Abstract
Aptamers are chemical antibodies possessing the capability of overcoming the limitations posed by conventional antibodies, particularly for diagnostic, therapeutic, and theranostic applications in cancer. The ease of chemical modifications or functionalization, including conjugations with nucleic acids, drug molecules, and nanoparticles, has made these aptamers to gain priorities in research. In this Mini-review, various reports on therapeutics with aptamer-functionalized nanomaterials for controlled or multistep drug release, targeted delivery, stimuli-responsive drug release, etc. are discussed. In the case of nucleic-acid-conjugated aptamers, DNA nanotrains and DNA beacons are discussed in terms of the possibility of multidrug loading for chemotherapy and gene therapy. Developments with electrochemical aptasensors and signal-enhanced immune aptasensors are also discussed. Further, the future scope of aptamer technology in cancer theranostics and the prevailing limitations are discussed.
Introduction
The emergence of new diseases and syndromes demands the development of highly efficient diagnostic and therapeutic strategies.1 Cancer, a heterogeneous and multifaceted disease that leads to anomalistic cell growth and proliferation by altering genomic and molecular characteristics, is one among them that has received much attention from researchers in recent years. The challenges posed by the rapidly increasing tumor-induced mortality rate from cancer are significantly due to inefficient diagnostic and therapeutic management.2 Currently available traditional methods include chemotherapy, radiotherapy, and surgery, but all these methods display limitations for practical clinical applications. In chemotherapy, therapeutic quantities of drugs are distributed throughout the body intravenously. Though this method is effective for treating cancer cells, it lacks selectivity, causing non-specific damage to normal cells. Moreover, reports have revealed that patients can develop resistance to these chemotherapeutic drugs, which is again considered as a major limitation of this method.
The most widely used and prescribed anticancer drug, cisplatin, is being replaced with numerous other platinum metal complexes in order to overcome the limitations posed by the former. The major limitations include (i) the rapid development of resistance against cisplatin and (ii) adverse side effects (nausea, nephrotoxicity, cardiotoxicity, hepatotoxicity, and neurotoxicity) when used at efficient dosage concentrations that might result in events like arrhythmias, congestive heart failure, electrocardiographic changes, and myocarditis. Further, the toxicity of cisplatin is altered by the generation of non-enzymatic molecules and antioxidant enzymes, reduction of glutathione, etc. In order to overcome the above limitations, combination therapies are employed to inhibit the growth of tumors and metastasis and are known to potentially decrease the systemic toxicity. Several synthetic and therapeutic strategies using platinum-based anticancer complexes in combination with gene editing technology, photoactivation, and therapies that include reactive oxygen species (ROS)-based, thermal, immune, etc. have been developed recently. Investigations have revealed the mode of inhibition to be induced via various processes that lead to apoptosis, DNA damage, and cell cycle arrest.3a In general, the cisplatin anticancer drugs act through the formation of intra-strand and inter-strand cross-links with DNA.3b,3c However, literature evidence has shown that a small percentage of the intra-cellular drug enters the nucleus of rapidly proliferating cancer cells and reacts with nuclear DNA to result in cell cycle arrest and apoptosis. Cisplatin acts by targeting enzymes involved in cell proliferation and growth, autophagy, etc. Evidence reveals that promotion of cell cycle arrest by cisplatin is through enzyme-mediated p53/p21 pathways.4a,4b Several other mechanisms are adopted by cisplatin drugs, including induced stress in the estrogen receptor, disruption of nucleic acid transcription, inhibition of oncogenic enzymes, acidification of cytoplasm, etc.4c−4e The further potential of these drugs as photodynamic therapeutics has also been proven.4f Many computational methods were also successfully employed to investigate the potential of numerous heterocyclic compounds as anticancer agents.4g,4h
On the other hand, radiotherapy often produces adverse reactions like radiation-induced osteonecrosis and pneumonia. Therefore, it has become a necessity to explore more efficient methods for diagnosis, early detection, and treatment of cancers to reduce the increasing mortality rate. With the available diagnostics, the detection of cancer cells at early stages still remains a challenge. Numerous strategies are being reported and proven to be better than the existing ones. The advances in aptamers have opened avenues toward cancer therapeutics. Recently, the use of aptamers in cancer therapy has been successfully proven to have better efficiencies than the existing counterparts.
Aptamers are short single-stranded DNA/RNA oligonucleotides (A, G, C, and T/U) possessing unique secondary and tertiary structures with high specificity for a selected target. Aptamers are also known as chemical antibodies. They have proven better efficiencies as compared to antibodies, as the application of antibodies is complicated by their macromolecular size, stability, high immunogenicity, etc.5 The unique binding properties of these aptamer drugs place them above the existing molecular drugs. With the evolution of aptamers and possible conjugates, the process of selective and targeted delivery of drugs has been made easy, with very high efficiencies. Moreover, the recent developments with SELEX (Systematic Evolution of Ligands through Exponential enrichment) procedures, from a library of 1013–1016 ssRNA or ssDNA molecules, have made it possible to screen and filter out the aptamer probes with very high specificity to bind the target. Once aptamers are introduced into a system for targeted drug delivery, their specificities are enhanced upon entering the target environment. This further increases their sensitivity toward several key components or biomarkers that are available in very limited quantities.6a Their behaviour is very similar to monoclonal antibodies with enhanced affinity and specificity. The small size of aptamers promotes their easy access into the system. The high stability of these probes favors their use in cell-free assembly, lower preparation cost, and quick and wide-scale manufacturing applications.
One of the most important problems in cancer therapeutics is enzymatic degradation, which can be overcome with chemically modified aptamers.6b With the current advancements, it is possible to ensure the aptamers’ chemical integrity and bioavailability when utilized under physiological conditions. The therapeutic efficiencies of aptamers are enhanced when they are chemically modified to hold a hydrophobic group.7a−7c
Cancer is considered as one of the hardest diseases to treat for various reasons, including metastasis, induced drug resistance via overexpression of drug efflux pumps, etc. The available targeted drug therapies display typical side effects induced by non-targeted dose-limiting cytotoxicity. Although antibody–drug conjugates (ADCs) have been extensively utilized for targeted cancer therapy, aptamer–drug conjugates (ApDCs) display several advantages. These ApDCs are relatively more stable/inert with no cytotoxic effect under physiological conditions.7d The release of drugs is initiated under diseased conditions wherein many enzymes and biomarkers promote the dissociation of aptamer-bound drugs via simple enzymatic or chemical reactions.5b In most of the cases, the released drugs get intercalated with cellular DNA, while care is taken to design linkers to attach cytotoxic payloads with aptamers following the familiar “click” chemistry.
