Abstract
The black rhinoceros (Diceros bicornis L.) is a critically endangered species historically distributed across sub-Saharan Africa. Hunting and habitat disturbance have diminished both its numbers and distribution since the 19th century, but a poaching crisis in the late 20th century drove them to the brink of extinction. Genetic and genomic assessments can greatly increase our knowledge of the species and inform management strategies. However, when a species has been severely reduced, with the extirpation and artificial admixture of several populations, it is extremely challenging to obtain an accurate understanding of historic population structure and evolutionary history from extant samples. Therefore, we generated and analyzed whole genomes from 63 black rhinoceros museum specimens collected between 1775 and 1981. Results showed that the black rhinoceros could be genetically structured into six major historic populations (Central Africa, East Africa, Northwestern Africa, Northeastern Africa, Ruvuma, and Southern Africa) within which were nested four further subpopulations (Maasailand, southwestern, eastern rift, and northern rift), largely mirroring geography, with a punctuated north–south cline. However, we detected varying degrees of admixture among groups and found that several geographical barriers, most prominently the Zambezi River, drove population discontinuities. Genomic diversity was high in the middle of the range and decayed toward the periphery. This comprehensive historic portrait also allowed us to ascertain the ancestry of 20 resequenced genomes from extant populations. Lastly, using insights gained from this unique temporal data set, we suggest management strategies, some of which require urgent implementation, for the conservation of the remaining black rhinoceros diversity.
Keywords: black rhinoceros, population genomics, conservation genomics, ancient DNA
Introduction
Next-generation DNA-sequencing technology is finding increasing application in conservation management (Shafer et al. 2015). Until recently, however, the majority of population-scale conservation genomic studies have utilized reduced representation methods, which call single nucleotide polymorphisms (SNPs) from a limited set of randomly amplified loci (Hohenlohe et al. 2021). By comparison, whole-genome sequences allow for precise estimates of mutation and recombination rates; for higher resolution insights into population diversity, structure, demography, and evolutionary history; and with the benefit of positional information that allows the detection and timing of introgression, inbreeding and, as sample sizes increase, local adaptation (Theissinger et al. 2023).
These attributes of whole-genome sequences make them an indispensable tool for managers entrusted with the conservation of the planet's remaining biodiversity. Conservation practice is reliant on all knowledge available for the biodiversity under protection, but as an important starting point, the species population structure, or at a minimum, a subspecies-level taxonomy is essential (Coates et al. 2018). Genetic data are, in fact, crucial when defining management units, such as evolutionarily significant units (ESUs), and estimating levels of genetic diversity, inbreeding, and gene flow, all of which guide conservation decisions (Barbosa et al. 2018).
In the present study, we applied whole-genome resequencing across a temporally distributed data set, with the aim of filling existing knowledge gaps related to a critically endangered African megaherbivore, the black rhinoceros (Diceros bicornis L.). Prior to 1960, the black rhinoceros had been the most abundant extant rhinoceros species, although its population had started to decline in the 19th century due to habitat clearance and unsustainable hunting (Emslie 2020). As of 2021, some 6,195 black rhinoceroses were left across the continent (Ferreira et al. 2022), reflecting a modest, yet positive demographic recovery after the lowest recorded census size of 2,354 animals in the early 1990s. This historic low was the result of an approximately 98% decline in the wild population between 1960 and 1995, owing principally to intense poaching for the rhinoceros horn trade (Emslie 2020).
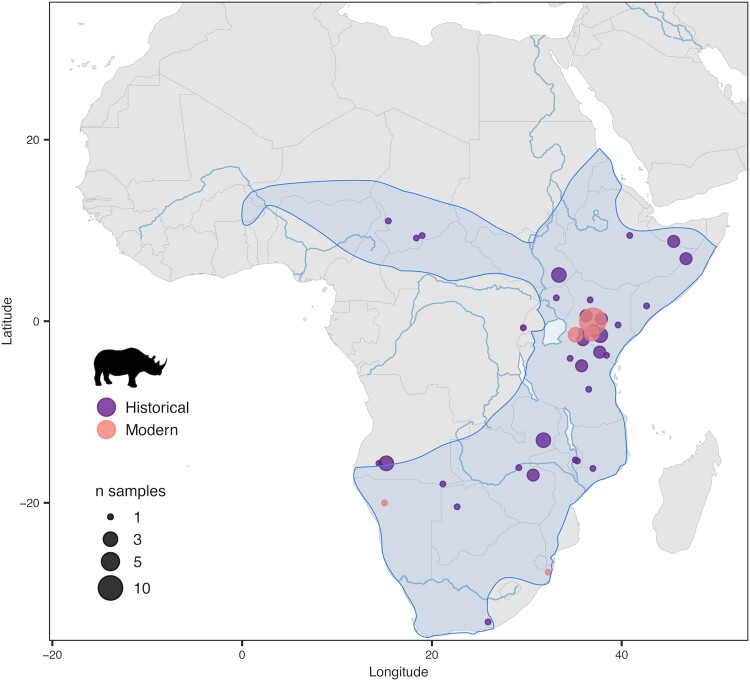
The historic distribution of the black rhinoceros encompassed a vast, continuous area across sub-Saharan Africa that spanned a broad range of habitats, from bushland and grassland to desert, only avoiding areas of dense tropical rainforest (Rookmaaker and Antoine 2012; fig. 1). Currently, outside of zoos, the species survives almost solely in a few protected areas, with large (>1,000 individuals) managed metapopulations only in South Africa and Namibia. In Kenya, the black rhinoceros has made a steady recovery from no more than 381 individuals in 1987 to 897 by 2021 (Kenya Wildlife Service 2021). In Zimbabwe, it has made a slower recovery from approximately 300 individuals in 1995 (Kotzé et al. 2014) to 616 in 2021 (Ferreira et al. 2022). Apart from these four countries, Tanzania is the only remaining country with aboriginal populations of black rhinoceros; however, the present-day population of no more than 160 is poorly managed, scattered across a handful of reserves and stem from a minimum estimate of 31 in 1995 (Emslie and Brooks 1999). Small satellite populations of black rhinoceros have been reestablished in some former range states, but these comprise a total population of about 212 (Ferreira et al. 2022) and have used animals mostly from South Africa. Therefore, the persistence of the black rhinoceros in what remains of its highly fragmented natural range is heavily dependent on active conservation efforts, which include population genetic management (Moodley et al. 2017).
Fig. 1.
The wide historic range of the black rhinoceros in sub-Saharan Africa and sampling locations. The shaded area indicates the historic range of distribution of the black rhinoceros (from Rookmaaker and Antoine 2012). Filled dots represent the sampling locations of 80 georeferenced samples in our data set, 53 historic and 27 modern. An additional eight samples lacked coordinates, but their country of origin was known. Two samples were of unknown origin (table 1; supplementary table S1, Supplementary Material online). Dot sizes represent the number of samples collected at each location.
The subspecies-level taxonomy of the black rhinoceros has been contentious among rhinoceros experts for over a century (Rookmaaker 2011). In the late 1980s, a pragmatic classification into four “ecotypes” was settled by the African Elephant and Rhino specialist group (AERSG; du Toit 1987) to aid conservation efforts. These were the southwestern black rhinoceros of Namibia, the south–central black rhinoceros ranging from South Africa to Tanzania, the eastern black rhinoceros of East Africa, and the now-extinct western black rhinoceros from West Africa. Despite a lack of supporting taxonomic evidence and ignoring other more detailed assessments by Groves (1967) and Zukowsky (1965), the AERSG classification has persisted until the present times. This is problematic because the four ecotypes are managed separately and are often incorrectly and misleadingly referred to as subspecies.
Genetic assessments are key sources of information for determining how populations are structured across the species distribution. In this regard, although a substantial body of prior work exists for the black rhinoceros, these have mostly focused on either single ecotypes or a subset of the managed populations (Harley et al. 2005; Karsten et al. 2011; Muya et al. 2011; Van Coeverden de Groot et al. 2011; Anderson-Lederer et al. 2012; Kotzé et al. 2014). The species- and range-wide understanding of the population structure and diversity has been less well explored. Furthermore, given the major recent population extirpations and bottlenecks that the black rhinoceros has experienced, its current population structure and diversity may not be an accurate reflection of what existed just half a century ago. Thus, an improved understanding of its predecline status will be essential for both expanding our knowledge of its ecology and evolution and informing future conservation efforts. In this regard, despite the extirpation of the species across much of its historic distribution, obtaining a representative range-wide genetic sample is possible thanks to the wealth of historic specimens preserved in museum collections.
This temporal sampling approach to study black rhinoceros genetics was explored for the first time by Moodley et al. (2017), who investigated the species-level population structure, phylogeny, and genetic erosion through time. However, their analyses were limited to molecular data from mitochondrial DNA and microsatellite markers. Although their data showed a major population genetic break on either side of the Zambezi River, most of their conclusions about genetic structure and diversity outside southern Africa were based on the history of the mitochondrial control region, since only a fraction of their historic samples yielded enough microsatellite data.
Therefore, in this study, we aimed to expand the resolution and scope of this previous work by taking advantage of palaeogenomic sequencing techniques. Specifically, we generated whole-genome resequencing data for a set of historic black rhinoceros specimens, representing most of the species’ historic distribution, and supplemented this with genomic data from a number of individuals from extant populations. Ultimately, our goal was to use these genomes to resolve the patterns of population structure, gene flow, and diversity in the black rhinoceros prior to their decline in order to better inform conservation management. In parallel, we aimed to evaluate whether modern individuals are still representative of historic individuals from the same geographic region and could therefore provide the basis for the recovery of historic populations by informing future range expansion efforts.
Results
A Black Rhinoceros Whole-Genome Temporal Data Set
We generated shotgun DNA-sequencing data for 98 individual black rhinoceroses originally sampled from 16 countries across the historic and contemporary range of the species. The historic specimens (n = 71) ranged in collection date between 1775 and 1981. The initial 27 modern samples derived from extant populations in natural reserves: one Namibian (Etosha National Park), three Kenyan (Maasai Mara Game Reserve, Nairobi National Park, and Ol Pejeta Conservancy), and two South African (iMfolozi and Mkhuze Game Reserves; fig. 1). However, based on our relatedness analysis (supplementary fig. S1, Supplementary Material online), we excluded seven individuals from our modern data set from further downstream analyses.
We mapped the raw sequence data against the publicly available whole-genome assembly for the black rhinoceros ASM1363453v1 (Genbank Assembly Accession: GCA_013634535.1; Moodley et al. 2020). We excluded individuals with depth of coverage < 1× from further analyses (n = 8), yielding a final data set consisting of 63 historic and 20 modern unrelated, resequenced whole genomes. Importantly, 53 of the historic genomes corresponded to samples whose associated metadata included coordinates indicating geographic origin (fig. 1).
Historic samples were named as follows: the alpha-2 code of the country of origin (see table 1), the year of collection, and an index number (to distinguish samples of identical country and year). Country of origin was unknown for two samples, which we indicated by replacing the country code with “un.” Modern samples were labeled with simpler identifiers that included the country code (for South Africa and Namibia) or reserve code (for Kenya) followed by an index number (see table 1 for further details on the distribution of samples across countries).
Table 1.
