Abstract
Objectives:
People living with HIV (PLWH) have elevated risk of non-Hodgkin lymphoma (NHL) and other diseases. Studying clonal hematopoiesis (CH), the clonal expansion of mutated hematopoietic stem cells, could provide insights regarding elevated NHL risk.
Design:
Cohort analysis of participants in the Multicenter AIDS Cohort Study (N=5,979).
Methods:
Mosaic chromosomal alterations (mCAs), a type of CH, were detected from genotyping array data using MoChA. We compared CH prevalence in men living with HIV (MLWH) to HIV-uninfected men using logistic regression, and among MLWH, assessed the associations of CH with NHL incidence and overall mortality using Poisson regression.
Results:
Comparing MLWH to HIV-uninfected men, we observed no difference in the frequency of autosomal mCAs (3.9% vs. 3.6%, p-value=0.09) or mosaic loss of the Y chromosome (mLOY) (1.4% vs. 2.9%, p-value=0.13). Autosomal mCAs involving copy-neutral loss of heterozygosity (CN-LOH) of chromosome 14q were more common in MLWH. Among MLWH, mCAs were not associated with subsequent NHL incidence (autosomal mCA p-value=0.65, mLOY p-value=0.48). However, two MLWH with diffuse large B-cell lymphoma had overlapping CN-LOH mCAs on chromosome 19 spanning U2AF2 (involved in RNA splicing), and one MLWH with Burkitt lymphoma had high-frequency mCAs involving chromosome 1 gain and chromosome 17 CN-LOH (cell fractions 22.1% and 25.0%, respectively). mCAs were not associated with mortality among MLWH (autosomal mCA p-value=0.52, mLOY p-value=0.93).
Conclusions:
We found limited evidence for a relationship between HIV infection and mCAs. Although mCAs were not significantly associated with NHL, mCAs detected in several NHL cases indicate a need for further investigation.
Keywords: clonal hematopoiesis, non-Hodgkin lymphoma, human immunodeficiency virus, genetic mosaicism, chromosomal alteration
Introduction
Despite improvements with effective antiretroviral therapy, HIV infection is associated with elevated risk of non-Hodgkin lymphoma (NHL) [1]. The two most common AIDS-associated NHLs (diffuse large B-cell lymphoma [DLBCL] and Burkitt lymphoma) manifest common genomic changes, including large segments of chromosomal loss or gain [2]. HIV infection is also associated with increased risk for infections, cardiovascular disease, and other conditions associated with chronic inflammation and aging [3,4].
Clonal hematopoiesis (CH) is the presence of a detectable clonal sub-fraction of circulating leukocytes that differs genetically from the inherited germline. CH expansion can be driven by small mutations of a few base pairs, referred to as clonal hematopoiesis of indeterminate potential (CHIP) [5,6], or can involve large structural mosaic chromosomal alterations (mCAs) such as gains, losses, or copy-neutral loss of heterozygosity (CN-LOH) [7,8]. CH increases in frequency with age and is associated with increased risk of infection and hematologic malignancies [5–9]. CHIP occurs more frequently among people living with HIV (PLWH) compared to HIV-free controls [10–14], and clone size is larger among PLWH [12]. However, studies of mCAs in PLWH have not been performed.
With improved survival, PLWH are aging, which translates into rising age-related comorbidity [15], promoting a need for markers that identify PLWH at elevated risk for NHL and other HIV-associated outcomes. Our study aimed to identify potential relationships between HIV infection, mCAs, NHL, and mortality in a population of 5,979 men living with HIV (MLWH) and men without HIV infection.
Methods
Study population
Starting in 1984, the Multicenter AIDS Cohort Study (MACS) enrolled men who have sex with men with HIV or at risk of acquiring HIV [16,17]. Participants were evaluated twice yearly for health assessment, blood collection, and HIV testing. NHL diagnoses were confirmed by medical records and cancer registry data. All participants provided informed consent.