Delivering suboptimal dosages of drugs and/or off-target delivery can induce inefficient therapeutic efficacy and drug resistance.8a Targeted drug delivery addresses the above limitations by reducing systemic toxicity and improving therapeutic efficiency. Numerous approaches for targeted drug delivery are known to date, and most of them employ antibodies as drug transport vehicles.8b
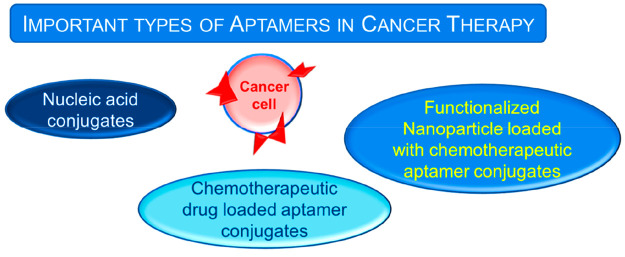
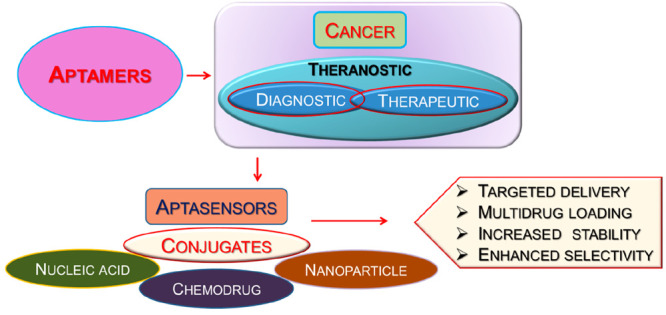
The high affinity and target specificity posed by aptamers display their great potential for therapeutic applications as drug delivery agents. These cost-effective, chemically synthesized aptamers can be easily hybridized to numerous other reagents like nucleic acids, functionalized nanoparticles, chemical agents, etc. that make them potent agents for therapeutic and diagnostic applications.9 The major types of aptamer conjugates that are available specifically for cancer therapies are shown in Figure 1.
Figure 1.
Different types of aptamer conjugates for anticancer applications.
Herein, the pioneering and more recent approaches to the development of aptamers for treating various types of cancers are reviewed. The major emphasis will be given to the aptamers that boost anticancer immunity and induce the expression of immunogenic antigens. The designs of aptamers that can overcome the existing limitations in cancer therapeutics are discussed. The aptamers that target and penetrate tumors and metastasis are also discussed. Herein, the role of aptamers in cancer theranostic along with various design approaches for targeted delivery will be discussed along with their recent developments. Further the future perspectives of aptamer-based tumor therapeutic and diagnostic approaches will also be discussed.
Chemical Modifications: Aptamer–Drug Conjugation
As aptamers are more susceptible to nuclease degradation and renal filtration, their stabilization via chemical modifications is very much necessary. These modifications focus on improving their interaction capabilities to widen their target spectrum and hence to expand their diagnostic and therapeutic applications. In particular with aptamer oligonucleotides, these chemical modifications can enhance their resistance to nuclease degradation and lower their renal filtration apart from increasing their binding affinity.10a,10b The most commonly employed chemical modification methods include click reaction, oxidation-induced coupling reaction, and avidin–biotin and thio–gold reactions.10c,10d The non-covalent coupling of drugs with aptamers is largely limited by its complicated and less efficient synthetic procedures, poor yields, low payloads, low spatiotemporal controllability, etc.
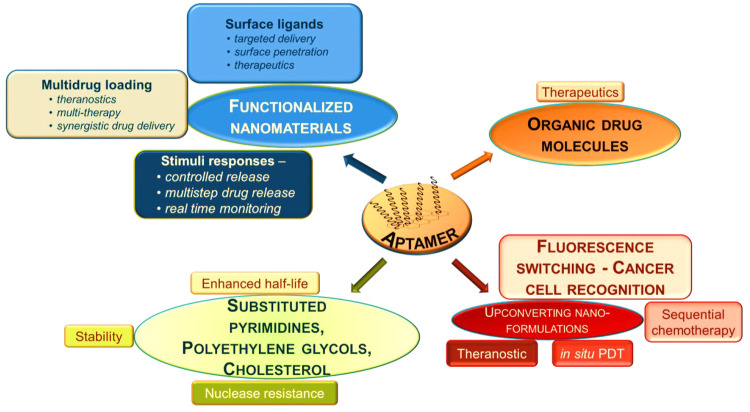
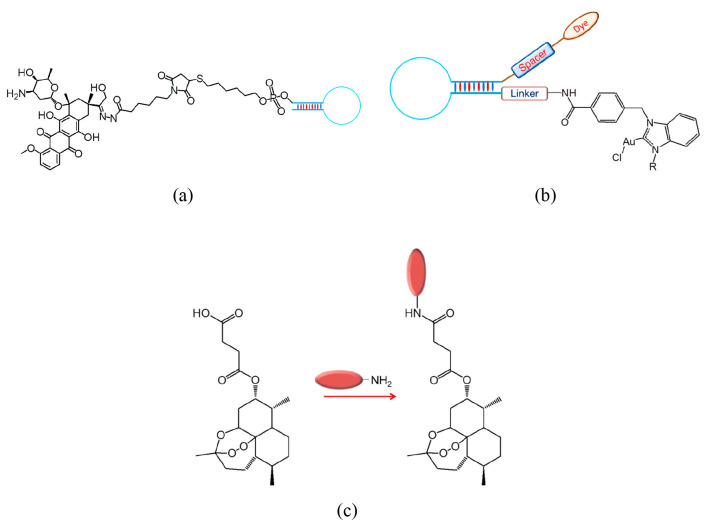
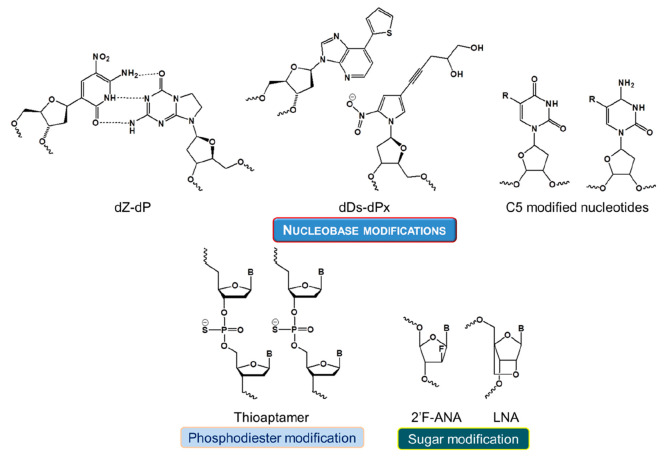
In general, aptamers have short life-spans in comparison to their modified counterparts.11 As they are processed by the kidneys and excreted from the body or degraded by nucleases, their physiological applications are very limited. Therefore, biochemical modifications of these aptamers are being attempted to ameliorate their pharmacodynamic and pharmacokinetic properties. These modifications include polymerization, truncation of sequences, functional optimization, etc., which are known to improve the binding efficiency and stability.12a−12c In particular, a variety of conjugation techniques like coupling methods and modification designs are available.12d−12f The potency and therapeutic impact of any aptamer-based drug delivery system greatly rely on a number of essential functional traits, including targeted accumulation, drug deployment, stability throughout its internal circulation, and drug loading capacity.12g These systems are designed to end with minimal or no residual components other than the target molecule. The various parameters that guide the aptamer–target interactions, such as van der Waals and hydrogen-bonding interactions, shape complementarity, hydrophobicity, etc., should dominate over other non-specific interactions with the surrounding molecules. Further, aptamer conjugations are classified into two types: non-covalent and covalent conjugation. The linker determines the effects of covalently conjugated aptamers, whereas the drug load embeds in between specific sequences of the non-covalent aptamer chains. Combinations of both were also reported to increase the therapeutic effects. The various types of possible chemical modifications in aptamers along with their functional roles in treating cancer are depicted in Figure 2.5,9,12h
Figure 2.
