Abstract
Background and purpose
Patients with brain metastases (BMs) are surviving longer and returning for multiple courses of stereotactic radiosurgery. BMs are monitored after radiation with follow-up magnetic resonance (MR) imaging every 2–3 months. This study investigated whether it is possible to automatically track BMs on longitudinal imaging and quantify the tumor response after radiotherapy.
Methods
The METRO process (MEtastasis Tracking with Repeated Observations was developed to automatically process patient data and track BMs. A longitudinal intrapatient registration method for T1 MR post-Gd was conceived and validated on 20 patients. Detections and volumetric measurements of BMs were obtained from a deep learning model. BM tracking was validated on 32 separate patients by comparing results with manual measurements of BM response and radiologists’ assessments of new BMs. Linear regression and residual analysis were used to assess accuracy in determining tumor response and size change.
Results
A total of 123 irradiated BMs and 38 new BMs were successfully tracked. 66 irradiated BMs were visible on follow-up imaging 3–9 months after radiotherapy. Comparing their longest diameter changes measured manually vs. METRO, the Pearson correlation coefficient was 0.88 (p < 0.001); the mean residual error was −8 ± 17%. The mean registration error was 1.5 ± 0.2 mm.
Conclusions
Automatic, longitudinal tracking of BMs using deep learning methods is feasible. In particular, the software system METRO fulfills a need to automatically track and quantify volumetric changes of BMs prior to, and in response to, radiation therapy.
Keywords: Longitudinal tumor tracking, Brain metastases, Image registration, T1 MR post-Gd, Deep learning, Stereotactic radiosurgery
1. Introduction
Brain metastases (BMs) are the most common form of brain tumors. It is estimated that 10–40% of all cancer patients will develop brain metastases, with an estimated 70,000–400,000 cases/year in the United States [1], [2], [3], [4]. Historically, the standard of care for patients with multiple BMs was whole brain radiation therapy (WBRT), which controls the disease for some time, but carries the possibility of cognitive side effects and reduced quality of life [5], [6], [7]. In addition, delivering additional courses radiation may increase the risk of radionecrosis [8], [9], [10]. Recent advances in technology have allowed treatment of multiple BMs with frameless single-fraction or hypofractionated stereotactic radiosurgery (SRS/HYPO) [11]. Evidence of reduced cognitive decline has led to the primacy of SRS/HYPO for treating BMs [12]. Moreover, there is an increasing trend to manage multiple or recurrent BMs with multiple courses of SRS/HYPO [13], [14], [15], [16], [17], [18], [19], [20], [21].
Patients with BMs are typically monitored with magnetic resonance (MR) imaging performed every two–three months [22], including T1 post-Gd (T1C+). The complexity of longitudinally tracking each BM and determining treatment response increases with each course of radiation or follow-up MR. Key challenges include classifying BMs as progressing, stable, or recurring, separating new BMs from treated BMs, and differentiating recurrent metastases from delayed radiation necrosis or post-treatment effects.
Limited prior work exists on automatic BM tracking. Shearkhani et al. applied a Jacobian operator field to detect size changes in brain metastases on longitudinal MRs, with significant challenges from new and resolved BMs and false positive detections from blood vessels [23]. Chitphakdithai et al. performed deformable intrapatient registration to estimate BM size changes, but relied on manual segmentation of the initial tumor [24]. Some commercially available software platforms can track BMs with significant manual user input, but do not provide a feasible or practical solution for patients with multiple BMs and multiple prior treatments. Recently, Petersen et al. evaluated a PACS-integrated tracking tool for tracking BMs [25], and demonstrated time savings and reduced manual clicks for physicians. However, there are currently no available tools which integrate automatic image registration, detection, segmentation, and tracking, and can provide information about all treated and untreated lesions in a timely manner to the radiologist and radiation oncologist.
The aim of this study was to investigate whether it is possible to automatically track size and radiation dose for multiple BMs, which could help clinicians make the optimal care decisions. We validated and investigated the efficacy of the METRO software (MEtastasis Tracking with Repeated Observations), which detects and segments all BMs using deep learning, tracks the BMs across co-registered MR timepoints, and reports the radiation dose from prior treatments.
