to pan out high-affinity Fab binders. After 3 rounds of panning, binding of 192 individual clones were analyzed in ELISA, and the selected clones after rescue of pCAT2 plasmid DNA, were sequenced. 1H5 Fab and IgG were purified as described previously [27].
Competitive ELISA to gauge relative bindings of 1H5 (human mAb) and 8C7 (murine mAb) to immobilized ADAM10 D+C domains construct (antigen):
The wells were coated with 100 ul of ADAM10 D+C (concentration 2 ug/ml). After three washes with PBS, the wells were blocked with 4 % non-fat dry milk. Varying concentrations of 1H5 (human) or 8C7 (murine) mAbs were added to the wells and incubated for 1 hr. Goat anti-human IgG cross-adsorbed secondary antibody conjugated to HRP, Invitrogen, (1:2000 dilution) was used to detect the binding of 1H5 to immobilized ADAM10 D+C antigen. Rabbit anti-mouse IgG cross-adsorbed secondary antibody conjugated to HRP, Invitrogen, (1:2000 dilution) was used to detect binding of 8C7 to ADAM10 D+C. Cross-adsorption ensured that the secondary antibodies maintained their desired species reactivity as shown in the graph (1H5/anti-murine secondary or 8C7/anti-human secondary). Color was developed using the TMB substrate kit (Thermo Scientific) and data was recorded at 450 nm. For the competition assay, we used a mixture of 1H5 and 8C7 at a 1:1 molar ratio. The final concentration of each mAb in the mixture was the same as when added individually. The mixture was added to the wells and incubated for 1 hr. Relative binding of 1H5 or 8C7 (in the mixture) to ADAM10 D+C was detected using goat anti-human IgG or rabbit anti-mouse IgG secondary (cross-adsorbed, 1:2000 dilution) conjugated to HRP.
Alamar Blue cell viability assay:
The cancer cells (colon/breast/ovarian/glioma/lung-adenocarcinoma) were harvested in the log phase of growth (after 3 days of culturing), adjusted to 5 × 104 cells/ml, and allowed to adhere and grow for 24 h in 96-well cell culture plates. The cells were then treated with test agent, in this case, the human anti-ADAM10 mAb 1H5 and allowed to grow for additional 38 h. Cells not treated with the mAb were used as a control. Alamar Blue (Bio-RAD Laboratories) (10 % of the well volume) was added aseptically. Cultures containing Alamar Blue were incubated for 6 hrs and cell proliferation was measured spectrophotometrically by absorbance at 570 and 600 nm. Cell viability was calculated using the following formula:
Percentage difference between treated and control cells.
= molar extinction coefficient (E) of oxidized alamarBlue® (Blue) at 570 nm, = E of oxidized alamarBlue® at 600 nm, = absorbance of test wells at 570 nm, = absorbance of test wells at 600 nm, = absorbance of positive growth control well (cells plus alamarBlue® but no test agent) at 570 nm, = absorbance of positive growth control well (cells plus alamarBlue® but no test agent) at 600 nm [28].
Sandwich ELISA to quantitate total Notch1 and NICD1 (Notch intracellular domain) in lysates of COLO205 cells untreated and treated with the human mAb 1H5:
The COLO205 cells were harvested in the log phase of growth (after 3 days of culturing), adjusted to 5 × 104 cells/ml, and allowed to adhere and grow for 24 h in 6-well cell culture plates (Greiner bio-one). The cells were treated with 5 μg/ml of 1H5 mAb and harvested after 10, 18 and 36 h of treatment. We used the PathScan® total Notch1 sandwich ELISA kit to measure total Notch1 according to the manufacturer’s protocol (Cell Signaling Technologies). Briefly, the cells harvested at different time points were resuspended in 1X lysis buffer containing 1 mM PMSF, sonicated on ice and centrifuged for 10 min (x14,000 rpm) at 4 °C. The supernatant, which is the cell lysate was used for further study. We added 100 μl of the lysates (diluted to 1 mg/ml) from the different time intervals to the microwells (in quadruplicate) coated with a Notch1 rat antibody. This antibody captures the total Notch1 (cleaved plus uncleaved) from the lysates. After extensive washing, a Notch1 rabbit antibody was added to detect the total captured Notch1 protein. To detect NICD1, we used PathScan® cleaved Notch1 (Val1744) sandwich ELISA Kit (Cell Signaling Technologies). Here, a cleaved-Notch1 rabbit detection antibody (Val1744) was used instead, to detect endogenous levels of Notch1 that is cleaved at Val1744. Anti-rabbit IgG, HRP-linked antibody was used in both cases to recognize the bound detection antibody. Color was developed using the TMB substrate. The data was recorded at 450 nm. Lysates from untreated cells served as controls. Comparison of Notch levels between treated and untreated groups was performed using independent t test. Total notch levels did not significantly differ between the two groups (A), p = 0.162. On the other hand, the mAb-treated group showed significant decrease of the cleaved notch levels when compared with the untreated control (B), p < 0.001. This demonstrates the unique capacity of the mAb to selectively inhibit notch cleavage.
In vivo antitumor efficacy studies:
The COLO205 cells were grown in monolayer culture, harvested by trypsinization, and implanted subcutaneously into the right flank of 6- to 8-week-old female athymic nude mice (n = 4). Approximately 10 million cells were injected per mouse. Mice were randomized into 4 mice per group (4 groups). When tumors reached 100–150 mm3, the anti-ADAM10 mAb 1H5, Irinotecan were injected intraperitoneally. Sterile PBS was used as a control. Tumor volume was determined by external caliper and calculated by the modified ellipsoidal formula: V = ½ (Length × Width2). Antitumor efficacy was calculated as (1-dT/dC) X100, where dT is the final tumor volume minus the starting tumor volume from the treatment group and dC is the final tumor volume minus the starting tumor volume of the control group [29]. Error bars were calculated as SEM. The mouse body weight and general health were monitored daily, and the experiments were carried out in accordance with Association for Assessment and Accreditation of Laboratory Animal Care and MSKCC Institutional Animal Care and Use Committee guidelines.