Different types of chemical modifications of aptamers and their roles in cancer therapies.
Covalent Conjugation
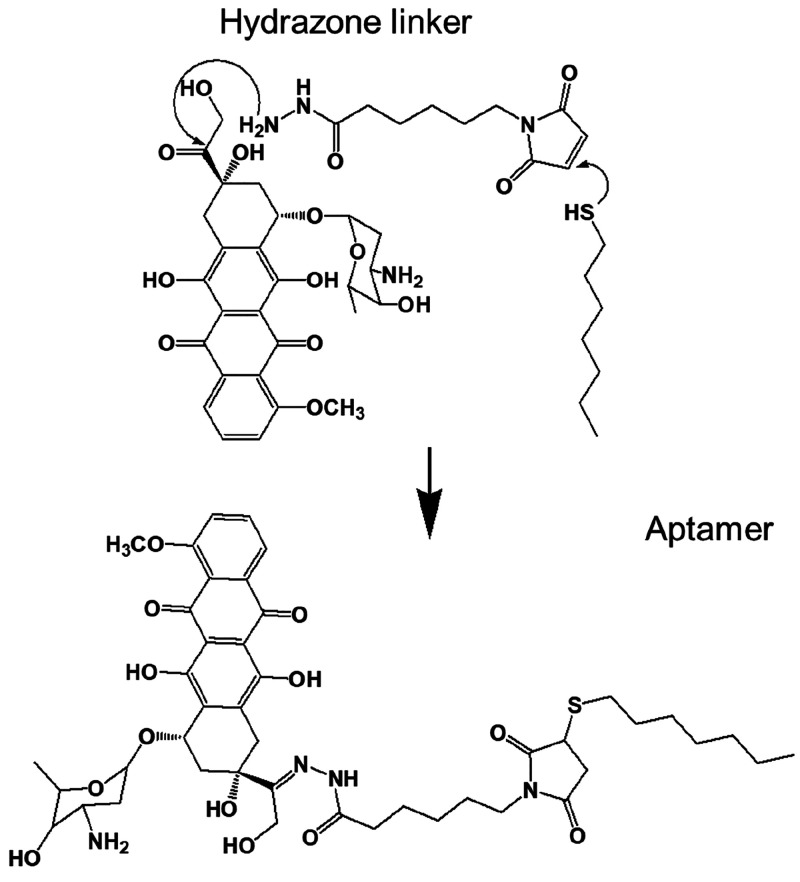
The first covalent conjugation of a chemotherapeutic drug (Figure 3) with an aptamer that specifically targets protein tyrosine kinase (PTK7) on the cellular membrane in lymphoblastic leukemia cells was successfully demonstrated by Huang et al.13a In this case, a hydrazone linker was used to conjugate to doxorubicin (DOX). The DOX C-13 hydrazone derivatives are known to display better cytotoxic effects than their unconjugated counterparts to release the drug at a pH of ∼5.0. Moreover, the fluorescence property of the above drug conjugate can aid in real-time monitoring of drug uptake by the target cancer cells. But the application of this conjugate was limited by the poor payload of drug conjugated to each aptamer.
Figure 3.
Synthetic scheme for aptamer–DOX conjugates that can bind to target cancer cells with high specificity and affinity.
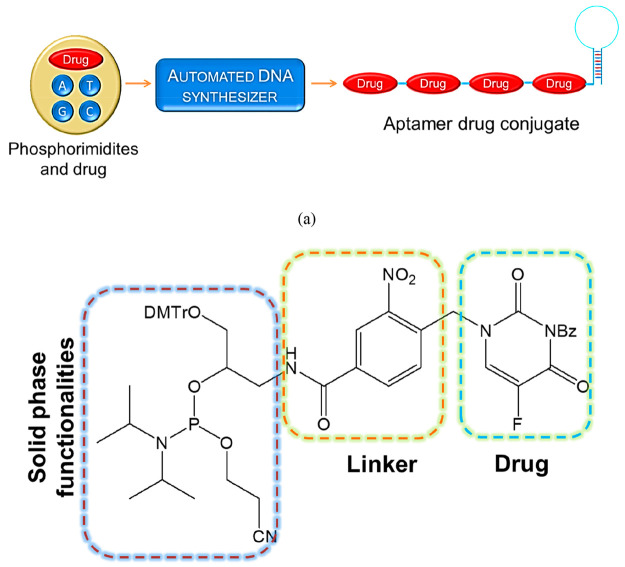
Wang et al. reported a new methodology for the preparation of automated and modular next-generation ApDCs employing solid-phase synthesis technology for targeted drug delivery applications.13b As shown in Figure 4, one end of the aptamers can attach to the drug molecular train. In order to release the drug into the therapeutic module, the spatiotemporal controllability was maintained through a photocleavable chemical linkage between the drugs and vehicles. Using this method, it is possible to conjugate multiple drug moieties onto a single aptamer with high drug-loading capacity. Moreover, this automated conjugation method is simple, with very high efficiency.
Figure 4.
(a) Schematic representation of ApDCs from phosphorimidites A, T, G, and C and drug moieties. (b) Structural features of a phosphorimidite drug.
The same aptamer was further modified to increase the payload by holding five copies of another well-known anticancer drug, 5-fluorouracil. Later modifications of the above included a photocleavable linker between the aptamer and the drug.13b Further modifications by Zhu et al. involved the construction of ApDCs with multiple drug copies that can inhibit cancer progression at different levels. Other representatives of covalently conjugated ApDCs are depicted in Figure 5.
Figure 5.
Representative examples of covalently coupled aptamer–drug conjugates: (a) DOX, (b) NHC–Au(I) complex, and (c) Artesunate.