2. Materials and methods
2.1. Overview of METRO process
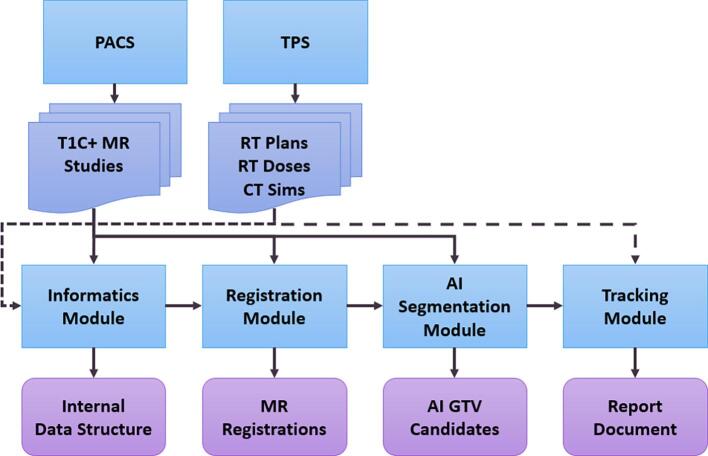
Fig. 1 shows an overview of METRO. The inputs are DICOM data (Digital Imaging and Communications in Medicine): longitudinal MR image series from the picture archiving and communication system (PACS) and DICOM-RT data from the treatment planning system (TPS). A recent MR series is chosen as the fixed image and rigid registration is performed with all the patient’s other MRs. After registration, artificial intelligence (AI) inference is performed at each MR series using a deep convolutional neural network to produce longitudinal maps of BM gross tumor volumes (GTVs) in the fixed MR frame of reference. Next, BM tracking is performed using the treatment plan structure sets and the AI segmentations. Depending on overlap with existing GTVs from treatment plans, the detected GTVs are associated with previously treated GTVs or classified as new AI GTV candidates. The size of each GTV is tracked over time. The calculated dose distributions from prior treatment plans are overlaid on the GTVs to track the physical dose over time. Finally, a report document is generated with separate pages for each tracked BM. Raw data is also saved for downstream analysis, including tracking information such as volume and 3D longest diameter.
Fig. 1.
Schematic showing an overview of the METRO software. Planning and follow-up MR scans are obtained from the PACS. Treatment plans with calculated dose and CT scans are obtained from the TPS.
2.2. Registration method
As months or years elapse, the patient’s brain may exhibit large changes over time, including tumor control and progression, edema, midline shift, and resection cavities. Furthermore, patients who survive longer have several follow-up imaging studies. METRO co-registers all longitudinal MR imaging using a six-degree-of-freedom (6DOF) rigid registration. A multiple-stage registration procedure is used, balancing speed and accuracy, while avoiding catastrophic failures requiring manual user intervention.
The choice of fixed image for the registration depends on the treatment plans. If no co-registered treatment plans are provided, the most recent T1C+ MR is the fixed image. Otherwise, it is the MR registered to the most recent treatment plan. The mutual information similarity metric was used as the cost function [26].
A brief description of the registration stages follows. The registration translation is initialized based on intensity moments, followed by a three-dimensional exhaustive search for initial rotation angle. Two successive stages align the entire head using gradient descent with 6DOF. Next, a fine 6DOF search is performed using only the brain parenchyma as the region of interest—this brain contour is obtained either from the structure sets of the co-registered treatment plan or an MR-based deep learning model [27]. Lastly, a fine-tuning 6DOF search is performed using a rectangular zone centered on the lateral ventricles. After registration, all images are resampled to the fixed image with isotropic 1 mm voxel spacing.
The registration method was validated by comparing with manually verified registrations performed by a medical physicist on 20 BM patients. Each patient had two T1C+ brain MR scans with a large time interval: median 392 days, interquartile range (IQR) 334–887. Seven anatomical points were chosen, and manually located on multi-planar views of the first scan. The XYZ position of each control point was passed through the spatial transformations from the manual and automatic registrations, and the registration error was quantified by differences in resulting position. Further details are given in Supplementary Materials.
2.3. AI segmentation
BM GTVs are automatically detected and segmented on co-registered T1C+ scans using a previously published AI model [27]. Using GTVs authored by the radiation oncologist as ground truths, this 3D V-Net convolutional neural network was trained (tested) on statistically independent samples of 409 (1 0 2) SRS/HYPO patients with 1345 (3 6 7) BMs. Dense evaluation of the neural network on each MR timepoint produces 3D BM probability maps in the fixed image frame of reference. To obtain discrete BM segmentations from the probability maps, a binary threshold is applied, followed by a connected-component analysis. The average patient sensitivity, false positive rate, and Dice coefficient were 95% ± 3%, 2.4 ± 0.5 per patient, and 0.76 ± 0.03, respectively (95% confidence).
A heuristic is applied to handle instances where two BMs are conjoined into one segmentation by a sandbar or isthmus of lower probability. A morphological opening operator is applied once to each initial segmentation blob of volume . Assuming the blob contains two spherical lesions each of volume roughly , an adaptive ball kernel radius of is chosen, where . If the result has two separate parts, they are now each counted as separate BM detections; otherwise, the original component is kept.
2.4. Brain metastasis tracking
After registration and AI segmentation, METRO tracks BMs over time. First, consider physician-authored GTVs with radiation prescriptions from prior treatment plans. Each expert GTV from the structure sets is rasterized, then expanded by a default 1 mm margin for longitudinal tracking. At each timepoint, any segmentation components that overlap with this expanded GTV are found; their union constitutes the tracked lesion on that scan. This search is applied to all scan timepoints—BMs are tracked and measured even at MRs preceding treatment. Measurements of (non-expanded) GTV volume and 3D longest diameter (3LD) at each timepoint are computed and saved. Longitudinal size changes are measured using AI segmentations.
BMs which are newly appeared or not prescribed treatment are also tracked. To mitigate false positive detections, an increased detection probability threshold (80%) was required to identify such lesions. METRO checks each MR timepoint and finds new or untargeted AI GTVs that do not overlap with AI GTVs found at previous timepoints. Like the expert GTV tracking, these AI GTVs are tracked on past and future timepoints, using the expansion margin to check for overlap with AI segmentations. False negative, true positive, and false positive detections were identified based on clinical radiology reports. The radiologist’s axial 2D diameter measurements were recorded for false negatives. False positives were measured using 3LD.
