Abstract
Chemical graph theory is a well-established discipline within chemistry that employs discrete mathematics to represent the physical and biological characteristics of chemical substances. In the realm of chemical compounds, graph theory-based topological indices are commonly employed to depict their geometric structure. The main aim of this paper is to investigate the degree-based topological indices of dominating David derived networks (DDDN) and assess their effectiveness. DDDNs are widely used in analyzing the structural and functional characteristics of complex networks in various fields such as biology, social sciences, and computer science. We considered the FN*, , and topological indices for DDDNs. Our computations' findings provide a clear understanding of the topology of networks that have received limited study. These computed indices exhibit a high level of accuracy when applied to the investigation of QSPRs and QSARs, as they demonstrate the strongest correlation with the acentric factor and entropy.
Subject terms: Mathematics and computing, Applied mathematics, Computational science
Introduction
In the field of graph theory, specifically in chemical graph theory, a chemical molecule is represented by a molecular graph, which is a simple graph. In this representation, vertices denote the atoms and edges represent bonds or connections. The edges goes beyond simple connectivity; it encompasses the type of bond as well. For instance, a single edge might denote a single covalent bond, while a double edge could represent a double bond involving the sharing of two pairs of electrons.
The emerging field of cheminformatics, which explores the relationship between quantitative structure–activity and structure–property, is gaining momentum as it aids in the prediction of biological activities. Topological indices are important invariants derived from graph theory that enable the characterization of a graph's topology. A topological index is a numerical value that provides information about the structure of a graph. Topological indices help in identifying various characteristics of a graph. Furthermore, the topology of a graph remains invariant under the automorphisms of graphs. Comperisons of the degree based toplogical indices hold a particularly significant place in research1, 2.
The first toplogical index introduced by Wiener, during the research of paraffin melting point. Initially termed “path number”, it was later renamed and has since become known as the Wiener index. Researchers have put a lot of effort into studying chemical graph theory. A key component of graph theory's work involves honeycomb networks. The honeycomb shape, with its hexagonal pattern of cells, finds a wide range of applications across various fields due to its unique structural and geometric properties. Some of the notable applications of the honeycomb shape include as: In structural engineering and architecture the honeycomb structure's hexagonal arrangement provides exceptional strength and stability while using minimal material. This makes it suitable for applications in construction, such as in lightweight yet strong support structures, building facades, and panels. On the other hands, in art and design, the visually appealing hexagonal pattern of the honeycomb has inspired artists, designers, and architects to incorporate it into their creations. From decorative elements in interior design to art installations, the honeycomb pattern adds a unique aesthetic.
In this article the notation E denotes an edge set and V denotes the vertex set of a graph G. The expression is the number of edges overall connected to a particular vertex v.
For the sake of simplicity, assume that a and b are two adjacent vertices and E is an edge between them, then the edge partition of E is denoted by and formulated as .
The degree-based topological indices shows a significant role in the field of mathematical chemistry3–7, and widely used to develop models that accurately predict the boiling points of alkanes with carbon atom8. Some current discovered degree-based neighborhood indices are presented in9, 10 and shown strong connections between entropy and the acentric factor.
In11–16, different chemical significant graphs' topological indices are considered. Baig et al.17 considered the topological indices for several silicates and oxide networks. Ullah et al.18, compared and examined the computational characteristics of two carbon nanosheets using some innovative topological indices. The topological characteristics of rhombus-type silicate and oxide networks were explored by Javaid et al.19. Recently, Koam et al.20, established the entropy measures of Y-junction based nanostructures. Ali et al.21 give some properties of ve-degree based topological indices for hex-derived networks. In this study, an examination was conducted on distance-based topological polynomials that are associated with zero-divisor graphs, as discussed in22. The authors of23 obtained the polynomials of degree-based indices of metal–organic networks. Zaman et al., determined the kemeny’s constant and spanning trees of hexagonal ring network24. Some upper bound and lower bound of graphs and also the spectral analysis of graphs are discussed in25–28. In this research, inspired by earlier studies, we establish some exact expressions of the different types of Dominating David derived networks and their comparisons.