Covalent coupling of aptamers with drug molecules is made possible by substituting reactive functional groups like dibenzocyclooctyne (DBCO), thiols, amines, etc.12 Recently, a cyclic bivalent ApDC (cb-ApDC) was reported by Zhou et al. that can selectively sense target cells and effectively localize itself between the cells. In this ApDC, release of the conjugated drug is initiated through cleavage mediated by an esterase.14a Moreover, these cb-ApDCs are reported to possess higher stability than single ApDCs. Also, with cb-ApDCs, it is possible to manage the medication ratio accurately through a simple, straightforward chemical reaction, which further paves way for the development of combinatorial cancer therapy and combinations for various medication quantities. Similarly, Yang et al. reported another ApDC aptamer–mitomycin C conjugate wherein an enhanced cytotoxicity was revealed to be induced by the linker.14b But this ApDC displayed limitations with internalization, specificity, and target recognition. In 2021, an aptamer–artesunate conjugate was designed by Li et al. where the aptamer enhanced the targeting activity by piling artesunate at the target site.14c Experimental evidence revealed that it possessed a specific target binding capacity with a longer localization time around the tumor site as compared with the control group. Besides, a nifty water-soluble aptamer, namely AS1411–paclitaxel conjugate (AS1411-PTX), was reported by Li et al. that can specifically target tumor location to deliver PTX.14d The 2-hydroxyl position of the drug was bound with the aptamer AS1411 via a dipeptide linker with cathepsin B. Upon recognizing the tumor site, the linker gets cleaved instantaneously, releasing the anticancer drug PTX. In the above, the reported drug combination was to selectively enhance the antitumor activity against ovarian tumor tissues. A modular aptamer–drug coupling model was proposed by Pusuluri et al.15 It is also evident that tumor-targeting aptamers along with selective drug combination in a synergistic ratio would result in efficient low-dose treatments. A precise combination of DOX and camptothecin (CPT) was coupled through a peptide scaffold that further linked to an aptamer to generate an aptamer–cooperative drug conjugate. Using the triple-negative breast cancer (TNBC) cell line MDA-MB-231 as a model, the probe revealed a relatively low IC50 (31.9 nmol/L). Moreover, recent reports have revealed that the specificity and selectivity of the aptamers conjugates of small molecules display negligible effects. Therefore, apart from the conventional methods, novel chemical strategies need to be developed for effective theranostic.
Non-covalent Conjugation
Non-covalent drug–aptamer conjugation happens through physical interactions like intercalative binding. The initial non-covalent ApDCs could not meet the required drug-carrying capacity for in vivo applications.16 In spite of several advantages, these non-covalently conjugated aptamer–drugs were highly unstable due to their reversible nature. A few examples of non-covalent modifications of aptamers are shown in Figure 6. The well-established anticancer drug DOX is known to be non-covalently conjugated to CG/GC-rich oligonucleotide sequences via intercalation. Recently, a 2-fluoro-modified RNA aptamer that was non-covalently conjugated to DOX was utilized to treat retinoblastoma with very high specificity and greatly enhanced therapeutic efficacy. Similarly, in an investigation of gene-mediated therapeutics, a 2-fluoro-modified RNA aptamer that was non-covalently conjugated to siRNA revealed an excellent inhibitory effect on an overexpressed B-cell activating factor receptor protein (BAFF-R) in B-cell malignancies. Further modification of the above was carried out with a multivalent aptamer, wherein the non-covalent conjugation was through a biotin–streptavidin linker.
Figure 6.
Chemical structures of non-covalent modifications of aptamers.
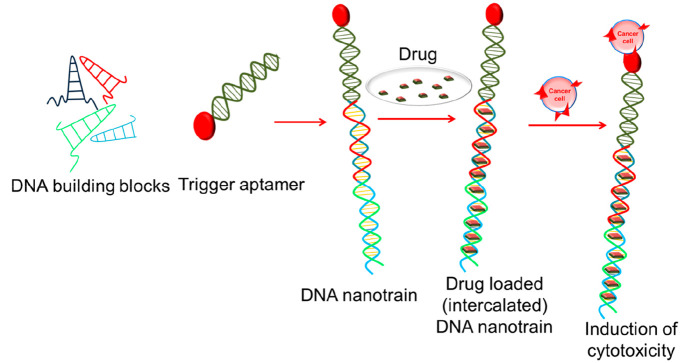
Further, high drug loading capacities were made possible with the development of DNA nanotrains by Tan et al.16 (Figure 7). These DNA nanostructures possessed the ability to serve as drug carriers for targeted delivery. In addition to increased payload capacity, the presence of multivalent aptamers increased the specificity to a large extent in DNA nanocentipedes.
Figure 7.
Self-assembly of nucleic acid building blocks into aptamer nanotrains for targeted therapeutics and theranostics.
In line with the above discussion, numerous reports are available in the literature. A diverse myeloma-cell-targeting ApDC with DOX drug was developed following hydrophobic interactions by Wen et al.17 The designed ApDC was then localized into the lysosome while exactly binding to the CD38 protein. Consequently, the drug molecule was released in the tumor cells with an acidic environment to further prohibit their growth. This method was reported with zero side effects. Further enhancement was made by Zeng et al. with 15 DOX molecules with trifurcated Newkome-type monomer structure (TNM-DOX) containing pH-dependent hydrazone bond. But, this approach was reported with propensity for drug leakage and induced toxic side effects. A more efficient system for the purpose is yet under investigations.