Cell-based ELISA assays to measure the binding of the anti-ADAM10 mAbs 1H5 and mAb1427 to cancer cell lines:
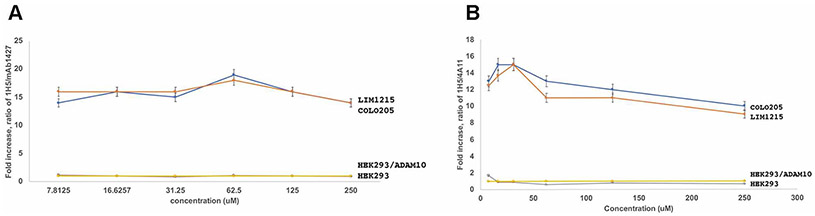
Cellular ELISA [30] was performed to measure the binding of the anti-ADAM10 human mAb 1H5, relative to the binding of the commercial anti-ADAM10 mAb1427 (monoclonal mouse IgG2B Clone # 163003, R&D), to ADAM10 expressed on the cell surface of colon cancer cell lines LIM1215 and COLO205 as well as to HEK293 cells and HEK293 cells transfected with full-length human ADAM10. Briefly, 5 × 104 cells/well were immobilized on 96-well ELISA plates (Greiner bio-one) with 1 % paraformaldehyde for 2hrs at 37° C. The plate was washed thrice with PBS and blocked for 2 hrs at room temperature with 4 % non-fat dry milk. The anti-ADAM10 mAbs were then added in varying concentrations. Mouse mAb conjugated to HRP and recognizing human IgG was used as a secondary antibody (Abcam) to detect 1H5 binding to ADAM10 expressed on the cells. For the commercial mAb1427 recognizing human ADAM10, we used goat anti-mouse IgG (H+L) secondary antibody conjugated to HRP (Thermofischer Scientific). Color was developed using the TMB substrate kit (Thermo Scientific). The data was recorded at 450 nm.
Fluorescent Peptide Cleavage Assay:
The purified anti-ADAM10 mAbs 1H5, 8C7 as well as the commercial anti-ADAM10 mAb1427 (R&D), were buffer-exchanged into 25 mM Tris, pH 9.0, containing 2 μM ZnCl2, and 0.005 % (w/v) Brij-35. ADAM10 ECD-antibody or ADAM10 ECD-inhibitor (GM6001) complexes were formed at a 1:1 molar ratio prior to the assay. The assay was carried out by mixing 50 μM of a fluorogenic peptide substrate Mca-PLAQAV-Dpa (R&D Systems Cat# ES003) with ADAMlO-antibody/inhibitor complexes at 37°C and monitoring the progress of the enzymatic reaction by fluorescence emission (excitation 320 nm and emission 405 nm) over a time course of 1 h using a SpectraMax M5. ADAM10 ECD (bovine or human) alone was used as positive control. The substrate peptide contains a highly fluorescent 7-methoxycoumarin group and a quencher group, 2,4-dinitrophenyl. ADAM10 cleaves the amide bond between the fluorescent and the quencher group causing an increase in fluorescence (R&D Systems, Cat Number ES003) [31,32]. The peptide was derived from TNFalpha.
Statistical Analysis:
The data from all in vitro and cell-based assays described, including Alamar blue, fluorescent peptide cleavage and ELISA-based assays, are representative of triplicate determinations and two independent experiments. Statistical analysis was performed using IBM SPSS version 29. P values were calculated using either one-way ANOVA with a Dunnett’s multiple comparison post hoc test, or one- or two-tailed independent t test analysis as indicated on each figure. A p value of < 0.05 was considered significant.
Crystallization and structure determination:
A bovine ADAM10 fragment containing the disintegrin and cysteine-rich domains (ADAM10(D+C), residues 455–646) was produced as described previously. [14] The 1H5 Fab fragment was prepared by digesting 1H5 with papain at pH 6.5 (enzyme/substrate ratio 1:100) for 2 h at room temperature. The final purification was performed using gel filtration chromatography (SD-200 column, 20 mM Hepes, and 150 mM NaCl, pH 7.5). The protein eluted as a monomer of ~50 kDa. For crystallization, ADAM10 (D+C) was mixed with the 1H5 Fab at 1:1 molar ratio (final concentration 22 mg/ml) in a buffer containing 20 mM HEPES, 150 mM NaCl, pH 7.4. The complex was crystallized in a hanging drop by vapor diffusion at room temperature against a reservoir containing 0.1 M BICINE, pH 8.5, 20 % PEG10,000. The initial thin plate like crystals were optimized using streak seeding. Sizeable crystals, in the space group P21221, were obtained and data was collected at Argonne National Laboratory, Chicago. The structure was determined using molecular replacement with the ADAM10(D+C)/8C7-F(ab′)2 structure as a search model (PDBID 5L0Q). The ADAM10/1H5 structure model was built with the program Coot and refined with PHENIX_Refine. The final structure was validated with PROCHECK. The accession number for the deposited structure is PDB 8GH4.
3. Results
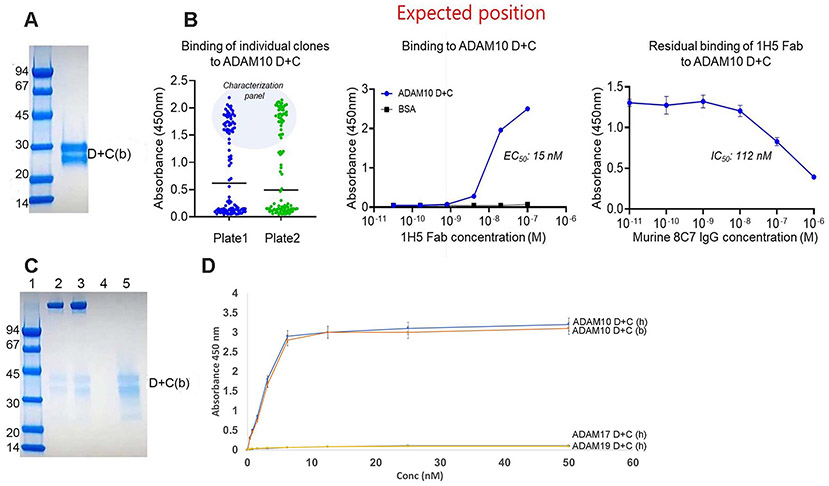
Generation of a fully human anti-ADAM10 mAb. We used a recombinant bovine ADAM10 D+C domain construct (ADAM10(D+C), Fig. 1A) in a phage displayed Fab library screen to identify an ADAM10 specific fully human antibody. We conducted three rounds of phage panning with ICAT5 and 5–1 Fab libraries constructed previously [27] and identified a dominant clone named 1H5 after 192 single clone analysis (Fig. 1B). After purification of 1H5 Fab, we confirmed that it bound to ADAM10(D+C) in a concentration dependent manner. In addition, competitive ELISA experiments with the murine 8C7 IgG demonstrated that 1H5 binds a similar epitope region in ADAM10 (Fig. 1B). Additional ELISA experiments (Fig. S1) document that both 1H5 and 8C7 efficiently bind to ADAM10 D+C when added alone, and that 1H5 outcompetes 8C7 when added at a 1:1 molar ratio mixture. Using cross-adsorbed secondary antibodies, we ascertained that there were no cross-species reactivity as evidenced from the 1H5/anti-murine secondary or 8C7/anti-human secondary plots.
Fig. 1.