We have calculated the forgotten index ()29, the second zargeb index ()30 and the Harmonic index ()31 for DDD networks. These topological indices are defined as , , .
Constructions of dominating David derived networks (DDDN)
In the field of chemistry, honeycomb networks are utilized as representations for benzoid hydrocarbons. Honeycomb networks find extensive applications in various domains, including graphics, such as cell phone base stations and image processing. The honeycomb network is formed by enclosing the boundaries with a layer of hexagons. Based on the honeycomb network, different types of Dominating David derived networks can be derived. One can follow the below steps to construct the DDDN (t dimension):
- Step 1:
Consider a t-dimension honeycomb network (see Fig. 1a).
- Step 2:
Add another vertex to divide each edge into two pieces (see Fig. 1b).
- Step 3:
In each hexagonal cell, connect the new vertices by an edge if they are at a distance of 4 within a hexagon (see Fig. 1c).
- Step 4:
Add new vertices at new edge intersections. (see Fig. 1d).
- Step 5:
Remove the starting vertices and edges of the honeycomb (see Fig. 1e).
- Step 6:
Divide each horizontal edge into two parts by addind a new vertex (see Fig. 1f).
Figure 1.
The steps to derive DDD (2).
Main results
Our key findings rely on the edge partitions of Figs. 2, 3 and 4 as given below. We have calculated these edge partitions based on the degrees of the end vertices of each edge. For instance, the first row of Fig. 1 shows the degrees of the end vertices of edges, while the second row illustrates the count of edges with those specific degrees. In the same way, we have obtained the other tables.
Figure 2.
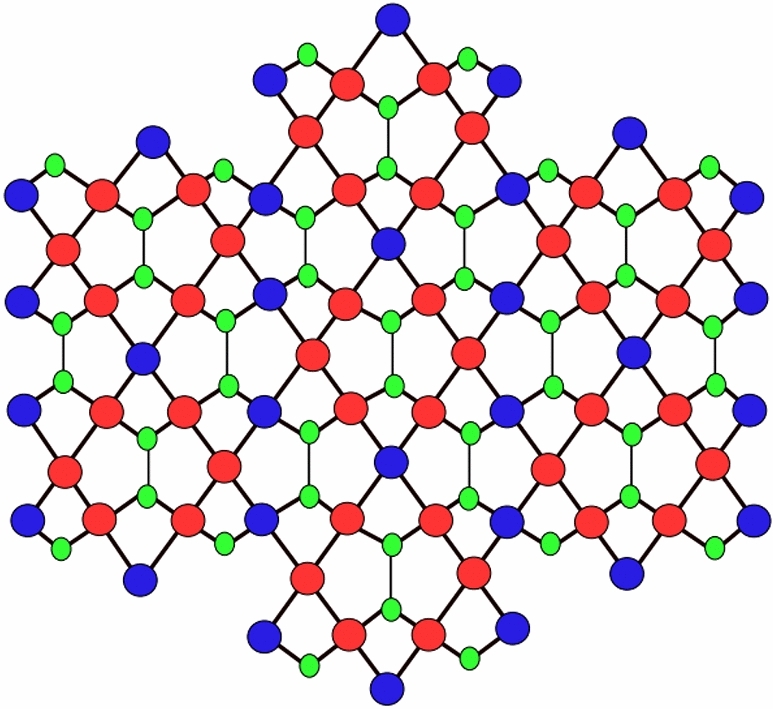
First type of D1(2) network.
Figure 3.

First type of D2(2) network.
Figure 4.