Aptamer–Drug Conjugates in Cancer Theranostics
Nucleic Acid Drugs
Nucleic acids are considered as versatile materials for the construction of nanodevices that allow a variety of functionalization for imaging applications, induced drug release, biovehicles, targeting entities, etc. The possibility of functional nucleic acids to alter and control gene expression in cells makes them as potential drug candidates.18 In order for them to exhibit their drug activity, nucleic acids need to be transported to target cells in an efficient manner. But the negative charges carried by the nucleic acids prevent their entry into the cell membrane or crossing the lipid bilayers. Therefore, to enhance their targeted delivery, aptamer conjugates were successfully employed. Recently many therapeutic oligonucleotides like siRNA and miRNA have proven their efficient roles in cancer therapy and other gene-related diseases that display undruggable targets for small-molecule drugs. These siRNA and miRNA drugs have shown excellent potential when used along with aptamer vehicles.19a Recently, a photocleavable aptamer–siRNA conjugate that can induce gene silencing upon irradiation was also reported.19b,19c
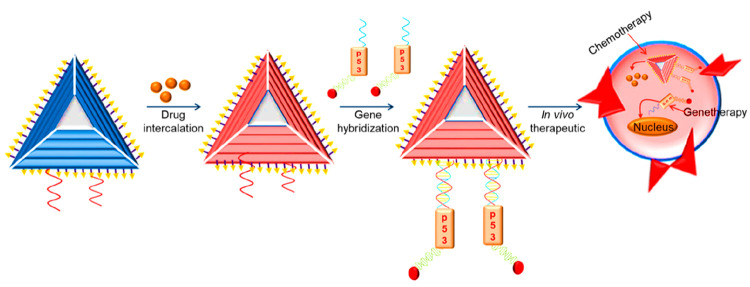
Similarly, Esposito et al. reported another tyrosine kinase binding aptamer that effectively inhibits transcription of STAT3 in glioblastoma cells.20a The same group also reported an aptamer–miRNA conjugate that could inhibit the proliferation of glioblastoma stem cells. As the overexpression of miRNAs indicates diseased conditions, they are considered as one of the important biomarkers in diagnostics, particularly in the case of various cancers. Garrido et al. reported an aptamer–nucleic acid conjugate that could effectively inhibit the growth of various tumors without displaying toxicity.20b Several DNA nanodevices were reported for the detection of important cancer biomarkers, employing gold nanoparticles, DNA beacons, etc. to understand tumor metastasis. A therapeutic multifunctional nanodevice (Figure 8) for the co-delivery of anticancer drug and multiple antisense oligonucleotides was developed for the targeted delivery of drug and nucleic acid sequences to combat the induced drug resistance. Such nanoassemblies display numerous advantages, including targeted delivery, increased drug loading, antisense oligonucleotides, etc.
Figure 8.
Multifunctional DNA nanostructure for combined gene therapy (p53 gene as tumor suppressor) and chemotherapy (anticancer drug).
Aptasensors
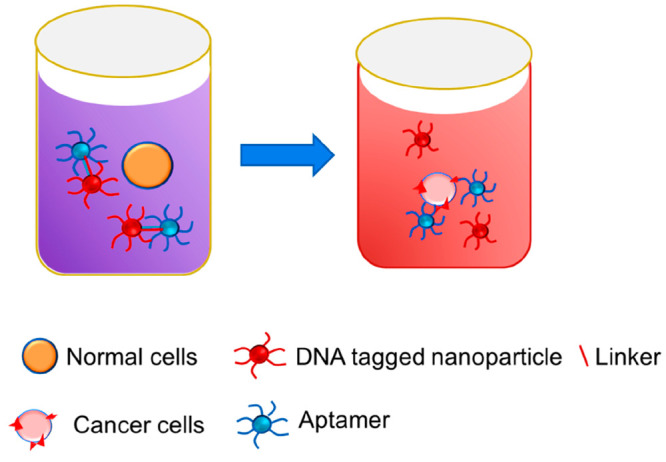
There are different types of aptasensors based on the mode of detection involved being electrochemical, colorimetric, fluorescence-based, etc.21a,21b In an electrochemical aptasensor, the response from the aptamer-immobilized electrodes is monitored. Based on the detection of response signals upon recognition of the target, they are further classified as cyclic voltammetry, impedance, and ampere analysis method-based sensors.21c,21d In the initial electrochemical aptasensors, the immobilization was done through electrostatic or affinity-oriented adsorption or covalent linking. Recently, a sensor was developed by electrodepositing gold–platinum nanoparticles on the electrode surface for signal amplification.22a Further activation was done with NHS-EDC to link streptomycin for depositing biotin-labeled aptamers.22b The response signals were monitored through differential pulse voltammetry (Figure 9).
Figure 9.
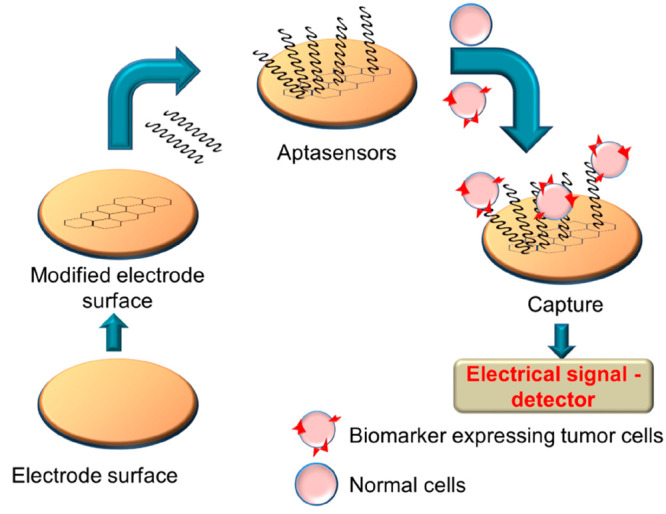
Mechanism of sensing of cancer cells via an electrochemical aptasensor.
Further attempts were made to increase the sensitivity of the electroaptasensors through sandwich aptasensors based on the principle of immunosensors: aptamer–aptamer, antibody–aptamer, and aptamer–antibody sandwich layers.23a,23b These kinds of aptasensors involve two steps: recognition of targets followed by the amplification of signals (Figure 10).
Figure 10.
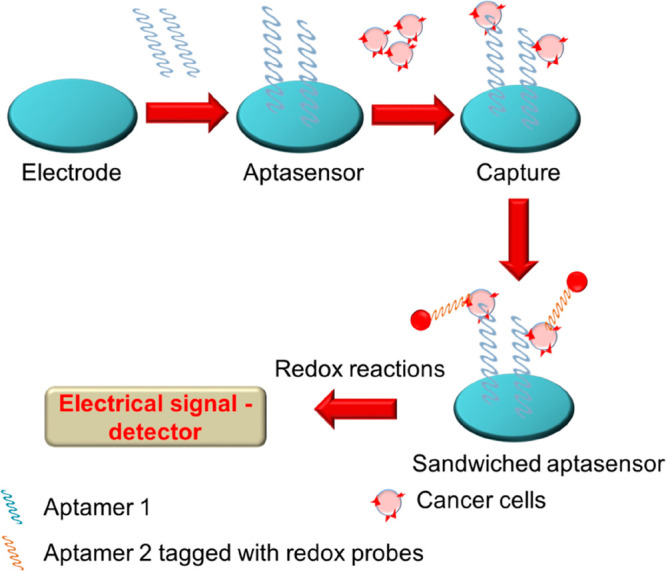
Mechanism of sensing of cancer cells via an electrochemical-based sandwich aptasensor.