Overview of the Number and Origin of Black Rhinoceros Samples in the Data Set.
| Country/Reserve | Code | Resequenced Genomes | Populations (K = 6) | Subpopulations (K = 10) |
|---|---|---|---|---|
| Historic | ||||
| Angola | AO | 5 | S | SW, SN/SE |
| Botswana | BW | 1 | S | SN/SE |
| Chad | TD | 3 | NW | NW |
| DRC | CD | 2 | EA, RU | EA, RU |
| Ethiopia | ET | 2 | NE | NE |
| Kenya | KE | 19 | EA, CE | CE, EA, MA, ER, NR |
| Malawi | MW | 2 | RU | RU |
| Mozambique | MZ | 1 | CE | ER |
| Nigeria | NG | 1 | NW | NW |
| Somalia | SO | 5 | NE | NE |
| South Africa | ZA | 2 | S | SN/SE |
| South Sudan | SS | 3 | EA | EA |
| Tanzania | TZ | 8 | CE, RU | MA, ER, RU |
| Uganda | UG | 1 | EA | NR |
| Unknown | un | 2 | CE, S | CE, SN/SE |
| Zambia | ZM | 3 | CE | CE |
| Zimbabwe | ZW | 3 | S | SN/SE |
| TOTAL | 63 | |||
| Modern | ||||
| Maasai Mara Game Reserve | MA | 7 | Modern CE-EA | MA |
| Nairobi National Park | NNP | 3 | Modern CE-EA | Modern CE-EA |
| Ol Pejeta Conservancy | OP | 14 | Modern CE-EA | Modern CE-EA |
| Etosha National Park, Namibia | NA | 1 | Modern S | SW |
| iMfolozi and Mkhuze, South Africa | ZA | 2 | Modern S | SN/SE |
| TOTAL | 27 | |||
For historic and modern samples separately, the countries of origin and their corresponding alpha-2 codes are specified, as well as the number of resequenced genomes and the historic populations present in each country.
As expected, the DNA-sequencing data from the historic specimens showed signals characteristic of ancient DNA, including cytosine deamination, shorter library insert sizes, and sizable fractions of nonendogenous DNA (supplementary fig. S2, Supplementary Material online). As such, the average depth of coverage for the nuclear genomes was generally lower and more variable among the historic specimens (ranging between 1.27× and 20.11×), compared with modern samples (ranging between 7.37× and 22.78×; supplementary table S1, Supplementary Material online). The endogenous DNA of the historic samples ranged from 5% to 62% (supplementary table S1 and fig. S2, Supplementary Material online), and levels of cytosine deamination ranged from 0.5% to 5%.
Black Rhinoceroses Exhibited Geography-Driven Population Structure
We used a range-wide data set comprising 63 historic genomes to determine the population structure of the black rhinoceros prior to its decline in the late 20th century. We used genotype likelihoods of variant transversions as input for the following population structure analyses (see Variant Site Identification in Materials and Methods and supplementary fig. S3, Supplementary Material online).
We first performed a principal component analysis (PCA) to explore historic population structure. The first principal component (PC) separated southern African (S) samples from the rest: Individuals from south and west of the Zambezi River, that is, southern Angola, Namibia, Botswana, Zimbabwe, and South Africa, were clearly grouped apart from central, eastern, and northern samples (fig. 2A). The second PC separated individuals sampled in Chad and Nigeria from others in Northeastern Africa (NE), East Africa (EA), and Central Africa (CE).
Fig. 2.
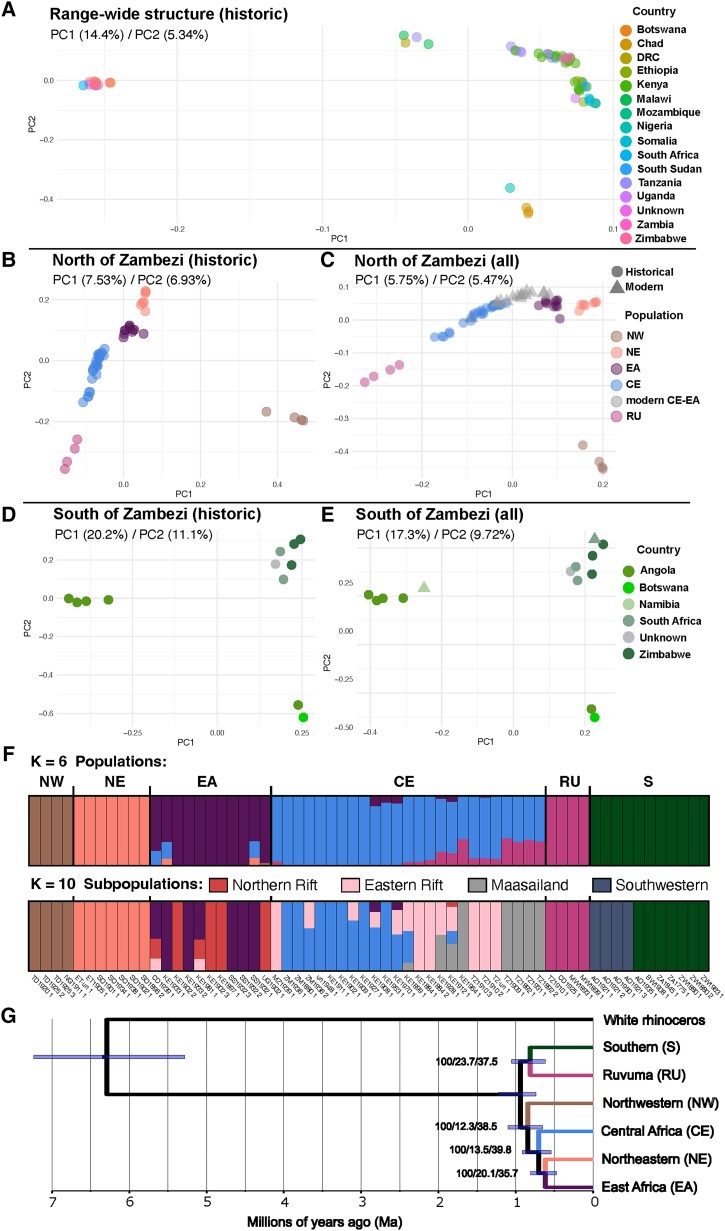
Range-wide population genomic structure of black rhinoceros historic and modern sample sets. (A) PCA of all 63 historic genomes colored by country of origin. (B) PCA of historic genomes sampled north of the Zambezi River. (C) PCA of historic and modern (all) genomes sampled north of the Zambezi River. (D) PCA of historic genomes sampled in southern Africa, south of the Zambezi River. (E) PCA of historic and modern (all) genomes sampled in southern Africa, south of the Zambezi River. (F) Admixture analysis of historic individuals showing range-wide population structure at K = 6 and subpopulation structure at (K = 10). Values of K 2 ≥ 10, including modern genomes, are available in supplementary figures S4 and S6, Supplementary Material online. (G) Fossil-calibrated phylogenomic tree using a single individual representative per population. Branch labels show bootstrap values, gene concordance factors, and site concordance factors, respectively.
To investigate these patterns of structure in finer detail, we conducted PCAs of the individuals on either side of the Zambezi River separately. We observed five major populations north and east of the river (fig. 2B), largely clustering according to geography: the northwestern (NW) population (Chad and Nigeria) observed in figure 2A; a NE population from Ethiopia and Somalia; an EA population including animals from South Sudan, Uganda, north Democratic Republic of Congo (DRC), and northwestern Kenya; a CE population from southern Kenya, northern and central Tanzania, Zambia, and Mozambique; and a more distinct population localized to Malawi, southeastern Tanzania, and putatively from the southern DRC, previously suggested by mtDNA and named Ruvuma (RU; Moodley et al. 2017).
The samples from south of the Zambezi River also displayed substructure but along an east–west axis. Individuals from South Africa (including our Cape rhinoceros sample), Zimbabwe, Botswana, and southeastern Angola were separated (along PC1) from those originating in southwestern Angola (fig. 2D). PC2 separated South Africa and Zimbabwe from Botswana and southeastern Angola (fig. 2D).
We conducted analogous PCAs including the genomes of 20 unrelated modern samples (see Relatedness Test in Materials and Methods and supplementary fig. S1, Supplementary Material online) on either side of the Zambezi River separately. Modern samples from the Kenyan reserves fell within and between the historic EA and CE samples, while the Namibian modern individual grouped with the historic southwestern Angola samples, and the South African modern genome grouped among the historic Zimbabwe–South Africa individuals. Therefore, the observed subpopulation groupings within southern Africa follow closely the three subpopulations, SW (Namibia and southwestern Angola), SN (southeastern Angola, Botswana, and Zimbabwe), and SE (South Africa and Zimbabwe) previously identified by Moodley et al. (2017).
To investigate historic population structure in more detail, we used the 63 historic genomes in an admixture proportion analysis and observed a pattern largely concordant with the results of the PCAs (fig. 2F; supplementary fig. S4, Supplementary Material online). The value of K = 6 was found to be the most likely for the data set using EvalAdmix (supplementary fig. S5, Supplementary Material online). At K = 2, as with PC1 in figure 2A, individuals from southern Africa separated from those north and east of the Zambezi River, although RU appeared to comprise a mixture of alleles from both populations. As K increased, NW, NE, and EA separated from CE and RU at K = 3, and NW was then distinguished from NE and EA at K = 4. Then, RU separated from CE at K = 5, while EA separated from NE at K = 6 (fig. 2F). Higher K models also yielded similar EvalAdmix results and geographically interesting and conservation-relevant subpopulation structure. At K = 7 (supplementary fig. S4, Supplementary Material online), a subpopulation, closely related to CE, could be distinguished among five genomes sampled in the Maasailand (MA) region in the rift valley of southern Kenya and northern Tanzania, previously identified from mtDNA as Chari-Victoria (CV; Moodley et al. 2017). However, the higher resolution offered by whole genomes placed the three individuals sampled on the Chari River into the NW population, making the name CV inappropriate for the Maasailand genomes. At K = 8, SW was delineated from SN/SE. K = 9 separated five further genomes from southern Kenya and Tanzania, but the range of this subpopulation did not overlap with MA; instead, these individuals were sampled in the relatively narrow gap to the east of the rift valley and to the west of the distribution of RU. Thus, while MA is characteristic of black rhinoceros in Maasailand and the southern rift valley, this new subpopulation is more associated with the area to the east of the rift valley, and so we name it here Eastern Rift (ER). Finally, at K = 10, four genomes from a region including Uganda, Lake Turkana, and Lake Baringo were differentiated from EA. We name the subpopulation of black rhinoceros inhabiting this arid landscape Northern Rift (NR), as it is dominated by volcanoes and lakes of the north of the rift valley.
The distinctive range-wide population structuring at K = 6 also allowed the detection of admixture (fig. 2F). Some individuals within EA and CE were not assigned fully to either population, instead appearing admixed. In Kenya and South Sudan, two EA genomes showed admixture with NE and CE, and one EA genome from Uganda appeared admixed with NW, while another in DRC was admixed with CE. Three CE genomes from Kenya were admixed with EA, two of which were the only samples from the valley of the Tana River in our data set (see supplementary table S1, Supplementary Material online). Fourteen CE individuals were admixed with RU at K = 6, but at K = 7, the most admixed of these were designated MA.
Analogously to our PCAs, we also conducted an admixture analysis including the 20 modern unrelated individuals (supplementary figs. S6 and S7, Supplementary Material online). At K = 6, modern individuals from two Kenyan reserves, Ol Pejeta Conservancy and Nairobi National Park, appeared either fully EA in ancestry, or as admixed between EA and CE (supplementary fig. S6, Supplementary Material online). On the other hand, our three samples from the Maasai Mara Game Reserve showed a high proportion of MA ancestry (K = 8; supplementary fig. S6, Supplementary Material online), which is geographically consistent because the reserve is situated within the Maasailand region, with some EA that is absent in historic MA genomes. The Namibian sample showed ancestry from the SW subpopulation, while the South African modern individual clustered with the historic SN/SE subpopulation (supplementary fig. S6, Supplementary Material online).
