Abstract
Enterococci with resistance to glycopeptides have recently emerged in Australia. We developed multiplex PCR assays for vanA, vanB, vanC1, and vanC2 or vanC3 in order to examine the genetic basis for vancomycin resistance in Australian isolates of vancomycin-resistant Enterococcus faecium and E. faecalis (VRE). The predominant genotype from human clinical E. faecium isolates was vanB. The PCR van genotype was consistent with the resistance phenotype in all but six cases. One vanA E. faecalis isolate had a VanB phenotype, one vanB E. faecium isolate had a VanA phenotype, and four E. faecalis isolates were consistently negative for vanA, vanB, vanC1, and vanC2 or vanC3, even though they exhibited a VanB phenotype. These four isolates were subsequently examined for the presence of vanD by published methods and were found to be negative. No vancomycin-susceptible strains produced a PCR product. On the basis of our findings the epidemiology of VRE in Australia appears to be different from that in either the United States or Europe. Our multiplex PCR assays gave a rapid and accurate method for determining the genotype and confirming the identification of glycopeptide-resistant enterococci. Rapid and accurate methods are essential, because laboratory-based surveillance is critical in programs for the detection, control, and prevention of the transmission of glycopeptide-resistant enterococci.
Vancomycin-resistant Enterococcus faecium and E. faecalis (VRE) were first described in Britain in 1988 (22) and soon afterward were reported from other European countries and the United States (14). In the United States they have become major nosocomial pathogens, rising in incidence from 0.3% in 1989 to 7.9% in 1993, as reported by the Centers for Disease Control and Prevention (5), and among patients in intensive care units they now represent 14% of isolates of enterococci retrieved from cultures of blood (5). Resistance to multiple antimicrobial agents as well as vancomycin, especially ampicillin and high levels of aminoglycosides, is typical of these isolates (15).
Two principal phenotypes of acquired vancomycin resistance have been described, VanA and VanB, encoded by two distinct gene clusters, the vanA and vanB clusters, respectively, which are carried on transposons Tn1546 and Tn1547, respectively (3). The VanA phenotype confers high-level resistance to both vancomycin and teicoplanin, while the VanB phenotype confers moderate to high-level resistance to vancomycin only. A third type of vancomycin resistance, termed VanC, has been known for many years to be a natural (intrinsic) vancomycin resistance found in the motile enterococci E. casseliflavus, E. gallinarum, and E. flavescens (3). VanC confers low-level resistance to vancomycin only. Unlike E. faecalis and E. faecium, the motile enterococci are infrequent pathogens in humans.
The troublesome features of enterococci with the VanA and VanB phenotypes include their high propensity for cross-infection and the resistance of many strains to all conventional agents. The demonstration that the vanA gene cluster can be transferred to Staphylococcus aureus in vitro and in vivo is further cause for concern (18).
The rapid emergence of VRE in the United States has been attributed to the intensive clinical use of vancomycin in both parenteral and oral forms in that country (13) on a background of high-level usage of cephalosporins, which promote enterococcal superinfection (16, 23). In Europe, investigators have postulated an additional role for the use of the glycopeptide avoparcin as a growth promoter in intensive animal industries, resulting in colonization with VanA E. faecium and subsequent transmission to humans via the food chain (1).
The first vancomycin-resistant E. faecium isolate in Australia was isolated from a liver transplant recipient in Melbourne in 1994 (12). Since March 1996 multiple isolates of vancomycin-resistant E. faecium and vancomycin-resistant E. faecalis have occurred throughout Australia. Only a few of these strains have been reported in the literature (4, 8, 9, 19).
The National Antimicrobial Resistance Surveillance Program at the Women’s and Children’s Hospital in Adelaide is a referral center for antimicrobial resistance in Australia, and we have collected isolates from virtually all patients known to have VRE infections that have occurred since 1994. In order to characterize these strains further we have developed multiplex PCR assays for vanA, vanB, vanC1, and vanC2 or vanC3 and have used these to examine the genetic basis for vancomycin resistance in Australian isolates of VRE. The results have been compared to those obtained by conventional susceptibility testing with glycopeptides.
