Abstract
Background/Aim
In recent years, individual patient cancer genomic profiling (CGP) has become more accessible, allowing determination of therapeutic strategies using driver gene mutations in cancer therapy. However, this precision oncology approach, tailored to specific patients, remains experimental. In this study, we verified the feasibility and benefit of using CGP to guide treatment of malignant head and neck tumors. We aimed to evaluate the profiling and clinical courses of patients with head and neck malignancies who underwent CGP and determine the extent to which CGP for head and neck malignancies has resulted in beneficial drug administration.
Patients and Methods
We analyzed CGP results, prognosis, and drug administration status in 27 patients. These patients had completed (or were expected to complete) standard therapy or had rare cancers without standard therapy.
Results
At least one somatic actionable gene alteration was seen in 25 (92.6%) patients, with a median number of actionable alterations per patient of 4 (range=0-11). Drugs in clinical trials were recommended to 22 (81.5%) patients, but none could participate. However, 3 patients (11.1%) could use approved drugs off-label based on CGP results. The most common genetic abnormality was TP53 (66.7%), with TP53 mutations leading to poor prognosis.
Conclusion
CGP is clinically useful and serves as a bridge to increase the number of therapeutic options. However, candidate drugs confirmed using CGP may be ineffective when administered. Therefore, oncologists should not blindly accept CGP therapeutic recommendations but should make recommendations that lead to optimal therapies after proper verification.
Keywords: Cancer genomic profiling, head and neck malignancies, precision oncology, TP53 mutation, immune checkpoint inhibitor
In 2003, the Human Genome Project decoded approximately 92% of the human genome sequence (1) and completely deciphered the entire sequence in 2022 (2). In addition, the Cancer Genome Atlas (http://cancergenome.nih.gov) and International Cancer Genome Consortium (http://www.icgc.org) published the cancer genome data that may be used for all cancer types based on this deciphering (3,4).
Therefore, in recent years, individual patient cancer genomic profiling (CGP) has become relatively easily available (3,4). Oncologists can specify molecular targetable cancer alterations and relevant cancer biomarkers for each patient using CGP (5,6). Hence, they can identify driver gene mutations that can serve as therapeutic targets. The medical technology called next-generation sequencing (NGS) has made this clinically feasible (5,6). NGS has dramatically improved the efficiency of genome analysis, enabling the timely detection and analysis of the mutation, amplification, loss, and fusion of hundreds of different gene types simultaneously (5,6). However, CGP results obtained using this remarkable technology can only lead to precision oncology when appropriately interpreted by oncologists and properly guided to available therapies. When driver gene mutations are discovered but not matched to therapies, the prognosis is the same as when driver gene mutations are not discovered. However, patients with driver gene mutations may have a better prognosis and can benefit from targeted therapies (7,8). Despite the discovery of target genes, only a small proportion of patients (6-20%) reach the treatment stage (8-10), and some problems still exist on the path to widely realizing precision oncology.
Moreover, these problems greatly differ depending on cancer type (11). The proportion of driver gene targeting drugs that are Food and Drug Administration (FDA)-approved and incorporated into standard therapy differs, with approximately 95%, 50%, 30%, and 25% for gastrointestinal stromal cancer, melanoma, non-small cell lung cancer, and breast cancer, respectively. Moreover, a significant difference in the realization rate of precision oncology exists between cancer types (11,12). The proportion of drugs for driver genes that are FDA-approved as standard therapy for head and neck cancer (HNC) is 0% (13), the difference from other cancers is obvious. Gradually, the genomic landscape of HNC is being clarified, the number of viable/druggable targets is increasing, and clinical trials are being conducted (14-17). However, they have not reached the stage of clinical application. Thus, whether precision oncology is feasible in the head and neck region and whether the role of CGP in precision oncology is clinically useful is unclear.
In this study, we aimed to evaluate the profiling and subsequent clinical courses of patients with malignant tumors of the head and neck region who underwent CGP at the Kyushu University Hospital and verified to what extent precision oncology was feasible using CGP. Further, we aimed to determine the extent to which CGP for head and neck malignancies has resulted in beneficial drug administration to patients. This verification will become the first step in making future precision oncology for malignant tumors of the head and neck region more effective.
Patients and Methods
We analyzed 27 Asian patients with malignant tumors of the head and neck who underwent CGP within 3 years, from December 2019 to December 2022, at the Kyushu University Hospital. These patients were patients with HNC who had completed (or were expected to complete) standard therapy or patients with rare cancers without standard therapy.
