Summary
Artificial intelligence (AI) models for automatic generation of narrative radiology reports from images have the potential to enhance efficiency and reduce the workload of radiologists. However, evaluating the correctness of these reports requires metrics that can capture clinically pertinent differences. In this study, we investigate the alignment between automated metrics and radiologists' scoring of errors in report generation. We address the limitations of existing metrics by proposing new metrics, RadGraph F1 and RadCliQ, which demonstrate stronger correlation with radiologists' evaluations. In addition, we analyze the failure modes of the metrics to understand their limitations and provide guidance for metric selection and interpretation. This study establishes RadGraph F1 and RadCliQ as meaningful metrics for guiding future research in radiology report generation.
Keywords: chest X-ray radiology report generation, automatic metrics, alignment with radiologists
Graphical abstract
Highlights
-
•
Examined correlation between automated metrics and scoring of reports by radiologists
-
•
Proposed metric based on overlap in clinical entities and relations named RadGraph F1
-
•
Proposed composite metric RadCliQ with better alignment with radiologists
-
•
Analyzed failure modes of automated metrics
The bigger picture
Artificial intelligence (AI) has made formidable progress in the interpretation of medical images, but its application has largely been limited to the identification of a handful of individual pathologies. In contrast, the generation of complete narrative radiology reports more closely matches how radiologists communicate diagnostic information. While recent progress on vision-language models has enabled the possibility of generating radiology reports, the task remains far from solved. Our work aims to tackle one of the most important bottlenecks for progress: the limited ability to meaningfully measure progress on the report generation task. We quantitatively examine the correlation between automated metrics and the scoring of reports by radiologists and investigate the failure modes of metrics. We also propose a metric based on overlap in clinical entities and relations extracted from reports and a composite metric, called RadCliQ, that is a combination of individual metrics.
Yu et al. quantitatively examine the correlation between automated metrics and the scoring of radiology reports by radiologists to understand how to meaningfully measure progress on automatic report generation. They propose RadGraph F1, a metric based on overlap in clinical entities and relations, and RadCliQ, a composite metric that combines individual metrics and aligns better with radiologists. They analyze the types of information metrics fail to capture to further understand metric usefulness and evaluate state-of-the-art report generation approaches.
Introduction
Artificial intelligence (AI) has been making great strides in tasks that require expert knowledge,1,2,3,4 including the interpretation of medical images.5 In recent years, medical AI models have been demonstrated to achieve expert-level performance,6 generalize to hospitals beyond which they were trained,3 and assist specialists in their interpretation.7 However, the application of AI to image interpretation tasks has often been limited to the identification of a handful of individual pathologies,8,9,10 representing an over-simplification of the image interpretation task. In contrast, the generation of complete narrative radiology reports11,12,13,14,15,16,17,18,19,20,21 moves past that simplification and is consistent with how radiologists communicate diagnostic information: the narrative report allows for highly diverse and nuanced findings, including association of findings with anatomic location, and expressions of uncertainty. Although the generation of radiology reports from medical images in their full complexity would signify a tremendous achievement for AI, the task remains far from solved. Our work aims to tackle one of the most important bottlenecks for progress: the limited ability to meaningfully measure progress on the report generation task.
Automatically measuring the quality of generated radiology reports is challenging. Most prior works have relied on a set of metrics inspired by similar setups in natural language generation, where radiology report text is treated as generic text.22 However, unlike generic text, radiology reports involve complex, domain-specific knowledge and critically depend on factual correctness. Even metrics that were designed to evaluate the correctness of radiology information by capturing domain-specific concepts do not align with radiologists.23 Therefore, improvement on existing metrics may not produce clinically meaningful progress or indicate the direction for further progress. This fundamental bottleneck hinders understanding of the quality of report generation methods thereby impeding work toward improvement of existing methods. We seek to remove this bottleneck by developing meaningful measures of progress in radiology report generation. The answer to this question is imperative to understanding which metrics can guide us toward generating reports that are clinically indistinguishable from those generated by radiologists.
In this study, we quantitatively examine the correlation between automated metrics and the scoring of reports by radiologists. We propose a new automatic metric that computes the overlap in clinical entities and relations between a machine-generated report and a radiologist-generated report, called RadGraph24 F1. We develop a methodology to predict a radiologist-determined error score from a combination of automated metrics, called RadCliQ. We analyze failure modes of the metrics, namely the types of information the metrics do not capture, to understand when to choose particular metrics and how to interpret metric scores. Finally, we measure the performance of state-of-the-art report generation models using the investigated metrics. The result is a quantitative understanding of radiology report generation metrics and clear guidance for metric selection to guide future research on radiology report generation.
Results
Alignment between automated metrics and radiologists
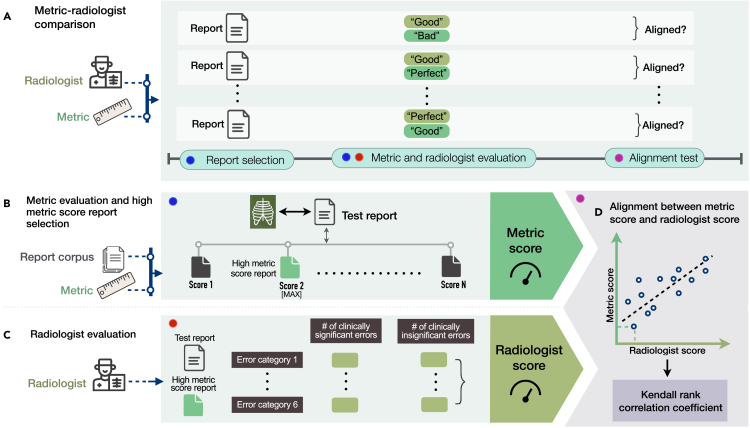
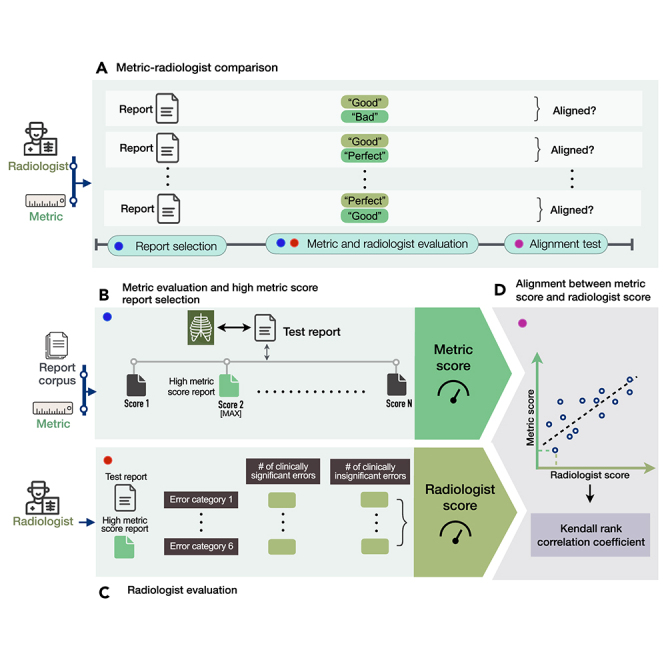
We study whether there is alignment between automated metric and radiologist scores assigned to radiology reports. An overview of our methodology is shown in Figure 1. Figure 1A shows the experimental design for determining alignment. Given a test report from MIMIC-CXR,25,26,27 we select a series of candidate reports from the MIMIC-CXR training set that score highly according to various metrics, including BLEU,28 BERTScore,29 CheXbert vector similarity (s_emb),9 and a novel metric RadGraph24 F1. Specifically, we select a candidate report by finding the test report’s metric-oracle: the highest-scoring report from the MIMIC-CXR training set with respect to a particular metric (Figure 1B). We choose this set of reasonably accurate reports so we can study their quality with more precision. An example study with a reference report and candidate metric-oracle reports is shown in Figure 2A.