MATERIALS AND METHODS
Bacterial strains.
Two hundred forty-eight isolates of Enterococcus spp. referred to the National Antimicrobial Resistance Surveillance Program were studied. Previously characterized VRE strains were used as controls. These included E. faecalis ATCC 51299 (vanB; vancomycin MIC, 6 μg/ml; teicoplanin MIC, 0.5 μg/ml), E. casseliflavus ATCC 25788 (vanC2; vancomycin MIC, 6 μg/ml; teicoplanin MIC, 0.75 μg/ml), E. gallinarum NCDO 2313 (vanC1; vancomycin MIC, 12 μg/ml; teicoplanin MIC, 1 μg/ml), E. faecalis ATCC 19433 (vancomycin MIC, 1 μg/ml; teicoplanin MIC, 0.19 μg/ml), E. faecium ATCC 19434 (vancomycin MIC, 1 μg/ml; teicoplanin MIC, 0.75 μg/ml), and E. faecalis E19 (12) (vanA; vancomycin MIC, >256 μg/ml; teicoplanin MIC, >256 μg/ml).
Identification and antimicrobial susceptibility testing.
Isolates were identified by a conventional test scheme (7). A multiplex PCR assay based on the specific detection of genes encoding d-alanine:d-alanine ligases (ddl) (6) was used to confirm the identification of E. faecalis and E. faecium.
The MICs of vancomycin and teicoplanin were determined for each isolate by the Etest (AB Biodisk, Solna, Sweden) method on Mueller-Hinton agar (2, 11). The interpretative criteria of the National Committee for Clinical Laboratory Standards (17) were used to determine the susceptibilities of the isolates.
Vancomycin resistance gene typing by PCR.
Enterococci were first grown overnight at 37°C in Todd-Hewitt broth, and then 1-ml volumes were microcentrifuged and the pellet was resuspended in 200 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). The suspensions were heated at 95°C for 20 min and then microcentrifuged for 2 min. Five-microliter volumes of the supernatant were subjected to PCR amplification in 50-μl reaction mixtures containing each deoxynucleoside triphosphate at a concentration of 200 μM, each primer at a concentration of approximately 1 μM, and 1 U of Taq polymerase (Boehringer Mannheim) in 10 mM Tris-HCl (pH 8.3)–50 mM KCl–2 mM MgCl2–0.1% gelatin–0.1% Tween 20–0.1% Nonidet P-40. The samples were subjected to 35 PCR cycles, each consisting of 1 min of denaturation at 94°C, 2 min of annealing at 60°C, and 2 min of elongation at 72°C. PCRs were analyzed by electrophoresis on 2% agarose gels and were stained with ethidium bromide. The oligonucleotide primers used for detection of vanA, vanB, vanC1, and vanC2 or vanC3 sequences were designed with reference to the sequences deposited in GenBank by P. Courvalin and colleagues under accession numbers X56895, L06138, M75132, L29638, and L29639, respectively. Primer sequences and specificities are presented in Table 1. For each sample, two PCRs were set up. One contained primers VanABF, VanAR, and VanBR, which direct amplification of 231- and 330-bp fragments from the vanA and vanB genes, respectively. The other contained primers VanC1F, VanC1R, VanC23F, and VanC23R, which direct amplification of 447- and 597-bp fragments from the vanC1 gene and either the vanC2 or the vanC3 gene, respectively. Multiplex PCRs were tested in duplicate. Known positive and negative controls were also included with each PCR run.
TABLE 1.