(A) SDS-PAGE profile of the purified bovine (b) ADAM10 disintegrin + cysteine-rich domain construct (D+C), used as an antigen to screen a large naïve human Fab library. (b) indicates bovine protein, (h) indicates human protein. (B) Characterization of fully human 1H5 Fab. Binding profile of individual Fab binders (left) and binding of 1H5 Fab (middle) to ADAM10 D+C. Competitive ELISA with the murine 8C7 antibody demonstrating that 1H5 Fab (15 nM fixed concentration) binds to a similar ADAM10 epitope (right). (C) Pull-down experiment showing that ADAM10 D+C (b) binds to protein A Sepharose bead-bound murine 8C7 and human 1H5. Lane 1: Low molecular weight standards (Bio-Rad). Lane 2: bead bound murine 8C7 IgG + ADAM10 D+C. Lane 3: bead bound human 1H5 IgG + ADAM10 D+C. Lane 4: bead bound ADAM10 D+C without any pre-bound mAb. Lane 5: ADAM10 D+C input. (D) Human 1H5 IgG binds specifically to immobilized human (h) and bovine (b) ADAM10 D+C, but not to human ADAM17 D+C or human ADAM19 D+C. Wells were coated with the ADAM D+C constructs and ELISA was performed as described in the Materials and Methods.
1H5 inhibits proliferation of colon cancer cell lines in Alamar Blue assays:
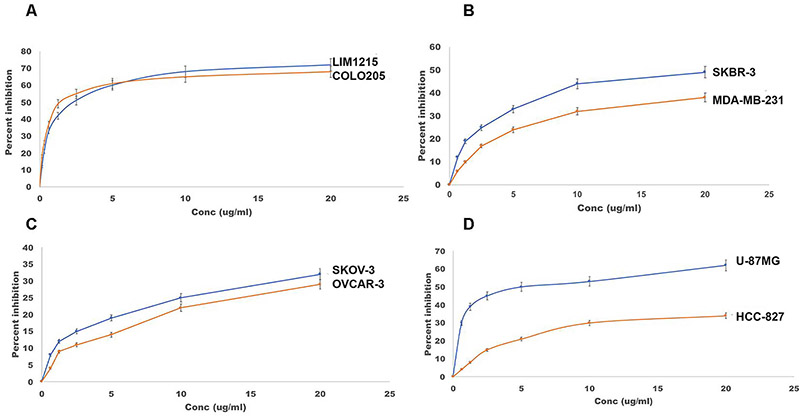
As ADAM10 is thought to control cell proliferation via activation of Notch and EGFR/HER2 signalling [2,24], we performed Alamar blue cell viability assays to evaluate the anti-proliferative potential of the fully human anti-ADAM10 1H5 mAb using a variety of cancer cell lines that are either Notch or EGFR/HER2 dependent. These included colon, breast, ovarian, glioma and lung adenocarcinoma (non-small cell lung cancer or NSCLC). The colon cancer cell lines LIM1215 and COLO205 display high levels of Notch receptors [33]. The triple-negative breast cancer cell line MDA-MB-231 has high EGFR expression, while SKBR-3 overexpresses HER2 [34]. The epithelial ovarian cancer cell line OVCAR-3 represent high-grade serous carcinoma (HGSC) type, while SKOV-3 belongs to the non-HGSC type [35]. Notch1 is known to be activated in glioblastoma and it has been shown that targeting Notch1 suppressed growth and proliferation of U-87 MG in vitro and in xenograft models [36]. EGFR signaling plays a crucial role in non-small cell lung cancer (NSCLC) occurrence and progression and is increased in over 45 % of the tumor lesions from NSCLC patients [37]. Our objective was to evaluate if the 1H5 mAb can inhibit the proliferation of these cancer cell lines. The Alamar blue assays show that 1H5 is more efficient in inhibiting Notch-dependent cancer cell lines, including LIM1215, COLO205 and U-87MG (60–70 % inhibition at 20 μg/ml), as compared to EGFR/HER2 dependent lines, such as MDA-MB-231, SKBR-3, OVCAR-3, SKOV-3 or HCC-827 (25–45 % inhibition at 20 μg/ml, Fig. 2).
Fig. 2.
Alamar blue cell viability assays with multiple cancer cell lines. Percent growth inhibition is shown for (A) colon (COLO205, LIM1215), (B) breast (MDA-MB-231, SKBR-3), (C) ovarian (SKOV-3, OVCAR-3) and (D) glioblastoma (U87-MG) and non-small cell lung cancer (HCC-827) tumor cell lines treated with human 1H5. The data represent mean of triplicate determinations and two independent experiments, and the bar plots show the effect of treatment of mAbs on cancer cells relative to the control, mean ± SEM (described in Materials and Methods).
1H5 inhibits Notch cleavage in COLO205 cells:
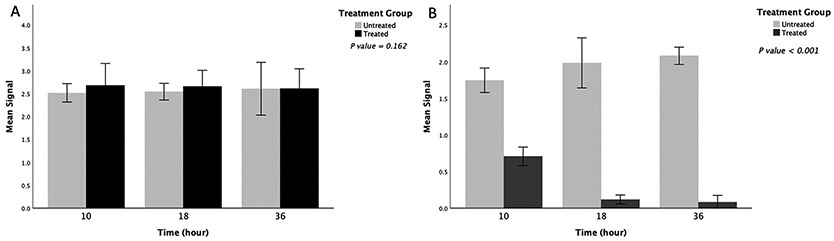
As previously mentioned, ligand-activated Notch signaling requires sequential cleavage of Notch by ADAM10 (which is the α-secretase for Notch) and by γ-secretase, releasing the Notch intracellular domain (NICD) in the cytoplasm [2]. To confirm that 1H5 blocks Notch processing, we evaluated the levels of cleaved and total (cleaved plus uncleaved) Notch1 in COLO205 cells using sandwich ELISA (PathScan®, Cell Signaling Technologies) following the manufacturer’s protocol (Materials and Methods). The detection antibody for cleaved notch specifically recognizes the cleaved (at Val1744) NICD1, while the detection antibody for total Notch1 binds the intracellular region in both cleaved and uncleaved Notch1. The results shown on Fig. 3 document that treatment with 1H5 significantly inhibits Notch1 cleavage without affecting the total Notch1 levels.
Fig. 3.
1H5 inhibits Notch cleavage. Sandwich ELISA was used to measure the levels of total (A) and cleaved (B) Notch1 in COLO205 cells upon treatment with 1H5. The data represent mean of triplicate experiments, and the bar plots show the effect of treatment with 1H5 relative to untreated control, mean ± SEM. Comparison of notch levels between treated and untreated groups was performed using independent t test. Total Notch1 levels did not significantly differ between the two groups (A), p = 0.162. On the other hand, the mAb-treated group showed significant decrease of the cleaved Notch1 levels when compared with the untreated control (B), p < 0.001.