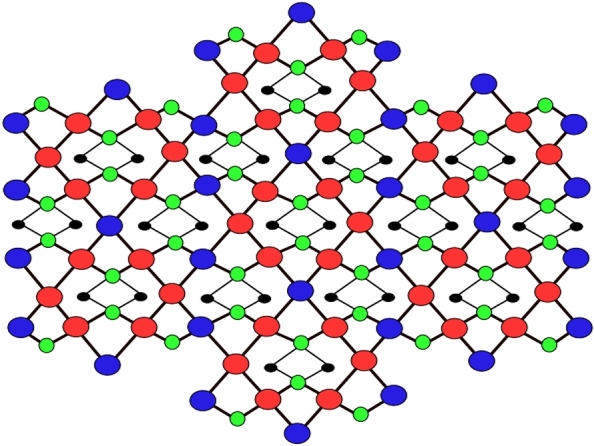
Third type of D3(2) network.
The FN* topological index for dominating David derived networks
Let G be a graph in D1(t), D2(t) and D3(t) then according to the definition of and Table 1, we have
Table 1.
Edges partition D1(t).
| Frequency |
Similarly, from Table 2, we have
Table 2.
Edge partition D2(t).
| Frequency |
And from Table 3, one has
Table 3.
Edge partition D3(t).
| Frequency |
The topological index for DDDN
Let G be a graph in D1(t), D2(t) and D3(t) then according to the definition of and Table 1 we have
Likewise, based on the information presented in Table 2, we obtain
And from Table 3, one has
The topological index for DDDN
Let G be a graph in D1(t), D2(t) and D3(t) then according to the definition of and Table 1, we have
Similarly, from Table 2, we have
And from Table 3, we have
Concluding Remarks
In this study, we have considered the , and topological indices. Our simulated results help for the better comprehend topology and enhance physical properties of the honeycomb structure. The computed indices, and above, as previously mentioned, have the most closely relates to the acentric factor and entropy consequently, they are extremely accurate in QSPR and QSAR analysis.
In Table 4, the topological indices computed are represented mathematically. As we can see, increasing the values of t, increases the value of the indices as well. We have precise analytical formulations for the D1, D2 and D3 networks, considering various topological indices. In the rapidly expanding fields of nanotechnology and applications, such as networks, our current discoveries and techniques can be applied to other, more complex structures. The utilization of distance-based topological indices poses greater challenges and complexity, but they can be employed alongside existing methods. Exploring these types of studies will be the focus of future research endeavors. In Table 4 and Fig. 5, we computed the numerical comparison of the certain topological indices for D1, D2 and D3 networks, which shows that when we increase t as a result the values of the topological indices also increases. These numerical comaprisons also shows that the inceasing rate of for D3 is greater than the other topological indices. Since, in graph theory, the is a mathematical concept used to describe the connectivity. Therefore, a higher reflects the more connectivity among the atoms of a molecule. This indicates that the D3 molecule has a greater potential for forming diverse interactions with other molecules and participating in a wider range of chemical reactions.
Table 4.
The comparison of , and for D1(t), D2(t) and D3(t) graphs.
| (D1) | (D2) | (D3) | (D1) | (D2) | (D3) | (D1) | (D2) | (D3) | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1221 | 526 | 608 | 233 | 236 | 272 | 2270 | 1493 | 1152 |
| 2 | 5912 | 4626 | 5856 | 2143 | 2188 | 2720 | 10,084 | 9992 | 11,296 |
| 3 | 15,031 | 13,298 | 17,152 | 6231 | 6372 | 8048 | 26,682 | 27,527 | 33,248 |
| 4 | 28,578 | 26,542 | 34,496 | 12,497 | 12,788 | 16,256 | 52,064 | 54,098 | 67,008 |
| 5 | 46,553 | 44,358 | 57,888 | 20,941 | 21,436 | 27,344 | 86,230 | 89,705 | 112,576 |
| 6 | 68,956 | 66,746 | 87,328 | 31,563 | 32,316 | 30,512 | 129,180 | 134,348 | 169,952 |
Figure 5.