Figure 1.
Method overview
(A) Experimental design for selecting radiology reports and comparing metrics and radiologists in evaluating reports.
(B) Given a test report, selecting the report with the highest metric score from the training report corpus with respect to the test report and a particular metric.
(C) Conducting radiologist evaluation on the high metric score report relative to the test report, where radiologists identify the number of clinically significant and insignificant errors in the high metric score report across six error categories.
(D) Determining the alignment between metric scores and radiologist scores assigned to the same reports using the Kendall rank correlation coefficient.
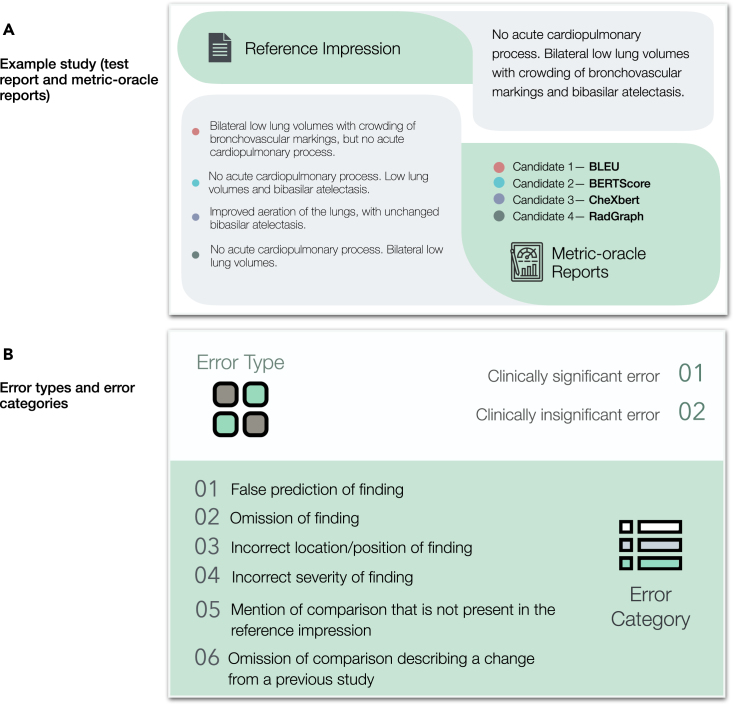
Figure 2.
Example study of reports, and error types and categories
(A) Example study of a test report and four metric-oracle reports corresponding to BLEU, BERTScore, CheXbert vector similarity, and RadGraph F1 that radiologists evaluate to identify errors.
(B) Two error types and six error categories that radiologists identify for each pair of test report and metric-oracle report.
Next, we have six board-certified radiologists score how well the candidates match the test report (Figure 1C). Radiologists scored the number of errors that various candidate reports make compared with the test report, and errors are categorized as clinically significant or insignificant. Radiologists subtyped every error into the following six categories: (1) false prediction of finding (i.e., false positive), (2) omission of finding (i.e., false negative), (3) incorrect location/position of finding, (4) incorrect severity of finding, (5) mention of comparison that is not present in the reference impression, and (6) omission of comparison describing a change from a previous study. The error types and error categories are summarized in Figure 2B. The instructions and interface presented to radiologists can be seen in Figures S1 and S2. The radiologist error scores on the 50 studies are shown in Figure S3.
We quantify metric-radiologist alignment using the Kendall rank correlation coefficient (tau b) between metric scores and number of radiologist-reported errors in the reports (Figure 1D). We determine the metric-radiologist alignment from metric-oracle generations from 50 chosen studies on both a total error and significant error level. The coverage of pathologies, as determined by the CheXpert8 labels in MIMIC-CXR, for the 50 randomly sampled reports is shown in Table S1. The per-radiologist Kendall rank correlation coefficients are listed in Table S2.
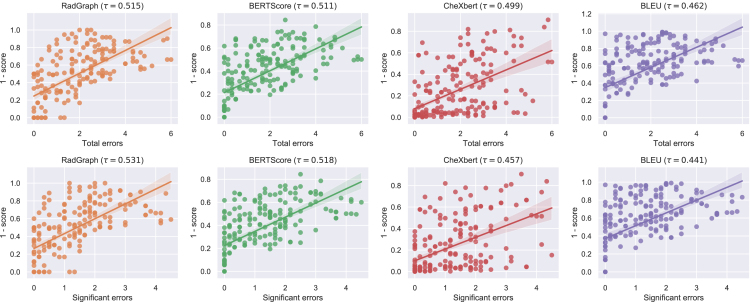
We find that RadGraph F1 and BERTScore are the metrics with the two highest alignments with radiologists. Specifically, RadGraph has a tau value of 0.515 (95% CI, 0.449 0.578) for total number of clinically significant and insignificant errors and 0.531 (95% CI, 0.465 0.584) for significant errors. BERTScore has a tau value of 0.511 (95% CI, 0.429 0.584)] for total number of clinically significant and insignificant errors and 0.518 (95% CI, 0.440 0.586) for significant errors. We find that CheXbert vector similarity is the third best metric under this evaluation with a 0.499 (95% CI, 0.417 0.576) tau value for total number of clinically significant and insignificant errors and 0.457 (95% CI, 0.370 0.538) for significant errors. Finally, BLEU has the worst alignment with a tau value of 0.462 (95% CI, 0.368 0.539) for total number of clinically significant and insignificant errors and 0.441 (95% CI, 0.350 0.521) for significant errors. From these results, we see that RadGraph and BERTScore are the metrics with closest alignment to radiologists. For the total number of clinically significant and insignificant errors, BERTScore has a significantly higher alignment than BLEU. Looking at significant errors, BERTScore and RadGraph have a significantly higher alignment than BLEU and, additionally, RadGraph has a significantly higher alignment than CheXbert. CheXbert, and BLEU have alignment with radiologists but are less concordant than the other two metrics. The metric-radiologist alignment graphs are shown in Figure 3.
Figure 3.
Correlations between metric scores and radiologist scores
Scatterplots and correlations between metric scores and radiologist scores of four metric-oracle generations from 50 studies, where radiologist scores are represented by the total number of clinically significant and insignificant errors (top row) and number of clinically significant errors (bottom row) identified by the radiologists. The translucent bands around the regression line represent 95% confidence intervals.