The samples were formalin-fixed paraffin-embedded tissues derived from major tissues of all 27 patients via surgery or biopsy, and gene profiling data were analyzed using FoundationOne® CDx platform [supplied by Foundation Medicine Inc. (FMI) Laboratories, Penzberg, Germany]. The detection analysis of the mutation, amplification, loss, and fusion of genes, and measurement of microsatellite instability (MSI) and tumor mutation burden (TMB) were performed using FoundationOne® CDx mounted with 324 genes. The actionable gene alterations and available therapies were discussed at the molecular tumor board meeting. The board members included medical oncologists, pathologists, genome researchers, and genetic counselors.
We examined the frequency of driver gene mutations, MSI and TMB, frequency of cases matched to therapies, and subsequent therapeutic courses of the 27 patients based on CGP results. The survival curve was computed using the period from the CGP detection date to the final survival confirmation date or date of death as the survival period.
This study was approved by the institutional review board of the Kyushu University (approval number: 2022-27). All patients had provided written informed consent for future research and genomic analysis. We followed the guidelines of the NHGRI Policy on Release of Human Genomic Sequence Data.
Statistical analyses. All calculations were performed using the SPSS statistics software program, ver. 22.0 (IBM Japan, Ltd., Tokyo, Japan). The overall survival (OS) was estimated using the Kaplan-Meier method and evaluated using the log-rank test. Statistical significance was set at p<0.05.
Results
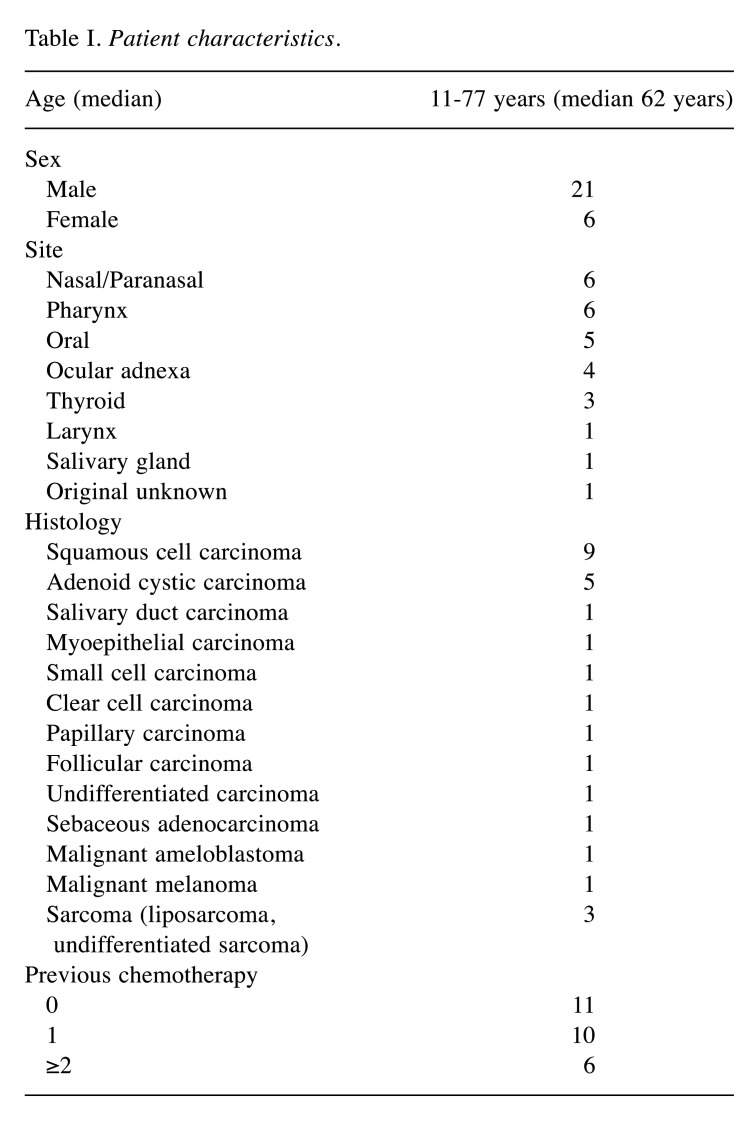
Patient characteristics. The patients included 21 men (77.8%) and 6 women (22.2%), with a median age of 62 years (11-77 years). The most common primary site was nasal/paranasal and pharynx in six patients (22.2%) each, followed by oral in five patients (18.5%) and ocular adnexa in four patients (14.8%). The most common histopathological type was squamous cell carcinoma in nine patients (33.3%), followed by adenoid cystic carcinoma in five patients (18.5%) (Table I).
Table I. Patient characteristics.