Genotyping and mosaicism detection
Peripheral blood DNA was genotyped using the Illumina Multi-Ethnic Genotyping Array at the MACS visit with the greatest amount of available DNA. Raw genotyping array intensity data were used to calculate B allele frequencies and log2 R ratios which were utilized by MoChA (v2020–08-14) to detect mCAs [7] (Supplemental Methods, Supplementary Digital Content). Mosaic loss of chromosome Y (mLOY) detection utilized probes in the pseudoautosomal region [18]. Included samples had a genotyping call rate ≥96%, and mCAs were restricted to >2 megabases (Mb) to minimize false positives.
Statistical Analyses
HIV status refers to when the genotyped blood sample was collected (baseline). A total of 6,170 samples were scanned for mCAs. We removed duplicate observations (n=1) and samples with unknown collection date/no follow-up time (n=165) or conflicting data on HIV status at sample collection (n=13), resulting in 5,979 included participants.
Logistic regression examined associations of autosomal mCA and mLOY with HIV, adjusting for established mCA risk factors: age per decade, age2 per decade2, and smoking status (never, former, current). For analyses where mCA was not the outcome of interest, we categorized age into groups (<35, 35–44, 45–54, and 55+ years). Fisher exact tests were used to identify differences in mCA distribution.
Poisson regression examined the relationship between mCAs and NHL incidence among MLWH, with follow-up starting at the baseline timepoint. The analysis was unadjusted due to the limited number of NHL events. Poisson regression was also used to assess the association between mCAs and mortality among MLWH, adjusting for age group, calendar year at entry, and smoking status. All statistical tests were two-sided (α=0.05).
Results
Fifty-three percent of the 5,979 included MACS participants had HIV infection at baseline (Table S1, Supplemental Digital Content). Most participants were non-Hispanic White (74%), and MLWH were significantly younger than HIV-uninfected participants. Approximately one-third (36%) of MLWH were currently smoking.
We identified 223 individuals (3.7%) with at least one detectable autosomal mCA and 127 (2.1%) with mLOY (Table S2, Supplemental Digital Content). Autosomal mCA frequency was similar in MLWH and HIV-uninfected participants (3.9% vs. 3.6%), and, although mLOY appeared less common in MLWH (1.4% vs. 2.9%), multivariable analysis indicated HIV status was not significantly associated with the presence of autosomal mCAs (odds ratio [OR]=1.28, 95%CI=0.96–1.71, p-value=0.09) or mLOY (OR=0.73, 95%CI=0.49–1.09, p-value=0.13) (Table 1).
Table 1.
Multivariable associations of mosaic chromosomal alterations with HIV infection, smoking, and age
| Autosomal mCA | mLOY | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Exposure | Subjects | N | % | Adjusted OR (95% CI) | P Value* | N | % | Adjusted OR (95% CI) | P-Value* |
|
|
|
|
|||||||
| HIV status | 0.09 | 0.13 | |||||||
| Without | 2840 | 102 | 3.6% | reference | 83 | 2.9% | reference | ||
| Living With | 3139 | 121 | 3.9% | 1.28 (0.96, 1.71) | 44 | 1.4% | 0.73 (0.49, 1.09) | ||
| Age, per decade | 5979 | 223 | 3.7% | 0.95 (0.49, 1.91) | 0.88 | 127 | 2.1% | 2.06 (0.64, 7.39) | 0.23 |
| Age2, per decade2 | 5979 | 223 | 3.7% | 1.05 (0.99, 1.12) | 0.13 | 127 | 2.1% | 1.04 (0.93, 1.15) | 0.46 |
| Smoking status | 0.54 | 0.29 | |||||||
| Never | 1817 | 63 | 3.5% | reference | 33 | 1.8% | reference | ||
| Former | 1988 | 83 | 4.2% | 1.01 (0.72, 1.42) | 65 | 3.3% | 1.30 (0.83, 2.05) | ||
| Current | 1863 | 64 | 3.4% | 1.20 (0.84, 1.73) | 25 | 1.3% | 1.54 (0.87, 2.69) | ||
Likelihood ratio test p-value
Abbreviations: CI confidence interval; mCA mosaic chromosomal alterations; OR odds ratio
In analyses restricted to MLWH and adjusted for age and smoking status (Table S3, Supplemental Digital Content), no HIV-specific risk factors were associated with autosomal mCAs. Relative to MLWH with CD4 counts above 500 cells/mm3, MLWH with CD4 counts below 350 cells/mm3 had elevated mLOY frequency (OR=2.63, 95%CI=1.25–5.48, p-value=9.8 × 10−3). Antiretroviral therapy and baseline HIV viral load were not associated with mCAs (Table S3, Supplemental Digital Content).