Lastly, to determine the relationships among populations and the timing of key divergence events, we reconstructed a fossil-calibrated genome-wide phylogeny using one, least admixed, individual for each of the populations identified at K = 6 (fig. 2G). We used a sliding window approach with 20-kb windows and a 1-Mb slide. As in the above analyses, the most supported topology featured an initial split between southern Africa and other regions. The only exception was that RU was a sister lineage with S and not to other genomes from East Africa. Within the eastern clade, NW branched before CE, while NE and EA were the most derived sister lineages. Although we recovered high bootstrap values (100) for all nodes, both the gene and site concordance factors were low, with maximum values of 27.3 and 39.8, respectively, suggesting high levels of phylogenetic discordance in our data set (fig. 2G). Using an estimate for the divergence of the black and white rhinoceros species from a common ancestor of between 5.3 and 7.3 Ma, we inferred the first population split to have occurred between 0.73 and 1.22 Ma, with all other major population subdivisions likely occurring before ∼500 Ka.
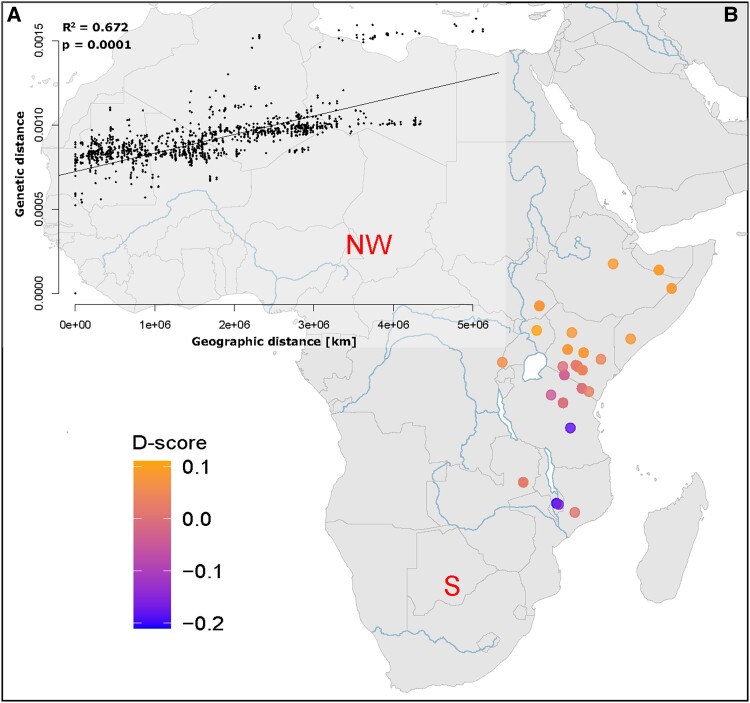
The observed levels of structuring at increasing values of K prompted a more explicit test of whether a model of isolation by distance (IBD) might have driven the population structure of the black rhinoceros in historic times. We therefore conducted a Mantel test (Mantel 1967) on our 53 georeferenced historic samples, which revealed a significant correlation between genomic and geographical distances, with geography potentially explaining up to 68% of the total variation in the data set (fig. 3A). The pairwise distances obtained when comparing our one individual from South Africa (ZA1775.1) to any other individuals in our data set were markedly higher relative to other comparisons. This individual was not only highly geographically isolated but also temporally isolated, as it was sampled from 1775, compared with 1845–1981 for other historic samples. However, this sample was also the lowest coverage (1.27×), which could have also driven relatively higher levels of divergence. We suspected that these higher pairwise values could increase the significance of our Mantel regression; however, the test remained significant even when this individual was removed from the analysis (supplementary fig. S8, Supplementary Material online). We further investigated the effect of sampling date with genetic distance but found only a very weak correlation (supplementary fig. S9, Supplementary Material online).
Fig. 3.
IBD across the historic range of the black rhinoceros. (A) Mantel regression showing the significant relationship between pairwise genetic and geographic distances for all georeferenced historic black rhinoceros. (B) Distribution of D-statistic values showing the relative genetic distance between the central and eastern populations (CE, RU or EA) to either the S or NW population. A negative D-score (blue) indicates a closer relationship to S, whereas a positive D-score indicates a closer relationship to NW (yellow).
We also calculated D-statistics to determine whether EA, CE or RU individuals, inhabiting the middle of the species range in central and East Africa, were closer to NW or to S, located at the extremes of the range. Under the IBD scenario, the expectation would be a linear decline in D-statistic values with distance from the center of the range. For this, we investigated shared derived polymorphisms using the topology (((S, NW), EA|CE|RU), Outgroup). The D-statistic is commonly used for assessments of gene flow; that is, assuming the input topology corresponds to the correct phylogenetic tree (fig. 2G). However, in this case, S and NW are not sister populations, and so elevated D-scores in this analysis will reflect shared polymorphisms due to closer common ancestry as opposed to gene flow (Westbury et al. 2018, 2021). Thus, a negative D-score would indicate a closer relationship of the test group (EA, CE or RU) to the S population, whereas a positive D-score would indicate a closer relationship to the NW population. We observed a decline in D-statistics as the distance to the NW end of the range increased (fig. 3B). Interestingly, however, the decline was not as linear as expected (fig. 3B). In fact, three groups of samples were identifiable through this approach, and they matched the EA, CE, and RU individuals as sorted by the PCA and admixture analyses (fig. 3B).
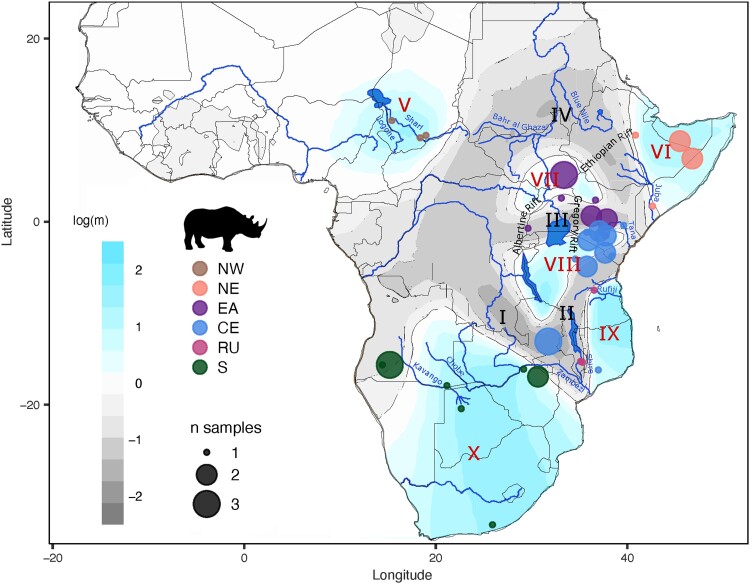
To further investigate these potentially varying degrees of connectivity among the historic black rhinoceros populations, we explored range-wide gene flow (or barriers to it) by computing effective migration rates with EEMS (Petkova et al. 2016) using the 53 georeferenced historic genomes. The resulting effective migration surface pinpointed areas where genetic differentiation decayed quickly with distance (higher than average effective migration, blue-shaded; fig. 4) versus areas where genetic differentiation remained high even in relatively close geographic space (lower than average effective migration, gray-shaded; fig. 4). Importantly, low effective migration might be due to an actual barrier to gene flow or to low population density in the area (Petkova et al. 2016).
Fig. 4.
Effective migration across the historic range of the black rhinoceros and summary of the inferred historic population structure. The effective migration surface was inferred with EEMS (Petkova et al. 2016) based on genome-wide data from 53 georeferenced historic samples. The color gradient represents effective migration rates in logarithmic scale; blue shades indicate rates higher than average, while gray shades represent migration rates lower than average. The six inferred historic populations were mapped onto the migration surface to determine their geographical distribution. Roman numerals denote regions of low (I–IV) and high (V–X) migration described in the text. Dot size represents the number of samples from each location.
We observed broad regions of low effective migration for the black rhinoceros across sub-Saharan Africa. These included the central Congo Basin, where the species never occurred, extending not only south approximately through the valleys of the Kafue and Lower Zambezi to the Indian Ocean (fig. 4, I) but also up the Shire valley into the basin of Lake Malawi and, from there, into the Kilombero and Rufiji valleys of south–central Tanzania (II). From the Congo Basin, this low effective migration surface also extended both east roughly through the basin of Lake Victoria, across the Gregory Rift and along the Tana River valley to the Indian Ocean (III), and north into Central African Republic and Sudan, along the Bahr-al-Ghazal and southeast through the valleys of the White and the Blue Nile, across the Ethiopian Rift and eventually reaching the Indian Ocean via the Juba River (IV). These complex patterns of low effective migration resulted in six pockets of relatively high effective migration: in western Central African Republic, southern Chad, and northern Cameroon (fig. 4, V); the Horn of Africa (VI); South Sudan, northern Uganda, and NW Kenya (VII); southern Kenya, northern and western Tanzania, and northern Zambia (VIII); southeastern Tanzania and northern Mozambique (IX); and finally southern Africa roughly south of the Zambezi basin to the Cape of Good Hope (X). We then overlaid the geographic distribution of the six putative historic populations in previous analyses and found that the distribution of high and low effective migration areas corresponds largely with major population boundaries (fig. 4).
Geographic Distribution of Genome-wide Diversity in the Black Rhinoceros
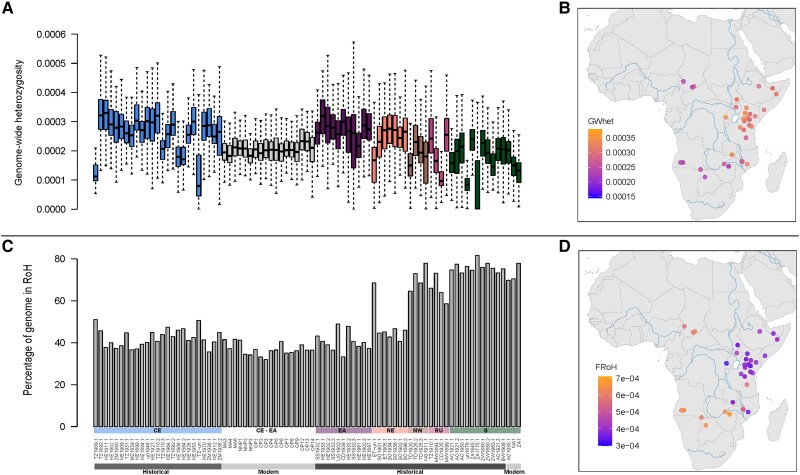
First, we jointly estimated the effective diversity surface for our georeferenced historic data set using EEMS for an initial idea of the geographic distribution of genome-wide diversity (supplementary fig. S10, Supplementary Material online). This analysis suggested a broad region of high diversity corresponding to EA and CE and two regions of low diversity in S and NW Africa. Then, we estimated the genome-wide heterozygosity (GWhet) per sample based on transversion sites. Historically, GWhet was highest in CE (median = 3.28 × 10−4) and EA (median = 3.25 × 10−4) and lowest in the S population (median = 2.36 × 10−4; fig. 5A and B). We also estimated levels of inbreeding among black rhinoceroses by calculating the average length of homozygous regions, known as runs of homozygosity (RoH), and divided it by the total length of the scaffolds considered (>14 Mb; see Variant Site Identification in Materials and Methods) to obtain individual inbreeding coefficients (FRoH; fig. 5C and D). Among historical samples, we found that FRoH was inversely related to GWhet, being lowest in CE and EA and highest in NW and S. Thus, both GWhet and the inverse of FRoH peaked in CE and EA, at the center of the species distribution, and decayed toward the northern and southern peripheries (fig. 5A–D). Both associations were significantly correlated with distance from the individual with the highest GWhet, thus explaining 73% (P < 0.001; supplementary fig. S11, Supplementary Material online) and 68% (P < 0.001; supplementary fig. S12, Supplementary Material online) of the variation in GWhet and FRoH, respectively. We did not find a significant association between sampling date and either GWhet or FRoH (supplementary figs. S13 and S14, Supplementary Material online, respectively).