Next, dose metrics are computed to each BM for patients with prior treatment data. This includes BMs not targeted for radiation by a given SRS/HYPO or WBRT plan, using the BM segmentations and the calculated dose images. Dose tracking is handled slightly differently for targeted and untargeted lesions. For linac-based SRS plans, a targeted lesion is defined as an expert GTV which has an associated planning target volume and a mean physical dose sum > 10 Gy. Otherwise, it is an untargeted BM which received spillage dose. Using the original GTV volume that initiated the tracking, physical dose metrics are recorded for each treatment, including fractionation, dose delivered to 99% volume (D99%), and mean dose.
2.5. Longitudinal patient dataset for clinical validation
32 patients with BMs were retrospectively identified under a patient consent waiver approved by our Institutional Review Board. They were treated in 2018–2021 with VMAT SRS/HYPO on linear accelerators using multiple arcs, couch rotations, and optical surface monitoring [11]. Patients were imaged with T1C+ MR prior to treatment and received regular follow-up imaging (183 total scans). METRO was used to longitudinally track each of the 187 lesions treated within this cohort. 53 lesions were excluded due to lack of follow-ups within a chosen time window of 90–270 days. Three resection cavities, one skull lesion, and seven missing PTVs were also excluded. For the remaining 123 lesions, the follow-up MR closest to 180 days post-treatment was chosen. Median time between treatment start date and the chosen follow-up was 185 days (IQR 149.5–196).
2.6. Brain metastasis response assessment
To evaluate the accuracy measuring the size changes of tracked BMs, the pre-treatment 3LD and volume for each lesion was compared to the size at follow-up. The percent changes of 3LD and equivalent sphere diameter (ESD) between pre-treatment and follow-up scans were calculated. To validate the tracking performance, a trained operator contoured the same 123 BMs on the pre-treatment and follow-up MR scans using MIM Maestro 6. Volume and longest diameter measurements were extracted from each manual contour. The ESD was derived from the manual and automatic volume measurements. 3LD and ESD were compared between METRO and manual measurements using linear regression and the Pearson correlation coefficient. For percent size changes, the residual error was defined as the software observation minus the human observation, with size changes determined either by 3LD or ESD. Distributions of the size change residual errors were analyzed for both size metrics.
3. Results
3.1. Registration accuracy