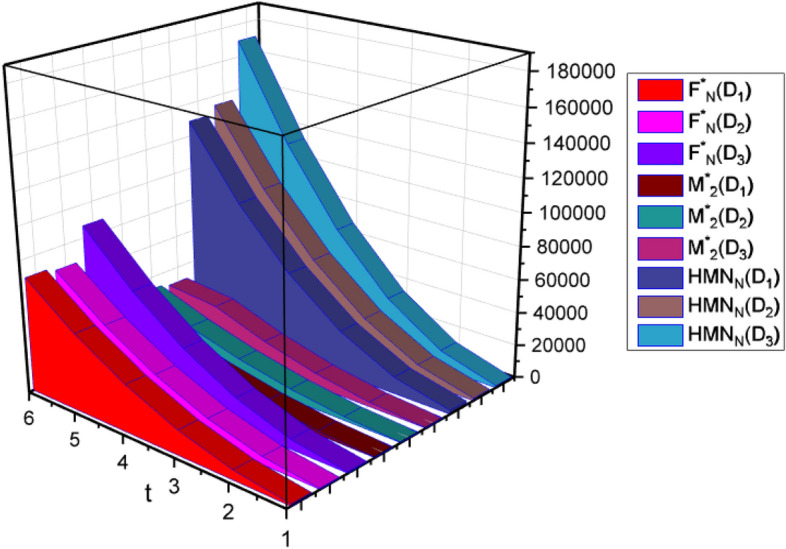
The comparison graph.
Author contributions
All authors have made equal contributions to this paper at every stage, including conceptualization and the final drafting process. The manuscript has been approved by all authors and consent for publication has been granted.
Data availability
All data generated or analysed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zaman S, et al. Mathematical analysis and molecular descriptors of two novel metal–organic models with chemical applications. Sci. Rep. 2023;13(1):5314. doi: 10.1038/s41598-023-32347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H. Comparison between Merrifield–Simmons index and some vertex-degree-based topological indices. Comput. Appl. Math. 2023;42(2):89. doi: 10.1007/s40314-023-02240-x. [DOI] [Google Scholar]
- 3.Babar U, et al. Multiplicative topological properties of graphs derived from honeycomb structure. Aims Math. 2020;5(2):1562–1587. doi: 10.3934/math.2020107. [DOI] [Google Scholar]
- 4.Ali H, et al. On the degree-based topological indices of some derived networks. Mathematics. 2019;7(7):612. doi: 10.3390/math7070612. [DOI] [Google Scholar]
- 5.Yang S, et al. Structure of Fejes Tóth cells in natural honey bee combs. Apidologie. 2022;53(1):6. doi: 10.1007/s13592-022-00915-8. [DOI] [Google Scholar]
- 6.Hayat S, Malik MA, Imran M. Computing topological indices of honeycomb derived networks. Rom. J. Inf. Sci. Technol. 2015;18(2):144–165. [Google Scholar]
- 7.Imran M, et al. On topological properties of poly honeycomb networks. Period. Math. Hung. 2016;73:100–119. doi: 10.1007/s10998-016-0132-5. [DOI] [Google Scholar]
- 8.Mukwembi S, Nyabadza F. A new model for predicting boiling points of alkanes. Sci. Rep. 2021;11(1):24261. doi: 10.1038/s41598-021-03541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad MS, et al. Calculating degree-based topological indices of dominating David derived networks. Open Phys. 2017;15(1):1015–1021. doi: 10.1515/phys-2017-0126. [DOI] [Google Scholar]
- 10.Imran M, Baig AQ, Ali H. On topological properties of dominating David derived networks. Can. J. Chem. 2016;94(2):137–148. doi: 10.1139/cjc-2015-0185. [DOI] [Google Scholar]
- 11.Aslam A, et al. On topological indices of certain dendrimer structures. Z. Naturforsch. A. 2017;72(6):559–566. doi: 10.1515/zna-2017-0081. [DOI] [Google Scholar]
- 12.Ullah A, Zeb A, Zaman S. A new perspective on the modeling and topological characterization of H-naphtalenic nanosheets with applications. J. Mol. Model. 2022;28(8):1–13. doi: 10.1007/s00894-022-05201-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, et al. The connective eccentricity index of graphs and its applications to octane isomers and benzenoid hydrocarbons. Int. J. Quantum Chem. 2020;120(18):e26334. doi: 10.1002/qua.26334. [DOI] [Google Scholar]
- 14.Mondal S, De N, Pal A. Topological properties of graphene using some novel neighborhood degree-based topological indices. Int. J. Math. Ind. 2019;11(01):1950006. doi: 10.1142/S2661335219500060. [DOI] [Google Scholar]