Fig. 5.
Individual genomic diversity across geographically informed populations of black rhinoceros. (A) Individual GWhet for 83 modern and historic samples. (B) GWhet based on geographical distribution for 53 georeferenced historic samples. (C) Distribution of individual FRoH values with a window size of 1 Mb and larger for 83 modern and historic samples per group is visualized. (D) The geographic distribution of FRoH with a window size of 1 Mb and larger is shown for 53 historic, georeferenced samples.
Modern individuals showed much lower GWhet. Although Kenyan samples (modern CE-EA) showed lower GWhet (median = 2.57 × 10−4) than their presumed historic sources, EA and CE, these levels were still within the range of some of the historic samples, being comparable with NE and RU. Conversely, modern individuals from South Africa and Namibia (modern S) featured much lower GWhet than that of any historic populations (fig. 5A). However, unlike GWhet, historical FRoH was not significantly different from levels in modern CE-EA and modern S (fig. 5C).
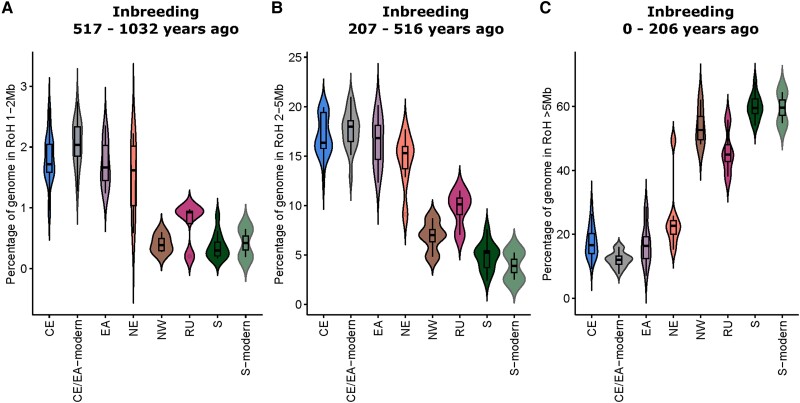
We explored this breakdown in the relationship between FRoH and GWhet among modern samples by dividing FRoH into three different size classes, with RoH between 1 and 2 Mb equating to inbreeding within the last 43 generations (fig. 6A), RoH between 2 and 5 Mb reflects inbreeding within the last 21.5 generations (fig. 6B) and RoH > 5 Mb equates to inbreeding within the last 8.6 generations (fig. 6C). By assuming a generation time of 24 years (Moodley et al. 2017), we estimated timeframes for historical inbreeding of 517–1,032, 207–516 years, and 0–206 years for the small, medium, and large FRoH size classes, respectively. While southern Africa expectedly showed considerably more recent inbreeding during the colonial period (17th to 20th centuries), along with NW and RU; populations in East Africa (CE, EA, and NE) displayed more inbreeding within the two older timeframes.
Fig. 6.
Inbreeding through time and space. Violin plots of individual percentages of genome in RoH across the six major black rhinoceros populations were divided into size classes to investigate inbreeding at three sequential timeframes of the recent past. Allowing for a generation time of 24 years equates to inbreeding between 517–1,032 (A), 207–516 (B), and 0–206 years (C) for the small, medium, and large FRoH size classes, respectively. CE, Central Africa; EA, East Africa; NE, Northeastern; NW, Northwestern, RU, Ruvuma; S, Southern.
The Burden of Inbreeding
The high levels of inbreeding observed in previous analyses necessitated an analysis of the genetic load borne by each population across the species range. We found differences in realized genetic load that is due to homozygous loss-of-function alleles and the masked genetic load of heterozygous loss-of-function alleles between populations as well as between historic and modern samples (supplementary fig. S15, Supplementary Material online). Similar to both GWhet and FRoH, southern Africa appears to suffer the highest burden in both realized and masked genetic loads. However, modern S had significantly lower realized genetic load than its historic counterpart (supplementary table S6, Supplementary Material online), with large variability between individuals, which could suggest efficient purging of deleterious alleles while masked genetic load between historic and modern S individuals overlapped. On the other hand, we observed no obvious differences in realized genetic load between historic and modern CE and EA populations (supplementary fig. S15, Supplementary Material online). The masked genetic load may be somewhat lower in the modern CE/EA population compared with its historic counterparts; however, differences appear only minor.
Discussion
The aim of this study was to characterize the population structure and the distribution of genomic diversity in the black rhinoceros before its range-wide collapse in the latter half of the 20th century. Today, the natural populations of black rhinoceros occurring in Kenya, Tanzania, Zimbabwe, Namibia, and South Africa are remnants of a much richer genetic diversity in the recent past. We therefore sourced and analyzed whole-genome data from 63 museum specimens representing the continental-scale historic distribution of the species.
Historic Populations of the Black Rhinoceros
Our PCA and admixture analyses indicated that historic genomic variation in the black rhinoceros was geographically structured into six major populations (S, RU, CE, EA, NE, and NW), with further substructuring in southern Africa of S into SW and SE/SN subpopulations and in East Africa of CE into MA and ER, and EA into NR. We were concerned that gaps in our sampling scheme, for example, in the Central African Republic, western South Sudan, southern Tanzania, and northern Mozambique, may have contributed to the observed population genomic structure. On the other hand, differentiation into distinct EA and CE populations was observed despite particularly dense sampling in their region of overlap.
Further exploration of the nature of population structure showed a significant pattern of IBD, where the genetic distance between pairs of individuals increased as a linear function of geographic distance between their sampling locations. However, we also observed that several of the geographically distant pairwise comparisons (>2,000 km) were more genetically distinct from each other than would be predicted from the distance between their sampling locations (fig. 3A). These outliers suggested genetic discontinuities in parts of the species range. A nonlinear decline in D-statistics from the center of the species range confirmed this observation. Finally, effective migration rates modeled using EEMS defined six regions of high migration that corresponded directly with the six major populations observed in PCA and admixture analyses (fig. 4B). Although this latter analysis was conducted only on our 53 georeferenced samples, and further data would help define these regions more precisely; taken together, we are confident that the genomic variation in black rhinoceros was structured as described above. Our observations of population and subpopulation structure largely corroborate the findings of Moodley et al. (2017). However, the increased resolution of our whole-genome data set enabled the detection of additional substructuring of populations ER and NR, to the east and north of the rift valley, respectively. Our whole-genome data did not retrieve the divergent mtDNA clade WW, despite sampling from west of the Chari–Logone Basin in Nigeria. We suggest that WW maternal lineage might be a relic of an ancient migration into West Africa that has become fixed west of the Chari–Logone by genetic drift. This discrepancy, and the high-resolution population structure observed above, highlights the growing necessity for the use of genome-scale data to infer intraspecific phylogeography.
Evolutionary History of the Historic Black Rhinoceros Populations
Central and East Africa, dominated by populations EA and CE, appear to have been the hotspots of black rhinoceros diversity, whereas individual diversity decayed with increasing latitude both northward (populations NW and NE) and southward (populations RU and S), toward the limits of the species range (fig. 5). Decreasing genetic diversity from the central parts of a species range is commonly observed in both plant and animal species (Eckert et al. 2008) and is thought to result from increasing isolation and smaller effective sizes. Genetic diversity is often, but not always, highest at or near the species origin (Liu et al. 2006), particularly in species exhibiting significant IBD, as do humans (Manica et al. 2005).
We have shown here that up to 67% of the heterogeneity in our whole-genome data set reflected IBD, and thus, we propose that central Africa and/or East Africa, east of the Congo Basin, as the putative region of origin for the black rhinoceros. This inference is supported by the fossil record, with the earliest emergence of modern D. bicornis at Koobi Fora in Kenya, 2.5 Ma. The species range then appears to have expanded rapidly, as it appears subsequently at Baard's Quarry in South Africa 2.0 Ma and in the Konso Formation in Ethiopia 1.8 Ma (Geraads 2010).
Regarding the isolation of the southern African populations, it is known that tectonic upliftment across the Kalahari sands of southern Africa resulted in a drainage depression that gave rise to enormous Lake Palaeo-Makgadikgadi between 1.4 and 0.5 Ma (Moore et al. 2012; Riedel et al. 2014). This event isolated the western and central parts of southern Africa from the basins of the Kavango, Chobe, and Upper Zambezi rivers through what is now central Botswana and ties in with the first split in our phylogeny, separating the ancestors of S and RU from the rest of Africa. The Upper Zambezi was eventually captured 125–150 Ka into its present-day course (Moore and Larkin 2001), effectively isolating all of southern Africa from RU and the rest of the continent. We observed the major genetic discontinuity in our range-wide data set across the axis of the Zambezi River and suggest that the series of geological events outlined above may have provided the strongest barrier to gene flow across the historic range of the black rhinoceros. In such a scenario, it seems most plausible that black rhinoceros inhabiting the area west of the central Kalahari were most isolated by Lake Palaeo-Makgadikgadi, potentially explaining why SW is well differentiated from SN/SE subpopulation, which would have inhabited the region to the east of the paleo-lake, and with possibly greater access to CE, prior to the capture of the Lower Zambezi River. Lake Palaeo-Makgadikgadi is known to have fluctuated greatly in size in the last 50 Ka (Riedel et al. 2014), eventually allowing the black rhinoceros to repopulate northern Botswana and southeastern Angola from Zimbabwe (SN).
Meanwhile, in the rest of the continent, the black rhinoceros had begun to diverge into populations firstly along the axis of the Albertine and Gregory rifts, with NW to the west of this system the first to differentiate (supplementary fig. S4, Supplementary Material online). CE then became differentiated from EA and NE along the axis of the Tana River and also potentially via admixture into the latter two populations from NW (supplementary fig. S4, Supplementary Material online). Although the upper Tana River altered its course during the Middle Pleistocene, it has flowed nevertheless across central Kenya to the Indian Ocean since the upliftment of the Aberdare Range and Mount Kenya in the Late Miocene and Pliocene (Baker et al. 1971; Veldkamp et al. 2012). Thus, the river and its mountain sources within the Gregory Rift system have likely been barriers to gene flow throughout the evolutionary history of the black rhinoceros in East Africa. At a finer scale, the geographic localization of populations MA, ER, and NR to the western, eastern, and northern parts of the Gregory Rift, respectively, provides further evidence that rifting and upliftment were major drivers of black rhinoceros differentiation in East Africa.
Admixture also appears to be a common feature of historical populations. Admixture profiles for K ≥ 2 (supplementary fig. S4, Supplementary Material online) indicate that while barriers to gene flow were important in isolating populations, introgression between populations was common once such barriers were removed. We postulate that admixture, followed by isolation, may have been responsible for the evolution of several populations including RU, EA, MA, and ER. Ancient introgression between CE and S explains why RU is closely affiliated to CE on admixture plots (fig. 2F; supplementary figs. S4 and S6, Supplementary Material online) and yet a sister taxon to S on the phylogenetic tree (fig. 2G). Such gene flow across the lower Zambezi between eastern southern Africa and southeastern parts of central Africa would only have been possible prior to the capture of the upper Zambezi when the river's flow might not have been as permanent as it is today. Likewise, EA is likely derived through admixture between NW and CE, MA from admixture between CE and RU, and ER through admixture between CE and MA. These signatures for admixture may also indicate shared ancestral polymorphisms rather than gene flow, and since neither is accounted for in a bifurcating tree approach, they are likely responsible for the low gene and site concordance values observed in our phylogenetic reconstruction. We are hopeful, however, that these evolutionary events can be teased apart in the future, through demographic modeling when greater sample sizes become available.