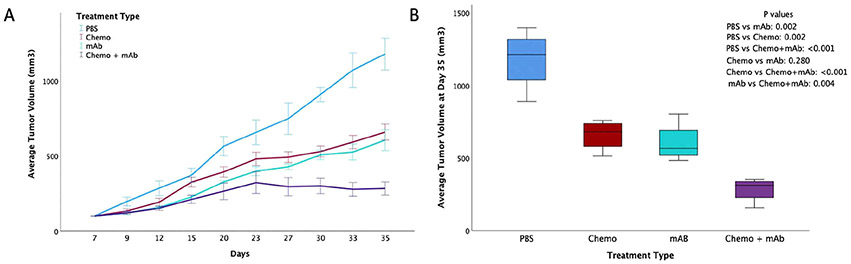
1H5 causes tumor growth inhibition of the colon cancer cell line COLO205 in vivo and is most effective in combination with chemotherapy. We determined the in vivo anti-tumor efficacy of human 1H5 mAb in a mouse model of CRC using xenografted COLO205 cells. Our previous results with the murine anti-ADAM10 mAb 8C7 showed that 8C7 inhibited tumor growth in combination with irinotecan, a chemotherapeutic drug used for the treatment of CRC [24]. In the same light we investigated the effect of the mAb 1H5 alone and in combination with Irinotecan. In the combination treatment (Fig. 4), we recorded 83 % tumor growth inhibition compared to 54 % (mAb alone) and 48 % (Irinotecan alone). There was no loss in mouse weight or presence of visible diarrhea. 1H5 also binds equally well to human and mouse ADAM10 in ELISA-based assays (data not shown). This indicates that despite ADAM10 manifesting ubiquitous expression profile, the 1H5 mAb selectively targets the tumor cells without any significant side effects or toxicity.
Fig. 4.
Anti-tumor effect of 1H5 in a colon cancer xenograft model. 6–8 weeks old female athymic nude mice were used. 5 million COLO205 cells per mouse were implanted (subcutaneous with Matrigel). Four groups were used (n = 4) for the experiment. When tumor volumes reached ~100 mm3, the four groups were injected as follows: (Group 1) sterile PBS (as a control). (Group 2) Irinotecan (i.p.) 20 mg/kg, three doses, once a week starting day 12. (Group 3) 1H5 (i.p.) at a dose of 30 mg/kg, biweekly (total 7 doses) starting day 7. (Group 4) Irinotecan (20 mg/kg, i.p., once a week starting day 12, total three doses) + continued 1H5 treatment (30 mg/kg, i.p., biweekly, total 7 doses). (A) Mean tumor volume ± 1 SD from day 7–35. 83 % tumor growth inhibition was recorded for the Group 4, treated with Irinotecan+ 1H5, 54 % for mAb alone (Group 3) and 48 % for Irinotecan alone (Group 2). No change in mouse weight or presence of diarrhea was observed in any of the groups. (B) Independent t test analysis showed significant difference in tumor volume reduction (day 35) between PBS and all treatments (with mAb and chemotherapy alone: p = 0.002; with combined treatment: p < 0.001). Significant tumor volume reduction differences were also observed between chemotherapy alone and combined treatment (p < 0.001), and between mAb treatment alone and combined treatment (p = 0.004). ANOVA with Dunnett multiple comparison post-hoc analysis likewise showed statistically significant tumor volume reduction between all treatment regimens vs. PBS control (all are p < 0.001), as well as between combination therapy and monotherapy (vs. chemotherapy p = 0.004; vs. mAB p = 0.011). In both analyses, the difference between mAb treatment and chemotherapy was not statistically significant. In the box and whiskers plot, the black horizontal lines indicate the average value, the top and bottom of the boxes, the interquartile range, and the whiskers, the range. Color coding on Panel B is the same as on Panel A.
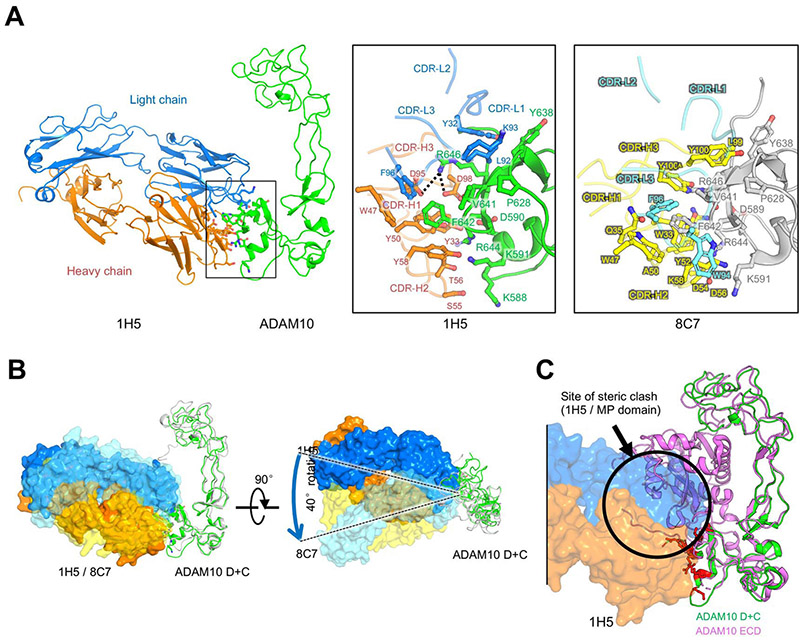
1H5 binds the ADAM10 C domain disrupting the ADAM10 MP-C domain autoinhibitory interactions. The crystal structure of the ADAM10 extracellular domain revealed its ‘closed’, auto-inhibited conformation, where the MP and C domains are in contact, partially obscuring the catalytic cleft, and the substrate-interacting C domain region [31]. Importantly, our previously determined crystal structure of the murine anti-ADAM10 mAb 8C7 bound to the isolated ADAM10 D+C domain region showed that 8C7 binds a defined epitope in the substrate-interacting C domain [24], which would be inaccessible in the auto-inhibited ADAM10 conformation. To address how the fully human mAb 1H5 binds ADAM10, and compare it to 8C7, we determined the structure of ADAM10(D+C) in complex with the isolated Fab fragment of 1H5 at 3.8 Å resolution (Fig. 5, Supplementary Table S1 and Fig. S2). The structure reveals that 1H5 binds ADAM10 at an epitope similar to 8C7 (Fig. S2) but with a distinct recognition strategy and approaching angle (Fig. 5A and B).
Fig. 5.
Crystal structure of the 1H5/ADAM10 (D+C) complex. (A). Overall structure of the ADAM10(D+C)/1H5 complex shown in ribbon view, as well as binding interface comparison (zoom-in insets) with the ADAM10(D+C)/8C7 structure showing a similar epitope region with distinct recognition strategies (see also Fig. S2 for epitope comparison). (B). Superimposition of the mAbs, 1H5 and 8C7, in complex with the ADAM10 D+C domain, illustrating the different antibody approaching angles. ADAM10 is shown as ribbons (green: 1H5 bound; grey: 8C7 bound). The antibodies are shown as molecular surfaces (1H5: nontransparent orange and blue; 8C7: semi-transparent yellow and cyan). (C) Overlay of the ADAM10 D+C (in green) / 1H5 (in orange and blue surface representation) complex and the ADAM10 ECD (in magenta) structures showing a partial overlay (indicated with the black circle) of 1H5 with the M domain in the autoinhibited ADAM10 conformation.