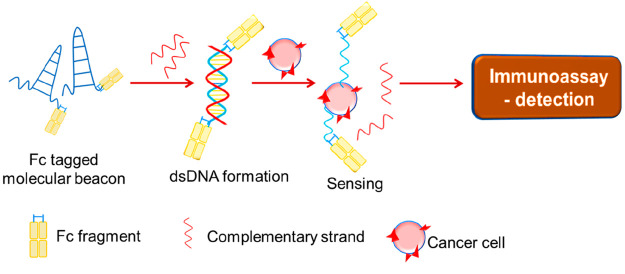
In the former two types of layers, the immobilization of aptamers was reported to hinder the target recognition in many cases. In order to overcome this limitation, an immobilization-free technique (Figure 11) was developed wherein a double-stranded DNA was formed using molecular beacons or antibody fragment Fc-labeled aptamers, which can further get adsorbed over modified electrodes. Using this method, many limitations posed by the earlier methods were overcome.24
Figure 11.
Immobilization-free Fc-tagged aptasensor.
The changes in the optical properties of gold nanoparticles upon aggregation and dispersion were utilized in the recognition of cancer biomarkers. By appropriately labeling the gold nanoparticles with ssDNA followed by a complementary aptamer probe that can link the adjacent gold nanoparticles, the aptasensor was developed. Upon recognizing the high-affinity target, this sensor modulates the optical properties of interlinked nanoparticles, as shown in Figure 12.25
Figure 12.
Gold-nanoparticle-tagged colorimetric aptasensor.
Fluorescence-based aptasensors are considered as better alternatives for colorimetric sensors because of their enhanced sensitivity and rapid detection capability. These systems, in combination with a variety of nanomaterials and other antitumor drugs, have been developed as an efficient approach. Generally fluorescence turn-on and -off approaches following FRET are employed. As depicted in Figure 13, a fluorophore is attached to a flexible aptasensor to which a quencher is linked appropriately to modulate the FRET activities. Upon sensing specific tumor biomarkers, conformation changes are induced that can further influence the fluorescence signals. Several such fluorescence aptasensors were successfully combined with photodynamic therapy, photothermal therapy, and chemotherapy techniques to diagnose and treat cancers.26
Figure 13.
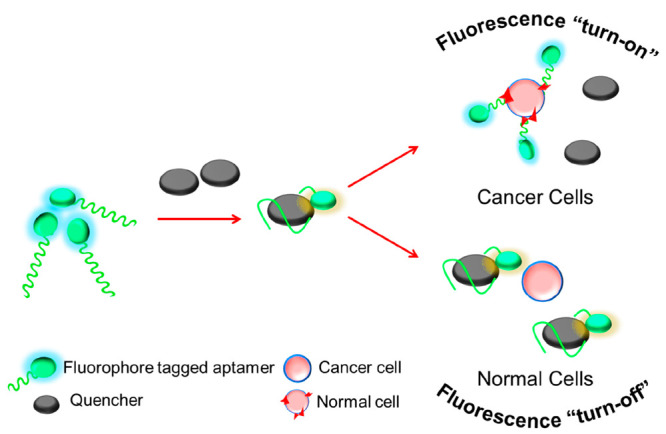
FRET-based fluorescence “turn-on” and “turn-off” aptasensor.
Functionalized Nanoparticles: Gold, Polymers, Liposomal, and Iron Oxide
Following the earlier discussions, targeting immune checkpoint blockades (ICBs) is considered as an important strategy for cancer treatment. Further strategies utilizing functionalized nanoparticles along with aptamers have been developed as novel approaches with enhanced therapeutic efficacies. There are many approaches to enhance the immunotherapy responses. The H1-antihistamine fexofenadine (FEXO) is known to induce an immunotherapy response via suppression of M2-like macrophages’ expression. This drug was very effective when loaded on self-assembled albumin nanoparticles that were further attached to PD-L1 aptamers (Figure 14). This strategy was successfully proven to display enhanced efficacy against ICBs.27
Figure 14.
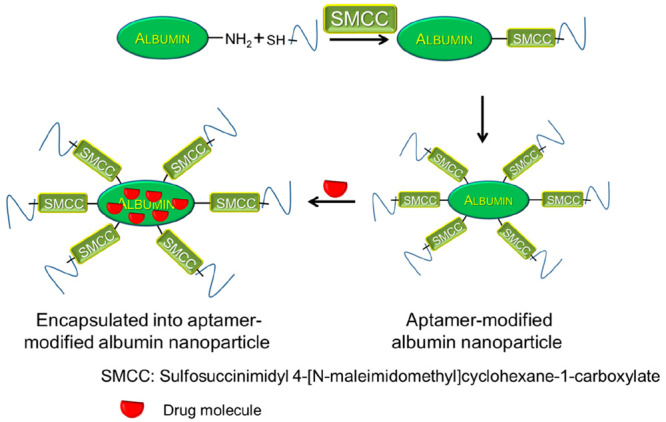
Schematic illustration of an aptamer-modified albumin nanoparticle.
Before the emergence of nanomedicines, several challenges for disease treatment and drug delivery were observed. More than 50 FDA- and EMA-approved nanodrugs were reported and successfully proven for clinical trials in 1995, with most of them being utilized for cancer therapy. This opened avenues in the field of nanomedicine with excellent translational values. Recently, an albumin-based nanoparticle was reported for PD-L1 aptamer delivery and the antihistamine drug FEXO.28 As revealed from dynamic light scattering (DLS) evaluations, the average size of the PD-L1 aptamer-modified nanoparticle without FEXO (PDL1-NP) was 135.5 nm, while that of PDL1-NP-FEXO was 154.6 nm. Moreover, like free PD-L1 aptamers, the PDL1-NP could effectively bind to tumor cells (MDA-MB-231) that express PD-L1. In vivo studies with mouse models have revealed that PDL1-NP could significantly boost the tumor inhibition as compared to free PD-L1 aptamer without enhancing the systemic toxicity. This method has proven to be a successful strategy to improve the efficacy of ICBs and may find potential application in cancer immunotherapy.
The emergence of reprogrammed immune cells has attracted the attention of researchers toward cancer immunotherapy. These genetically programmed cells display high expression of highly specific tumor-recognizing antibodies, enabling them with better cancer cell recognition and therapeutic efficiency. The application of this method is currently limited by the highly active killing nature of the immune cells, leading to continuous cytokine secretion. In order to enhance the cell recognition ability and controlled spatiotemporal activation of the killing effect by the immune cells, recently macrophages engineered with gold-based polyvalent spherical aptamers were developed.29a,29b In these, the cell membranes of the natural macrophages were modified with the above-mentioned aptamers that were further irradiated with X-rays to generate ROS.
Immunotherapy
The immune system is known to play significant roles toward inhibiting tumor development and progression. This paved way for researchers to open up avenues in cancer immunotherapies. In general, cancer immunotherapy boosts the immune system to specifically target only the cancer cells to avoid damage caused to the normal cells. More specifically, the binding of programmed death ligand 1 (PDL1) to the programmed cell death protein 1 (PD1) localized on the T-lymphocytes is blocked by an ICB (Figure 15). In order to widen the therapeutic scope, recently an alternate approach involving multiple markers was identified through Boolean logic. This led to the development of several logic-based methods to identify multiple biomarkers.30 Further, by coupling the above logic-based methods with photosensitizers or radioisotope labels on aptamers, such logic computing reactions could be extended precisely for photodynamic therapy.