In conclusion, although we demonstrate distinct population structure across its range, we also show that the evolutionary history of the black rhinoceros was likely driven not just by allopatric separation of populations owing to the species inability, or reluctance, to cross large and permanent water bodies and mountains but also by secondary contact followed by isolation, in cases where barriers to gene flow were temporarily removed. Thus, the overlaying of these various evolutionary events upon each other has led to a significant pattern of IBD across the wide sub-Saharan range of the black rhinoceros.
Historic Levels of Inbreeding Vary with Geography
Assessing the structure of modern samples from Kenya, Namibia, and South Africa allowed insight into how population declines have compressed and distributed the remaining historical diversity. The breakdown in relationship between GWhet and FRoH among modern samples is intriguing. While among southern African individuals, where the colonial period began in the 1600s and is known to have heralded the onset of habitat destruction and trophy hunting on a vast scale, it might be expected that historical samples from 1776 (ZA1776.1) and 1845 (ZA1845.1) may already have been subjected to inbreeding at the time of sampling, and so their FRoH values appear similar to levels in modern samples from the same region. However, in East Africa, similarly scaled interventions by Europeans began much later, in the mid to late 1800s, and so most historical samples in our data set were expected to have significantly lower levels of RoH. By dividing RoH into size (and hence time) classes, we show that East African black rhinoceros (CE, EA, and NE), while possessing fewer large RoH tracts than S, RU, and NW, still contained appreciable levels of colonial-period inbreeding (fig. 6C), showing, for the first time, the negative genetic consequences of the ubiquitous European hunting safari on black rhinoceros diversity in East Africa. However, the same three East African populations had significantly more RoH at medium and small size classes (supplementary table S1B, Supplementary Material online). This provides evidence of precolonial inbreeding among black rhinoceros in East Africa, whereas populations outside this region, in West Africa, southern East Africa, and southern Africa, show inbreeding mainly during the colonial period. A similar result was shown for white rhinoceros, where effective population sizes among southern white rhinoceros of South Africa were lowest during the colonial period, whereas values for the northern white rhinoceros were lowest during Bantu migrations into East Africa (Moodley et al. 2018). Thus, independent genetic data from both African rhinoceros species point to geographically distinct patterns of inbreeding between southern and East Africa, suggesting that anthropogenic pressures on African rhinoceros date back to antiquity, and may have been, as it is today, associated with rhinoceros horn. This view is corroborated by the fact that rhinoceros horn and other wildlife products from East Africa were already being traded along the Arabian coast and further east by 100 AD (Boeyens and van der Ryst 2014).
Conservation Implications
Our whole-genome data set provides the first resolution of nuclear DNA populations NW, NE, RU, and MA, which were previously only suggested by mtDNA, plus two entirely unknown populations ER and NR. Moreover, our genome-scale, georeferenced data set allowed the more precise localization of all black rhinoceros populations and subpopulations across the species range than was previously possible with spatial modeling of low-resolution traditional markers (Moodley et al. 2017). One such example occurred in East Africa, where variation neither at microsatellites nor at mtDNA was able to resolve the geographic ranges of populations EA and CE. Here, we show that the distribution of EA is clearly distinguishable from CE, with the former ranging in suitable habitat between the Albertine and Gregory rifts and the latter distributed from about the Tana River south to the Zambezi River, with a zone of secondary contact between EA and CE in southern Kenya. Our genome data also identified distinct black rhinoceros subpopulations MA, ER, and NR that we localized to different regions within the rift valley. Thus, the additional structure and better geographic localization of populations offered by whole-genome data have major implications for conservation-oriented management.
Unfortunately, both NW and NE have been extirpated, with no known record of animals from those populations ever successfully contributing to ex situ populations. On the other hand, confirmation of the existence of RU places enormous conservation value on any of its remaining individuals in the wild. Its historic range, covering the eastern part of Central Africa from the Zambezi River in the south to the Rufiji in the north, contains only two possible options for the persistence of RU individuals: Selous Game Reserve in Tanzania and Niassa Game Reserve in Mozambique. With no recent reports of black rhinoceros activity in either reserve and with local authorities incapable of providing the necessary protection, the future of RU and its unique portion of black rhinoceros diversity look bleak. Similarly bleak prognosis can be made about the existence of ER and NR among modern populations.
The modern samples highlight the devastating effect of population contractions and subsequent genetic drift. This observation was shown to be worst among modern southern African individuals, which featured the lowest heterozygosity and highest inbreeding across all populations, descending from a limited number of founding individuals in Damaraland and Kaokoland, Namibia (SW) (Endangered Wildlife Trust 1984), Zululand, South Africa (SE), and the Zambezi Valley, Zimbabwe (SN; Emslie and Brooks 1999).
In Kenya, despite sustaining precolonial inbreeding associated with Bantu migrations, colonial inbreeding associated with European hunting, and finally the heavy population contractions from the 1970s to the 1990s, modern Kenyan black rhinoceros still maintain much higher levels of present-day variation than modern southern African populations (fig. 5). At one stage, the plight of the Kenyan black rhinoceros was so serious that local authorities located, caught, and translocated the last animals from the dwindling populations scattered across that country, in a desperate effort to consolidate the national metapopulation into intensive protection zones (IPZs). In the absence of genetic knowledge at that time, the origin of each animal was not considered, and so EA, NR, CE, and ER individuals were inadvertently placed within the same IPZs. However, two IPZs in Kenya never received introductions from elsewhere, and these were the Maasai Mara Game Reserve and Chyulu National Park.
These management decisions have resulted in the admixture of EA and CE in much of the present-day Kenyan metapopulation, as is clear from the intermediate PC space occupied by most modern-day Kenyan samples (fig. 2C), and in contrast to separately managed southern African populations (fig. 2E), where modern and historic samples cluster together. In our data set, individuals with highly admixed EA/CE profiles were typically from Nairobi National Park and the Ol Pejeta Conservancy (supplementary fig. S6, Supplementary Material online). Although it is possible that typically NR and ER individuals may also have contributed to the diversity of the present-day Kenyan metapopulation, our restricted modern sample from Kenya did not allow for their detection. The conservation benefit of the consolidation of the Kenyan metapopulation was thus the maintenance of high genetic diversity in the face of population collapse.
On the other hand, our modern samples from the Maasai Mara possess lower genomic diversity compared with other Kenyan populations, but because no translocations ever entered this IPZ, they reveal the original mix of population ancestries that would have been present in Maasailand in historical times. Therefore, all three genomes sampled in the Maasai Mara were typical of MA ancestry and probably represent the last place in Africa, together with the adjoining Serengeti, where the MA population still exists. It is unlikely that MA would have survived in the Massai Mara had this reserve been part of the original translocation plans to protect the Kenyan black rhinoceros, and so our results vindicate the original decision to manage this reserve separately from others in Kenya.
A similar situation may exist among the nonadmixed black rhinoceros population of Chyulu National Park. Although this population was reduced to only two individuals in 1992, it had grown to 21 by 2011 (Muya et al. 2011). Chyulu is in southern Kenya and, importantly, to the east of the rift valley. It may therefore still harbor individuals with ER ancestry, although the national park has never been sampled. From a conservation perspective, this possibility alone elevates Chyulu National Park to a similar level to that of the Maasai Mara as it may be the last place in Africa where ER might exist. Another interesting possibility is that ER and CE may exist in ex situ black rhinoceros populations that were removed from southern Kenya, east of the rift valley during the 1960s (Moodley et al. 2017). These ex situ populations can be found at Thabo Tholo Reserve in South Africa, where unfortunately, many have been admixed with S individuals and thus unsuitable for reintroduction anywhere in East Africa. Another possibility for the existence of ER and CE is in European and American zoos, particularly Dvůr Králové Zoo in the Czech Republic, whose black rhinoceros collection stems directly from Tsavo National Park, also to the east of the rift valley (Moodley et al. 2017).
Based on these results, we suggest strictly separate management for the Maasai Mara-Serengeti and Chyulu National Park from each other and the rest of the Kenyan metapopulation. We suggest local authorities step up measures to genetically profile all remaining black rhinoceroses in Kenya, particularly for those populations with little or no genetic data. The overarching goal for the long-term management of the Kenyan black rhinoceros would be to maintain MA in the Maasai Mara, potentially ER and CE in Chyulu, and EA/CE within the remaining metapopulation, with regular monitoring to sustain levels of diversity, attenuate genetic drift, and limit inbreeding. Similarly, but more urgently, we recommend that authorities in Tanzania obtain genetic data for all their remaining black rhinoceros, with their top priorities to maintain RU, MA, ER, and CE populations, wherever they might still occur in that country.
In southern Africa, our results confirmed previous findings, and we therefore recommend a continuation of the current management scheme, where SW (the Namibian black rhinoceros) is managed separately from subpopulation SN/SE. Our results also confirm the close relationship between SN and SE, which were previously managed separately. We suggest, as did Moodley et al (2017), that new reserves established anywhere in eastern southern Africa from the Cape to the Zambezi consider founders from both SN and SE when available. As both the realized and masked genetic loads were highest among southern African black rhinoceros, we recommend measures to avoid further inbreeding, such as the movement of males between reserves and population monitoring using a studbook, be implemented in all facilities with small populations, whether wild or captive. We also caution that although individual numbers are highest in southern Africa, these populations represent but a small fraction of the remaining species diversity and a conservation management focus on maintaining as many different genetic populations is now required, rather than simply increasing numbers and growth rates of southern African black rhinoceros.
Beyond these conclusions, having genome-wide data available opens promising new avenues for conservation-related research on the black rhinoceros. Our map of black rhinoceros genomic diversity could be leveraged to develop more sophisticated molecular tools to identify the provenance of black rhinoceros material seized from the illegal market. Also, with genomic information, we could venture into the potential phenotypic effects of the intraspecies diversity observed in order to guide management actions. For instance, gaining insight into local adaptation, inbreeding, and outbreeding depression might greatly enhance the success of breeding programs. Overall, our results support and highlight the importance of improving the resolution of traditional molecular markers by carrying out population level, whole-genome studies, and by sampling widely across the species range to better understand population structure and evolutionary history and, ultimately, to better inform conservation management.
Materials and Methods
Whole-Genome Data Generation
Our historic sample collection included material obtained from 71 museum specimens. Collection dates ranged between 1775 and 1981, with the oldest sample a bona fide representative of the Cape rhinoceros (D. b. bicornis), which was thought to be extinct. All samples consisted of keratinous material (pieces of skin, horn powder, or hairs), except for ZA1845.1, which was a piece of bone from a skull, and ZA1775.1, which was a molar tooth. Samples from historic specimens were stored and processed in facilities dedicated to ancient DNA work at the Swedish Museum of Natural History (Stockholm) and the Natural History Museum of Denmark (Copenhagen).
We followed Sánchez-Barreiro et al. (2021) for keratinous tissue processing. The skin pieces were manually cut and then hydrated for 2–3 h at 4 °C in 0.5–1 ml of molecular biology-grade water. The tissue was then briefly washed with 0.5 ml of a 1% bleach solution, followed by two rinsing steps with molecular biology-grade water (Sánchez-Barreiro et al. 2021). Bone material was crushed with a small hammer, and small pieces amounting to 150–200 mg were used for extraction after a brief washing with a 1% bleach solution, and two rounds of rinsing with molecular biology-grade water. Our collection also included 27 modern samples in the form of keratinous material either preserved in ethanol or dry. Dry samples were hydrated with molecular biology-grade water prior to manipulation, and then each piece of skin was cut with a disposable scalpel. For extraction, 20 mg of material was used.