PCR primer sequences
| Primer | Sequence | Specificity | Location within gene (strand)a |
|---|---|---|---|
| VanABF | GTAGGCTGCGATATTCAAAGC | vanA | 358–378 (+) |
| vanB | 355–375 (+) | ||
| VanAR | CGATTCAATTGCGTAGTCCAA | vanA | 568–588 (−) |
| VanBR | GCCGACAATCAAATCATCCTC | vanB | 664–684 (−) |
| VanC1F | TGGTATTGGTATCAAGGAAACC | vanC1 | 139–160 (+) |
| VanC1R | AGATTGGAGCGCTGTTTTGTC | vanC1 | 565–585 (−) |
| VanC23F | CAGCAGCCATTGGCGTACAA | vanC2 and vanC3 | 431–450 (+) |
| VanC23R | CAAGCAGTTTTTGTAGTAGTTC | vanC2 and vanC3 | 1006–1027 (−) |
Position in nucleotide sequence relative to the initiation codon. +, positive strand; −, negative strand.
Genotype-negative VRE isolates, for which vancomycin MICs were >4 μg/ml, were also tested for the presence of vanD by using the primers described by Perichon et al. (20).
RESULTS
Characterization of multiplex PCR assays for vanA, vanB, vanC1, and vanC2 or vanC3.
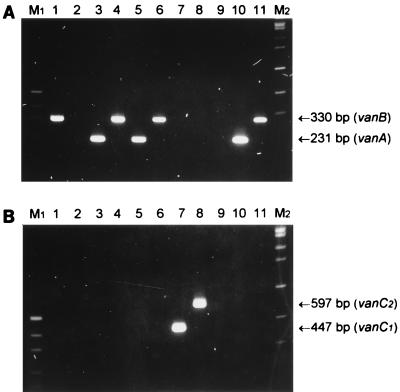
In order to confirm the specificities of the various PCR primers listed in Table 1, representative reference VRE isolates and vancomycin-susceptible enterococci were analyzed as described in Materials and Methods (Fig. 1). Clear PCR products of the expected size (231, 330, 447, and 597 bp for vanA, vanB, vanC1 and vanC2 or vanC3, respectively) were obtained. There was 100% agreement between the PCR results and the previously published genotypes and phenotypes.
FIG. 1.
PCR analysis of VRE. Enterococci were subjected to PCR analysis, as described in Materials and Methods, with primers VanABF, VanAR, and VanBR (A) or VanC1F, VanC1R, VanC23F, and VanC23R (B). The PCR mixtures were electrophoresed on 2% agarose gels and stained with ethidium bromide. Lanes: 1, E. faecalis ATCC 51299; 2, E. faecium ATCC 19434; 3, E. faecalis 91; 4, E. faecalis 3; 5, E. faecium 143; 6, E. faecium 135; 7, E. gallinarum 129; 8, E. casseliflavus 38; 9, E. faecalis 26; 10, E. faecalis 21; 11, E. faecium 30. Lane M1, pUC19 DNA digested with HpaII (fragments of 501 and 489, 404, 331, 242, 190, and 147 bp are visible); lane M2, bacteriophage SPP1 DNA digested with EcoRI (fragments of 1,950, 1,860, 1,510, 1,390, 1,160, 980, 720, 480, and 360 bp are visible.
PCR analysis of Australian enterococci.
A total of 139 VRE isolates from 14 institutions in seven cities (Adelaide, Brisbane, Darwin, Melbourne, Newcastle, Perth, and Sydney) throughout all mainland states of Australia were obtained. The VRE were either human clinical isolates (n = 41) or isolates from the contacts of index patients (n = 63), the environment (n = 33), or animals (n = 2). Other referred Enterococcus spp. were clinical isolates (n = 89) or were from animals (n = 20). The results of PCR analysis of the van genotype are presented in Table 2. PCR van genotype results for E. faecalis and E. faecium isolates were consistent with the resistance phenotype for all but six isolates. The discrepancies were as follows: one vanA E. faecalis isolate with the VanB phenotype, one vanB E. faecium isolate with the VanA phenotype, and four E. faecalis isolates consistently negative for vanA, vanB, vanC1, vanC2 or vanC3, and vanD, even though they exhibited a VanB phenotype. No vancomycin-susceptible strains produced a PCR product. All E. faecium (n = 133) and E. faecalis (n = 60) isolates were correctly identified by PCR with the ddl primers. Using the vanC1 and vanC2 or vanC3 primers, we further confirmed the identification of 42 E. gallinarum and 9 E. casseliflavus isolates. Of the 41 VRE isolates noted as index isolates by the sending institution, 23 (56%) were E. faecium vanB, 7 (17%) were E. faecium vanA, 6 (15%) were E. faecalis vanB, 3 were van-negative E. faecalis, and 2 were E. faecalis vanA.