Failure modes of metrics
In addition to evaluating the clinical relevance of metrics in terms of the total number of clinically significant and insignificant errors, we also examine the particular error categories of metric-oracles to develop a granular understanding of the failure modes of different metrics, as shown in Figure 4. We use the following six error categories as described earlier:
-
1.
false prediction of finding
-
2.
omission of finding
-
3.
incorrect location/position of finding
-
4.
incorrect severity of finding
-
5.
mention of comparison that is not present in the reference impression
-
6.
omission of comparison describing a change from a previous study and analyze the total number of errors and the number of clinically significant errors within each error category
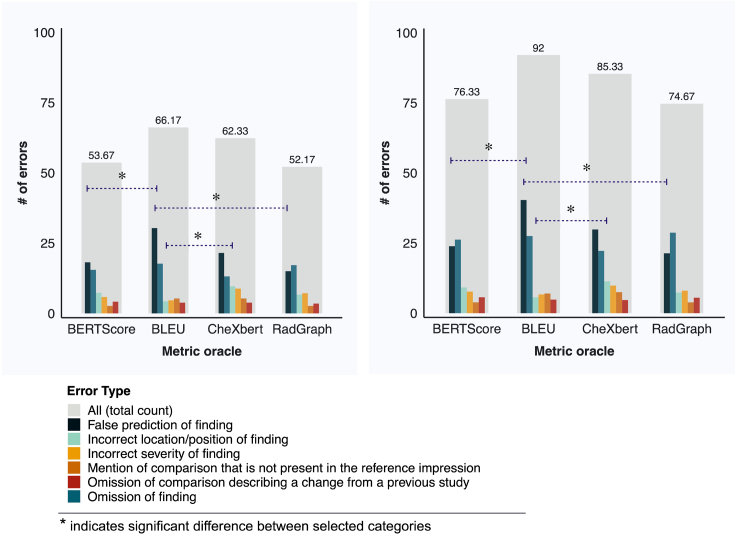
Figure 4.
Distribution of errors across error categories for metric-oracle reports
Distribution of errors across six error categories for metric-oracle reports corresponding to BERTScore, BLEU, CheXbert vector similarity, and RadGraph F1, in terms of the number of clinically significant errors (left) and the total number of clinically significant and insignificant errors (right). Statistical significance is determined using the Benjamini-Hochberg procedure with a false discovery rate (FDR) of 1% to correct for multiple-hypothesis testing.
BLEU exhibits a prominent failure mode in identifying false predictions of finding in reports. Metric-oracle reports with respect to BLEU produce more false predictions of finding than BERTScore and RadGraph in terms of both the total number of clinically significant and insignificant errors (0.807 average number of errors per report versus 0.477 and 0.427 for BERTScore and RadGraph) and the number of clinically significant errors (0.607 average number of errors per report versus 0.363 and 0.300 for BERTScore and RadGraph). BLEU exhibits a less prominent failure mode in identifying incorrect locations/positions of finding compared with CheXbert vector similarity. Metric-oracle reports with respect to BLEU have fewer incorrect locations/positions of finding than CheXbert in terms of both the total number of clinically significant and insignificant errors (0.113 average number of errors per report versus 0.227 for CheXbert) and the number of clinically significant errors (0.087 average number of errors per report versus 0.193 for CheXbert). These differences are statistically significant after accounting for multiple-hypothesis testing. Metric-oracle reports of the four metrics exhibit similar behavior in the other error categories, as the differences in number of errors are not statistically significant. The raw error counts and the statistics testing results for two-sample t tests and the Benjamini-Hochberg procedure for accounting for multiple-hypothesis testing are shown in Tables S3 and S4.
Measuring progress of prior methods in report generation
Using the four metrics, we evaluated the following state-of-the-art radiology report generation methods: M2 Trans,11 R2Gen,12 CXR-RePaiR,13 WCL,14 and CvT2DistilGPT2.15 As a baseline, we also implemented a random radiology report generation model, which retrieves a random report from the training set for each test report. The prior methods were trained to generate different sections of radiology reports: CXR-RePaiR generates the impression section, M2 Trans the findings section, and R2Gen, WCL, and CvT2DistilGPT2 jointly the findings and impression sections. For each method, we compute metric values using the corresponding section(s) of radiology reports it generates as the ground-truth report to ensure accurate evaluation of the method. We also generated three versions of random baselines that retrieved different sections of the reports and compared each method with its corresponding random baseline. Because the impression section of radiology reports is an interpretation of the findings section, we can assume that both sections use the same medical vocabulary and style, which the report metrics evaluate. Conclusions about the report metrics drawn from the radiologist experiment and associated analyses, which used the impression section, can carry over to the evaluation of different report sections.
The performances of metric-oracle selection models, prior models, and random retrieval baselines on the MIMIC-CXR test set are shown in Tables 1, 2, and 3, grouped by the report sections they generate. Note that the results are comparable within each table, but not across. With respect to the most radiologist-aligned metric RadGraph F1, among impression-generating models, metric-oracle models significantly outperform real report generation models, achieving a maximum score of 0.677. Among findings-generating models, M2 Trans performs the best (0.244). Among models that jointly generate the findings and impression sections, CvT2DistilGPT2 performs the best (0.154).
Table 1.
Metric scores of impression-generating models, including metric-oracle models, CXR-RePaiR, and the random retrieval baseline model
| BLEU | BERTScore | CheXbert vector similarity | RadGraph F1 | |
|---|---|---|---|---|
| BLEU metric-oracle | 0.557∗ | 0.661 | 0.689 | 0.476 |
| BERTScore metric-oracle | 0.491 | 0.721∗ | 0.738 | 0.498 |
| CheXbert metric-oracle | 0.381 | 0.573 | 0.954∗ | 0.403 |
| RadGraph metric-oracle | 0.366 | 0.541 | 0.739 | 0.677∗ |
| CXR-RePaiR | 0.055 | 0.193 | 0.379 | 0.090 |
| Random retrieval of impression | 0.048 | 0.222 | 0.269 | 0.050 |
The 95% confidence interval and range of metric scores are available in Table S5.
indicates the best-performing model.
Table 2.
Metric scores of findings-generating models, including M2 Trans, and the random retrieval baseline model
| BLEU | BERTScore | CheXbert vector similarity | RadGraph F1 | |
|---|---|---|---|---|
| M2 Trans | 0.220∗ | 0.386∗ | 0.452∗ | 0.244∗ |
| Random retrieval of findings | 0.123 | 0.323 | 0.235 | 0.105 |
The 95% confidence interval and range of metric scores are available in Table S6.
indicates the best-performing model.
Table 3.
Metric scores of models that jointly generate findings and impression sections, including R2Gen, WCL, CvT2DistilGPT2, and the random retrieval baseline model
| BLEU | BERTScore | CheXbert vector similarity | RadGraph F1 | |
|---|---|---|---|---|
| R2Gen | 0.137 | 0.271 | 0.286 | 0.134 |
| WCL | 0.144∗ | 0.275 | 0.309 | 0.143 |
| CvT2DistilGPT2 | 0.143 | 0.280∗ | 0.335∗ | 0.154∗ |
| Random retrieval of jointly the findings and impression | 0.100 | 0.256 | 0.190 | 0.090 |
The 95% confidence interval and range of metric scores are available in Table S7.
indicates the best-performing model.
Composite metric RadCliQ
To improve upon individual metrics, we propose a novel composite metric RadCliQ (radiology report clinical quality) that combines the four investigated metrics. We trained a model to predict the total number of clinically significant and insignificant errors that radiologists would assign to a report. The model input consisted of the four metric scores computed for each report. We applied zero-mean unit-variance normalization on the scores of each type of metric before passing the scores as model input. Prediction of the trained model therefore combines evaluations of BLEU, BERTScore, CheXbert vector similarity, and RadGraph F1.