We extracted DNA from the historic keratinous samples with the DNeasy Blood and Tissue Kit (Qiagen), but introducing two modifications to the manufacturer's guidelines, as indicated in Sánchez-Barreiro et al. (2021): on one hand, “adding of DTT (dithiothreitol) 1 M to a final concentration of 40 mM to the lysis buffer” and also “the substitution of the purification columns in the kit by MinElute silica columns (Qiagen) to favor retention of small fragments.” DNA extraction from the bone and the tooth samples was carried out following (Gilbert et al. 2007) with the modifications detailed in Dabney et al. (2013) to enhance the retrieval of small DNA molecules. We assessed the concentration and fragment size distribution in each extract using a TapeStation 2200 (Agilent).
Extraction of DNA from the modern samples was carried out with the KingFisher Duo Prime instrument and its associated Cell and Tissue DNA Kit, following the manufacturer's guidelines. The concentration of DNA extracts was measured with a Thermo Scientific Qubit dsDNA high-sensitivity (HS) assay. A 20-μl aliquot of each extract was fragmented in a Covaris-focused ultrasonicator with a customized program to reduce fragment length to ∼400 bp. Size distribution upon fragmentation was assessed with a TapeStation 2200 (Agilent, Santa Clara, CA, USA).
Sequencing library preparation followed the procedure described in Sánchez-Barreiro et al. (2021), using the BEST protocol (Carøe et al. 2018). We used 100 ng of extracted DNA to which we ligated adapter sequences compatible with BGISEQ 500 sequencing (Mak et al. 2017). Libraries were polymerase chain reaction amplified and single indexed following strictly the protocol described in Sánchez-Barreiro et al. (2021). The resulting indexed libraries were distributed in pools containing equimolar proportions of eight indexed libraries each. Each of these pools was given one lane of BGISEQ 500 PE150 sequencing.
For samples ZA1, ZA2, and NA1, sequencing libraries were built using the Illumina TruSeq Nano DNA Library Prep Kit for NeoPrep on DNA inserts that were 350 bp in length and following the manufacturer's guidelines. Libraries were then sequenced on an Illumina HiSeq X platform, giving 0.5 lanes per sample in PE150 mode.
Bioinformatic Processing of Raw Data
Quality Assessment and Mapping of DNA-Sequencing Data
We generated shotgun-sequencing data for a total of 98 black rhinoceros samples, 71 historic and 27 modern. We conducted a quality check per sample with FastQC v0.11.7 (Andrews 2010). Subsequently, we ran the pipeline PALEOMIX v1.2.13.2 (Schubert et al. 2014) on each sample separately to remove sequencing adapters and exclude reads shorter than 25 bp with AdapterRemoval v2.2.2 (Schubert et al. 2016); align the raw reads against the D. bicornis assembly ASM1363453v1 (Genbank Assembly Accession: GCA_013634535.1; Moodley et al. 2020) using bwa v0.7.16a and its backtrack algorithm (Li and Durbin 2009) setting minimum base quality filtering to 0 to maximize reads retained; filter out duplicates with Picard MarkDuplicates (Broad Institute 2019); and calculate the level of ancient DNA damage with mapDamage v2.0.6 (Jónsson et al. 2013). From the total 98 samples, eight samples were excluded from whole-genome analyses due to low depth of coverage (<1×; see supplementary table S1, Supplementary Material online) or systematic failure to align against the whole-genome assembly. The resulting 90 aligned genomes were divided into 63 historic and 27 modern.
Variant Site Identification
To optimize computational memory usage and omit potentially poorly assembled regions of the reference assembly, we restricted variant site finding to scaffolds > 14 Mb (n = 47), which represent 72.83% of the total length of the assembly. We verified that none of these scaffolds belonged to sex chromosomes by evaluating if male samples showed a 0.5× normalized depth of coverage, indicative of X chromosome regions (supplementary fig. S3, Supplementary Material online). We identified biallelic variant sites that were transversions and computed their genotype likelihoods using the GATK genotype likelihood model (-GL 2) within ANGSD v0.921 (Korneliussen et al. 2014). Transitions were excluded with the -rmTrans option, and the minimum number of individuals in which a variant site must be present (-minInd) was 95%. Minimum and maximum global depths per site were based on a global depth assessment with ANGSD -doDepth: 500 and 1,500 respectively when including 63 or more genomes and 200 and 1,500 when including fewer than 63 genomes. Additionally, the following quality filtering and output choice parameters were set: -remove_bads 1 -uniqueOnly 1 -baq 1 -C 50 -minMapQ 30 -minQ 20 -doCounts 1 -GL 2 -doGlf 2 -doMajorMinor 1 -doMaf 1 -doHWE 1 -dosnpstat 1 -HWE_pval 1e-2 -SNP_pval 1e-6.
Statistical Analyses of Genomic Data
Relatedness Test
We ran a pairwise analysis of relatedness based on genotype likelihoods with ngsRelate v2 (Hanghøj et al. 2019). The computation of this panel of genotype likelihoods followed the procedure detailed above, except for the parameters -setMaxDepth and -setMinDepth, which were set to 900 and 200, respectively. The files containing the genotype likelihoods and allele frequencies were reformatted with commands in bash language to match the input requirements of ngsRelate v2. As per Waples et al. (2019), the degree of relatedness between each pair of samples was assessed qualitatively based on the relative values of coefficient of relatedness R1 versus coefficients KING and R0.
We found 11 pairs of individuals showing a relatedness signal among the modern samples (supplementary fig. S1, Supplementary Material online). Seven of those samples were therefore excluded from the analyses of population structure: MA1, MA2, MA5, MA7, OP10, OP11, and ZA2. As a criterion to exclude samples from a related pair, the sample of the lowest depth of coverage was discarded.
PCA
We used PCAngsd 0.973 (Meisner and Albrechtsen 2018) to compute covariance matrices from genotype likelihoods for different sets of samples: all historic genomes, those north and south of the Zambezi River separately, and the latter plus the unrelated modern genomes stemming from those respective regions. Standard packages in R v3.4.4 (R Core Team 2022) were used for decomposition of each matrix in eigenvectors and eigenvalues and ggplot2 (Wickham 2016) for visualization of PCs.
Admixture
Assessment of admixture proportions across individuals was conducted with NGSadmix v32 (Skotte et al. 2013). We used the genotype likelihoods of transversion variant sites for the 63 historic genomes as input. Values of ancestral clusters, K, ranged between 2 and 10, and for each value of K, we ran NGSadmix 100 times. We repeated the analyses with the inclusion of the modern individuals for K values 2–10. For each value of K, the run of the highest log-likelihood was chosen for visualization with the software Pong (Behr et al. 2016) (supplementary figs. S4 and S5, Supplementary Material online). We used EvalAdmix (Garcia-Erill and Albrechtsen 2020) to evaluate the goodness of fit of the clustering for each K value (supplementary figs. S6 and S7, Supplementary Material online).
Phylogenetic Tree
We selected a single individual per population (EA, CE, NE, NW, RU, and S) that showed the low levels of mixed ancestry based on K = 6 in the admixture analysis: KE1911.1, KE1933.1, TZ1910.1, TD1925.2, SO1896.2, and ZW1880.1. We additionally mapped a white rhinoceros individual (P9109_108) to the black rhinoceros genome to act as an outgroup. The white rhinoceros individual was mapped to the black rhinoceros reference genome using PALEOMIX, following the same protocol described above for the black rhinoceros data. We generated consensus FASTA files from each of the individuals using ANGSD and a consensus haploid call (-doFasta 2) and the following filters: -remove_bads 1 -uniqueOnly 1 -minMapQ 30 -minQ 20 -setmindepthind 5. We limited this to scaffolds > 14 Mb. We generated a bed file containing sliding windows of 20 kb in size with 1-Mb slides using BEDTools v2.29.1 (Quinlan and Hall 2010) and extracted each window from the individual specific consensus file using SAMtools. We built a phylogenetic tree for each window (gene tree) using IQ-TREE v2.2.0.3 (Minh et al. 2020) with the GTR substitution model + six gamma distribution rate categories (R6) and 1,000 bootstrap replicates. We also concatenated all windows into a single sequence and built a phylogenetic tree using IQ-TREE. We calculated gene concordance factors (percentage of gene trees supporting a given node) and site concordance factors (percentage of sites supporting a given node) based on the topology from the concatenated data and the individual gene trees in IQ-TREE (--gcf and --scf). We dated the concatenated tree using MCMCTree from the PAML package (Yang 2007) and specified a root age (split between black and white rhinoceros) between 5.3 and 7.3 Ma. This range is based on records of Diceros in Upper Miocene deposits (>5.3 Ma) at Lothagam (Kenya, 6.54–5.2 Ma; Brown and McDougall 2011) and Albertine (Uganda, 7.25–5.3 Ma; Pickford et al. 1993).
Factors Influencing Genetic Distance
To perform a Mantel test for IBD, we generated two distance matrices: One based on genetic distance, and one based on geographic distance. We calculated the genome-wide pairwise distance between either all 53 georeferenced historical black rhinoceros or 52 (we excluded one southern African individual [ZA1775.1] due to elevated putative genetic distances caused by low coverage data [1.27×]) using ANGSD with a consensus base call (-doIBS 2) and the following parameters: -rmtrans 1 -minind 53 -remove_bads 1 -uniqueOnly 1 -minMapQ 30 -minQ 20 -GL 1 -doMajorMinor 1 -minMinor 0 -makeMatrix 1. Similar to the other analyses, we limited our analysis to scaffolds > 14 Mb in length. We generated the geographic distance matrix for the same individuals using their GPS coordinates (supplementary table S1, Supplementary Material online) and R using the geodist library. We ran the Mantel test in R specifying the two distance matrices as input and 9,999 permutations. We also performed a regression test by comparing pairwise differences between dates and pairwise genetic distance of the same individuals to assess whether there was a temporal factor driving the genetic differences between samples. The correlation coefficient was calculated using R v4.2.1 (R Core Team 2022).
D-statistics
To estimate the relatedness of the individuals found in the central range (CE, RU or EA) of the species to either the S or northwestern (NW) populations, we used D-statistics in ANGSD. We used a random base call (-doabbababa 1), specified the white rhinoceros (BioSample accession: SAMEA8896056) as the outgroup, only used scaffolds > 14 Mb in length, excluded repeat regions, and chose the following parameters: -remove_bads 1 -uniqueOnly 1 -baq 1 -C 50 -minMapQ 30 -minQ 20 -setMaxDepth 1500 -setMinDepth 500 -rmTrans 1. The output was parsed through the jackKnife.R script, which is part of the ANGSD toolsuite, to make it into a more readable format. ANGSD calculates D-statistics for all possible triplet combinations. However, we only extracted comparisons following the defined topology of (((S, NW), central population), Outgroup). Based on this topology, a negative D-score would indicate a closer relationship to the S population, whereas a positive D-score would indicate a closer relationship to the NW population. As we have multiple individuals from S and NW, we took the average of all possible combinations of S/NW.
Estimation of Effective Migration and Diversity Surfaces with EEMS
We employed EEMS (Petkova et al. 2016) to link genetic and geographic data and estimated the effective migration and diversity surfaces along the black rhinoceros range of distribution using 53 georeferenced historic genomes. As input, EEMS takes a pairwise distance matrix which we calculated with PLINK using an input file generated using ANGSD (-doplink 2) across the 47 largest scaffolds of the assembly and the following parameters: -rmtrans 1 -minind 51 -remove_bads 1 -uniqueOnly 1 -minMapQ 30 -minQ 20 -GL 1 -doMajorMinor 1 -doPlink 2 -doGeno -4 -doPost 1 -postCutoff 0.95 -SNP_pval 1e-6 -doMaf 1 -minMaf 0.05. Using PLINKv1.90b6.2, we converted the resultant tped and tfam to map/pedfiles using --recode and then converted those to bed/fam files using --make-bed. From the bed file, we generated a distance matrix as input for EEMS using bed2diffs_v1, part of the EEMS toolsuite. The matrix was fed as input to EEMS with an MCMC chain of 2,000,000 iterations and assuming 1,000 underlying demes (a specification of grid size). The geographic area of interest was outlined by hand with the online tool Google Maps API v3 Tool (Scharning). Visualization of the estimated migration (m) and effective diversity (q) surfaces was conducted in R v3.4.4 (R Core Team 2022).