Formation of the 1H5 Fab/ADAM10 (D+C) complex buries ~791 Å2 of surface area in each molecule. The antibody complementarity determining regions (CDRs) target the C domain of ADAM10 as expected, via residues on the first and third CDR of the light chain (CDR-L1 and L3) and heavy-chain CDR-H1–3. The center of this interface is formed by embedding of three hydrophobic ADAM10 residues, V641 and F642 and P628, into a hydrophobic groove defined by 1H5 CDR-L1, L3 residues (Y32, L92, K93, and F96) and CDR-H1, H3 and framework residues (W33, W47, Y50 and Y58). Adjacent to this hydrophobic core, CDR-H3 residues D95 and D98 form salt-bridges with ADAM10 residue R646. There are multiple hydrogen bonds in the surrounding regions that further stabilize the interaction, including hydrogen bonds between R644 (ADAM10) and Y33, Y50 and T56 (CDR-H1 and H2); between K59 (ADAM10) and Y58 (CDR-H2); and between D590 (ADAM10) and Y33 (CDR-H1). The data also reveals that the ADAM10(D+C) structure in the 1H5/ADAM10 complex is very similar to that in the 8C7/ADAM10 complex structure [24], as well as in the structures of the unbound ADAM10(D+C) [14] and ADAM10(ECD) [31]. Indeed, the 1H5-bound and 8C7-bound ADAM10 D+C structures can be superimposed with a root-mean-square deviation (r.m.s.d.) of 1.675 Å between 185 Cα atoms (Fig. 5B).
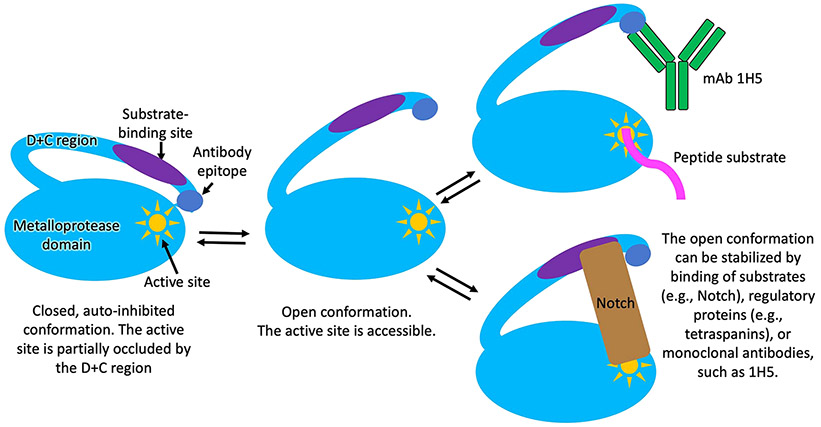
Importantly, binding of 1H5, like 8C7, would not be compatible with the autoinhibited ADAM10 conformation, as it would sterically clash with the D+C region-interacting M domain in the autoinhibited state [31] (Fig. 5C, see also Fig. 8). Consistent with these observations, 1H5 binds the isolated recombinant ADAM10 D+C region with a of 3.3 nM, and the full recombinant ADAM10 ECD, which predominantly adopts the autoinhibited conformation in solution [31], with a KD of ~200 nM (as measured by biolayer interferometry, data not shown). For comparison, the murine 8C7, binds the ADAM10 D+C region with a KD of 14 nM and the ADAM10 ECD, with a KD of ~100 nM [23].
Fig. 8.
Schematic representation of a proposed mechanism for ADAM10 activation and interactions with substrates and the 1H5 antibody. In the autoinhibited conformation, which is the predominant conformation of the ADAM10 ectodomain observed in solution [31], the cysteine rich domain partially occludes the ADAM10 active site, hindering substrate binding. In the open ADAM10 conformation, the active site is fully accessible for substrate binding [39]. In addition to interacting with the active site, ADAM10 cell-surface substrates also interact with the D+C region of the molecule, which is required for substrate selection and/or proper substrate positioning for productive cleavage. The open ADAM10 conformation can be stabilized by binding of substrates, such as Notch, or other regulatory molecules, such as tetraspanins. The latter not only regulate the activity of ADAM10 by stabilizing the open conformation, but also selectively enhance the cleavage of certain substrate, while downregulating the cleavage of other substrates [2,3,14,31,39]. The 1H5 mAb, in this regard, acts in a similar fashion to the tetraspannins: it (i) selectively binds and stabilizes the activated, open ADAM10 conformation; and (ii) it selectively downregulates the cleavage of certain ADAM10 substates (Notch), while upregulating (or not affecting) the cleavage of other substrates (e.g., peptides, APP).
1H5 recognizes an activated ADAM10 conformation present on cancer cells. To verify, the human 1H5 preferentially recognizes the tumor specific ADAM10 conformation, we employed an ELISA-based assay. The cell-based ELISA experiments were performed as previously reported for the anti-ADAM17 mAb D8P1C1 [38], which provides more details for this approach. We measured human 1H5 binding to two human colon cancer cell lines relative to the binding of a commercial mAb (mAb1427). MAb1427 was shown previously to not be conformation specific [23,24]. The cell-based ELISA results clearly document that 1H5 binds to tumor-expressed ADAM10 approximately fourteen-fold better than to ADAM10 expressed on HEK293 cells (Fig. 6).
Fig. 6.
1H5 preferentially binds to the activated, tumor cell-specific conformation of ADAM10. Cellular ELISA was performed to gauge the binding of 1H5, relative to the binding of the control anti-ADAM10 mAb1427 (commercial, R&D systems) to ADAM10 expressed on the cell surface of colon cancer cell lines LIM1215, COLO205, as well as HEK293 cells and HEK293 cells transfected with human ADAM10. MAb1427 bind equally well to the activated (tumor-associated) and the autoinhibited conformation of ADAM1023,24. The graph show the 1H5/mAb1427 signal ratio observed for the noted cell line relative to the 1H5/mAb1427 signal ratio observed for the untransfected HEK293 cells. Specifically, on the Y axes is plotted the value of: (A(1H5)/A(mAb1427))/(A(1H5-HEK)/A(mAb1427-HEK)) where A(1H5-HEK) is the signal for 1H5 using the untransfected HEK293 cells; A(mAb1427-HEK) is the signal for the mAb1427 using the untransfected HEK293 cells; A (1H5) is the signal for 1H5 using the cells that are being evaluated; A(mAb1427) is the signal for mAb1427 using the cells being evaluated. 1H5 binds to ADAM10 on tumors approximately fourteen-folds better than to ADAM10 on HEK2923 cells. The data represent triplicate determinations and two independent experiments, mean ± SEM; P < 0.001 by unpaired two-tailed independent t test (cancer cell lines vs. HEK293 cells).