We had 200 metric-oracle reports that were evaluated by radiologists, containing 50 metric-oracle reports corresponding to each of the four investigated metrics. These training data correspond to a subset of 50 studies from the MIMIC-CXR test set. We split our dataset by 8:2 into a development set (160 data points) and a test set (40 data points). On the development set, we conducted a cross-validation of 10-fold with 16 data points per validation fold to experiment with different model formulations for RadCliQ and build a fair comparison between RadCliQ and existing metrics.
Specifically, for each cross-validation setup, we built a normalizer with zero-mean and unit-variance and a linear regression model that took in the normalized metric values, on the cross-validation training set (144 data points). We then used the normalizer and regression model to normalize and generate predictions on the held-out validation set (16 data points). Finally, we computed the Kendall tau b correlation on the held-out set predictions with respect to the held-out set ground-truth radiologist total number of errors. We also computed the Kendall tau b correlation for each existing metric. Across the 10 cross-validation setups, we computed the mean Kendall tau b correlations for the composite metric and existing metrics, and verified that the composite metric had stronger alignment with radiologists.
Our proposed model builds upon the standard linear regression by introducing constraints that improve its performance. Specifically, we require the negative of the coefficients to be non-negative and sum up to 1, resulting in a well-defined and interpretable convex function. Thus, we ensure that, when one metric score increases, while the others remain constant, the predicted number of errors will decrease. This property makes our model more sensitive to changes in individual metrics and thus more accurate in predicting error rates. Furthermore, the constraint makes the coefficients interpretable as weights, providing insights into the relative importance of each metric in predicting errors. To obtain the constrained coefficients, we use the convex optimization solver CVXPY, which guarantees global optimality and fast convergence. With this approach, we can effectively balance the trade-off between accuracy and interpretability, and obtain a robust and reliable model for error prediction.
After finalizing the model formulation, we fit the normalizer and composite metric model on the full development set and obtain RadCliQ. The coefficients were 0.000 for BLEU, −0.370 for BERTScore, −0.253 for CheXbert, and −0.377 for RadGraph F1. The intercept value for the regression model was 0.000. Finally, on the held-out test set, the composite metric (RadCliQ) has higher Kendall tau b correlations than the other metrics, as shown in Table 4. This result indicates that RadCliQ has the strongest alignment with radiologists than any individual metric.
Table 4.
Kendall tau b correlations of individual metrics and the composite metric (RadCliQ) on the held-out test set of 40 data points with radiologist error annotations
| Kendall tau b correlation | |
|---|---|
| BLEU | 0.414 (95% CI, 0.156 0.635) |
| BERTScore | 0.505 (95% CI, 0.273 0.671) |
| CheXbert vector similarity | 0.537 (95% CI, 0.330 0.717) |
| RadGraph F1 | 0.528 (95% CI, 0.357 0.687) |
| Composite metric (RadCliQ) | 0.615 (95% CI, 0.450 0.749)∗ |
indicates the best-aligned metric.
We used RadCliQ to evaluate all generations of metric-oracle models, prior models, and random retrieval baselines for the MIMIC-CXR test set. The metric scores are shown in Tables 5, 6, and 7. Among impression-generating models, the BERTScore metric-oracle model performs the best (−0.095). CXR-RePaiR (1.642) outperforms the random retrieval baseline (1.755). Among findings-generating models, M2 Trans performs the best (1.059). Among models that jointly generate findings and impression sections, CvT2DistilGPT2 performs the best (1.463).
Table 5.
RadCliQ scores of impression-generating models, including metric-oracle models, CXR-RePaiR, and the random retrieval baseline model
| RadCliQ | |
|---|---|
| BLEU metric-oracle | 0.081 |
| BERTScore metric-oracle | −0.095∗ |
| CheXbert metric-oracle | 0.052 |
| RadGraph metric-oracle | −0.020 |
| CXR-RePaiR | 1.642 |
| Random retrieval of impression | 1.755 |
Lower is better. The 95% confidence interval and range of metric scores are available in Table S5.
indicates the best-performing model.
Table 6.
RadCliQ scores of findings-generating models, including M2 Trans and the random retrieval baseline model
| RadCliQ | |
|---|---|
| M2 Trans | 1.059∗ |
| Random retrieval of findings | 1.553 |
Lower is better. The 95% confidence interval and range of metric scores are available in Table S6.
indicates the best-performing model.
Table 7.
RadCliQ scores of models that jointly generate findings and impression sections, including R2Gen, WCL, CvT2DistilGPT2, and the random retrieval baseline model
| RadCliQ | |
|---|---|
| R2Gen | 1.552 |
| WCL | 1.511 |
| CvT2DistilGPT2 | 1.463∗ |
| Random retrieval of jointly the findings and impression | 1.726 |
Lower is better. The 95% confidence interval and range of metric scores are available in Table S7.
indicates the best-performing model.
Discussion
The purpose of this study was to investigate how to meaningfully measure progress in radiology report generation. We studied popular existing automated metrics and designed novel metrics, the RadGraph graph overlap metric and the composite metric RadCliQ, for report evaluation. We quantitatively determined the alignment of metrics with clinical radiologists and the reliability of metrics against specific failure modes, clarifying whether metrics meaningfully evaluate radiology reports and therefore can guide future research in report generation. We also showed that selecting the best-match report from a large corpus performs better on most metrics that the current state-of-the-art radiology report generation methods. Although the best-match method is unlikely to be clinically viable, it served as a useful tool to derive the RadCliQ composite metric developed in this study and could serve as a useful benchmark against which to evaluate report generation algorithms developed in the future.
The design of automated evaluation metrics that are aligned with manual expert evaluation has been a challenge for research in radiology report generation as well as medical report generation as a whole. Prior works have used metrics designed to improve upon n-gram matching28,29,30,31,32 or include clinical awareness,8,9,11,13,24 such as with BLEU28 and CheXpert labels.8 However, these evaluations nevertheless poorly approximate radiologists’ evaluation of reports. The expressivity of prior metrics is often restricted to a curated set of medical conditions. Therefore, the quantitative investigation of metric-radiologist alignment conducted in this study is necessary for understanding whether these metrics meaningfully evaluate reports. Prior works have investigated the alignment between metrics and human judgment.23,33 However, to the best of our knowledge, these works pose one of two limitations for radiology report evaluation: (1) they study metric alignment with humans for general image captioning, which does not involve radiology-specific terminology, a high prevalence of negation, or expert human evaluators, and (2) they do not create a leveled comparison between metrics and radiologists, where metrics and radiologists assign scores to reports in identical experimental settings, or a granular understanding of metric behavior beyond the overall metric score. Our work builds a fair comparison between general natural language and clinically aware metrics and radiologists by providing them with the same set of information that is the reports and goes beyond metric scores to examine six granular failure modes of each metric. In addition, our work proposes a novel composite metric, RadCliQ, that aligns more strongly than any individual metric. We also show that current radiology report generation algorithms exhibit relatively low performance by all of these metrics.
To study metric-radiologist alignment, we designed metric-oracles: the reports selected from a large corpus with the highest metric score with respect to test reports. We had metrics and radiologists assign scores to the metric-oracles based on how well the metric-oracles match their respective test reports, and computed the alignment between metric and radiologist scores on the same reports. Pairing metric-oracles with test reports produces a narrower distribution of scores than using random reports. However, metric-oracles are necessary because comparisons with test reports are only reliable when the differences are small. If a random report, rather than a high-scoring report, was paired with the test report, the two reports could diverge to the extent that they were difficult to compare directly. In contrast, metric-oracles are comparable with test reports and therefore allow a meaningful evaluation of errors.