Figure 15.
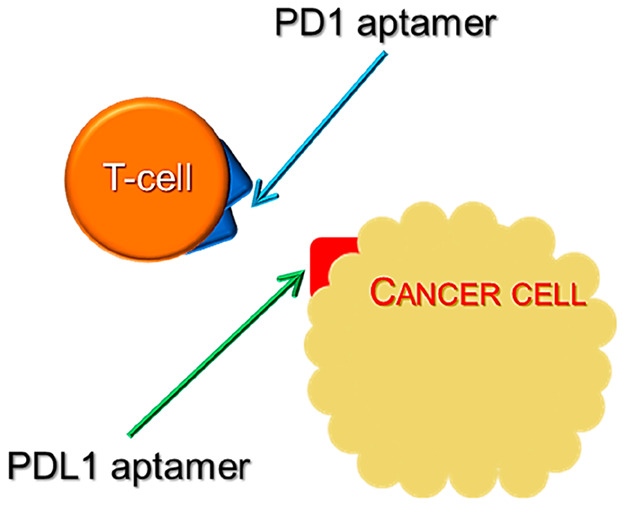
Schematic representing the inhibition of immune checkpoints through aptamers for PD1 and PDL1.
The major challenge faced with cancer immunotherapies—particularly with those which are highly immunogenic, like TNBC—is that most of the tumors are capable of evading the immune system. Therefore, it is necessary to develop strategies that aim to probe the tumor microenvironment and further promote antitumor immunity. Two such immunotherapies are available for tumors that express PDL1. But the challenges to predict their efficacy still exist due to the heterogeneity displayed in clinical trials.31a Further developments toward improving the stability and specificity of this strategy were achieved with aptamers. Recently, tremendous advancements have been made toward developing immunotherapeutic targeting strategies that aim to block specific molecules that can evade the immune system. In recent years, numerous aptamer-based strategies have been investigated to induce immune system responses against various tumors.31b
In the treatment of cancer, blocking the immune checkpoint is the most commonly adopted technique that could replace the conventional use of immune checkpoint inhibitors that display adverse side effects. Along this line, a strategy for photoimmunotherapy for cancer with an aptamer-based spherical nucleic acid (SNA), along with an anticancer drug encapsulated in a metal–organic framework nanoparticle core, was reported by Zhang et al.32a that can initialize concomitant photodynamic therapy, chemotherapy, and immunotherapy to inhibit tumors in mouse models.
In ICB therapy, the inhibitors for PDL1 and its receptor PD1 function by releasing the brake on T-cell antitumor activity. Sun et al. developed an aptamer labeled with DBCO that can bring about covalent conjugation with PD1 and PDL1 upon cell recognition, which can further induce potent immunological antitumor effects.32b
Platelet-derived growth factor (PDGF) is another target in immunotherapy that is found to be overexpressed in malignant cells. Recently, a DNA aptamer that can inhibit the binding of PDGF to its receptor was reported. Later, several modifications of this aptamer were carried out to enhance the binding affinity. Following this, a RNA aptamer was reported that can specifically bind to the ectodomain of human PDGF receptor-β and inhibit the activation of receptor and other downstream signaling processes in human glioblastoma cells. In addition to this, the above RNA aptamer was able to induce the differentiation of tumor cells, inhibit cell migration and proliferation, and impede cancer progression. Further, in the case of TNBC, it is reported that the aggressiveness of bone-marrow-derived mesenchymal stem cells (BM-MSCs) could be significantly enhanced. Therefore, treating BM-MSCs with the above RNA aptamer could inhibit the receptor-dependent signaling pathways further to block the uptake of BM-MSCs by TNBC cells.
Recently, Li et al. reported a novel method for the quantification and imaging of exosomal PDL1 that follows a cascade of primer exchange reaction-induced DNA nanostructures.32b This assay is recognized as a diagnostic toolkit to detect cancer and monitor immunotherapy.
Conclusions
Aptamers are widely used to treat numerous diseases. In addition to their application in therapeutics, they are also employed for drug delivery via covalent or non-covalent attachments of drugs. With the display of exceptional binding affinity and specificity, these aptamers are more widely utilized in diagnosis, imaging, and apta-sensing. Development of aptamer drugs is one of the most interesting investigations toward the inhibition of cancer progression. Herein we have summarized various aptamers that were successfully screened for targets in different types of tumors. Further development and commercialization of these aptamer drugs will require additional experiments in live models and clinical trials.
Aptamers are chemically synthesized analogues, with numerous benefits over conventional methodologies in drug development. Their distinctive characteristics have promoted their design strategies for numerous applications. Numerous strategies are offered, including in vitro diagnosis, drug delivery, biomarker development and screening, and molecular imaging. Regardless of the remarkable advantages, aptamer technology is still developing in the fields of detection and treatment of cancers.
Several of the existing flaws and limitations are being investigated for them to be resolved. To date, no therapeutic aptamers are available as medicines to treat cancers, though many of them have entered but failed at the level of clinical trials. To overcome the above limitations, the aptamers are being chemically modified further to lower their toxicity and prolong their half-life. In this regard, ApDCs serve as promising candidates for numerous clinical applications. To obtain appropriate tumor suppression activity, it is necessary to develop accurate, controlled, and optimized ApDCs. Further investigations must focus on improving the existing ApDC systems, the linkers that can store a large payload and cleave the drug molecules upon reaching the target sites. ApDCs have more promising benefits, as they possess high stability, simple conjugation, ease of customization, and high payload that make them suitable for applications in chemotherapy, photodynamic therapy, and photokinetic therapy and in developing fluorescent probes, drug delivery systems, and bioimaging.
Acknowledgments
The authors thank the Chancellor and Vice Chancellor of Vellore Institute of Technology for providing the opportunity to carry out this study. Further the authors wish to thank the management of this institute for providing seed money as a research grant.
Glossary
Abbreviations
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- PTK7
protein tyrosine kinase
- A
adenine
- T/U
thymine/uracil
- G
guanine
- C
cytosine
- ssRNA
single-stranded RNA
- ssDNA
single-stranded DNA
- DOX
doxorubicin
- CPT
camptothecin
- ApDC
aptamer–drug conjugate
- PTX
paclitaxel
- DBCO
dibenzocyclooctyne
- siRNA
small interfering RNA
- miRNA
microRNA
- NHS
N-hydroxysuccinimide
- EDC
1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide
- FRET
Förster resonance energy transfer
- PDL
programmed death ligand
- PD1
death protein 1
- ICB
immune checkpoint blockade
- TNBC
triple-negative breast cancer
- SNA
spherical nucleic acid
- PDGF
platelet-derived growth factor
Biographies

Swathi Venkatesan was awarded a Master’s degree in chemistry by Hindustan Institute of Technology & Science, Tamil Nadu, India, in 2020. She is currently, pursuing her Ph.D. at Vellore Institute of Technology, Chennai campus. under the guidance of Dr. Balamurali MM. Her research interests include biophysical chemistry, biosensors, and photophysical chemistry.