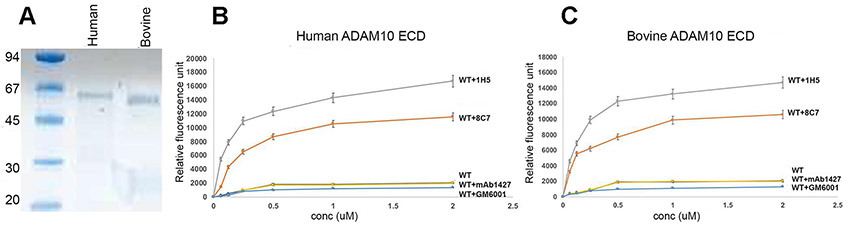
1H5 stabilizes the activated ADAM10 conformation and augments the cleavage of peptide substrates. Consistent with its binding to activated ADAM10, the murine mAb 8C7 was previously shown to enhance the ADAM10 catalytic activity in a fluorogenic peptide cleavage assay [31]. Using the same assay, we evaluated the effect of the human mAb 1H5. We used purified active human and bovine ADAM10 ECDs for these assays. The pro-domains of the human and bovine ADAM10 are removed during secretion of the proteins into the culture medium and the final recombinant products consist of the MP, D and C domains (confirmed by N-terminal sequencing). We included the commercial, conformation non-specific, anti-ADAM10 mAb1427, the murine anti-ADAM10 mAb 8C7 (that recognizes both human and bovine ADAM10), and the broad-spectrum metalloprotease inhibitor GM6001, in our assays. These results establish that: (i) 1H5 binding stabilizes activated ADAM10 and promotes the ADAM10 enzymatic activity (human or bovine); (ii) 1H5 is more efficient in augmenting the cleavage of substrate peptides than 8C7 (approximately two-fold); (iii) the commercial mAb1427 has no effect on substrate cleavage; (iv) the broad-spectrum inhibitor that is known to bind to the protease domain provides marginal inhibition (Fig. 7). Overall, these results validate our structural observations with ADAM10 D+C bound to either 1H5 or 8C7, confirming that 1H5 binding relieves ADAM10 autoinhibition thus enhancing the catalytic activity of the enzyme towards peptides in vitro (Fig. 8).
Fig. 7.
(A) SDS-PAGE profile of purified human (h) and bovine (b) catalytically active ADAM10 extracellular domains (ECD). (B and C) FRET-based peptide cleavage assays. The data shows that 1H5, similar 8C7 (but even more efficiently), promotes cleavage of a short peptide substrate by ADAM10. The data represent mean of triplicate determinations and two independent experiments (mean fluorescence ± SEM). Maximum dispersion was within 10 % of the mean value. Bovine or human ADAM10(ECD)-antibody complexes were formed at 1:1 molar ratio prior to the assay, and the assay was carried out in the presence of 50 μM of a fluorogenic peptide as described in the Materials and Methods section. GM6001 is a MMP inhibitor, which was also used as a control. Statistical analysis revealed significant difference/increase in peptide cleavage (fluorescence) between the 1H5 and 8C7 treated and untreated ADAM10 samples (p < 0.001, for both 1H5 and 8C7 treatment, one way ANOVA with a Dunnett’s multiple comparison post hoc test), as well as between the 1H5-treated and 8C7-treated ADAM10 samples (p = 0.003, two-tailed independent t test). Treatment with the control mAb1427 or the GM6001 inhibitor did not significantly change the ADAM10 enzymatic activity (p > 0.05).
4. Discussion
ADAM10 overexpression correlates with aberrant signaling from Notch, erbBs, and other receptors, as well as a more aggressive, metastatic phenotype in a range of cancers including colon, gastric, prostate, breast, ovarian, uterine, and leukemia [40-42]. As such, ADAM10 overexpression, has been shown to induce metastases of human HCT116 CRC cells in mice [40], suggesting that its inhibition may have potent antitumor effects. Current anti-colorectal treatments, predominantly relying on surgery, radiation and conventional chemotherapeutics, are not curative and commonly result in drug-resistant disease relapse. Almost half of all colorectal cancer patients will develop recurrent disease. Surgically resected cases of colorectal cancer (CRC) are known to have a 40 %–60 % recurrence rate in the first 3 years after surgery with the majority in the second year. Lymph node metastasis and/or adjacent organ involvement in stage II is said to have a recurrence of 20 %–30 % and stage III 50 %–80 % recurrence after surgery [43].
We set out to target the metalloprotease ADAM10, which is not only essential for ligand-activated Notch signaling but also controls other pathways deregulated in cancer, including signaling from erbB receptor tyrosine kinases. Attempts to inhibit ADAM10 with small-molecule inhibitors directed at its catalytic site failed clinical trials due to lack of specificity and efficacy, as well as dose-limiting toxicity. We previously proposed a novel, alternative strategy based on our discovery of a unique ADAM10 substrate recognition module outside the catalytic domain (thus avoiding side-effects and toxicity associated with catalytic-site targeting) [14] and raised murine ADAM10 specific monoclonal antibodies (mAbs) [23]. One of these mAbs, 8C7, recognized an activated conformation of ADAM10 present in the cancer stem cells that are characterized by elevated Notch activity. Consequently, 8C7 treatment inhibited Notch signaling and tumor relapse in pre-clinical models without exhibiting any dose-limiting toxicity in animals.
As part of our venture to translate anti-ADAM10 mAbs into clinic, we now report a fully human mAb, isolated from a new phage displayed Fab library screen, named 1H5, which binds to a similar epitope on the C substrate-binding domain of ADAM10 as the murine mAb 8C7. The data reported here shows that 1H5 preferentially recognizes ADAM10 present on cancer cells by binding to its activated conformation, and not to the autoinhibited conformation present in normal tissues [24,31]. Consequently, 1H5 activates the ADAM10 ECD in vitro and promotes its enzymatic activity towards small peptides in a FRET-based assay. The reason that 1H5 binding inhibits Notch cleavage, but activates small-peptide cleavage, is presumably due to its interference with the ADAM10-Notch interactions required for the formation of a productive enzyme-substrate complex, which also involve substrate/C-domain contacts. It should be noted that while the ADAM10 D and C domains (D+C) are considered important for interactions with, and cleavage of, many ADAM10 cell-surface substrates [23,24,31,39,44] (similar observations have also been reported for other ADAMs [15,45,46]), the exact substrate-binding region on ADAM10 is not well defined for any substrate and might be different for different substrates. Indeed, while 1H5 inhibits Notch cleavage (Fig. 3), it does not inhibit the cleavage of another ADAM10 substrate, APP, in cell-based assays (Fig. S3), indicating that 1H5 binding affects cleavage of different cell-surface-attached ADAM10 substrates, differently. This selective inhibition of oncogenic Notch signaling highlights the rationale behind targeting the substrate-binding ADAM10 D+C domains for therapeutic intervention (vs. targeting the proteinase domain) and likely contributes to the lack of side effects and low toxicity of both 1H5 and 8C7 in animal studies. In a sense, 1H5 acts similarly to the tetraspannins [39] in that it binds and stabilizes the activated ADAM10 conformation, while also selectively modulating (downregulates or upregulates) the cleavage of specific substates (Fig. 8).