TABLE 2.
Characteristics of Australian enterococcia
| Species | Phenotype | No. of isolates with the following van genotype:
|
|||||
|---|---|---|---|---|---|---|---|
| vanA | vanB | vanC1 | vanC2 or vanC3 | Negative | Total | ||
| E. faecalis | Sensitiveb | 44 | 44 | ||||
| VanA | 1 | 1 | |||||
| VanB | 1 | 10 | 4 | 15 | |||
| E. faecium | Sensitiveb | 10 | 10 | ||||
| VanA | 82 | 1 | 83 | ||||
| VanB | 40 | 40 | |||||
| E. gallinarum | VanC | 42 | 42 | ||||
| E. casseliflavus | VanC | 9 | 9 | ||||
| Other speciesc | 4 | 4 | |||||
A total of 248 isolates were tested; VRE (n = 139) comprised human clinical isolates (n = 41) and isolates from contacts of index patients (n = 63), the environment (n = 33), and animals (n = 2); other Enterococcus spp. (n = 109) included clinical isolates (n = 89) and isolates from animals (n = 20). Refer to text for discussion on the prevalence of VRE from clinical isolates.
Vancomycin MIC, ≤4 μg/ml.
E. durans (n = 2) and E. hirae (n = 2).
The vancomycin and teicoplanin susceptibility results are presented in Table 3. Interestingly, one E. faecium isolate and three E. faecalis isolates with intermediate resistance to vancomycin (MICs, 8 to 16 μg/ml) were PCR positive for vanB. For the E. faecalis vanA isolate with the VanB phenotype referred to above, the teicoplanin MIC was 4 μg/ml. For the VanA E. faecium isolate which was vanB positive, the teicoplanin MIC was ≥256 μg/ml. For the four VRE isolates which were van negative, the vancomycin MICs were in the range of 12 to 16 μg/ml.
TABLE 3.
Glycopeptide susceptibility
| Gene | Species | MIC (μg/ml)
|
No. of isolates | |
|---|---|---|---|---|
| Vancomycin | Teicoplanin | |||
| vanA | E. faecium (n = 82) | 256 | >32 | 53 |
| 256 | 32 | 19 | ||
| 256 | 24 | 7 | ||
| 256 | 16 | 2 | ||
| 256 | 12 | 1 | ||
| E. faecalis (n = 2) | 256 | 256 | 1 | |
| 256 | 4 | 1 | ||
| vanB | E. faecium (n = 41) | 256 | 256 | 1 |
| 256 | 0.25–1.5 | 33 | ||
| 128 | 1 | 1 | ||
| 64 | 0.75–1 | 3 | ||
| 24 | 0.5–0.75 | 2 | ||
| 12 | 0.5 | 1 | ||
| E. faecalis (n = 10) | 256 | 0.25 | 1 | |
| 128 | 0.38 | 1 | ||
| 48 | 0.5–0.75 | 2 | ||
| 32 | 0.5 | 2 | ||
| 24 | 0.38 | 1 | ||
| 16 | 0.5 | 1 | ||
| 12 | 0.25 | 1 | ||
| 8 | 0.25 | 1 | ||
| vanC1 | E. gallinarum (n = 42) | 12 | 1 | 1 |
| 8 | 0.5–1.5 | 20 | ||
| 6 | 0.38–1 | 18 | ||
| 4 | 0.75–1 | 2 | ||
| 3 | 0.5 | 1 | ||
| vanC2 or vanC3 | E. casseliflavus (n = 9) | 6 | 0.5–1 | 4 |
| 4 | 0.5–1 | 5 | ||
| None | E. faecium (n = 10) | 1.5 | 0.75–1 | 3 |
| 1 | 0.75–1 | 5 | ||
| 0.75 | 0.125–1 | 2 | ||
| E. faecalis (n = 48) | 16 | 0.38 | 2 | |
| 12 | 0.5 | 2 | ||
| 4 | 0.38–0.75 | 4 | ||
| 3 | 0.25–1 | 13 | ||
| 2 | 0.25–0.75 | 19 | ||
| 1.5 | 0.125–0.5 | 8 | ||
| E. durans (n = 2) | 6 | 0.38 | 1 | |
| 4 | 0.125 | 1 | ||
| E. hirae (n = 2) | 1 | 0.094 | 1 | |
| 0.75 | 0.094 | 1 | ||
DISCUSSION
The VRE isolated in Australia to date show considerable diversity in their phenotypes, genotypes, and geographic locations. All four combinations of genotype and species have been found, with the commonest being E. faecium vanB. While the clinical profiles of VRE-affected patients appear to be similar to those recorded in the United States and elsewhere (13), the predominance of E. faecium vanB rather than E. faecium vanA suggests an epidemiology different from that in either Europe or the United States.