We observed differences in the distribution of mCAs comparing MLWH to HIV-uninfected men. Chromosome 14 CN-LOH mCAs were more common in MLWH (N=12 events vs. N=1 in HIV-uninfected men, p-value=0.003) (Figure 1; Table S4, Supplemental Digital Content), frequently impacting the q arm spanning chr14:102,902,329–107,349,540 (GRCh37) (Figures S1 and S2, Supplemental Digital Content). In addition, chromosome 1 gains and chromosome 11 losses were present only among HIV-uninfected men (p-values 0.028 and 0.028, respectively, based on N=5 and N=5 events) (Table S4, Supplemental Digital Content).
Figure 1.
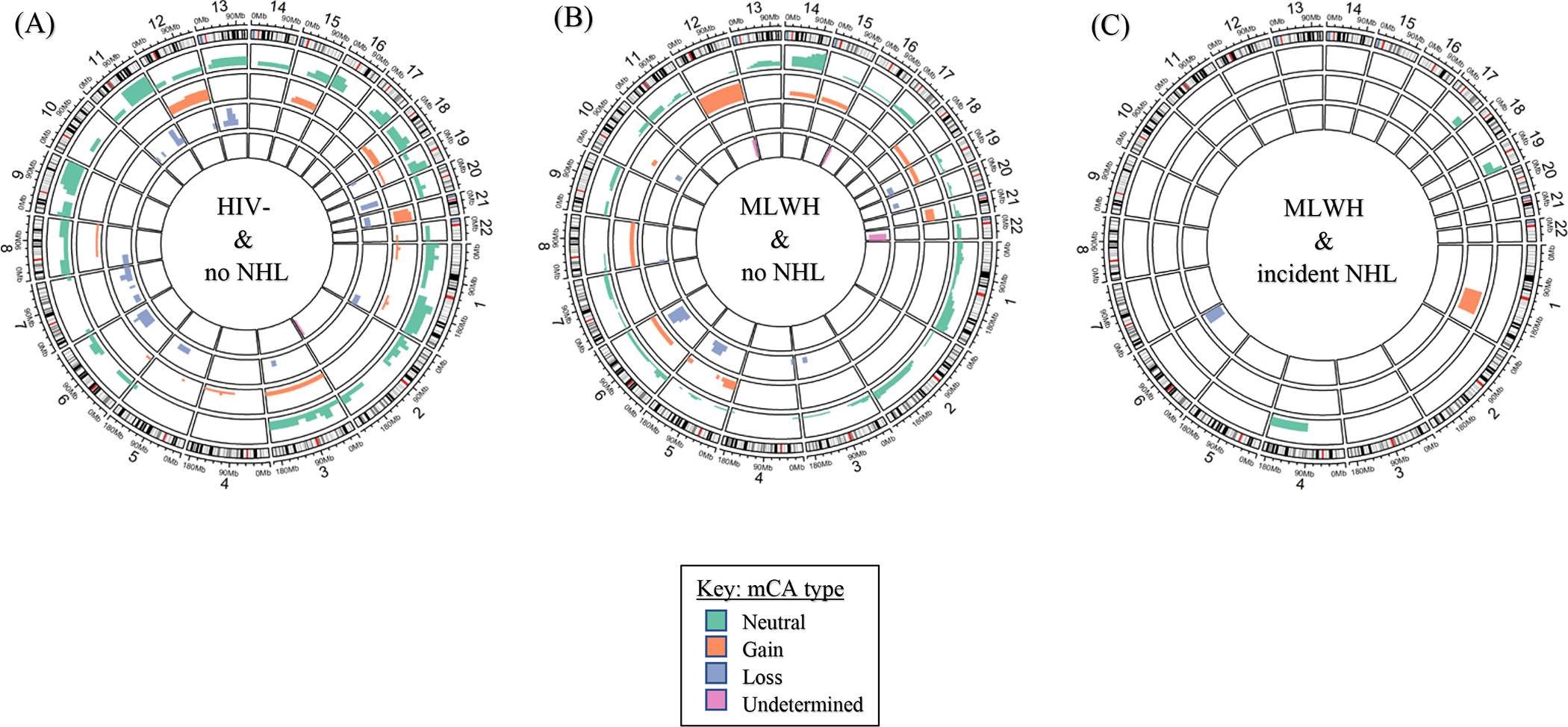
Circos plots displaying the frequency and type of autosomal mosaic chromosomal alteration (mCA) by HIV and non-Hodgkin lymphoma (NHL) status. Participants with NHL diagnosed before the blood draw are not included in these analyses. (A) HIV-uninfected participants with no record of incident or prevalent NHL, (B) Men living with HIV (MLWH) with no record of incident or prevalent NHL, and (C) MLWH with incident NHL. The height of the colored bars indicates the number of participants with mCA in the specified chromosomal region
NHL was diagnosed in 203 participants (16 prevalent cases and 187 incident cases), most of whom were MLWH (N=187, 92.1%). Incident NHL was diagnosed in 177 MLWH (5.7%), with DLBCL being the most common subtype (N=64) followed by central nervous system lymphoma (N=51), Burkitt lymphoma (N=18), and other/unspecified lymphoma (N=54).
Nine autosomal mCAs (7 copy-neutral events, 2 gains) and 4 mLOY events were detected in 11 participants with prevalent or incident NHL (Table S5, Supplemental Digital Content). Among MLWH with incident NHL, 8 had an mCA at baseline (5 autosomal mCA, 3 mLOY, Table S5, Supplemental Digital Content). The presence of mCAs was not significantly associated with NHL incidence among MLWH (p=0.65 for autosomal mCAs, p=0.48 for mLOY; Table S6, Supplemental Digital Content). However, the time from baseline until NHL diagnosis was shorter for MLWH with detectable mCAs (median 0.46 years, interquartile range [IQR] 0.22–0.60, range=0.02–1.02) than for those with no detectable mCA (median 3.76 years, IQR=1.28–6.15 years, range=0.06–21; p-value=7×10−5).