Most importantly, in combination with chemotherapy, 1H5 prevents tumor growth in a CRC preclinical model without any discernible toxicity effects. 1H5 has only human antibody sequences in its framework and complementarity determining regions (CDRs), rendering it more favorable for clinical trials in human patients. We envisage that the mAb-mediated ADAM10 inhibition represents a novel and effective way to specifically inhibit drug-resistance and metastasis in CRC. Interestingly, it was recently reported that ADAM10 cleavage of PD-L1 is a mechanism for developing resistance to immunotherapy [47], suggesting that, in the future, 1H5 could also be evaluated in combination with PD-(L)1 inhibitors.
Supplementary Material
Acknowledgements and Funding
The authors thank the staff of the NE-CAT beamline ID-24 at the Advanced Photon Source of Argonne National Laboratory, for assistance with crystallographic data collection. This work was supported by the Functional Genomics Initiative of Memorial Sloan Kettering Cancer Center to D.B.N., NIH/NCI Cancer Center Support Grant P30 CA008748 to Memorial Sloan Kettering Cancer Center, and internal funding support from University of Pittsburgh to D.D.
Abbreviations:
- mAb
monoclonal antibody
- NICD
Notch intracellular domain
- CSC
Cancer stem cells
- HRP
Horseradish peroxidase
- NSCLC
Non-small cell lung cancer
- TNF
Tumor necrosis factor
Footnotes
CRediT authorship contribution statement
Nayanendu Saha: Conceptualization, Methodology, Investigation, Validation, Writing – review & editing. Du-San Baek: Methodology, Investigation, Data curation, Validation. Rachelle P. Mendoza: Resources, Data curation, Investigation. Dorothea Robev: Investigation, Methodology. Yan Xu: Investigation, Methodology. Yehuda Goldgur: Methodology, Software, Validation. M. Jason De La Cruz: Resources, Data curation. Elisa de Stanchina: Supervision, Methodology, Investigation. Peter W. Janes: Formal analysis. Kai Xu: Methodology, Software, Validation. Dimiter S. Dimitrov: Supervision, Formal analysis, Funding acquisition. Dimitar B. Nikolov: Formal analysis, Funding acquisition, Writing – original draft, Supervision, Project administration, Validation.
Conflict of Interest Statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2023.114494.
References
- [1].Munro MJ, Wickremesekera SK, Peng L, Tan ST, Itinteang T, Cancer stem cells in colorectal cancer: a review, J. Clin. Pathol 71 (2) (2018) 110–116. [DOI] [PubMed] [Google Scholar]
- [2].Hartmann DB, De Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P, The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts, Hum. Mol. Genet 11 (2002) 2615–2624. [DOI] [PubMed] [Google Scholar]
- [3].Murphy G, The ADAMs: signalling scissors in the tumour microenvironment, Nat. Rev. Cancer 8 (2008) 929–941. [DOI] [PubMed] [Google Scholar]
- [4].Fischer M, Yen WC, Kapoun AM, Wang M, O’Young G, et al. , Anti-DLL4 inhibits growth and reduces tumor-initiating cell frequency in colorectal tumors with oncogenic KRAS mutations, Cancer Res. 71 (2011) 1520–1525. [DOI] [PubMed] [Google Scholar]
- [5].Li JL, Swanson RC, Oon CE, Turley H, Leek R, Sheldon H, Bridges E, et al. , DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo, Cancer Res. 71 (2011) 6073–6083. [DOI] [PubMed] [Google Scholar]
- [6].Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Marti M, Quinn SA SA, et al. , Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells, Cancer Cell 22 (2012) 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McAuliffe SM, Morgan SL, Wyant GA, Tran LT, Muto KW, et al. , Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy, Proc. Natl. Acad. Sci 109 (2012) E2939–E2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Timme CR, Gruidi M, Yeatman TJ, Gamma-secretase inhibition attenuates oxaliplatin-induced apoptosis through increased Mcl-1 and/or Bcl-xL in human colon cancer cells, Apoptosis: Int. J. Program. Cell Death 18 (2013) 1163–1174. [DOI] [PubMed] [Google Scholar]
- [9].Meng RD, Shelton CC, Li YM, Qin LX, Notterman D, Paty PB, Schwartz GK, Gamma-Secretase inhibitors abrogate oxaliplatin-induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity, Cancer Res. 69 (2009) 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Groth C, Fortini ME, Therapeutic approaches to modulating Notch signaling: current challenges and future prospects, Semin Cell Dev. Biol 23 (4) (2012) 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dikic I, Schmidt MH, Notch: Implications of endogenous inhibitors for therapy, Bioessays 32 (2010) 481–487. [DOI] [PubMed] [Google Scholar]
- [12].Wu Y, Cain -Hom C, Choy L, Hagenbeek TJ, De Leon GP, Therapeutic antibody targeting of individual Notch receptors, Nature 464 (2010) 1052–1057. [DOI] [PubMed] [Google Scholar]
- [13].Seals DF, Courtneidge SA, The ADAMs family of metalloproteases: multidomain proteins with multiple functions, Genes Dev. 17 (1) (2003) 7–30. [DOI] [PubMed] [Google Scholar]
- [14].Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB, Adam Meets Eph: An ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans, Cell 123 (2005) (2005) 291–330. [DOI] [PubMed] [Google Scholar]
- [15].Smith KM, Gaultier KM,A, Cousin H, Alfandari D, White JM, DeSimone DW, The cysteine-rich domain regulates ADAM protease function in vivo, J. Cell Biol 159 (2002) 893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Janes PW, Wimmer-Kleikamp SH, Frangakis AS, Treble K, Griesshaber B, Sabet O, Grabenbauer M, Ting AY, Saftig P, Bastiaens PI, Lackmann M, Cytoplasmic relaxation of active Eph controls ephrin shedding by ADAM10, PLoS Biol. 7 (2009), e1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Andersson ER, Lendahl U, Therapeutic modulation of Notch signalling–are we there yet? Nat. Rev. Drug Discov 13 (2014) 357–378. [DOI] [PubMed] [Google Scholar]
- [18].DasGupta S, Murumkar PR, Giridhar R, Yadav MR, Current perspective of TACE inhibitors: a review, Bioorg. Med. Chem 17 (2009) (2009) 444–459. [DOI] [PubMed] [Google Scholar]
- [19].Saftig P, Reiss K, The "A Disintegrin And Metalloproteases" ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur. J. Cell Biol 90 (2011) 527–535. [DOI] [PubMed] [Google Scholar]