The origin of VRE in Australia remains unclear. No strains appear to have been imported, although one occurred in a liver transplant recipient who was a New Zealand-born resident of Taiwan. This patient had entered Australia specifically for transplantation a few days prior to the procedure. E. faecalis of the VanB phenotype was initially isolated from blood cultures after surgery. The patient was treated with teicoplanin, but several days later a vancomycin-resistant enterococcus was again isolated from blood cultures, with the isolate identified as E. faecalis of the VanA phenotype. Genotyping showed that both isolates possessed the vanB gene, and subsequent ribotyping confirmed that the strains were identical. The emergence of resistance to teicoplanin has been recorded previously, albeit rarely (10).
The level of vancomycin use in Australia is relatively high and has been increasing over the last decade. There is significant regional variation in its use due to the variation in prevalence of multidrug-resistant S. aureus (21). Australia is also a high-level user of avoparcin as a growth promoter in the intensive animal industries. It is possible that the novel epidemiology of VRE in Australia may result from a combination of the high rates of use of vancomycin and avoparcin in humans and animals, respectively.
PCR methods have previously been used for the rapid identification of the vancomycin resistance genotype (6). In the present study we designed a set of PCR primers that provides for the simultaneous identification of all the major van genotypes under identical amplification conditions. Our multiplex van PCR assays were rapid and simple, giving clear-cut answers within 6 h. On the basis of phenotypic analysis, no false-positive results were generated by this test. PCR analysis also indicated that MIC determination alone is not sufficient for the unambiguous classification of isolates of VRE. Also, difficulties continue to occur with commercial identification systems, and the not infrequent occurrence of nonmotile E. gallinarum and E. casseliflavus isolates and nonpigmented E. casseliflavus isolates compounds the problem. The ddl PCR was extremely useful for the identification of E. faecalis and E. faecium and, in combination with vanC1 and vanC2 or vanC3 PCR, for the identification of E. gallinarum and E. casseliflavus. It is essential to have a rapid and accurate method for determination of the genotype and for confirmation of the identification of glycopeptide-resistant enterococci, especially during an outbreak or when performing surveillance for VRE. It is unlikely that the four strains with the VanB resistance phenotype that appeared to lack vanA, vanB, vanC1, and vanC2 or vanC3 or even the recently described vanD (20) were false-negative isolates (e.g., had the VanB resistance phenotype due to minor sequence variations in the primer annealing sites), because PCR analysis with independent vanA and vanB primers (6) was also negative. All four strains (from three patients) came from a single institution and gave two distinct pulsed-field gel electrophoresis patterns. Our results are consistent with either the existence of a significant variant of a current van genotype or a novel one. The van loci of these strains are undergoing further analysis.