- 15.De N, et al. On some degree based topological indices of mk-graph. J. Discrete Math. Sci. Cryptogr. 2020;23(6):1183–1194. doi: 10.1080/09720529.2020.1809112. [DOI] [Google Scholar]
- 16.Zaman S, Ali A. On connected graphs having the maximum connective eccentricity index. J. Appl. Math. Comput. 2021;67(1):131–142. doi: 10.1007/s12190-020-01489-3. [DOI] [Google Scholar]
- 17.Baig AQ, Imran M, Ali H. On topological indices of poly oxide, poly silicate, DOX, and DSL networks. Can. J. Chem. 2015;93(7):730–739. doi: 10.1139/cjc-2014-0490. [DOI] [Google Scholar]
- 18.Ullah A, et al. Computational and comparative aspects of two carbon nanosheets with respect to some novel topological indices. Ain Shams Eng. J. 2022;13(4):101672. doi: 10.1016/j.asej.2021.101672. [DOI] [Google Scholar]
- 19.Javaid M, Rehman MU, Cao J. Topological indices of rhombus type silicate and oxide networks. Can. J. Chem. 2017;95(2):134–143. doi: 10.1139/cjc-2016-0486. [DOI] [Google Scholar]
- 20.Koam AN, et al. Entropy measures of Y-junction based nanostructures. Ain Shams Eng. J. 1913;2022:10. [Google Scholar]
- 21.Ahmad A, Imran MJC. Vertex-edge-degree-based topological properties for hex-derived networks. Complexity. 2022;2022:1–13. [Google Scholar]
- 22.Ahmad A, et al. Analysis of distance-based topological polynomials associated with zero-divisor graphs. Comput. Mater. Contin. 2022;70(2):2898–2904. [Google Scholar]
- 23.Ahmad A, et al. Polynomials of degree-based indices of metal-organic networks. Comb. Chem. High Throughput Screen. 2022;25(3):510–518. doi: 10.2174/1386207323666201229152828. [DOI] [PubMed] [Google Scholar]
- 24.Zaman, S., et al., The Kemeny’s Constant and Spanning Trees of Hexagonal Ring Network.
- 25.Khabyah AA, et al. Minimum zagreb eccentricity indices of two-mode network with applications in boiling point and benzenoid hydrocarbons. Mathematics. 2022;10(9):1393. doi: 10.3390/math10091393. [DOI] [Google Scholar]
- 26.Zaman S, et al. Maximum H-index of bipartite network with some given parameters. AIMS Math. 2021;6(5):5165–5175. doi: 10.3934/math.2021306. [DOI] [Google Scholar]
- 27.Zaman S. Cacti with maximal general sum-connectivity index. J. Appl. Math. Comput. 2021;65(1):147–160. doi: 10.1007/s12190-020-01385-w. [DOI] [Google Scholar]
- 28.Zaman S. Spectral analysis of three invariants associated to random walks on rounded networks with 2 n-pentagons. Int. J. Comput. Math. 2022;99(3):465–485. doi: 10.1080/00207160.2021.1919303. [DOI] [Google Scholar]
- 29.Gao W, et al. Forgotten topological index of chemical structure in drugs. Saudi Pharm. J. 2016;24(3):258–264. doi: 10.1016/j.jsps.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalifeh M, Yousefi-Azari H, Ashrafi AR. The first and second zagreb indices of some graph operations. Discrete Appl. Math. 2009;157(4):804–811. doi: 10.1016/j.dam.2008.06.015. [DOI] [Google Scholar]
- 31.Zhong L. The harmonic index for graphs. Appl. Math. Lett. 2012;25(3):561–566. doi: 10.1016/j.aml.2011.09.059. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.