Evaluating the registrations qualitatively, differences between scan timepoints were handled well by the registration method, including ventricle size, edema, tumor progression, tumor response, and motion and susceptibility artifacts. No BM tracking failures were caused by registration uncertainty. The spatial shifts between manual and automatic registration at each anatomical control point were assessed to quantify the registration accuracy. The average shift per point in millimeters was: Point A, 1.8 ± 0.7; Point B, 1.2 ± 0.5; Point C, 1.3 ± 0.5; Point D, 1.6 ± 0.6; Point E, 1.7 ± 0.7; Point F: 1.2 ± 0.5; Point G: 1.3 ± 0.5 (95% confidence). The average shift across all points was 1.5 ± 0.2 mm (95% confidence) with median 1.1 mm (IQR 0.6–1.6). Registration examples for three patients with time intervals of about three years are shown in Figure S2.
3.2. Report document
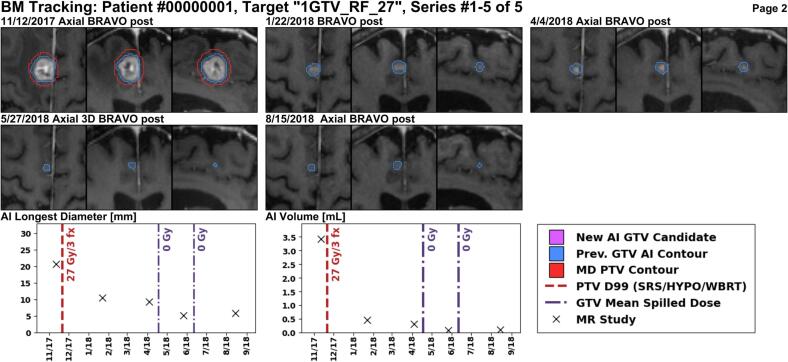
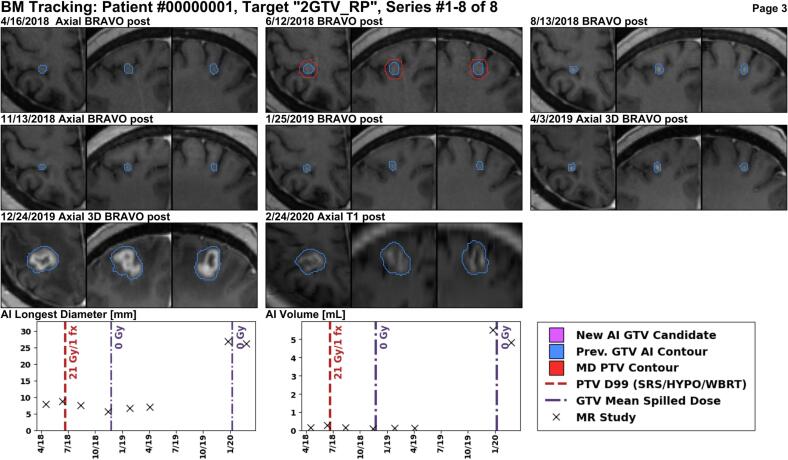
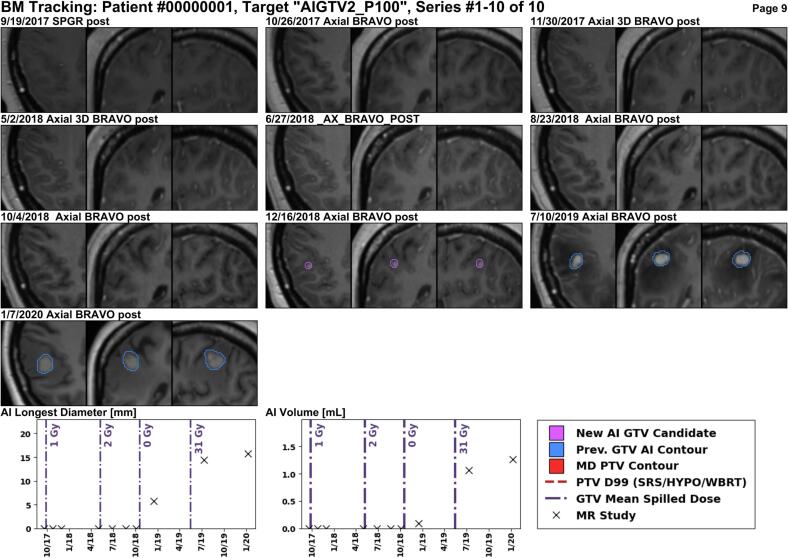
Several example reports are shown for anonymized patients. In Fig. 4, a larger enhancing lesion in the right frontal lobe was treated with 27 Gy over three fractions, and the report correctly identifies that the lesion responded to treatment on follow-up images. Fig. 5 shows a BM in the right parietal lobe which was treated with 21 Gy (single-fraction) and is stable for some time, but exhibits an increase in size over one year later. Lastly, Fig. 6 shows a new BM identified by METRO in the right parietal lobe after four previous courses of radiation treatments to other BMs. See Figures S3 and S4 for additional examples.
Fig. 4.
This 2 cm enhancing lesion in the right frontal lobe was treated with 27 Gy in three fractions using the MR scan from 11/12/2017 for planning. The planning target volume (PTV) is shown in red, and the longitudinal AI segmentations in blue. The lesion size is decreasing at all follow-up MR images with the last MR from 8/15/2018. The METRO report shows both the 3D longest diameter and the volume based on the AI segmentation at each MR. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
A lesion in the right parietal lobe is targeted with single-fraction SRS using the MR from 6/12/2018 for treatment planning. There is not much size change in the first nine months after treatment, but a year and a half after treatment, at the MR scan from 12/24/2019, the lesion size is increasing. The increase in size must be evaluated for potential progression or necrosis.
Fig. 6.
This patient received three courses of SRS elsewhere in the brain, followed by WBRT (Rx 30 Gy). Then, the METRO workflow identified a new lesion in the right parietal lobe at the 7/10/2019 MR, which is increasing in size at subsequent follow-up on 1/7/2020.
3.3. Detection
72% (38/54) of new or unirradiated BMs were detected in the longitudinal tracking dataset. 92 false positive new lesions were tracked (0.5/scan). The median sizes of true positive, false negative, and false positive detections of unirradiated BMs were respectively: 0.8 cm (IQR 0.6–1.2), 0.5 cm (IQR 0.3–0.7), and 0.7 cm (IQR 0.6–0.9). The median initial size of all tracked lesions was 0.9 cm (IQR 0.6–1.3). The isthmus rule was applied 64 times (0.3/scan) and affected measurements of 15 irradiated BMs. For example, two nearby BMs in the left cerebellum and temporal lobe were measured at 7.58 and 0.73 cm3 pre-treatment, after an isthmus was removed between their conjoined segmentations.