Metrics of Individual Genomic Diversity
We estimated the GWhet of each genome, based on transversion biallelic sites within the scaffolds > 14 Mb, following strictly the approach described in Sánchez-Barreiro et al. (2021). Briefly, for each sample, we first calculated the site allele frequency likelihood of there being 0, one or two alternative alleles with the -doSaf 1 option of ANGSD (Korneliussen et al. 2014) and the folded option (-fold 1). Both the reference (-ref) and the ancestral (-anc) genome used were the black rhinoceros assembly. We only included transversion sites (-noTrans 1), and sites of a depth of coverage of at least 5× (-setMinDepth 5). Identical quality filtering parameters as for computing genotype likelihoods were set. Then, we used RealSFS, within ANGSD, to compute the folded site frequency spectrum (SFS) for each sample using the output of the previous step. To investigate the variance of heterozygosity across the genome, we calculated the SFS in 10-Mb windows of covered bases (-nSites). The count of heterozygous sites was divided by the total count of sites to obtain the individual estimate of GWhet.
RoH were also estimated for each genome in our data set with >5× coverage using PLINK based on the approach used by Foote et al (2021). We generated a PLINK file from the scaffolds > 14 Mb in length from all individuals using ANGSD (-doPlink 2) and the following parameters: -rmtrans 1 -minind 83 -remove_bads 1 -uniqueOnly 1 -minMapQ 30 -minQ 20 -GL 1 -doGlf 2 -doMajorMinor 1 -doPlink 2 -doGeno -4 -doPost 1 -postCutoff 0.95 -SNP_pval 1e-6 -doMaf 1 -minMaf 0.05. We ran the resultant PLINK file in PLINK to calculate the RoH using the following parameters: --homozyg-snp 50 --homozyg-kb 1000 --homozyg-density 50 --homozyg-gap 1000 --homozyg-window-snp 50 --homozyg-window-het 5 --homozyg-window-missing 5 --homozyg-window-threshold 0.05 --allow-extra-chr. Individual inbreeding coefficients (FRoH) were calculated by dividing the total length within RoH > 1 Mb by the total number of base pairs found in the scaffolds > 14 Mb in length (1,698,121,211 bp).
We also filtered the output into three different RoH categories: 1–2, 2–5, and >5 Mb. We estimated the number of generations since inbreeding occurred using the calculation g = 100/(2rL; Kardos et al. 2018), where r is the recombination rate, L is the length of RoH in Mb, and g is the number of generations. As genome-wide recombination rates for black rhinoceros are unavailable, we present results based on the horse (Equus caballus, 1.16 cM/Mb; Beeson et al. 2020). Given this calculation, RoH > 1 Mb equates to inbreeding occurring within the last 43 generations, RoH > 2 Mb equates to inbreeding occurring within the last 21.5 generations, and RoH > 5 Mb equates to inbreeding occurring within the last 8.6 generations.
We performed regressions of the original sampling date of the individual and GWhet and FRoH as well as the geographic distance to the central population and GWhet and FRoH. For the latter, we picked distance to the individual with the highest mean GWhet with GPS coordinates (TZ1910.2) as the central point of the species and calculated distance from that individual taken from the geographic distance matrix calculated above. We limited our analyses to the 52 georeferenced historic black rhinoceros individuals with the exclusion of one S African individual (ZA1775.1) due to low coverage (1.27×). The correlation coefficients were calculated using R v4.2.1 (R Core Team 2022).
Genetic Load
The genetic load was estimated to explore the potential consequence of genomic erosion for each individual with a sequencing depth of >5× following the approach described in Sánchez-Barreiro et al. (2021). BCFtools v1.15 (Danecek et al. 2021) was used to call genotypes within scaffolds > 14 Mb in length. We masked the individual genotype as missing for samples with sequencing depth lower than 5× or samples showing heterozygous genotype with either allele having less than 3 reads of coverage. We excluded transition sites and SNPs with fewer than 2 allele counts or having over 20% missing information. We used SnpEff v5.1d to annotate the function of each variation. For simplicity, we considered the major allele of our black rhinoceros samples as the ancestral state. We then counted the total number of nonsynonymous and loss-of-function homozygous and heterozygous sites separately for each sample to estimate the realized and masked genetic loads (Bertorelle et al. 2022).
Visualizations
All visualizations were produced in R v3.4.4 (R Core Team 2022) using standard packages and ggplot2 (Wickham 2016). Visualization of maps and geographical data required the packages maps (Becker et al. 2018), mapdata (Becker and Brownrigg 2018), maptools (Bivand and Lewin-Koh 2019), rgdal (Bivand et al. 2019), and sp (Pebesma and Bivand 2005; Bivand et al. 2013).
Supplementary Material
Acknowledgments
This work was supported by ERC Consolidator Grant 681396 “Extinction Genomics” to M.T.P.G. and by EMBO Short-Term Fellowship 7578 to F.S.-B. The authors would like to acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure (NGI), Sweden, the Knut and Alice Wallenberg Foundation, and UPPMAX for providing assistance in massively parallel DNA sequencing and computational infrastructure. T.M.-B. is supported by funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No. 864203), BFU2017-86471-P (MINECO/FEDER, UE), “Unidad de Excelencia María de Maeztu”, funded by the AEI (CEX2018-000792-M), and NIH 1R01HG010898-01A1. Y.M. acknowledges support from the National Research Foundation of the Republic of South Africa. The authors are very grateful to all the museums who contributed samples to this study: the Natural History Museum London, the Museum of Natural History Berlin, the Powell-Cotton Museum, the Natural History Museum Vienna, the Natural History Museum at the National Museum Praha, the Natural History Museum of Zimbabwe, the Swedish Museum of Natural History, the Royal Museum for Central Africa Tervuren, the Senckenberg Museum Frankfurt, Smithsonian National Museum of Natural History and the Bavarian State Collection of Zoology. The authors would also like to thank Professor Alfred L. Roca (Department of Animal Sciences, University of Illinois Urbana-Champaign) for insightful comments on the preliminary results of this manuscript. Lastly, it is with great sadness that we acknowledge the death of our friend, colleague, mentor, and coauthor Michael W. Bruford, who succumbed to illness during the resubmission phase of this manuscript. He will be sorely missed far beyond the bounds of just the conservation genetics community.
Contributor Information
Fátima Sánchez-Barreiro, Globe Institute, University of Copenhagen, Copenhagen, Denmark.
Binia De Cahsan, Globe Institute, University of Copenhagen, Copenhagen, Denmark.
Michael V Westbury, Globe Institute, University of Copenhagen, Copenhagen, Denmark.
Xin Sun, Globe Institute, University of Copenhagen, Copenhagen, Denmark.
Ashot Margaryan, Globe Institute, University of Copenhagen, Copenhagen, Denmark.
Claudia Fontsere, Globe Institute, University of Copenhagen, Copenhagen, Denmark; Institut de Biologia Evolutiva (Consejo Superior de Investigaciones Científicas–Universitat Pompeu Fabra), Barcelona Biomedical Research Park, Barcelona, Catalonia, Spain.
Michael W Bruford, Cardiff School of Biosciences, Cardiff University, Cardiff, UK.
Isa-Rita M Russo, Cardiff School of Biosciences, Cardiff University, Cardiff, UK.
Daniela C Kalthoff, Department of Zoology, Swedish Museum of Natural History, Stockholm, Sweden.
Thomas Sicheritz-Pontén, Globe Institute, University of Copenhagen, Copenhagen, Denmark; Centre of Excellence for Omics-Driven Computational Biodiscovery (COMBio), Faculty of Applied Sciences, AIMST University, Kedah, Malaysia.
Bent Petersen, Globe Institute, University of Copenhagen, Copenhagen, Denmark; Centre of Excellence for Omics-Driven Computational Biodiscovery (COMBio), Faculty of Applied Sciences, AIMST University, Kedah, Malaysia.
Love Dalén, Department of Zoology, Centre for Palaeogenetics, Stockholm University, Stockholm, Sweden; Department of Bioinformatics and Genetics, Swedish Museum of Natural History, Stockholm, Sweden.
Guojie Zhang, Section for Ecology and Evolution, Department of Biology, University of Copenhagen, Copenhagen, Denmark; State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, People's Republic of China; Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, People's Republic of China; BGI Research, BGI-Shenzhen, Shenzhen, People's Republic of China.
Tomás Marquès-Bonet, Institut de Biologia Evolutiva (Consejo Superior de Investigaciones Científicas–Universitat Pompeu Fabra), Barcelona Biomedical Research Park, Barcelona, Catalonia, Spain; National Centre for Genomic Analysis–Centre for Genomic Regulation, Barcelona Institute of Science and Technology, Barcelona, Spain; Life & Medical Sciences, Institucio Catalana de Recerca i Estudis Avançats (ICREA), Barcelona, Catalonia, Spain.
M Thomas P Gilbert, Globe Institute, University of Copenhagen, Copenhagen, Denmark; Department of Natural History, NTNU University Museum, Trondheim, Norway.
Yoshan Moodley, Department of Biological Sciences, University of Venda, Thohoyandou, Republic of South Africa.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The sequencing data underlying this article is available on SRA under BioProject Number PRJNA1002571.
References
- Anderson-Lederer RM, Linklater WL, Ritchie PA. 2012. Limited mitochondrial DNA variation within South Africa's black rhino (Diceros bicornis minor) population and implications for management. Afr J Ecol. 50:404–413. [Google Scholar]
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.