Two MLWH with incident DLBCL (participants 2 and 6) had CN-LOH mCAs that involved an overlapping region on chromosome 19 that included U2AF2 (Table S5, Supplemental Digital Content), a gene involved in RNA splicing [19]. The highest mCA cell fractions were observed in participant 1, a 25-year-old MLWH with Burkitt lymphoma diagnosed 8 days after sample collection who had a chromosome 1 gain (22.1% of cells) and chromosome 17 CN-LOH event (25.0%) that spanned SETDB1 and SRSF2, respectively. SETDB1 encodes a histone methyltransferase involved in transcriptional repression, and SRSF2 encodes a pre-mRNA splicing factor involved in protein translation [20,21].
A total of 1,732 deaths were observed in MLWH, among whom 67 (2.1%) had autosomal mCAs and 16 (0.5%) had mLOY at baseline. In multivariable models adjusted for age, calendar year, and smoking status (Table S7, Supplemental Digital Content), mortality was not associated with presence of autosomal mCAs (mortality rate ratio 1.16, 95%CI 0.90–1.47, p-value=0.23) or mLOY (1.19, 0.69–1.91, p-value=0.49).
Discussion
In this large study of MLWH and HIV-uninfected men, the similarity in the frequency of mCAs by HIV status suggests that HIV infection does not notably impact overall clonal expansion of leukocytes with autosomal mCAs and mLOY. Chromosome 14 CN-LOH events were more common among MLWH, although we could not identify any genes plausibly related to HIV, CH, or myeloid or lymphoid malignancies in the minimally impacted region [10,22]. In addition, while no overall association between HIV infection and mLOY was observed, MLWH with the lowest CD4 cell counts (<350 cells/mm2) had a higher frequency of mLOY compared to those with CD4 cell counts of 500+ cells/mm2. Future studies may identify important genes in these regions relevant to clonal expansion in the presence of HIV.