3.4. Treatment response
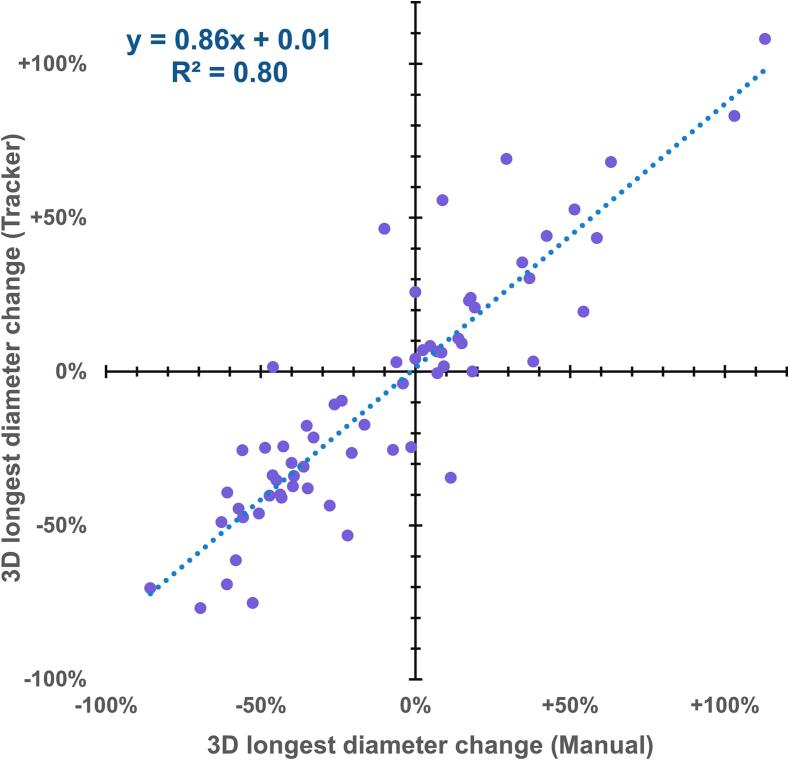
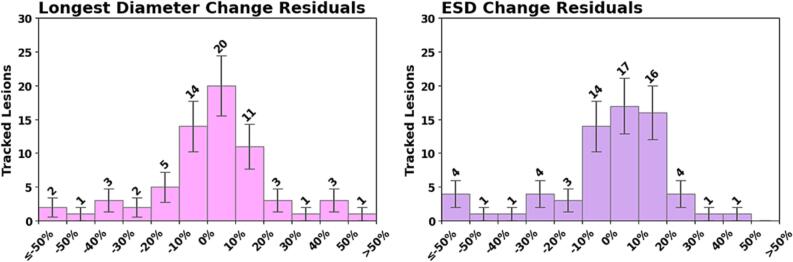
With complete BM disappearance as the quantity of interest, compared to the human observer, METRO produced 48 true positives (i.e. truly disappeared BMs), 66 true negatives, four false positives, and five false negatives. For the 66/123 BMs still visible on follow-up, linear regression (Fig. 2) shows a correlation (R2 = 0.80) between the size responses measured by human versus METRO. Comparing the size changes of non-disappeared lesions measured by both observers, the Pearson correlation coefficient was 0.88 for 3LD and 0.86 for ESD (p < 0.001). Distributions of the size change residual errors are shown in Fig. 3. The mean residual of 3LD changes was −8 ± 17% (95% confidence), with standard deviation 70%, median 2%, IQR (-7%, 12%). The mean residual of ESD changes was −12% ± 17% (95% confidence) with standard deviation 81%, median 2%, and IQR (-7%, 11%). The residual distributions were characterized by a central mode plus large outliers.
Fig. 2.
Linear regression of size response for treated BMs that were still visible on follow-up, as measured automatically by METRO versus manually. One outlier with diameter change greater than + 200% is suppressed.
Fig. 3.
Distributions of residual errors for the size changes measured by automatic METRO method for the BMs still visible at follow-up. The residual is the difference of the percent change measured with METRO minus that measured manually. Left: 3D longest diameter. Right: equivalent sphere diameter.
4. Discussion
The METRO process was capable of reading the patient data and tracking BMs over time. The longitudinal intrapatient registration method was reliable when tested on numerous clinical cases, including those with significant time intervals and notable changes in brain anatomy, and its accuracy was sufficient for longitudinal tracking. The information gathered from each patient’s imaging and treatment history is compiled in a concise, lightweight report document accessible to clinicians, and data are organized for downstream analysis.
We observed a strong correlation between the BM size changes measured by METRO and the human operator. Furthermore, positive versus negative size changes were well-differentiated by METRO—this can be observed by the lack of points in the upper-left and lower-right quadrants of Fig. 2. The distribution of residuals for size changes measured by human versus METRO (Fig. 3) were well-centered at zero, but highly non-normal with large variance due to outliers. We observed a strong correlation coefficient of 88% for the measured change in longest diameter, but there is room for improvement. One potential avenue is a longitudinal segmentation model trained using multiple annotated longitudinal scans. We noticed several cases with non-spherical BMs where the 3LD remains stable, but the BM volume is increasing. This indicates that automatic measurements of BM volume could be more clinically relevant to tumor burden than the manually-measured 2D longest diameter frequently employed in radiology practice.
The accuracy for tracking new sub-centimeter BMs was limited; several false positives and false negatives were observed. The sample of unirradiated BMs was also biased towards smaller size, since larger, growing BMs are more likely to be targeted with radiation. The driving factor is the imperfect detection accuracy of the AI model used to analyze individual MR scans. Unfortunately, this problem is omnipresent in the BM AI literature [27], [28], [29], [30], [31], [32], [33], [34]. There is ongoing work studying how to mitigate false positives [35], [36].
Varying imaging protocols and quality standards can hamper the generalizability of AI-based work such as this across different centers. Consensus guidelines are emerging for BM studies [37], [38]. MR imaging parameters such as pixel spacing and slice thickness can be automatically detected, but other quality aspects require standards developed by experts.