ACKNOWLEDGMENTS
We thank all the contributing laboratories throughout Australia that provided enterococci for this study; the Gram-Positive Bacteria Typing and Research Unit at Royal Perth Hospital and Curtin University (Geoff Coombs, Cheryll McCullough, Todd Pryce, Ian Kay, and Frances O’Brien) for providing some of the type strains, ribotyping, and confirming the genotype of non-vanA, non-vanB, and non-vanC E. faecalis; and Barrie Mayall for providing strain E19.
REFERENCES
- 1.Aarestrup F M, Ahrens P, Madsen M, Pallesen L V, Poulsen R L, Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob Agents Chemother. 1996;40:1938–1940. doi: 10.1128/aac.40.8.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AB Biodisk. Etest technical guide 3B. Solna, Sweden: AB Biodisk; 1995. [Google Scholar]
- 3.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 4.Branley J, Yan B, Benn R A V. Vancomycin-resistant Enterococcus faecalis. Med J Aust. 1996;165:292. doi: 10.5694/j.1326-5377.1996.tb124971.x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Nosocomial enterococci resistant to vancomycin—United States, 1989–1993. Morbid Mortal Weekly Rep. 1993;42:597–599. [PubMed] [Google Scholar]
- 6.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. . (Erratum, 33:1434.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faoagali J, Bodman J, Geary A. Isolation of vancomycin-resistant enterococci in Queensland, case 2. Communicable Dis Intell Aust. 1996;20:402–403. [Google Scholar]
- 9.Ferguson J, Butt H, Johnson C, Boyle M. Vancomycin-resistant Enterococcus faecium colonisations. Med J Aust. 1996;165:292–293. doi: 10.5694/j.1326-5377.1996.tb124972.x. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 10.Hayden M K, Trenholme G M, Schultz J E, Sahm D F. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis. 1993;167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 11.Jones R N, Erwin M E, Anderson S C. Emerging multiply resistant enterococci among clinical isolates. II. Validation of the Etest to recognize glycopeptide-resistant strains. Diagn Microbiol Infect Dis. 1995;21:95–100. doi: 10.1016/0732-8893(94)00146-n. [DOI] [PubMed] [Google Scholar]
- 12.Kamarulzaman A, Tosolini F A, Boquest A L, Geddes J E, Richards M J. Vancomycin-resistant Enterococcus faecium in a liver transplant recipient. Aust N Z J Med. 1995;25:560. . (Abstract.) [Google Scholar]
- 13.Leclercq R, Courvalin P. Resistance to glycopeptides in enterococci. Clin Infect Dis. 1997;24:545–546. doi: 10.1093/clind/24.4.545. [DOI] [PubMed] [Google Scholar]
- 14.LeClercq R, Perlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 15.Moellering R C., Jr Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14:1173–1178. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 16.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 18.Noble W C, Virani Z, Cree R G A. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Newsl. 1992;93:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 19.Paterson D, Jennings A, Allen A, Sherlock K, Whitby M. Isolation of vancomycin-resistant enterococci in Queensland, case 1. Communicable Dis Intell Aust. 1996;20:400–401. [Google Scholar]
- 20.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnidge J D, Nimmo G R, Francis G. Evolution of resistance in Staphylococcus aureus in Australian teaching hospitals. Med J Aust. 1996;164:68–71. [PubMed] [Google Scholar]
- 22.Uttley A H C, Collins C H, Naidoo J, George R C. Vancomycin-resistant enterococci. Lancet. 1988;i:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 23.Woodford N, Johnson A P, Morrison D, Speller D C. Current perspectives on glycopeptide resistance. Clin Microbiol Rev. 1995;8:585–615. doi: 10.1128/cmr.8.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]