Previous work identified an association between HIV and an increased frequency of CHIP, in which single base pair alterations occur primarily in myeloid driver genes [10–13]. However, CHIP and mCAs differentially impact protein products and gene expression, potentially explaining observed differences of association. Likewise, the null association of mCAs with mortality among MLWH is contrary to previous reports from larger CH studies in the general population [23]. Future studies that investigate both types of CH jointly will be beneficial in understanding how CH is related to HIV and HIV-associated outcomes.
We did not observe an association between mCAs and subsequent NHL incidence among MLWH. In interpreting this null association, one possible explanation is that mCAs are not etiologically relevant. However, an alternative interpretation is that the time interval between the baseline blood sample and NHL diagnosis was too long for most NHL cases. Of note, the time between sample collection and NHL diagnosis was a year or less among the incident NHL cases who had an autosomal mCA or mLOY, suggesting that mCAs arise late in the development of NHL and that the sampling timeframe could be a critical component for detecting emerging neoplastic clones.
If mCAs arise late in the course of NHL development, expanded cell clones would likely include mutations in the same genomic regions found in NHL tumors; however, incident NHL cases among MLWH had no genomic regions that were generally elevated in mCA frequency. None of the mCAs among MLWH diagnosed with DLBCL encompassed genes previously identified as chromosomal translocation targets in HIV-related DLBCL (e.g., BLC2, BLC6, and MYC) [2]. Among the 8 DLBCL cases with autosomal mCAs, two had mCAs in a region that includes the chromosome 19 gene U2AF2. U2AF2 mutations have been associated with hematological malignancies, but not specifically NHL [24–27]. In addition, the highest mCA cell fraction was observed in a man who developed Burkitt lymphoma within days of baseline. His mCAs encompassed SETDB1 and SRSF2, both of which are mutated in other hematologic malignancies [28–30].
The present study is the first to examine the relationships of mCAs with NHL and mortality among MLWH. We used prospectively collected blood samples and employed robust methods that detected mCAs in the MACS study down to cell fractions of 0.6% and 2.6% for autosomal and mLOY events, respectively. As analyses were restricted to men, the findings may not be generalizable to women living with HIV. In addition, the low frequency of mCAs could be due to the age of the MACS study population and the timing of blood sample collection relative to NHL diagnosis.
In conclusion, we found limited evidence for a relationship between HIV infection and mCAs, although the enrichment among MLWH of copy-neutral mCAs on chromosome 14, mCAs encompassing U2AF2 in two DLBCL cases, and mCAs spanning SETDB1 and SRSF2 in one Burkitt lymphoma case may warrant further investigation. Future studies examining the relationship between mCAs and NHL risk should examine blood samples collected closer in time to NHL diagnosis.
Supplementary Material
Table S1. Comparison of 5,979 MACS participants by HIV status.
Table S2. Unadjusted associations of mosaic chromosomal alterations (mCA) with demographic characteristics and other factors.
Table S3. Multivariable associations between mosaic chromosomal alterations (mCAs) and HIV-related factors among 3,139 men living with HIV.
Table S4. Distribution and frequency of autosomal mosaic chromosomal alterations (mCAs) among MACS participants without prevalent non-Hodgkin lymphoma at the time of blood collection.
Table S5. Detected mosaic chromosomal alterations (mCAs) among 11 MACS participants with non-Hodgkin lymphoma.
Table S6. Unadjusted associations between mosaic chromosomal alterations and non-Hodgkin lymphoma among men living with HIV.
Table S7. Multivariable associations of mortality with mosaic chromosomal alterations, age group, smoking status, and sample collection period among men living with HIV.
Figure S1. Circos plot of chromosome 14 copy-number neutral mosaic chromosome alterations among 12 men living with HIV with no history of non-Hodgkin lymphoma.
Figure S2. The minimally shared region of chromosome 14q neutral events among 12 men living with HIV, with no history of non-Hodgkin lymphoma. The tracks display genes included in this region that could be important for increased frequencies of 14q CNLOH among MLWH.