It is currently difficult to differentiate post-treatment tumor volumes from radionecrosis treatment effects. Despite the current ambiguity, post-treatment tracking of abnormal volumes associated with tumors could be an important tool to help distinguish recurrences from radionecrotic volumes.
Manually identifying multiple, potentially irradiated BMs on longitudinal imaging is time-consuming and prone to human error [39], [40], [41], [42]. Measuring or segmenting primary and metastatic brain tumors is similarly challenging [31], [43]. With the aim of aiding clinical practice, we are currently working to integrate this tracking workflow with our existing clinical systems. To address potential inaccuracies, a radiologist or trained operator could view and modify the results, while referencing other synchronized image series from the MR studies. The user would delete false positives and adjust inaccurate longitudinal segmentations, then an approved report would be generated. The approved structure sets would be available if further SRS treatment is indicated, and confirmed new BM appearances would be monitored automatically. These data would be invaluable not only for longitudinal retrospective studies, but also as a continuous source of BM annotations for developing better longitudinal AI models. While manual corrections do cost time, there is great potential for time savings and clinical insights as the tracking accuracy is further improved.
In conclusion, we show that automatic longitudinal tracking of brain metastases using deep learning methods is feasible. In particular, the software system METRO fulfills a need to automatically track and quantify volumetric changes of brain metastases prior to, and in response to, radiation therapy. The accuracy achieved in detecting and tracking tumor volumes, excepting tumors smaller than 1 cm, appears to be adequate to support clinical workflows. Further development is needed to improve the detection of small BMs and the response measurement accuracy. Future work may explore how to distinguish post-treatment tumor volumes from radionecrosis treatment effects. We invite interested researchers to discuss potential multicenter collaborations. In the future, the addition of interactivity for clinicians can make this a routine part of the patient’s medical record.
Funding: MSKCC Support Grant/Core Grant P30 CA008748.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2023.100452.
Contributor Information
Dylan G. Hsu, Email: hsud3@mskcc.org.
Åse Ballangrud, Email: ballanga@mskcc.org.
Kayla Prezelski, Email: kayla.prezelski@health.slu.edu.
Nathaniel C. Swinburne, Email: swinburn@mskcc.org.
Robert Young, Email: youngr@mskcc.org.
Kathryn Beal, Email: kab4027@med.cornell.edu.
Joseph O. Deasy, Email: deasyj@mskcc.org.
Laura Cerviño, Email: cervinol@mskcc.org.
Michalis Aristophanous, Email: aristopm@mskcc.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lamba N., Wen P.Y., Aizer A.A. Epidemiology of brain metastases and leptomeningeal disease. Neuro Oncol. 2021;9:1447–1456. doi: 10.1093/neuonc/noab101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frisk G., Svensson T., Bäcklund L.M., Lidbrink E., Blomqvist P., Smedby K.E. Incidence and time trends of brain metastases admissions among breast cancer patients in Sweden. Br J Cancer. 2012;11:1850–1853. doi: 10.1038/bjc.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nayak L., Lee E.Q., Wen P.Y. Epidemiology of brain metastases. Curr Oncol Rep. 2012;1:48–54. doi: 10.1007/s11912-011-0203-y. [DOI] [PubMed] [Google Scholar]
- 4.Steeg P.S., Camphausen K.A., Smith Q.R. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;5:352–363. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown P.D., Ahluwalia M.S., Khan O.H., Asher A.L., Wefel J.S., Gondi V. Whole-Brain Radiotherapy for Brain Metastases: Evolution or Revolution? J Clin Oncol. 2018;5:483–491. doi: 10.1200/jco.2017.75.9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang E.L., Wefel J.S., Hess K.R., Allen P.K., Lang F.F., Kornguth D.G., et al. Neurocognition in patients with brain metastases treated with radiosurgery or radiosurgery plus whole-brain irradiation: a randomised controlled trial. Lancet Oncol. 2009;11:1037–1044. doi: 10.1016/s1470-2045(09)70263-3. [DOI] [PubMed] [Google Scholar]