To generate metric-oracles, any report generation model is theoretically feasible. There are three main categories: the first generates free text based on semantics extracted from input chest X-ray images,16,34,35 the second retrieves existing text that best matches input images from a report corpus,13,36 and the third selects curated templates corresponding to a predefined set of abnormalities.10,37 We chose to use retrieval-based models to generate metric-oracles because retrieval from a training report corpus produces a controlled output space, instead of an unpredictable one produced by models that generate free text. Retrieval-based models also improve upon templating-based models in terms of flexibility and generalizability because the report corpus better captures real-world occurring conditions, combinations of conditions, and uncertainty. Furthermore, retrieval-based metric-oracle models outperformed existing report generation methods by a large margin.
By investigating the different categories of errors that radiologists identified in metric-oracle reports, we also uncovered specific metric failure modes that valuably inform the choice of metrics and interpretation of metric scores for evaluating generated reports. We find that BLEU performs worse than BERTScore and RadGraph in evaluating false prediction of finding. Yet, BLEU performs better than CheXbert vector similarity in evaluating incorrect position/location of finding. Therefore, RadGraph and BERTScore, which offer the strongest radiologist-alignment, also have better overall reliability against failure modes.
Using the individual metrics and RadCliQ, we also measured the progress of prior state-of-the-art models. Among impression-generating models, we find a significant performance gap between real report generation models and metric-oracle models, which represent the theoretical performance ceiling of retrieval-based methods on MIMIC-CXR for a given metric. This gap suggests that prior models in report generation still have significant room for improvement in creating high-quality reports that are useful to radiologists. We identify M2 Trans to be the best findings-generating model and CvT2DistilGPT2 to be the best model that jointly generates findings and impression sections. Overall, RadGraph is the best individual metric to use for its strong alignment with radiologists and reliability across failure modes. RadCliQ, a composite metric, offers the strongest alignment with radiologists.
This study has several important limitations. A main limitation is the inter-observer variability in radiologist evaluation. Although the evaluation scheme—the separation of clinically significant and insignificant errors, and the six error categories—was designed to be objective and consistent across radiologist evaluation, the same report often received varying scores between radiologists, a common occurrence in experiments that employ subjective ratings from clinicians. This suggests a potential limitation of the evaluation scheme used, but may also present an intrinsic problem with objective evaluation of radiology reports. Another limitation is the coverage of metrics. Although a variety of general and clinical natural language metrics are investigated, there exist other metrics in these two categories that may have different behaviors than the four investigated metrics. For instance, other text overlap-based metrics are commonly used in natural language generation beyond BLEU, such as CIDEr,31 METEOR,30 and ROUGE,32 which may have better or worse radiologist-alignment and reliability than BLEU in report generation.
In this study, we determined that the novel metrics RadGraph F1 and RadCliQ meaningfully measure progress in radiology report generation and hence can guide future report generation models in becoming clinically indistinguishable from radiologists. We have open-sourced the code for computing the individual metrics and RadCliQ on reports in the hope of facilitating future research in radiology report generation.
Experimental procedures
Resource availability
Lead contact
The lead contact for this work is Pranav Rajpurkar (pranav_rajpurkar@hms.harvard.edu).
Materials availability
Does not apply.
Datasets
We used the MIMIC-CXR dataset to conduct our study. The MIMIC-CXR dataset25,26,27 is a de-identified and publicly available dataset containing chest X-ray images and semi-structured radiology reports from the Beth Israel Deaconess Medical Center Emergency Department. There are 227,835 studies with 177,110 images conducted on 65,379 patients. We used the recommended train/validation/test split. We pooled the train and validation splits as the training report corpus from which metric-oracles are retrieved and used the test split as the set of ground-truth reports. We preprocessed the reports by filtering nan reports and extracting the impression and findings sections of reports, which contain key observations and conclusions drawn by radiologists. We follow the section extraction code provided in the MIMIC-CXR repository. In the training set, 187,383 impression reports, 153,415 findings reports, and 214,344 findings and impression joint reports are available. In the test set, 2,191 impression reports, 1,597 findings reports, and 2,192 findings and impression joint reports are available. We refer to the impression section when discussing reports for the metric-oracle reports and failure modes. When evaluating prior models, we use either the impression section, the findings section, or jointly the findings and impression sections based on what the prior model generates.
Metric-oracle reports
We constructed metric-oracle reports for four metrics. These include BLEU,28 BERTScore,29 CheXbert vector similarity (s_emb),9 and a novel metric RadGraph24 F1. BLEU and BERTScore are general natural language metrics for measuring the similarity between machine-generated and human-generated texts. BLEU computes n-gram overlap and is representative for the family of text overlap-based natural language generation metrics such as CIDEr,31 METEOR,30 and ROUGE.32 BERTScore has been proposed for capturing contextual similarity beyond exact textual matches. CheXbert vector similarity and RadGraph F1 are metrics designed to measure the correctness of clinical information. CheXbert vector similarity computes the cosine similarity between the CheXbert model embeddings for machine-generated and human-generated radiology reports. The CheXbert model is designed to evaluate radiology-specific information but its training supervision was limited to 14 pathologies. To address this limitation, we propose the use of the knowledge graph of the report to represent arbitrarily diverse radiology-specific information. We design a novel metric, RadGraph F1, that computes the overlap in clinical entities and relations that RadGraph extracts from machine- and human-generated reports. The four metrics are detailed in the “textual based and natural language generation performance metrics” subsection and the “clinically aware performance metrics” subsection.
For every test report, we generated the matching metric-oracle report by selecting the highest scoring report according to each of the four investigated metrics from the training set. We specifically used the impression section of the report. As an example of our setup, for the test report of “No acute cardiopulmonary process. Bilateral low lung volumes with crowding of bronchovascular markings and bibasilar atelectasis,” the metric-oracle retrieved with respect to BERTScore was: “No acute cardiopulmonary process. Low lung volumes and bibasilar atelectasis,” while the metric-oracle retrieved with respect to RadGraph F1 was: “No acute cardiopulmonary process. Bilateral low lung volumes.”
Using metric-oracles as the candidate reports as opposed to using other strategies such as randomly sampling reports offers two primary advantages: (1) metric-oracles are sufficiently accurate for radiologists to pinpoint specific errors and not be bogged down by candidate reports that are not remotely similar to the test reports, and (2) metric-oracles allow us to analyze where certain metrics fail since the reports are the hypothetical top retrievals.
Radiologist scoring criteria
In this work, we develop a scoring system for radiologists to evaluate the quality of candidate reports. The goals of our scoring system are to be objective, limit radiologist bias, and change linearly with report quality. To this end, scores are determined by counting the number of errors that candidate reports make where types of errors are broken down into six different categories. By explicitly defining each error category, we clarify what should be classified as an error. Following ACR’s RADPEER40 program for peer review, we differentiate between clinically significant and clinically insignificant errors. The detailed scoring criteria allow us to analyze report quality based on the accuracy of its findings and the clinical impact of its mistakes.