Acknowledgements
Shu-Hong Lin – performed statistical analyses, drafted the manuscript
Sairah M. Khan – performed statistical analyses, drafted the manuscript
Weiyin Zhou – performed mCA detection
Derek W. Brown – contributed to analytic design
Candelaria Vergara – managed the MACS data
Steven M. Wolinsky – performed MACS genotyping
Otoniel Martínez-Maza – provided subject matter expertise and critically revised the manuscript
Joseph B. Margolick – provided subject matter expertise and critically revised the manuscript
Jeremy J. Martinson – provided subject matter expertise and critically revised the manuscript
Shehnaz K. Hussain – provided subject matter expertise and critically revised the manuscript
Eric A. Engels – conceived and designed the study; secured funding for the study; supervised the analysis; drafted the manuscript
Mitchell J. Machiela – conceived and designed the study; secured funding for the study; supervised the analysis; drafted the manuscript
All authors reviewed and approved the final version of the manuscript.
This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. Data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centers at Baltimore (U01-AI35042): The Johns Hopkins University Bloomberg School of Public Health: Joseph B. Margolick (PI), Todd Brown (PI), Jay Bream, Adrian Dobs, Michelle Estrella, W. David Hardy, Lisette Johnson-Hill, Sean Leng, Anne Monroe, Cynthia Munro, Michael W. Plankey, Wendy Post, Ned Sacktor, Jennifer Schrack, Chloe Thio; Chicago (U01-AI35039): Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services: Steven M. Wolinsky (PI), Sheila Badri, Dana Gabuzda, Frank J. Palella, Jr., Sudhir Penugonda, John P. Phair, Susheel Reddy, Matthew Stephens, Linda Teplin; Los Angeles (U01-AI35040): University of California, UCLA Schools of Public Health and Medicine: Roger Detels (PI), Otoniel Martínez-Maza (PI), Otto Yang (Co-PI), Peter Anton, Robert Bolan, Elizabeth Breen, Anthony Butch, Shehnaz Hussain, Beth Jamieson, John Oishi, Harry Vinters, Dorothy Wiley, Mallory Witt, Stephen Young, Zuo Feng Zhang; Pittsburgh (U01-AI35041): University of Pittsburgh, Graduate School of Public Health: Charles R. Rinaldo (PI), Lawrence A. Kingsley (PI), Jeremy J. Martinson (PI), James T. Becker, Phalguni Gupta, Kenneth Ho, Susan Koletar, John W. Mellors, Anthony J. Silvestre, Ronald D. Stall; Data Coordinating Center (UM1-AI35043): The Johns Hopkins University Bloomberg School of Public Health: Lisa P. Jacobson (PI), Gypsyamber D’Souza (PI), Alison Abraham, Keri Althoff, Michael Collaco, Priya Duggal, Sabina Haberlen, Eithne Keelaghan, Heather McKay, Alvaro Muñoz, Derek Ng, Anne Rostich, Eric C. Seaberg, Sol Su, Pamela Surkan, Nicholas Wada. Institute of Allergy and Infectious Diseases: Robin E. Huebner; National Cancer Institute: Geraldina Dominguez. The MACS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA), and the National Institute of Mental Health (NIMH). Targeted supplemental funding for specific projects was also provided by the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute on Deafness and Communication Disorders (NIDCD). MACS data collection is also supported by UL1-TR001079 (JHU ICTR) from the National Center for Advancing Translational Sciences (NCATS) a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH), Johns Hopkins ICTR, or NCATS. The MACS website is located at http://aidscohortstudy.org/.
Cancer incidence data were provided by the following state agencies: 1) Maryland Cancer Registry, Center for Cancer Prevention and Control, Department of Health and Mental Hygiene, Baltimore, MD 21201; 2) Illinois Department of Public Health, Illinois State Cancer Registry; 3) Bureau of Health Statistics & Research, Pennsylvania Department of Health, Harrisburg, Pennsylvania; 4) Ohio Cancer Incidence Surveillance System (OCISS), Ohio Department of Health (ODH), a cancer registry partially supported in the National Program of Cancer Registries at the Centers for Disease Control and Prevention (CDC) through Cooperative Agreement # 5U58DP000795–05; and 5) California Department of Public Health pursuant to California Health and Safety Code Section 103885; CDC’s National Program of Cancer Registries, under cooperative agreement 5NU58DP003862–04/DP003862; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute. We acknowledge the State of Maryland, the Maryland Cigarette Restitution Fund, and the National Program of Cancer Registries of the CDC for the funds that support the collection and availability of the cancer registry data. The analyses, findings, interpretations, and conclusions of this report are those of the authors. No endorsement by any of the states providing data, the National Cancer Institute, the CDC or their Contractors and Subcontractors is intended nor should be inferred.