- 7.McTyre E., Scott J., Chinnaiyan P. Whole brain radiotherapy for brain metastasis. Surg Neurol Int. 2013;Suppl 4 doi: 10.4103/2152-7806.111301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berber T., Raturi V., Aksaray F., Hojo H., Fujisawa T., Ohyoshi H. Clinical outcome after CyberKnife® radiosurgery re-irradiation for recurrent brain metastases. Cancer Radiother. 2021;5:457–462. doi: 10.1016/j.canrad.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Nieder C., Yobuta R., Mannsåker B. Second Re-irradiation of Brain Metastases: A Review of Studies Involving Stereotactic Radiosurgery. Cureus. 2018;12:e3712. doi: 10.7759/cureus.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milano M.T., Grimm J., Niemierko A., Soltys S.G., Moiseenko V., Redmond K.J., et al. Single- and Multifraction Stereotactic Radiosurgery Dose/Volume Tolerances of the Brain. Int J Radiat Oncol Biol Phys. 2021;1:68–86. doi: 10.1016/j.ijrobp.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballangrud Å., Kuo L.C., Happersett L., Lim S.B., Beal K., Yamada Y., et al. Institutional experience with SRS VMAT planning for multiple cranial metastases. J Appl Clin Med Phys. 2018;2:176–183. doi: 10.1002/acm2.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinde A, Akhavan D, Sedrak M, Glaser S, Amini A. Shifting paradigms: whole brain radiation therapy versus stereotactic radiosurgery for brain metastases. CNS Oncol. 2019;1:Cns27. 10.2217/cns-2018-0016. [DOI] [PMC free article] [PubMed]
- 13.Kotecha R., Damico N., Miller J.A., Suh J.H., Murphy E.S., Reddy C.A., et al. Three or More Courses of Stereotactic Radiosurgery for Patients with Multiply Recurrent Brain Metastases. Neurosurgery. 2017;6:871–879. doi: 10.1093/neuros/nyw147. [DOI] [PubMed] [Google Scholar]
- 14.Yuan J., Lee R., Dusenbery K.E., Lee C.K., Mathew D.C., Sperduto P.W., et al. Cumulative doses to brain and other critical structures after multisession gamma knife stereotactic radiosurgery for treatment of multiple metastatic tumors. Front Oncol. 2018::65. doi: 10.3389/fonc.2018.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuntz L., Le Fèvre C., Jarnet D., Keller A., Meyer P., Bund C., et al. Local recurrence and cerebral progression-free survival after multiple sessions of stereotactic radiotherapy of brain metastases: a retrospective study of 184 patients: statistical analysis. Strahlenther Onkol. 2022;6:527–536. doi: 10.1007/s00066-022-01913-6. [DOI] [PubMed] [Google Scholar]
- 16.Kowalchuk R.O., Niranjan A., Lee C.C., Yang H.C., Liscak R., Guseynova K., et al. Reirradiation With Stereotactic Radiosurgery After Local or Marginal Recurrence of Brain Metastases From Previous Radiosurgery. Int J Radiat Oncol Biol Phys. 2022;3:726–734. doi: 10.1016/j.ijrobp.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Lee W.J., Choi J.W., Kong D.S., Seol H.J., Nam D.H., Lee J.I. Clinical outcomes of patients with multiple courses of radiosurgery for brain metastases from non-small cell lung cancer. Sci Rep. 2022;1:10712. doi: 10.1038/s41598-022-13853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamin C.G., Gurewitz J., Kavi A., Bernstein K., Silverman J., Mureb M., et al. Survival and outcomes in patients with ≥ 25 cumulative brain metastases treated with stereotactic radiosurgery. J Neurosurg. 2021 doi: 10.3171/2021.9.Jns21882. [DOI] [PubMed] [Google Scholar]
- 19.Robin T.P., Camidge D.R., Stuhr K., Nath S.K., Breeze R.E., Pacheco J.M., et al. Excellent outcomes with radiosurgery for multiple brain metastases in ALK and EGFR driven non-small cell lung cancer. J Thorac Oncol. 2018;5:715–720. doi: 10.1016/j.jtho.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Alongi F., Nicosia L., Figlia V., Giaj-Levra N., Cuccia F., Mazzola R., et al. Long-term disease outcome and volume-based decision strategy in a large cohort of multiple brain metastases treated with a mono-isocentric linac-based Stereotactic Radiosurgery technique. Clin Transl Oncol. 2021;8:1561–1570. doi: 10.1007/s12094-020-02550-0. [DOI] [PubMed] [Google Scholar]
- 21.Nicosia L., Figlia V., Mazzola R., Napoli G., Giaj-Levra N., Ricchetti F., et al. Repeated stereotactic radiosurgery (SRS) using a non-coplanar mono-isocenter (HyperArc™) technique versus upfront whole-brain radiotherapy (WBRT): a matched-pair analysis. Clin Exp Metastasis. 2020;1:77–83. doi: 10.1007/s10585-019-10004-3. [DOI] [PubMed] [Google Scholar]
- 22.Eggen A.C., Wind T.T., Bosma I., Kramer M.C.A., van Laar P.J., van der Weide H.L., et al. Value of screening and follow-up brain MRI scans in patients with metastatic melanoma. Cancer Med. 2021;23:8395–8404. doi: 10.1002/cam4.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shearkhani O., Khademi A., Eilaghi A., Hojjat S.P., Symons S.P., Heyn C., et al. Detection of Volume-Changing Metastatic Brain Tumors on Longitudinal MRI Using a Semiautomated Algorithm Based on the Jacobian Operator Field. AJNR Am J Neuroradiol. 2017;11:2059–2066. doi: 10.3174/ajnr.A5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chitphakdithai N., Chiang V.L., Duncan J.S. Tracking Metastatic Brain Tumors in Longitudinal Scans via Joint Image Registration and Labeling. Spatiotemporal Image Anal Longitud Time Ser Image Data. 2012;2012:124–136. doi: 10.1007/978-3-642-33555-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassinelli Petersen G., Bousabarah K., Verma T., von Reppert M., Jekel L., Gordem A., et al. Real-time PACS-integrated longitudinal brain metastasis tracking tool provides comprehensive assessment of treatment response to radiosurgery. Neurooncol Adv. 2022;1: vdac116 doi: 10.1093/noajnl/vdac116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattes D., Haynor D.R., Vesselle H., Lewellen T.K., Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Trans Med Imaging. 2003;1:120–128. doi: 10.1109/tmi.2003.809072. [DOI] [PubMed] [Google Scholar]