Textual-based and natural language generation performance metrics
In this study we make use of two natural language generation metrics: BLEU and BERTScore. The BLEU scores were computed as BLEU-2 bigrams with the fast_bleu library for parallel scoring. BERTScore uses the contextual embeddings from a BERT model to compute similarity of two text sequences. We used the bert_score library directly and used the “distilroberta-base” version of the model. We used the unscaled scores for metric-oracle retrieval and the baseline-scaled scores for all other analyses.
Clinically aware performance metrics
In addition to traditional natural language generation metrics, we also investigated metrics that were designed to capture clinical information in radiology reports. Since radiology reports are a special form of structured text that communicate diagnostics information, their quality depends highly on the correctness of clinical objects and descriptions, which is not a focus of traditional natural language metrics. To address this gap, the CheXbert labeler (which is improved from the CheXpert labeler)8,9 and RadGraph,24 were developed to parse radiology reports. We investigated whether they could be used as clinically aware metrics. We defined a metric as the cosine similarity between CheXbert model embeddings of the generated report and test report. We extracted the CLS token output embeddings before the final dropout layer and prediction heads. We used the implementation here: https://github.com/stanfordmlgroup/CheXbert. In prior literature, a common way of comparing generated reports against ground-truth reports is to compute the micro- and/or macro-F1/precision/recall scores averaged over 14 observation labels outputted by CheXpert/CheXbert. For instance, CXR-RePaiR computes the macro-average F1 over 14 observations to evaluate generations. Positive observation labels are treated as positive, while other labels, including negative, uncertain, and blank labels, are treated as negative. However, this approach limits the evaluation of generated reports to 14 observations and discrete outputs. Because radiology reports can reference diverse observations beyond the 14 and contain more nuanced semantics about the observations, we decided to use the CheXbert model embedding before the final classifiers, which produce 14 outputs to capture a more accurate representation of the report. Our design is supported by a prior work that uses the same CheXbert model embeddings as deep representations of radiology reports for heart failure patient mortality prediction.41 In their experiments, they also found that these hidden features led to better prediction performance than the features of 14 observations extracted by CheXpert. This suggests that the model embeddings may preserve more information about the reports than the final model output of observation labels. CXR-RePaiR also adopts the same formulation of CheXbert vector similarity as a report evaluation metric. We propose a novel metric as the overlap in parsed RadGraph graph structures: the RadGraph entity and relation F1 score. RadGraph is an approach for parsing radiology reports into knowledge graphs containing entities (nodes) and relations (edges), which can capture radiology concept dependencies and semantic meaning. We used the model checkpoint as provided here: https://physionet.org/content/radgraph/1.0.0/,27 and inference code as provided here: https://github.com/dwadden/dygiepp,42 to generate RadGraph entities and relations on generated and test reports.
Retrieval-based metric-oracle models
To generate metric-oracle reports, the most immediate attempt is to adopt methods akin to those for multi-label classification tasks. Namely, we can curate a set of medical conditions and obtain radiologist annotations for each condition over a training set of reports. Then, we can train a classifier that outputs the likelihood of having each condition given an X-ray image, and proceed to select the corresponding report templates for conditions with high likelihood.10 Some more nuanced approaches paraphrase the curated templates after selection.37 The attempt at templating for report generation is well-grounded in abundant experience in multi-label image classification as well as its highly controlled output space. However, its flaw is also prominent, in that it is restricted to a manually curated predefined set of medical conditions and report templates. It does not generalize to unseen or complex conditions, express combinations of conditions, or capture uncertainty in diagnoses. The CheXbert labeler, for instance, can classify 13 conditions and the no-finding observation.9 This set is representative of common medical observations but not comprehensive. Therefore, while we may define a larger set of conditions with the help of radiologists, manual curation and templating are nevertheless too inflexible for optimizing with respect to automated metrics. To generate reports of higher quality, we consider matching reports more closely onto test reports. We can do so by either generating new text from scratch or retrieving free text from an existing corpus of reports written by radiologists, given an X-ray image.34,36 Out of the two approaches, retrieval-based methods have the advantage of a controlled output space that is the set of training report corpus. Therefore, in this study, we use retrieval-based methods to generate metric-oracle reports.
RadGraph metric-oracle model entities and relations match
The RadGraph F1 metric-oracle model retrieves reports with the highest F1 score match in terms of entities and relations. Specifically, we treat two entities as matched if their tokens (words in the original report) and labels (entity type) match. We treat two relations as matched if their start and end entities match and the relation type matches. These criteria are consistent with what the RadGraph authors have done. For combining entities and relations, we take the average of F1 score of entity match and relation match, respectively. We generated RadGraph entities and relations for each report in the training and test corpora. We implemented the metric-oracle model by finding, for each report in the test set, which report in the training set is the best match based on the average of entity and relation F1 scores. For reports without nonzero F1 score matches, we used the most frequent report in the training set, “No acute cardiopulmonary process,” as the metric-oracle report in the radiologist experiment.
Statistical analysis
Metric-radiologist alignment
The alignment of metrics with radiologists’ scoring was determined using the Kendall tau b correlation coefficient. We construct 95% bootstrap confidence intervals by creating 1,000 resamples with replacement where each resample size is the number of studies (50). In this calculation, the number of errors is the mean number across all raters. We additionally test for the difference in correlations between two metrics by counting the number of positive correlation differences computed on 1,000 resamples of metric scores. The fraction of positive correlation differences indicate the p value for the null hypothesis that there is zero difference in correlation between the metrics.
Metric failure modes
We conduct one-sided two-sample t tests on pairs of metrics’ error counts for total number of clinically significant and insignificant errors and clinically significant errors within each of the six error categories. We assume equal population variances for the t tests. We take the error count of one radiologist and one study as one data point. Because there are 6 radiologists and 50 studies, we have 300 data points per metric for either total number of clinically significant and insignificant errors or clinically significant errors and for 1 error category. With 4 metrics, there are 12 unique pairs of 2 different metrics for one-sided two-sample t tests with (300 + 300 – 2 = 598) degrees of freedom. We use the Benjamini-Hochberg procedure with a false discovery rate of 1% to account for multiple-hypothesis testing on 12 tests within an error type and an error category, and determine the significance of a metric having a more-/less-prominent failure mode compared with other metrics.
Prior models evaluation
To evaluate performance of metric-oracle models and prior state-of-the-art models, we construct 95% bootstrap confidence intervals by taking 5,000 resamples with replacement of metric scores assigned to generated reports.
Composite metric RadCliQ
The composite metric model used to predict the total number of errors was evaluated using the Kendall tau b statistical test. This test produces a tau value correlation coefficient and a corresponding p value, which was used to determine the significance of the result (p < 0.01). The same statistical comparison procedure described in metric-radiologist alignment with 5,000 resmaples was used to compare the correlation of RadCliQ with that of other metrics.
The analyses were performed using statsmodels, scikit-learn, and SciPy packages in Python.