This study has been granted an exception from the NIH Genomic Data Sharing Policy and has no individual level phenotype and molecular data (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000506.v1.p1)
This study was funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics of the National Cancer Institute; National Institute of Allergy and Infectious Diseases; National Institute on Drug Abuse; National Institute of Mental Health; National Heart, Lung, and Blood Institute; National Institute of Deafness and Communication Disorders; National Center for Advancing Translational Sciences (UL1-TR001079); and the NIH Roadmap for Medical Research.
Footnotes
Conflicts of Interest & Sources of Funding
The authors declare no conflicts of interest.
Disclaimers
The analyses, findings, interpretations, and conclusions of this report are those of the authors. No endorsement by the NCI, NIH, or their contractors and subcontractors is intended nor should be inferred.
References
- 1.Gibson TM, Morton LM, Shiels MS, Clarke CA, Engels EA. Risk of non-Hodgkin lymphoma subtypes in HIV-infected people during the HAART era: a population-based study. AIDS Lond Engl 2014; 28:2313–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capello D, Scandurra M, Poretti G, Rancoita PMV, Mian M, Gloghini A, et al. Genome wide DNA-profiling of HIV-related B-cell lymphomas. Br J Haematol 2010; 148:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Triant VA. Cardiovascular Disease and HIV Infection. Curr HIV/AIDS Rep 2013; 10:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeks SG, Overbaugh J, Phillips A, Buchbinder S. HIV infection. Nat Rev Dis Primer 2015; 1:1–22. [DOI] [PubMed] [Google Scholar]
- 5.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal Hematopoiesis and Blood-Cancer Risk Inferred from Blood DNA Sequence. N Engl J Med 2014; 371:2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-Related Clonal Hematopoiesis Associated with Adverse Outcomes. N Engl J Med 2014; 371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh P-R, Genovese G, Handsaker RE, Finucane HK, Reshef YA, Palamara PF, et al. Insights about clonal hematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018; 559:350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machiela MJ, Zhou W, Sampson JN, Dean MC, Jacobs KB, Black A, et al. Characterization of Large Structural Genetic Mosaicism in Human Autosomes. Am J Hum Genet 2015; 96:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin S-H, Brown DW, Rose B, Day F, Lee OW, Khan SM, et al. Incident disease associations with mosaic chromosomal alterations on autosomes, X and Y chromosomes: insights from a phenome-wide association study in the UK Biobank. Cell Biosci 2021; 11:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dharan NJ, Yeh P, Bloch M, Yeung MM, Baker D, Guinto J, et al. HIV is associated with an increased risk of age-related clonal hematopoiesis among older adults. Nat Med 2021; 27:1006–1011. [DOI] [PubMed] [Google Scholar]
- 11.Bick AG, Popadin K, Thorball CW, Uddin MM, Zanni MV, Yu B, et al. Increased prevalence of clonal hematopoiesis of indeterminate potential amongst people living with HIV. Sci Rep 2022; 12:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Heijden WA, van Deuren RC, van de Wijer L, van den Munckhof ICL, Steehouwer M, Riksen NP, et al. Clonal Hematopoiesis Is Associated With Low CD4 Nadir and Increased Residual HIV Transcriptional Activity in Virally Suppressed Individuals With HIV. J Infect Dis 2021; :jiab419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Pasca S, Post WS, Langan S, Pallavajjala A, Haley L, et al. Clonal hematopoiesis in men living with HIV and association with subclinical atherosclerosis. AIDS 2022; 36:1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiley B, Parsons TM, Burkart S, Young AL, Erlandson KM, Tassiopoulos KK, et al. Effect of Clonal Hematopoiesis on Cardiovascular Disease in People Living with HIV. Exp Hematol 2022; 114:18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiels MS, Islam JY, Rosenberg PS, Hall HI, Jacobson E, Engels EA. Projected Cancer Incidence Rates and Burden of Incident Cancer Cases in HIV-Infected Adults in the United States Through 2030. Ann Intern Med 2018; 168:866–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Souza G, Bhondoekhan F, Benning L, Margolick JB, Adedimeji AA, Adimora AA, et al. Characteristics of the MACS/WIHS Combined Cohort Study: Opportunities for Research on Aging With HIV in the Longest US Observational Study of HIV. Am J Epidemiol 2021; 190:1457–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Detels R, Jacobson L, Margolick J, Martinez-Maza O, Muñoz A, Phair J, et al. The multicenter AIDS Cohort Study, 1983 to …. Public Health 2012; 126:196–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh P-R, Danecek P, Palamara PF, Fuchsberger C, A Reshef Y, K Finucane H, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016; 48:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U2AF2 U2 small nuclear RNA auxiliary factor 2 [Homo sapiens (human)] - Gene - NCBI. https://www.ncbi.nlm.nih.gov/gene/11338 (accessed 3 Aug2022).