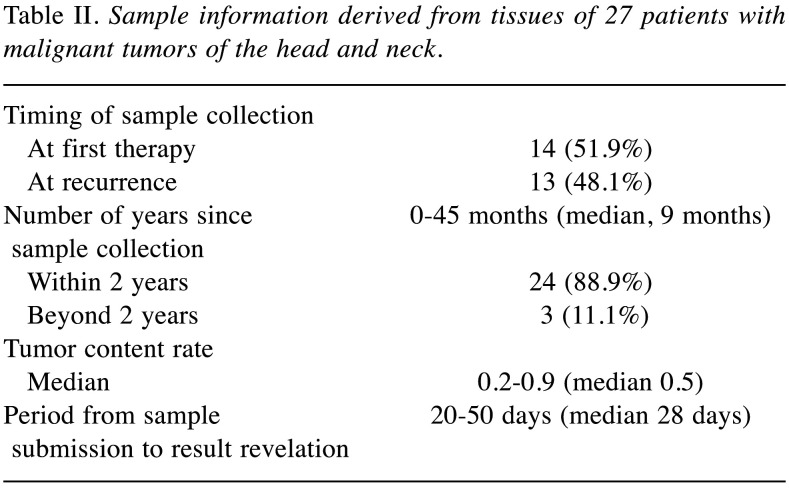
Sample condition. The median period from sample collection to submission to FoundationOne® CDx was nine months (0-45 months), and samples were collected within 2 years in 24 patients (88.9%). Samples were collected during the first therapy in 14 patients, and at recurrence in 13 patients, with a median tumor content rate of 50% (range=20-90%). All 27 patients were analyzed. The median number of days from submission to FoundationOne® CDx to results analysis and submission to oncologists was 28 days (20-50 days) (Table II).
Table II. Sample information derived from tissues of 27 patients with malignant tumors of the head and neck.
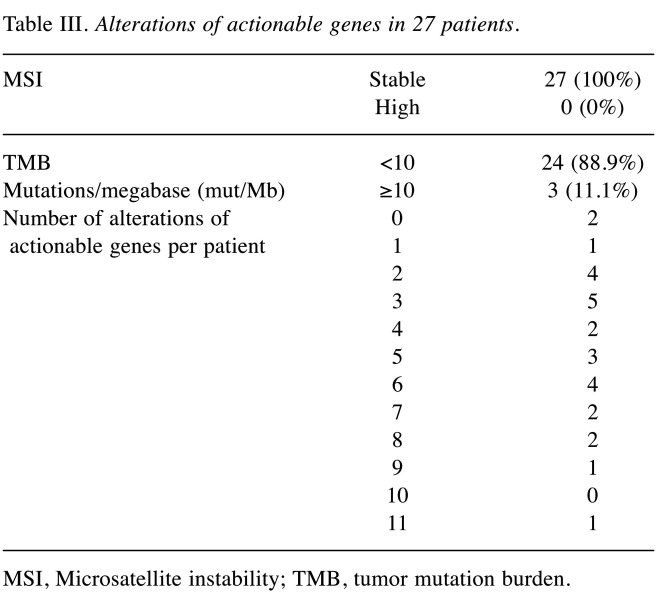
Alterations of actionable genes in 27 patients. All 27 patients were MSI-stable, with no confirmed patient with high MSI. Regarding TMB, three patients (11.1%) had ≥10 mutations/megabase (mut/Mb), and 24 patients (88.9%) had <10 mut/Mb. Secondary findings of germline mutation were observed in one patient (3.7%) (Table III).
Table III. Alterations of actionable genes in 27 patients.
MSI, Microsatellite instability; TMB, tumor mutation burden.
Among the 27 patients, 25 (92.6%) had at least one somatic actionable gene alteration. The median number of actionable alterations per patient was four (range=0-11). TP53 had the most genetic abnormalities (66.7%, 18/27), followed by FGF (55.6%, 15/27), CDKN2A/B (29.6%, 8/27), PTEN (14.8%, 4/27), TERT (14.8%, 4/27), NOTCH1 (14.8%, 4/27), and ERBB2 (14.8%, 4/27).
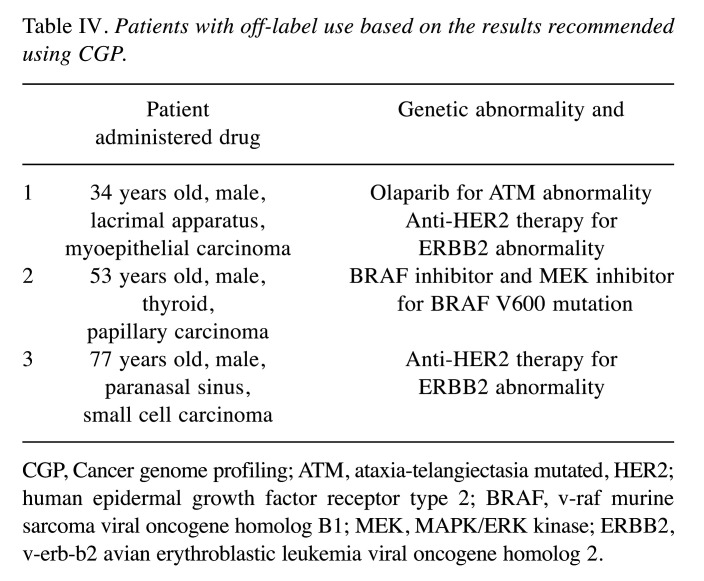
Clinical course. Drugs in clinical trials were recommended to 22 of the 27 patients (81.5%), but no patient could participate in the clinical trials. However, based on CGP results, three patients (11.1%) purchased FDA-approved drugs at their expense and used them off-label. One out of three patients could maintain stable disease (SD) for 7 months but developed progressive disease (PD) subsequently and died 10 months after the CGP results were revealed. The remaining two patients have recently commenced drug administration; therefore, the effects are unknown (Table IV).