- Baker BH, Williams LAJ, Miller JA, Fitch FJ. 1971. Sequence and geochronology of the Kenya rift volcanics. Tectonophysics 11:191–215. [Google Scholar]
- Barbosa S, Mestre F, White TA, Paupério J, Alves PC, Searle JB. 2018. Integrative approaches to guide conservation decisions: using genomics to define conservation units and functional corridors. Mol Ecol. 27:3452–3465. [DOI] [PubMed] [Google Scholar]
- Becker RA, OSC, by Brownrigg R, ARWRV. 2018. mapdata: Extra Map Databases. Available from: https://CRAN.R-project.org/package=mapdata
- Becker RA, OSC, by Minka TP, ARWRV by RBE, Deckmyn A. 2018. maps: Draw Geographical Maps. Available from: https://CRAN.R-project.org/package=maps
- Beeson SK, Mickelson JR, McCue ME. 2020. Equine recombination map updated to EquCab3.0. Anim Genet. 51:341–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr AA, Liu KZ, Liu-Fang G, Nakka P, Ramachandran S. 2016. Pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32:2817–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Raffini F, Bosse M, Bortoluzzi C, Iannucci A, Trucchi E, Morales HE, van Oosterhout C. 2022. Genetic load: genomic estimates and applications in non-model animals. Nat Rev Genet. 23:492–503. [DOI] [PubMed] [Google Scholar]
- Bivand R, Keitt T, Rowlingson B. 2019. rgdal: Bindings for the “Geospatial” Data Abstraction Library. Available from: https://CRAN.R-project.org/package=rgdal
- Bivand R, Lewin-Koh N. 2019. maptools: Tools for Handling Spatial Objects. Available from: https://CRAN.R-project.org/package=maptools
- Bivand RS, Pebesma E, Gomez-Rubio V. 2013. Applied spatial data analysis with R, 2nd edition. NY: Springer. [Google Scholar]
- Boeyens JC, Van der Ryst MM. 2014. The cultural and symbolic significance of the African rhinoceros: a review of the traditional beliefs, perceptions and practices of agropastoralist societies in Southern Africa. Southern African Humanities 26(1):21–55. [Google Scholar]
- Broad Institute . 2019. Picard Toolkit. Available from: https://broadinstitute.github.io/picard/
- Brown FH, McDougall I. 2011. Geochronology of the Turkana depression of northern Kenya and southern Ethiopia. Evol Anthropol. 20:217–227. [DOI] [PubMed] [Google Scholar]
- Carøe C, Gopalakrishnan S, Vinner L, Mak SST, Sinding MHS, Samaniego JA, Wales N, Sicheritz-Pontén T, Gilbert MTP. 2018. Single-tube library preparation for degraded DNA. Methods Ecol Evol. 9:410–419. [Google Scholar]
- Coates DJ, Byrne M, Moritz C. 2018. Genetic diversity and conservation units: dealing with the Species-population continuum in the age of genomics. Front Ecol Evol. 6:165. [Google Scholar]
- Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, et al. 2013. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci U S A. 110:15758–15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience [Internet] 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Toit R. 1987. African rhino systematics—the existing basis for subspecies classification of black and white rhinos. Pachyderm 9:3–7. [Google Scholar]
- Eckert CG, Samis KE, Lougheed SC. 2008. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol. 17(5):1170–1188. [DOI] [PubMed] [Google Scholar]
- Emslie R. 2020. IUCN Red List of Threatened Species: Black Rhino. IUCN Available from: https://www.iucnredlist.org/species/6557/152728945#assessment-information
- Emslie R, Brooks M. 1999. African rhino: status survey and conservation action plan. IUCN/SSC African Rhino Specialist Group, editor. Gland, Switzerland and Cambridge, UK: IUCN. [Google Scholar]
- Endangered Wildlife Trust . 1984. Rhinoceros in South and South West Africa. p. 1–25.
- Ferreira SM, Ellis S, Burgess G, Baruch-Mordo S, Talukdar B, Knight MH. 2022. The African and Asian rhinoceroses—status, conservation and trade: a report from the IUCN Species Survival Commission (IUCN/SSC) African and Asian Rhino Specialist Groups and TRAFFIC to the CITES Secretariat pursuant to Resolution Conf. 9.14 (Rev. CoP15). CoP19 Doc. 75 (Rev. 1), CITES Secretariat, Geneva, Switzerland.
- Foote AD, Hooper R, Alexander A, Baird RW, Baker CS, Ballance L, Barlow J, Brownlow A, Collins T, Constantine R, et al. 2021. Runs of homozygosity in killer whale genomes provide a global record of demographic histories. Mol Ecol. 30:6162–6177. [DOI] [PubMed] [Google Scholar]
- Garcia-Erill G, Albrechtsen A. 2020. Evaluation of model fit of inferred admixture proportions. Mol Ecol Resour. 20:936–949. [DOI] [PubMed] [Google Scholar]
- Geraads D. 2010. Rhinocerotidae, editors. Cenozoic mammals of Africa: University of California Press. [Google Scholar]
- Gilbert MTP, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van Marck E, Worobey M. 2007. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One 2(6):e537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves CP. 1967. Geographic variation in the black rhinoceros Diceros bicornis (L., 1758). Zeitschrift für Säugetierkd. 32:267–276. [Google Scholar]
- Hanghøj K, Moltke I, Andersen PA, Manica A, Korneliussen TS. 2019. Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. Gigascience [Internet] 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley EH, Baumgarten I, Cunningham J, O’Ryan C. 2005. Genetic variation and population structure in remnant populations of black rhinoceros, Diceros bicornis, in Africa. Mol Ecol. 14:2981–2990. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Funk WC, Rajora OP. 2021. Population genomics for wildlife conservation and management. Mol Ecol. 30:62–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. Mapdamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29:1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M, Åkesson M, Fountain T, Flagstad Ø, Liberg O, Olason P, Sand H, Wabakken P, Wikenros C, Ellegren H. 2018. Genomic consequences of intensive inbreeding in an isolated wolf population. Nat Ecol Evol. 2:124–131. [DOI] [PubMed] [Google Scholar]
- Karsten M, van Vuuren BJ, Goodman P, Barnaud A. 2011. The history and management of black rhino in KwaZulu-Natal: a population genetic approach to assess the past and guide the future. Anim Conserv. 14:363–370. [Google Scholar]
- Kenya Wildlife Service . 2021. National Wildlife Census 2021 Report. Available from: https://kws.go.ke/content/national-wildlife-census-2021-report.
- Korneliussen TS, Albrechtsen A, Nielsen R. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzé A, Dalton DL, du Toit R, Anderson N, Moodley Y. 2014. Genetic structure of the black rhinoceros (Diceros bicornis) in south-eastern Africa. Conserv Genet. 15:1479–1489. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Prugnolle F, Manica A, Balloux F. 2006. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 79:230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SST, Gopalakrishnan S, Carøe C, Geng C, Liu S, Sinding M-HS, Kuderna LFK, Zhang W, Fu S, Vieira FG, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience 6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A, Prugnolle F, Balloux F. 2005. Geography is a better determinant of human genetic differentiation than ethnicity. Hum Genet. 118:366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220. [PubMed] [Google Scholar]
- Meisner J, Albrechtsen A. 2018. Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210:719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Russo I-RM, Dalton DL, Kotzé A, Muya S, Haubensak P, Bálint B, Munimanda GK, Deimel C, Setzer A, et al. 2017. Extinctions, genetic erosion and conservation options for the black rhinoceros (Diceros bicornis). Sci Rep. 7:41417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Russo IR, Robovský J, Dalton DL, Kotzé A, Smith S, Stejskal J, Ryder OA, Hermes R, Walzer C, et al. 2018. Contrasting evolutionary history, anthropogenic declines and genetic contact in the northern and southern white rhinoceros (Ceratotherium simum). Proc Biol Sci R Soc. 285(1890). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Westbury MV, Russo I-RM, Gopalakrishnan S, Rakotoarivelo A, Olsen R-A, Prost S, Tunstall T, Ryder OA, Dalén L, et al. 2020. Interspecific gene flow and the evolution of specialization in black and white rhinoceros. Mol Biol Evol. 37:3105–3117. [DOI] [PubMed] [Google Scholar]
- Moore AE, Cotterill FP, Eckardt FD. 2012. The evolution and ages of Makgadikgadi palaeo-lakes: consilient evidence from Kalahari drainage evolution South–Central Africa. South Afr J Geol. 115:385–413. [Google Scholar]
- Moore AE, Larkin PA. 2001. Drainage evolution in South–Central Africa since the breakup of Gondwana. South Afr J Geol. 104:47–68. [Google Scholar]
- Muya SM, Bruford MW, Muigai A W-T, Osiemo ZB, Mwachiro E, Okita-Ouma B, Goossens B. 2011. Substantial molecular variation and low genetic structure in Kenya's black rhinoceros: implications for conservation. Conserv Genet. 12:1575–1588. [Google Scholar]
- Pebesma EJ, Bivand RS. 2005. Classes and methods for spatial data in R. Available from: https://cran.r-project.org/doc/Rnews/.
- Petkova D, Novembre J, Stephens M. 2016. Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet. 48:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford M, Senut B, Hadoto D. 1993. Geology and palaeobiology of the albertine rift valley, Uganda-Zaire. Volume I: geology. Publication occasionnelle—Centre international pour la formation et les échanges géologiques [Internet]. Available from: https://pascal-francis.inist.fr/vibad/index.php?action=getRecordDetail&idt=6369901 [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2022. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: https://www.r-project.org/ [Google Scholar]
- Riedel F, Henderson ACG, Heußner K-U, Kaufmann G, Kossler A, Leipe C, Shemang E, Taft L. 2014. Dynamics of a Kalahari long-lived mega-lake system: hydromorphological and limnological changes in the Makgadikgadi Basin (Botswana) during the terminal 50 Ka. Hydrobiologia 739:25–53. [Google Scholar]
- Rookmaaker K. 2011. A review of black rhino systematics proposed in ungulate taxonomy by Groves and Grubb (2011) and its implications for rhino conservation. Pachyderm 50:72–76. [Google Scholar]
- Rookmaaker K, Antoine P-O. 2012. New maps representing the historical and recent distribution of the African species of rhinoceros: Diceros bicornis, Ceratotherium simum and Ceratotherium cottoni. Pachyderm 52:91–96. [Google Scholar]
- Sánchez-Barreiro F, Gopalakrishnan S, Ramos-Madrigal J, Westbury MV, de Manuel M, Margaryan A, Ciucani MM, Vieira FG, Patramanis Y, Kalthoff DC, et al. 2021. Historical population declines prompted significant genomic erosion in the northern and southern white rhinoceros (Ceratotherium simum). Mol Ecol. 30:6355–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharning K. Google Maps API v3 Tool. www.birdtheme.org/useful/v3tool.html [Internet]. Available from: http://www.birdtheme.org/useful/v3tool.html
- Schubert M, Ermini L, Der Sarkissian C, Jónsson H, Ginolhac A, Schaefer R, Martin MD, Fernández R, Kircher M, McCue M, et al. 2014. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc. 9:1056–1082. [DOI] [PubMed] [Google Scholar]
- Schubert M, Lindgreen S, Orlando L. 2016. Adapterremoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer ABA, Wolf JBW, Alves PC, Bergström L, Bruford MW, Brännström I, Colling G, Dalén L, De Meester L, Ekblom R, et al. 2015. Genomics and the challenging translation into conservation practice. Trends Ecol Evol. 30:78–87. [DOI] [PubMed] [Google Scholar]
- Skotte L, Korneliussen TS, Albrechtsen A. 2013. Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissinger K, Fernandes C, Formenti G, Bista I, Berg PR, Bleidorn C, Bombarely A, Crottini A, Gallo GR, Godoy JA, et al. 2023. How genomics can help biodiversity conservation. Trends Genet. 39(7):545–559. [DOI] [PubMed] [Google Scholar]
- Van Coeverden de Groot PJ, Putnam AS, Erb P, Scott C, Melnick D, O’Ryan C, Boag PT. 2011. Conservation genetics of the black rhinoceros, Diceros bicornis bicornis, in Namibia. Conserv Genet. 12:783–792. [Google Scholar]
- Veldkamp A, Schoorl JM, Wijbrans JR, Claessens L. 2012. Mount Kenya volcanic activity and the Late Cenozoic landscape reorganisation in the upper Tana fluvial system. Geomorphology 145–146:19–31. [Google Scholar]
- Waples RK, Albrechtsen A, Moltke I. 2019. Allele frequency-free inference of close familial relationships from genotypes or low-depth sequencing data. Mol Ecol. 28:35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbury MV, Hartmann S, Barlow A, Wiesel I, Leo V, Welch R, Parker DM, Sicks F, Ludwig A, Dalén L, et al. 2018. Extended and continuous decline in effective population size results in low genomic diversity in the world's rarest hyena species, the brown hyena. Mol Biol Evol. 35:1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbury MV, Thompson KF, Louis M, Cabrera AA, Skovrind M, Castruita JAS, Constantine R, Stevens JR, Lorenzen ED. 2021. Ocean-wide genomic variation in Gray's beaked whales, Mesoplodon grayi. R Soc Open Sci [Internet]. 8:201788.201788. Available from: https://royalsocietypublishing.org/doi/10.1098/rsos.201788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Available from: https://ggplot2.tidyverse.org
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zukowsky L. 1965. Die systematik der gattung Diceros gray, 1821. Zool Gart. 30:1–178. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data underlying this article is available on SRA under BioProject Number PRJNA1002571.