- 20.SETDB1 SET domain bifurcated histone lysine methyltransferase 1. Bethesda(MD): National Library of Medicine (US), National Center for Biotechnology Information; 2004. https://www.ncbi.nlm.nih.gov/gene/9869 (accessed 29 Dec2022). [Google Scholar]
- 21.SRSF2 serine and arginine rich splicing factor 2. https://www.ncbi.nlm.nih.gov/gene/6427 (accessed 29 Dec2022).
- 22.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 2020; 586:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loftfield E, Zhou W, Graubard BI, Yeager M, Chanock SJ, Freedman ND, et al. Predictors of mosaic chromosome Y loss and associations with mortality in the UK Biobank. Sci Rep 2018; 8:12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albacker LA, Buonamici S, Frampton GM, Smith P, Stephens PJ, Warmuth M, et al. Abstract 3406: Comprehensive genomic profiling of hematologic malignancies identifies recurrent somatic splicing factor mutations in non-Hodgkin’s lymphoma (NHL) and multiple myeloma (MM). Cancer Res 2018; 78:3406. [Google Scholar]
- 25.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011; 478:64–69. [DOI] [PubMed] [Google Scholar]
- 26.Glasser E, Agrawal AA, Jenkins JL, Kielkopf CL. Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs. Biochemistry 2017; 56:4757–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maji D, Glasser E, Henderson S, Galardi J, Pulvino MJ, Jenkins JL, et al. Representative cancer-associated U2AF2 mutations alter RNA interactions and splicing. J Biol Chem 2020; 295:17148–17157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thol F, Kade S, Schlarmann C, Löffeld P, Morgan M, Krauter J, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 2012; 119:3578–3584. [DOI] [PubMed] [Google Scholar]
- 29.Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 2012; 119:3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang J, Chen X, Chen M, Hou J. Increased Expression of SETDB1 Predicts Poor Prognosis in Multiple Myeloma. BioMed Res Int 2022; 2022:3307873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Comparison of 5,979 MACS participants by HIV status.
Table S2. Unadjusted associations of mosaic chromosomal alterations (mCA) with demographic characteristics and other factors.
Table S3. Multivariable associations between mosaic chromosomal alterations (mCAs) and HIV-related factors among 3,139 men living with HIV.
Table S4. Distribution and frequency of autosomal mosaic chromosomal alterations (mCAs) among MACS participants without prevalent non-Hodgkin lymphoma at the time of blood collection.
Table S5. Detected mosaic chromosomal alterations (mCAs) among 11 MACS participants with non-Hodgkin lymphoma.
Table S6. Unadjusted associations between mosaic chromosomal alterations and non-Hodgkin lymphoma among men living with HIV.
Table S7. Multivariable associations of mortality with mosaic chromosomal alterations, age group, smoking status, and sample collection period among men living with HIV.
Figure S1. Circos plot of chromosome 14 copy-number neutral mosaic chromosome alterations among 12 men living with HIV with no history of non-Hodgkin lymphoma.
Figure S2. The minimally shared region of chromosome 14q neutral events among 12 men living with HIV, with no history of non-Hodgkin lymphoma. The tracks display genes included in this region that could be important for increased frequencies of 14q CNLOH among MLWH.