Table IV. Patients with off-label use based on the results recommended using CGP.
CGP, Cancer genome profiling; ATM, ataxia-telangiectasia mutated, HER2; human epidermal growth factor receptor type 2; BRAF, v-raf murine sarcoma viral oncogene homolog B1; MEK, MAPK/ERK kinase; ERBB2, v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2.
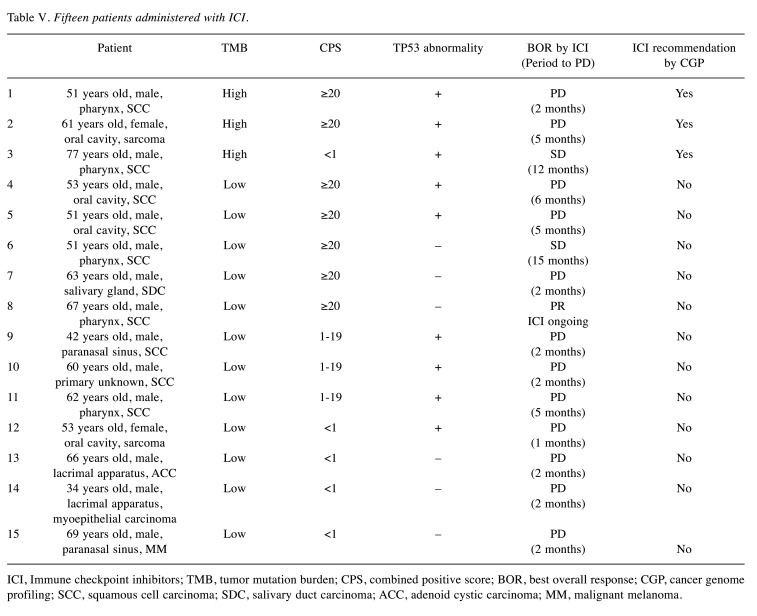
Immune checkpoint inhibitors (ICI) were administered to 15 of the 27 patients before CGP, and programmed death-ligand 1 (PD-L1) was measured using the combined positive score (CPS) before ICI administration. Table V shows the CPS results, TMB and MSI status, and ICI effect examination results. According to the molecular tumor board meeting based on CGP, ICI, such as anti- PD-L1 or PD-L1 antibodies, were recommended for three patients with high TMB. Two of them had high CPS, but the effects of ICI were not observed. TP53 gene mutations were observed in these two patients. The remaining patient had CPS <1 and TP53 gene mutations; however, he developed PD after maintaining the SD effect for 12 months following ICI administration (Table V).
Table V. Fifteen patients administered with ICI.
ICI, Immune checkpoint inhibitors; TMB, tumor mutation burden; CPS, combined positive score; BOR, best overall response; CGP, cancer genome profiling; SCC, squamous cell carcinoma; SDC, salivary duct carcinoma; ACC, adenoid cystic carcinoma; MM, malignant melanoma.
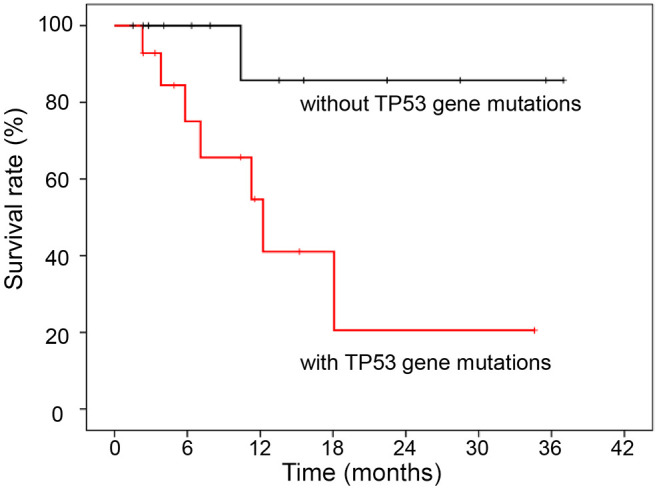
The 1-year survival rate of the 27 patients was 68%. The survival rate was examined based on TP53 gene mutations, and it was discovered that patients with TP53 mutation had a significantly poorer prognosis than patients without TP53 mutations (p=0.022, Figure 1).