- 27.Hsu DG, Ballangrud Å, Shamseddine A, Deasy JO, Veeraraghavan H, Cervino L, et al. Automatic segmentation of brain metastases using T1 magnetic resonance and computed tomography images. Phys Med Biol 2021;1710.1088/1361-6560/ac1835. [DOI] [PMC free article] [PubMed]
- 28.Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med 2018:43-54. 10.1016/j.compbiomed.2018.02.004. [DOI] [PubMed]
- 29.Dikici E., Ryu J.L., Demirer M., Bigelow M., White R.D., Slone W., et al. Automated Brain Metastases Detection Framework for T1-Weighted Contrast-Enhanced 3D MRI. IEEE J Biomed Health Inform. 2020;10:2883–2893. doi: 10.1109/jbhi.2020.2982103. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Stojadinovic S., Hrycushko B., Wardak Z., Lau S., Lu W., et al. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One. 2017;10:e0185844. doi: 10.1371/journal.pone.0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudie J.D., Weiss D.A., Colby J.B., Rauschecker A.M., Laguna B., Braunstein S., et al. Three-dimensional U-Net Convolutional Neural Network for Detection and Segmentation of Intracranial Metastases. Radiol Artif Intell. 2021;3:e200204. doi: 10.1148/ryai.2021200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue J., Wang B., Ming Y., Liu X., Jiang Z., Wang C., et al. Deep learning-based detection and segmentation-assisted management of brain metastases. Neuro Oncol. 2020;4:505–514. doi: 10.1093/neuonc/noz234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M., Young G.S., Chen H., Li J., Qin L., McFaline-Figueroa J.R., et al. Deep-Learning Detection of Cancer Metastases to the Brain on MRI. J Magn Reson Imaging. 2020;4:1227–1236. doi: 10.1002/jmri.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z, Sanders JW, Johnson JM, Gule-Monroe M, Chen M, Briere TM, et al. MetNet: Computer-aided segmentation of brain metastases in post-contrast T1-weighted magnetic resonance imaging. Radiother Oncol; 2020:189-196. 10.1016/j.radonc.2020.09.016. [DOI] [PubMed]
- 35.Chartrand G., Emiliani R.D., Pawlowski S.A., Markel D.A., Bahig H., Cengarle-Samak A., et al. Automated Detection of Brain Metastases on T1-Weighted MRI Using a Convolutional Neural Network: Impact of Volume Aware Loss and Sampling Strategy. J Magn Reson Imaging. 2022;6:1885–1898. doi: 10.1002/jmri.28274. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Chen M, Kazemimoghadam M, Ma L, Stojadinovic S, Timmerman R, et al. Deep-learning and radiomics ensemble classifier for false positive reduction in brain metastases segmentation. Phys Med Biol 2022; 210.1088/1361-6560/ac4667. [DOI] [PMC free article] [PubMed]
- 37.Kaufmann T.J., Smits M., Boxerman J., Huang R., Barboriak D.P., Weller M., et al. Consensus recommendations for a standardized brain tumor imaging protocol for clinical trials in brain metastases. Neuro Oncol. 2020;6:757–772. doi: 10.1093/neuonc/noaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin N.U., Lee E.Q., Aoyama H., Barani I.J., Barboriak D.P., Baumert B.G., et al. Response assessment criteria for brain metastases: proposal from the RANO group. Lancet Oncol. 2015;6:e270–e278. doi: 10.1016/s1470-2045(15)70057-4. [DOI] [PubMed] [Google Scholar]
- 39.Zopfs D., Laukamp K., Reimer R., Grosse Hokamp N., Kabbasch C., Borggrefe J., et al. Automated Color-Coding of Lesion Changes in Contrast-Enhanced 3D T1-Weighted Sequences for MRI Follow-up of Brain Metastases. AJNR Am J Neuroradiol. 2022;2:188–194. doi: 10.3174/ajnr.A7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al Yassin A., Salehi Sadaghiani M., Mohan S., Bryan R.N., Nasrallah I. It is About “Time”: Academic Neuroradiologist Time Distribution for Interpreting Brain MRIs. Acad Radiol. 2018;12:1521–1525. doi: 10.1016/j.acra.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Ganesan A., Alakhras M., Brennan P.C., Mello-Thoms C. A review of factors influencing radiologists' visual search behaviour. J Med Imaging Radiat Oncol. 2018;6:747–757. doi: 10.1111/1754-9485.12798. [DOI] [PubMed] [Google Scholar]
- 42.Berbaum K.S., Franken E.A., Jr., Dorfman D.D., Rooholamini S.A., Kathol M.H., Barloon T.J., et al. Satisfaction of search in diagnostic radiology. Invest Radiol. 1990;2:133–140. doi: 10.1097/00004424-199002000-00006. [DOI] [PubMed] [Google Scholar]
- 43.Egger J., Kapur T., Fedorov A., Pieper S., Miller J.V., Veeraraghavan H., et al. GBM volumetry using the 3D Slicer medical image computing platform. Sci Rep. 2013:1364. doi: 10.1038/srep01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.