Implementation of prior report generation methods
We used the following implementations of prior methods in radiology report generation: M2 Trans, https://github.com/ysmiura/ifcc11,42; R2Gen, https://github.com/cuhksz-nlp/R2Gen12; CXR-RePaiR, https://github.com/rajpurkarlab/CXR-RePaiR13; WCL, https://github.com/zzxslp/WCL14; CvT2DistilGPT2, https://github.com/aehrc/cvt2distilgpt2.15 CXR-RePaiR was trained to generate the impression section through retrieval. M2 Trans and CvT2DistilGPT2 were trained to generate the findings section, with maximum sequence lengths of 128 and 60, respectively. R2Gen and WCL were trained to jointly generate the findings and impression sections, with maximum sequence lengths of 60 and 100, respectively. We did not shorten or cut off any part of the actual reports when evaluating our report generation method to avoid creating a problem in our evaluation process. If we had shortened the reports, it could have allowed a generation method to be trained to only produce very short reports that lack important information, but still receive good evaluation scores. Thus, to ensure accurate evaluation, we did not truncate the ground-truth reports. We used these prior methods to generate reports for all available studies in the test set. For impression only and findings only generations, there are fewer test reports and metric outputs. This is considered acceptable, because there are still sufficiently large numbers of reports for a reliable estimate of model performance. For each study ID, if the model generated multiple reports corresponding to different X-ray images for the same study, we used the generated report corresponding to the anterior-posterior or posterior-anterior views if any were present. If both were present, we randomly chose a report out of the two. If neither was present, we randomly chose a report out of the available reports corresponding to other views. Among variations of CXR-RePaiR, we chose CXR-RePaiR-2 to be consistent with their original study.13
Acknowledgments
We thank M.A. Endo MD for helpful review and feedback on the radiologist evaluation survey design and the manuscript. Support for this work was provided in part by the Medical Imaging Data Resource Center (MIDRC) under contracts 75N92020C00008 and 75N92020C00021 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health.
Author contributions
P.R. conceived the study. F.Y., M.E., and R.K. contributed to the design, implementation, and analyses of all aspects of this study. I.P., A.T., E.P.R., E.K.U.N.F., H.M.H.L., and V.K.V. provided suggestions on the setup of the radiologist evaluation survey and provided annotations in the radiologist evaluation process. Z.S.H.A. contributed to the design of the illustrations and figures. A.Y.N., C.P.L., and V.K.V. provided guidance on the study. P.R. supervised the study. All authors approved the final version.
Declaration of interests
The authors declare no competing non-financial interests but the following competing financial interests: I.P. is a consultant for MD.ai and Diagnosticos da America (Dasa). C.P.L. serves on the board of directors and is a shareholder of Bunkerhill Health. He is an advisor and option holder for GalileoCDS, Sirona Medical, Adra, and Kheiron. He is an advisor to Sixth Street and an option holder in whiterabbit.ai. His research program has received grant or gift support from Carestream, Clairity, GE Healthcare, Google Cloud, IBM, IDEXX, Hospital Israelita Albert Einstein, Kheiron, Lambda, Lunit, Microsoft, Nightingale Open Science, Nines, Philips, Subtle Medical, VinBrain, Whiterabbit.ai, the Paustenbach Fund, the Lowenstein Foundation, and the Gordon and Betty Moore Foundation.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2023.100802.
Supplemental information
Data and code availability
-
•
Original data for the radiologist error annotations have been deposited to the Radiology Report Expert Evaluation (ReXVal) Dataset38 with credentialed access at https://physionet.org/content/rexval-dataset/1.0.0/ (https://doi.org/10.13026/2fp8-qr71). The radiology report data used in the study are available with credentialed access at: https://physionet.org/content/mimic-cxr-jpg/2.0.0/ (https://doi.org/10.13026/8360-t248). Credentialed access can be obtained via an application to PhysioNet.
-
•
The code for computing the composite metric RadCliQ and individual metrics is made publicly available at: https://doi.org/10.5281/zenodo.7579952.39
References
- 1.Rajpurkar P., Chen E., Banerjee O., Topol E.J. AI in health and medicine. Nat. Med. 2022;28:31–38. doi: 10.1038/s41591-021-01614-0. [DOI] [PubMed] [Google Scholar]
- 2.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajpurkar P., Joshi A., Pareek A., Ng A.Y., Lungren M.P. Proceedings of the Conference on Health, Inference, and Learning. Association for Computing Machinery; 2021. CheXternal: Generalization of Deep Learning Models for Chest X-ray Interpretation to Photos of Chest X-rays and External Clinical Settings; pp. 125–132. [Google Scholar]
- 4.Jin B.T., Palleti R., Shi S., Ng A.Y., Quinn J.V., Rajpurkar P., Kim D. Transfer learning enables prediction of myocardial injury from continuous single-lead electrocardiography. J. Am. Med. Inf. Assoc. 2022;29:1908–1918. doi: 10.1093/jamia/ocac135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajpurkar P., Lungren M.P. The Current and Future State of AI Interpretation of Medical Images. N. Engl. J. Med. Overseas. Ed. 2023;388:1981–1990. doi: 10.1056/NEJMra2301725. [DOI] [PubMed] [Google Scholar]
- 6.Tiu E., Talius E., Patel P., Langlotz C.P., Ng A.Y., Rajpurkar P. Expert-level detection of pathologies from unannotated chest X-ray images via self-supervised learning. Nat. Biomed. Eng. 2022;6:1399–1406. doi: 10.1038/s41551-022-00936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarwal N., Moehring A., Pranav R., Salz T. 2023. Combining Human Expertise with Artificial Intelligence: Experimental Evidence from Radiology. [Google Scholar]
- 8.Irvin J., Rajpurkar P., Ko M., Yu Y., Ciurea-Ilcus S., Chute C., Marklund H., Haghgoo B., Ball R., Shpanskaya K., et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv. 2019 doi: 10.48550/arXiv.1901.07031. Preprint at. [DOI] [Google Scholar]
- 9.Smit A., Jain S., Rajpurkar P., Pareek A., Ng A.Y., Lungren M.P. CheXbert: Combining Automatic Labelers and Expert Annotations for Accurate Radiology Report Labeling Using BERT. arXiv. 2020 doi: 10.48550/arXiv.2004.09167. Preprint at. [DOI] [Google Scholar]
- 10.Pino P., Parra D., Besa C., Lagos C. Machine Learning in Medical Imaging. Springer; 2021. Clinically Correct Report Generation from Chest X-Rays Using Templates; pp. 654–663. [Google Scholar]
- 11.Miura Y., Zhang Y., Tsai E.B., Langlotz C.P., Jurafsky D. Improving Factual Completeness and Consistency of Image-to-Text Radiology Report Generation. arXiv. 2020 doi: 10.48550/arXiv.2010.10042. Preprint at. [DOI] [Google Scholar]
- 12.Chen Z., Song Y., Chang T.-H., Wan X. Generating Radiology Reports via Memory-driven Transformer. arXiv. 2020 doi: 10.48550/arXiv.2010.16056. Preprint at. [DOI] [Google Scholar]