Figure 1. Survival curve of 27 patients who underwent cancer genome profiling (CGP) with and without TP53 gene mutations. There was no effect of CGP implementation on survival. However, CGP did reveal a relationship between TP53 mutation, which can only be detected by CGP, and survival of patients with malignant tumors of the head and neck.
Discussion
Recurrent/metastatic head and neck squamous cell carcinoma (R/M HNSCC) has a poor prognosis with an approximately 1-year median survival period (18). Furthermore, while the number of existing therapeutic options has increased in recent years, they are still limited, and their effectiveness is also limited (19,20). Malignant tumors of the head and neck region, other than HNSCC, are very rare, and there is currently no standard therapy (21,22). In addition, the therapeutic contents for such cases are determined based on tumor biology and tumor stage classification (13).
Advancement in NGS has increased the number of explorable molecular druggable targets. Currently, approximately 3000 drug discoveries are possible, with approximately 10% being small-molecule drug targets (23,24). In addition, clinical trials evaluating these targets have increased drastically in recent years because CGP has become easily available in clinical practice (6,7). Furthermore, the path to biomarker-driven precision oncology is opening gradually for malignant tumors of the head and neck region, including R/M HNSCC (17). CGP, which offers access to clinical trials, is a test that might benefit patients with rare cancers that have few or no standard therapeutic options.
However, the usefulness of CGP is dependent on the interpretation of NGS results and the realization rate of drug access, with a limited number of patients experiencing the effects (25). In previous large-scale studies, the probability of discovering actionable mutations in all cancer types is approximately 40-60% overseas (8,9), while it ranges from 60 to 90% in Japan (25,26). The possibility that these results are linked to some therapy is only 6-11% in reports from overseas (8,9,11) and approximately 8-22% in reports from Japan (25,26). In this study, drugs accessible in Japan were discovered for approximately 81.5% of patients, but none could participate in clinical trials. This was mainly because most available drugs were in Phase 1 clinical trials, limiting the possible number of entries. Moreover, hospitals conducting the clinical trials were far distant, making visits difficult. A previous study identified the following shortcomings of genomic medicine: genome analysis processing takes a long time (approximately 2 months), clinical trial requirements are seldom met, and clinical trial slots are insufficient, even though patients undergoing CGP often progress rapidly (10). Thus, despite great efforts by doctors and patients, the effects of molecular targeted therapy for tumor genomic alterations are limited (10). Difficulties in accessing therapeutic drugs have become a big obstacle to realizing precision oncology using CGP in the head and neck region, and obtaining the effects of developing therapeutic drugs is challenging. However, based on CGP results and considering the three patients (11.1%) to whom drugs could be administered off-label, even though at their expense, the path to precision oncology is gradually advancing. The National Comprehensive Cancer Network (NCCN) guidelines also approve the benefits of off-label use for patients to some extent (27), and when the path to clinical trials is difficult, this approach should be actively recommended to patients as a useful option for realizing precision oncology.
The frequency of known genomic abnormalities in HNC that cause cancer is relatively high; thus, many viable/druggable effective drug candidates should exist. However, HNC is a carcinoma that is difficult to treat using cancer genome medicine (17,28). Apart from the insufficient number of cases and difficulties in establishing evidence due to the small number of HNC cases, this difficulty in treatment is caused by the intratumor heterogeneity of HNC at the molecular level, which hinders the identification of specific targets and development of targeted therapeutic drugs. Furthermore, new targeted therapies have not improved clinical outcomes (29,30).
In addition, the high TP53 mutation rate in HNC plays a significant role. HNC has high mutation rate mainly in TP53, CDKN2A, CCND1, EGFR, PIK3CA, PTEN, and NOTCH1 (14-17). Among them, mutations of the tumor suppressor protein TP53 are the most common gene mutations in HNSCC, accounting for approximately 70%, leading to a loss of tumor suppressor activity and genomic instability (15,31,32). Furthermore, the high frequency of TP53 mutations becomes more striking after metastasis and recurrence (18), and the mutation rate was high (66.7%) in this study. Moreover, TP53 mutations have a poor prognosis in HNC (31,32), which is consistent with our results. Additionally, even though TP53 is the most frequently altered gene in all cancer types (n=10,336), the frequency is 41% (9), and whether TP53 mutations are correlated with prognosis differs by cancer type (33,34). Furthermore, TP53 mutations affect the therapeutic effects of ICI (18,34). The effects of ICI should be expected in high TMB and PD-L1 positive (20,28,35,36); however, despite high TMB scores, there is essentially no response to therapy in HNC with TP53 mutations (18). In this study, despite the high TMB, ICI recommendation, and high PD-L1 status, patients with poor ICI effects accounted for two-thirds of patients in clinical practice, and TP53 mutations were confirmed in all patients. Moreover, TP53 mutations are more likely to promote responsiveness to immunotherapy in other cancer types (34). Hence, a part of the cancer genome gene information obtained from CGP should be interpreted based on each cancer type despite the same abnormal findings. CGP should now be a test that presents drug therapies across organs, although it has aspects that should be approached with an organ-specific mindset.