- [20].Huang Y, Benaich N, Tape C, Kwok HF, Murphy G, Targeting the sheddase activity of ADAM17 by an anti-ADAM17 antibody D1 (A12) inhibits head and neck squamous cell carcinoma cell proliferation and motility via blockage of bradykinin induced HERs transactivation, Int. J. Biol. Sci 10 (2014) 702–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yamamoto K, Trad A, Baumgart A, Huske L, Lorenzen I, Chalaris A, et al. , A novel bispecific single-chain antibody for ADAM17 and CD3 induces T-cell-mediated lysis of prostate cancer cells, Biochem J. 445 (2012) 135–144. [DOI] [PubMed] [Google Scholar]
- [22].Rios-Doria J, Sabol D, Chesebrough J, Stewart D, Xu L, et al. , A monoclonal antibody to ADAM17 inhibits tumor growth by inhibiting EGFR and Non-EGFR-mediated pathways, Mol. Cancer Ther 14 (2015) 1637–1649. [DOI] [PubMed] [Google Scholar]
- [23].Atapattu L, Saha N, Llerena C, Vail ME, Scott AM, Nikolov DB, Lackmann M, Janes PW, Antibodies binding the ADAM10 substrate recognition pocket inhibit Eph function, J. Cell Sci 125 (2012) 6084–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Atapattu L, Saha N, Chheang C, Eissman MF, Xu K, Vail ME, Hi L, Llerena C, Liu Z, Horvay K, Abud HE, Kusebauch U, Moritz RL, Ding BS, Cao Z, Rafii S, Ernst M, Scott AM, Nikolov DB, Lackman M, Janes PW, An activated form of ADAM10 is tumor selective and regulates cancer stem-like cells and tumor growth, J. Exp. Med 213 (9) (2016) 1741–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Whitehead RH, Macrae FA, St John DJ, Ma J, A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer, J. Natl. Cancer Inst 74 (4) (1985) 759–765. [PubMed] [Google Scholar]
- [26].Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB, Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3, Proc. Natl. Acad. Sci 105 (29) (2008) 9953–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Baek DS, Kim YJ, Vergara S, Conard A, Adams C, Calero G, Ishima R, Mellors JW, Dimitrov DS, Cancer Lett. 525 (2022) 97–107. [DOI] [PubMed] [Google Scholar]
- [28].O’Brien J, Wilson I, Orton T, Pognan F, Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cytotoxicity, Eur. J. Biochem 267 (17) (2000) 5421–5426. [DOI] [PubMed] [Google Scholar]
- [29].Rios-Doria J, Sabol D, Chesebrough J, Stewart D, Xu L, Tammali R, Cheng L, Du Q, Schifferli K, Rothstein R, Leow CC, Heidbrink-Thompson J, Jin X X, Gao C, Friedman J, Wilkinson B, Damschroder M, Pierce AJ, Hollingsworth RE, Tice DA, Michelotti EF, A monoclonal antibody to ADAM17 inhibits tumor growth by inhibiting EGFR and non-EGFR-mediated pathways, Mol. Cancer Ther 14 (2015) 1637–1649. [DOI] [PubMed] [Google Scholar]
- [30].Smith D D, Cohick CB, Lindsley HB, Optimization of cellular ELISA for assay of surface antigens on human synoviocytes, BioTechniques 22 (1997) 952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, Janes PW, Rubinstein E, Nikolov DB, Skiniotis G, Kruse AC, Blacklow SC, Structural basis for regulated proteolysis by the α-Secretase ADAM10, Cell 171 (7) (2017) 1638–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Black RA, Becherer JD, Tumor Necrosis Factor alpha -Converting Enzyme, in: Barret AJ, et al. (Eds.), Handbook of Proteolytic Enzymes, Academic Press, San Diego, 1998, p. 1315. [Google Scholar]
- [33].Pal S, Konkimalla VB, Kathawate L, et al. , Targeting a chemorefractory COLO205 (BRAF V600E) cell line using substituted benzo[α]phenoxazines, RCS Adv. 5 (2015) 82549–82563. [Google Scholar]
- [34].Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Xing L, Hung MC, Bonfiglio T, Hicks DG, Tang P, The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines, Breast Cancer (Auckl.) 4 (2010) 35–41. [PMC free article] [PubMed] [Google Scholar]
- [35].Potts AH, Dawson JC, Herrington CS, Ovarian cancer cell lines derived from non-serous carcinomas migrate and invade more aggressively than those derived from high-grade serous carcinomas, Sci. Rep 9 (2019) article number: 5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Long H, Zhang C, Li T, Zhou X, Liu B, Li S, Zhu M, Lin Y, Yu S, Zhang K, Ren B, et al. , Notch1 is a prognostic factor that is distinctly activated in the classical and proneural subtype of glioblastoma and that promotes glioma cell survival via the NF-κB(p65) pathway, Cell Death Dis. 9 (2018) 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rusch V, Baselga J, Cordon-Cardo C, Orazem J, Zaman M, Hoda S, McIntosh J, Kurie J, Dmitrovsky E, Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung, Cancer Res. 53 (1993) 2379–2385. [PubMed] [Google Scholar]
- [38].Saha N, Xu K, Zhu Z, Robev D, Kalidindi T, Xu Y, Himanen J, de Stanchina E, Pillarsetty NVK, Dimitrov DS, Nikolov DB, Inhibitory monoclonal antibody targeting ADAM17 expressed on cancer cells, Transl. Oncol 15 (1) (2022), 101265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lipper CH, Egan ED, Gabriel KK, Blacklow SC. Structural Basis for Selective Proteolysis of ADAM10 Substrates at Membrane-Proximal Sites. BioRxiv. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gavert N, Scheffer M, Raveh S, Spdarena S, Shtutman M, et al. , Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis, Cancer Res. 67 (2007) 7703–7712. [DOI] [PubMed] [Google Scholar]
- [41].Wang YY, Ye ZY, Li L, Zhao ZS, Shao QS, Tao HQ, ADAM 10 is associated with gastric cancer progression and prognosis of patients, J. Surg. Oncol 103 (2011) 116–123. [DOI] [PubMed] [Google Scholar]
- [42].Smith TM, Tharakan A, Martin RK, Targeting ADAM10 in cancer and autoimmunity, Front Immunol. 11 (2020). Article 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Young PE, Womeldorph CM, Johnson EK, Maykel JA, et al. , Early detection of colorectal cancer recurrence in patients undergoing surgery with curative intent: current status and challenges, J. Cancer 5 (4) (2014) 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science., 327(5971): 1380–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stawikowska R, Cudic M, Giulianotti M, Houghten RA, Fields G, Minond D, Activity of ADAM17 (a disintegrin and metalloprotease 17) is regulated by its noncatalytic domains and secondary structure of its substrates, J. Biol. Chem 288 (31) (2013) 22871–22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Saha N, Robev D, Himanen JP, Nikolov DB, ADAM proteases: emerging role and targeting of the non-catalytic domains, Cancer Lett. 467 (2019) 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Orme JJ, Jazieh KA, Xie T, Harrington S, Lin X, et al. , ADAM10 and ADAM17 cleave PD-L1 to mediate PD-(L)1 inhibitor resistance, Oncoimmunology 9 (1) (2020), e1744980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.