- 13.Endo M., Krishnan R., Krishna V., Ng A.Y., Rajpurkar P. Machine Learning for Health. PMLR; 2021. Retrieval-Based Chest X-Ray Report Generation Using a Pre-trained Contrastive Language-Image Model; pp. 209–219. [Google Scholar]
- 14.Yan A., He Z., Lu X., Du J., Chang E., Gentili A., McAuley J., Hsu C.-N. Weakly Supervised Contrastive Learning for Chest X-Ray Report Generation. arXiv. 2021 doi: 10.48550/arXiv.2109.12242. Preprint at. [DOI] [Google Scholar]
- 15.Nicolson A., Dowling J., Koopman B. Improving Chest X-Ray Report Generation by Leveraging Warm-Starting. arXiv. 2022 doi: 10.48550/arXiv.2201.09405. Preprint at. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H.-Y., Chen X., Zhang Y., Luo R., Wang L., Yu Y. Generalized radiograph representation learning via cross-supervision between images and free-text radiology reports. Nat. Mach. Intell. 2022;4:32–40. [Google Scholar]
- 17.Ramesh V., Chi N.A., Rajpurkar P. Proceedings of Machine Learning for Health. PMLR; 2022. Improving Radiology Report Generation Systems by Removing Hallucinated References to Non-existent Priors; pp. 456–473. [Google Scholar]
- 18.Jeong J., Tian K., Li A., Hartung S., Behzadi F., Calle J., Osayande D., Pohlen M., Adithan S., Rajpurkar P. Proceedings of Medical Imaging with Deep Learning. MIDI; 2023. Multimodal Image-Text Matching Improves Retrieval-based Chest X-Ray Report Generation. [Google Scholar]
- 19.Li M., Lin B., Chen Z., Lin H., Liang X., Chang X. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2023. Dynamic Graph Enhanced Contrastive Learning for Chest X-Ray Report Generation; pp. 3334–3343. [Google Scholar]
- 20.Yang S., Wu X., Ge S., Zhou S.K., Xiao L. Knowledge matters: Chest radiology report generation with general and specific knowledge. Med. Image Anal. 2022;80 doi: 10.1016/j.media.2022.102510. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen H., Nie D., Badamdorj T., Liu Y., Zhu Y., Truong J., Cheng L. Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. 2021. Automated Generation of Accurate & Fluent Medical X-ray Reports; pp. 3552–3569. [Google Scholar]
- 22.Hossain M.Z., Sohel F., Shiratuddin M.F., Laga H. A Comprehensive Survey of Deep Learning for Image Captioning. arXiv. 2018 doi: 10.48550/arXiv.1810.04020. Preprint at. [DOI] [Google Scholar]
- 23.Boag W., Kané H., Rawat S., Wei J., Goehler A. Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency. 2021. A Pilot Study in Surveying Clinical Judgments to Evaluate Radiology Report Generation. [Google Scholar]
- 24.Jain S., Agrawal A., Saporta A., Truong S.Q.H., Duong D.N., Bui T., Chambon P., Zhang Y., Lungren M.P., Ng A.Y., et al. RadGraph: Extracting Clinical Entities and Relations from Radiology Reports. arXiv. 2021 doi: 10.48550/arXiv.2106.14463. Preprint at. [DOI] [Google Scholar]
- 25.Johnson A.E.W., Pollard T.J., Berkowitz S.J., Greenbaum N.R., Lungren M.P., Deng C.-Y., Mark R.G., Horng S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data. 2019;6:317–318. doi: 10.1038/s41597-019-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson A.E.W., Pollard T.J., Greenbaum N.R., Lungren M.P., Deng C.-Y., Peng Y., Lu Z., Mark R.G., Berkowitz S.J., Horng S. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv. 2019 doi: 10.48550/arXiv.1901.07042. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberger A.L., Amaral L.A., Glass L., Hausdorff J.M., Ivanov P.C., Mark R.G., Mietus J.E., Moody G.B., Peng C.-K., Stanley H.E. PhysioBank, PhysioToolkit, and PhysioNet. Circulation. 2000;101:E215–E220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 28.Papineni K., Roukos S., Ward T., Zhu W.J. Proceedings of the 40th Annual Meeting on Association for Computational Linguistics. 2002. BLEU: a method for automatic evaluation of machine translation. [Google Scholar]
- 29.Zhang T., Kishore V., Wu F., Weinberger K.Q., Artzi Y. BERTScore: Evaluating Text Generation with BERT. arXiv. 2019 doi: 10.48550/arXiv.1904.09675. Preprint at. [DOI] [Google Scholar]
- 30.Lavie A., Agarwal . Proceedings of the Second Workshop on Statistical Machine Translation. 2007. Meteor: an automatic metric for MT evaluation with high levels of correlation with human judgments. [Google Scholar]
- 31.Vedantam R., Zitnick C.L., Parikh D. CIDEr: Consensus-based Image Description Evaluation. arXiv. 2014 doi: 10.48550/arXiv.1411.5726. Preprint at. [DOI] [Google Scholar]
- 32.Lin C.-Y. Text Summarization Branches Out. 2004. ROUGE: A Package for Automatic Evaluation of Summaries; pp. 74–81. [Google Scholar]
- 33.Anderson P., Fernando B., Johnson M., Gould S. SPICE: Semantic Propositional Image Caption Evaluation. arXiv. 2016 doi: 10.48550/arXiv.1607.08822. Preprint at. [DOI] [Google Scholar]
- 34.Monshi M.M.A., Poon J., Chung V. Deep learning in generating radiology reports: A survey. Artif. Intell. Med. 2020;106 doi: 10.1016/j.artmed.2020.101878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y., Huang L., Zhou T., Fu H., Shao L. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021. Visual-Textual Attentive Semantic Consistency for Medical Report Generation; pp. 3985–3994. [Google Scholar]
- 36.Wang X., Zhang Y., Guo Z., Li J. 2018. ImageSem at ImageCLEF 2018 Caption Task: Image Retrieval and Transfer Learning. [Google Scholar]
- 37.Li C.Y., Liang X., Hu Z., Xing E.P. Knowledge-Driven Encode, Retrieve, Paraphrase for Medical Image Report Generation. AAAI. 2019;33:6666–6673. [Google Scholar]
- 38.Yu F., Endo M., Krishnan R., Pan I., Tsai A., Reis E.P., Fonseca E.K.U., Lee H., Shakeri Z., Ng A., et al. 2023. Radiology Report Expert Evaluation (ReXVal) Dataset. [DOI] [Google Scholar]
- 39.Yu K., Rayan-Krishnan . 2023. rajpurkarlab/CXR-Report-Metric: v1.1.0. [DOI] [Google Scholar]
- 40.Goldberg-Stein S., Frigini L.A., Long S., Metwalli Z., Nguyen X.V., Parker M., Abujudeh H. ACR RADPEER Committee White Paper with 2016 Updates: Revised Scoring System, New Classifications, Self-Review, and Subspecialized Reports. J. Am. Coll. Radiol. 2017;14:1080–1086. doi: 10.1016/j.jacr.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Lee H.G., Sholle E., Beecy A., Al’Aref S., Peng Y. Leveraging Deep Representations of Radiology Reports in Survival Analysis for Predicting Heart Failure Patient Mortality. Proc. Conf. 2021;2021:4533–4538. doi: 10.18653/v1/2021.naacl-main.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadden D., Wennberg U., Luan Y., Hajishirzi H. 2019. Entity, Relation, and Event Extraction with Contextualized Span Representations. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original data for the radiologist error annotations have been deposited to the Radiology Report Expert Evaluation (ReXVal) Dataset38 with credentialed access at https://physionet.org/content/rexval-dataset/1.0.0/ (https://doi.org/10.13026/2fp8-qr71). The radiology report data used in the study are available with credentialed access at: https://physionet.org/content/mimic-cxr-jpg/2.0.0/ (https://doi.org/10.13026/8360-t248). Credentialed access can be obtained via an application to PhysioNet.
-
•
The code for computing the composite metric RadCliQ and individual metrics is made publicly available at: https://doi.org/10.5281/zenodo.7579952.39