One of this study’s limitations is the small number of cases. Hence, more cases must be accumulated in the future.
Conclusion
Standard therapy, the pillar of current therapies, or evidence-based medicine is established based on fully validated biomarkers and high-evidence drugs with improved effectiveness. However, the therapeutic option based on CGP, called precision oncology, is still experimental, with no established benefit. Nonetheless, it serves as a bridge to increasing the number of therapeutic options for future patients. Encouraging participation in clinical trials for molecular targeted therapies for driver genes and proving their effects is important to increasing the number of future drug therapy options for malignant tumors of the head and neck region. In addition, CGP, which may lead to therapeutic drugs, contributing to clinical usefulness for existing patients. However, our study proved that the candidate drugs judged to be effective in CGP, especially ICI are often ineffective when administered in practice. Thus, doctors should not blindly accept therapeutic recommendations from CGP but make recommendations for optimal therapy for patients after proper verification. This is true precision oncology.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
Mioko Matsuo, MD, Ph.D.: Writing - original draft, conceptualization, methodology, investigation; Kazuki Hashimoto, MD, Ph.D.: Validation; Ryunosuke Kogo, MD, Ph.D.: Resources; Rina Jiromaru, MD, Ph.D.: Data collection; Takahiro Hongo MD, Ph.D.: Data collection; Tomomi Manako, MD: Data collection; Takashi Nakagawa, MD, Ph.D.: Supervision.
Acknowledgements
We would like to thank Editage (www.editage.com) for English language editing.
References
- 1.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O’Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM. The complete sequence of a human genome. Science. 2022;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills GB. An emerging toolkit for targeted cancer therapies. Genome Res. 2012;22(2):177–182. doi: 10.1101/gr.136044.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohno T. Implementation of “clinical sequencing” in cancer genome medicine in Japan. Cancer Sci. 2018;109(3):507–512. doi: 10.1111/cas.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kou T, Kanai M, Yamamoto Y, Kamada M, Nakatsui M, Sakuma T, Mochizuki H, Hiroshima A, Sugiyama A, Nakamura E, Miyake H, Minamiguchi S, Takaori K, Matsumoto S, Haga H, Seno H, Kosugi S, Okuno Y, Muto M. Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. 2017;108(7):1440–1446. doi: 10.1111/cas.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, Shyr Y, Camidge DR, Sequist LV, Glisson BS, Khuri FR, Garon EB, Pao W, Rudin C, Schiller J, Haura EB, Socinski M, Shirai K, Chen H, Giaccone G, Ladanyi M, Kugler K, Minna JD, Bunn PA. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsimberidou AM, Hong DS, Wheler JJ, Falchook GS, Janku F, Naing A, Fu S, Piha-Paul S, Cartwright C, Broaddus RR, Nogueras Gonzalez GM, Hwu P, Kurzrock R. Long-term overall survival and prognostic score predicting survival: the IMPACT study in precision medicine. J Hematol Oncol. 2019;12(1):145. doi: 10.1186/s13045-019-0835-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Cheng DT, Chandramohan R, Mohanty AS, Ptashkin RN, Jayakumaran G, Prasad M, Syed MH, Rema AB, Liu ZY, Nafa K, Borsu L, Sadowska J, Casanova J, Bacares R, Kiecka IJ, Razumova A, Son JB, Stewart L, Baldi T, Mullaney KA, Al-Ahmadie H, Vakiani E, Abeshouse AA, Penson AV, Jonsson P, Camacho N, Chang MT, Won HH, Gross BE, Kundra R, Heins ZJ, Chen HW, Phillips S, Zhang H, Wang J, Ochoa A, Wills J, Eubank M, Thomas SB, Gardos SM, Reales DN, Galle J, Durany R, Cambria R, Abida W, Cercek A, Feldman DR, Gounder MM, Hakimi AA, Harding JJ, Iyer G, Janjigian YY, Jordan EJ, Kelly CM, Lowery MA, Morris LGT, Omuro AM, Raj N, Razavi P, Shoushtari AN, Shukla N, Soumerai TE, Varghese AM, Yaeger R, Coleman J, Bochner B, Riely GJ, Saltz LB, Scher HI, Sabbatini PJ, Robson ME, Klimstra DS, Taylor BS, Baselga J, Schultz N, Hyman DM, Arcila ME, Solit DB, Ladanyi M, Berger MF. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trédan O, Wang Q, Pissaloux D, Cassier P, De La Fouchardière A, Fayette J, Desseigne F, Ray-coquard I, De La Fouchardière C, Frappaz D, Heudel P, Bonneville-levard A, Fléchon A, Sarabi M, Guibert P, Bachelot T, Pérol M, You B, Bonnin N, Collard O, Leyronnas C, Attignon V, Baudet C, Sohier E, Villemin J, Viari A, Boyault S, Lantuejoul S, Paindavoine S, Treillleux I, Rodriguez C, Agrapart V, Corset V, Garin G, Chabaud S, Pérol D, Blay J. Molecular screening program to select molecular-based recommended therapies for metastatic cancer patients: analysis from the ProfiLER trial. Ann Oncol. 2019;30(5):757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 11.Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017;168(4):584–599. doi: 10.1016/j.cell.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D’Amico TA, DeCamp M, Dilling TJ, Dowell J, Gettinger S, Grotz TE, Gubens MA, Hegde A, Lackner RP, Lanuti M, Lin J, Loo BW, Lovly CM, Maldonado F, Massarelli E, Morgensztern D, Ng T, Otterson GA, Pacheco JM, Patel SP, Riely GJ, Riess J, Schild SE, Shapiro TA, Singh AP, Stevenson J, Tam A, Tanvetyanon T, Yanagawa J, Yang SC, Yau E, Gregory k, Hughes M. Non-small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20:497–530. doi: 10.6004/jnccn.2022.0025. [DOI] [PubMed] [Google Scholar]
- 13.Caudell JJ, Gillison ML, Maghami E, Spencer S, Pfister DG, Adkins D, Birkeland AC, Brizel DM, Busse PM, Cmelak AJ, Colevas AD, Eisele DW, Galloway T, Geiger JL, Haddad RI, Hicks WL, Hitchcock YJ, Jimeno A, Leizman D, Mell LK, Mittal BB, Pinto HA, Rocco JW, Rodriguez CP, Savvides PS, Schwartz D, Shah JP, Sher D, St. John M, Weber RS, Weinstein G, Worden F, Yang Bruce J, Yom SS, Zhen W, Burns JL, Darlow SD. NCCN guidelines® insights: Head and neck cancers, Version 1.2022. J Natl Compr Canc Netw. 2022;20(3):224–234. doi: 10.6004/jnccn.2022.0016. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, Zhang N, El-Naggar AK, Jasser SA, Weinstein JN, Treviño L, Drummond JA, Muzny DM, Wu Y, Wood LD, Hruban RH, Westra WH, Koch WM, Califano JA, Gibbs RA, Sidransky D, Vogelstein B, Velculescu VE, Papadopoulos N, Wheeler DA, Kinzler KW, Myers JN. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333(6046):1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortés ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareño C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farah CS. Molecular landscape of head and neck cancer and implications for therapy. Ann Transl Med. 2021;9(10):915. doi: 10.21037/atm-20-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinakis A, Rampias T. TP53 mutational landscape of metastatic head and neck cancer reveals patterns of mutation selection. EBioMedicine. 2020;58:102905. doi: 10.1016/j.ebiom.2020.102905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, Worden F, Saba NF, Iglesias Docampo LC, Haddad R, Rordorf T, Kiyota N, Tahara M, Monga M, Lynch M, Geese WJ, Kopit J, Shaw JW, Gillison ML. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, Psyrri A, Basté N, Neupane P, Bratland Å, Fuereder T, Hughes BGM, Mesía R, Ngamphaiboon N, Rordorf T, Ishak WZW, Hong RL, Mendoza RG, Roy A, Zhang Y, Gumuscu B, Cheng JD, Jin F, Rischin D. KEYNOTE-048 Investigators: Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 21.Morfouace M, Stevovic A, Vinches M, Golfinopoulos V, Jin DX, Holmes O, Erlich R, Fayette J, Croce S, Ray-Coquard I, Girard N, Blay JY. First results of the EORTC-SPECTA/Arcagen study exploring the genomics of rare cancers in collaboration with the European reference network EURACAN. ESMO Open. 2020;5(6):e001075. doi: 10.1136/esmoopen-2020-001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu N, Oshitari T, Yotsukura J, Yokouchi H, Baba T, Yamamoto S. Ten-year epidemiological study of ocular and orbital tumors in Chiba University Hospital. BMC Ophthalmol. 2021;21(1):344. doi: 10.1186/s12886-021-02108-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 24.Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J, Galver L, Kelley R, Karlsson A, Santos R, Overington JP, Hingorani AD, Casas JP. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9(383) doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunami K, Ichikawa H, Kubo T, Kato M, Fujiwara Y, Shimomura A, Koyama T, Kakishima H, Kitami M, Matsushita H, Furukawa E, Narushima D, Nagai M, Taniguchi H, Motoi N, Sekine S, Maeshima A, Mori T, Watanabe R, Yoshida M, Yoshida A, Yoshida H, Satomi K, Sukeda A, Hashimoto T, Shimizu T, Iwasa S, Yonemori K, Kato K, Morizane C, Ogawa C, Tanabe N, Sugano K, Hiraoka N, Tamura K, Yoshida T, Fujiwara Y, Ochiai A, Yamamoto N, Kohno T. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 2019;110(4):1480–1490. doi: 10.1111/cas.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoyagi Y, Kano Y, Tohyama K, Matsudera S, Kumaki Y, Takahashi K, Mitsumura T, Harada Y, Sato A, Nakamura H, Sueoka E, Aragane N, Kimura K, Onishi I, Takemoto A, Akahoshi K, Ono H, Ishikawa T, Tokunaga M, Nakagawa T, Oshima N, Nakamura R, Takagi M, Asakage T, Uetake H, Tanabe M, Miyake S, Kinugasa Y, Ikeda S. Clinical utility of comprehensive genomic profiling in Japan: Result of PROFILE-F study. PLoS One. 2022;17(3):e0266112. doi: 10.1371/journal.pone.0266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurzrock R, Gurski LA, Carlson RW, Ettinger DS, Horwitz SM, Kumar SK, Million L, von Mehren M, Benson AB 3rd. Level of evidence used in recommendations by the National Comprehensive Cancer Network (NCCN) guidelines beyond Food and Drug Administration approvals. Ann Oncol. 2019;30(10):1647–1652. doi: 10.1093/annonc/mdz232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon B, Young RJ, Rischin D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol. 2018;52:228–240. doi: 10.1016/j.semcancer.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 29.Alsahafi E, Begg K, Amelio I, Raulf N, Lucarelli P, Sauter T, Tavassoli M. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death Dis. 2019;10(8):540. doi: 10.1038/s41419-019-1769-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antra, Parashar P, Hungyo H, Jain A, Ahmad S, Tandon V. Unraveling molecular mechanisms of head and neck cancer. Crit Rev Oncol Hematol. 2022;178:103778. doi: 10.1016/j.critrevonc.2022.103778. [DOI] [PubMed] [Google Scholar]
- 31.Zhou G, Liu Z, Myers JN. TP53 Mutations in head and neck squamous cell carcinoma and their impact on disease progression and treatment response. J Cell Biochem. 2016;117(12):2682–2692. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dubot C, Bernard V, Sablin M, Vacher S, Chemlali W, Schnitzler A, Pierron G, Ait Rais K, Bessoltane N, Jeannot E, Klijanienko J, Mariani O, Jouffroy T, Calugaru V, Hoffmann C, Lesnik M, Badois N, Berger F, Le Tourneau C, Kamal M, Bieche I. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur J Cancer. 2018;91:47–55. doi: 10.1016/j.ejca.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, Li X, Babur O, Hsu TK, Lichtarge O, Weinstein JN, Akbani R, Wheeler DA. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 2019;28(5):1370–1384.e5. doi: 10.1016/j.celrep.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, Li M, Wang X. Cancer type-dependent correlations between TP53 mutations and antitumor immunity. DNA Repair (Amst) 2020;88:102785. doi: 10.1016/j.dnarep.2020.102785. [DOI] [PubMed] [Google Scholar]
- 35.Filipovic A, Miller G, Bolen J. Progress toward identifying exact proxies for predicting response to immunotherapies. Front Cell Dev Biol. 2020;8:155. doi: 10.3389/fcell.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, Kaley TJ, Kendall SM, Motzer RJ, Hakimi AA, Voss MH, Russo P, Rosenberg J, Iyer G, Bochner BH, Bajorin DF, Al-Ahmadie HA, Chaft JE, Rudin CM, Riely GJ, Baxi S, Ho AL, Wong RJ, Pfister DG, Wolchok JD, Barker CA, Gutin PH, Brennan CW, Tabar V, Mellinghoff IK, DeAngelis LM, Ariyan CE, Lee N, Tap WD, Gounder MM, D’Angelo SP, Saltz L, Stadler ZK, Scher HI, Baselga J, Razavi P, Klebanoff CA, Yaeger R, Segal NH, Ku GY, DeMatteo RP, Ladanyi M, Rizvi NA, Berger MF, Riaz N, Solit DB, Chan TA, Morris LGT. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]