Kaushik Chanda obtained his M.Sc. in organic chemistry from Gauhati University, Assam, India, in 2001. He subsequently worked as a Senior Research Fellow in an ICAR-NATP-funded project at St Anthonys College, Shillong, India, from 2002 to 2005. In 2006, he moved to Taiwan to pursuing a Ph.D. in applied chemistry from National Chiao Tung University under the guidance of Prof. Chung Ming Sun on the topic of combinatorial chemistry. After finishing his Ph.D. in 2010, he moved to the Department of Chemistry, National Tsing Hua University, Taiwan, for an NSC-Postdoctoral Fellowship in facet-dependent organic catalysis with Prof. Michael H. Y. Huang. Now he is working as an Associate Professor in the Department of Chemistry, Vellore Institute of Technology, Vellore. He has received funding from CSIR-India, ICMR-India, and DST-SERB. He is a member of the Royal Society of Chemistry and American Chemical Society. His research interests include diversity-oriented green synthesis, computational chemistry, anticancer drug design, drug delivery, and nanocatalysis.

Musuvathi Motilal Balamurali was awarded a Master’s degree in chemistry by Madurai Kamaraj University, Tamil Nadu, India, in 1999. He pursued doctoral research in photophysical chemistry at the Indian Institute of Technology, Kanpur, under the supervision of Prof. Sneh Kumar Dogra. In 2005, he was awarded a CSIR research associate fellowship and continued his postdoctoral research in Prof. Raghavan Varadarajan’s group at the Indian Institute of Science Bangalore. In 2007, he moved to the University of British Columbia, Canada, and in 2009 he joined Prof. Debkumar Pain’s research group at New Jersey Medical School, USA, to continue his interdisciplinary research in biophysical chemistry and biochemistry. In 2011, he served as a research scientist in DST India, funded by the National Hub for Healthcare Instrumentation Development, Anna University, Chennai, India. Since 2013, he continues to serve Vellore Institute of Technology, India, as an Associate Professor in the Department of Chemistry. His research interests include protein engineering and biophysical chemistry.
The authors declare no competing financial interest.
References
- Meganck R.-M.; Baric R.-S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat. Med. 2021, 27, 401–410. 10.1038/s41591-021-01282-0. [DOI] [PubMed] [Google Scholar]
- a Gillespie M.-S.; Ward C.-M.; Davies C.-C. DNA.Repair and Therapeutic Strategies in Cancer Stem Cells. Cancers (Basel) 2023, 15, 1897. 10.3390/cancers15061897. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kandasamy G.; Karuppasamy Y.; Krishnan U.-M. Emerging Trends in Nano-Driven Immunotherapy for Treatment of Cancer. Vaccines (Basel) 2023, 11, 458. 10.3390/vaccines11020458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Prathima T.-S.; Choudhury B.; Ahmad M.-G.; Chanda K.; Balamurali M.-M Recent developments on other platinum metal complexes as target-specific anticancer therapeutics. Coord. Chem. Rev. 2023, 490, 215231 10.1016/j.ccr.2023.215231. [DOI] [Google Scholar]; b Yimit A.; Adebali O.; Sancar A.; Jiang Y. Differential damage and repair of DNA-adducts induced by anti-cancer drug cisplatin across mouse organs. Nat. Commun. 2019, 10, 309. 10.1038/s41467-019-08290-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Hu D.; Yang C.; Lok C.-N.; Xing F.; Lee P.-Y.; Fung Y. M. E.; Jiang H.; Che C.-M. An Antitumor Bis(N-Heterocyclic Carbene)Platinum(II) Complex That Engages Asparagine Synthetase as an Anticancer Target. Angew. Chem., Int. Ed. Engl. 2019, 58, 10914–10918. 10.1002/anie.201904131. [DOI] [PubMed] [Google Scholar]
- a Huang B. P.; Lin C. H.; Chen H. M.; Lin J. T.; Cheng Y. F.; Kao S. H. AMPK activation inhibits expression of proinflammatory mediators through downregulation of PI3K/p38 MAPK and NF-kappaB signaling in murine macrophages. DNA Cell Biol. 2015, 34, 133–141. 10.1089/dna.2014.2630. [DOI] [PubMed] [Google Scholar]; b Chen S. Y.; Lin C. H.; Lin J. T.; Cheng Y. F.; Chen H. M.; Kao S. H. Adenine causes cell cycle arrest and autophagy of chronic myelogenous leukemia K562 cells via AMP-activated protein kinase signalling. Oncol. Lett. 2017, 14, 5575–5580. 10.3892/ol.2017.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rashid H. O.; Yadav R. K.; Kim H. R.; Chae H. J. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015, 11, 1956–1977. 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Raudenska M.; Kratochvilova M.; Vicar T.; Gumulec J.; Balvan J.; Polanska H.; Pribyl J.; Masarik M. Cisplatin enhances cell stiffness and decreases invasiveness rate in prostate cancer cells by actin accumulation. Sci. Rep. 2022, 12, 1660. 10.1038/s41598-022-23540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Liang Z. D.; Long Y.; Chen H. H.; Savaraj N.; Kuo M. T. Regulation of the high-affinity copper transporter (hCtr1) expression by cisplatin and heavy metals. J. Biol. Inorg. Chem. 2014, 19, 17–27. 10.1007/s00775-013-1051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Balamurali M. M.; Chanda K. Light emitting probes – approaches for interdisciplinary applications. Chem. Soc. Rev. 2021, 50, 3706–3719. 10.1039/D0CS01444C. [DOI] [PubMed] [Google Scholar]; g Dasmahapatra U.; Kiran K.-C.; Das S.; Prathima T.-S.; Poornimaa M.; Emerson I.-A.; Balamurali M.-M.; Chanda K. In-silico molecular modelling, MM/GBSA binding free energy and molecular dynamics simulation study of novel pyrido fused imidazo[4,5-c]quinolines as potential anti-tumor agents. Front. Chem. 2022, 10, 991369 10.3389/fchem.2022.991369. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Panchangam R.-L.; Rao R.-N.; Balamurali M.-M; Hingamire T.-B.; Shanmugam D.; Manickam V.; Chanda K. Antitumor Effects of Ir(III)-2 H-Indazole Complexes for Triple Negative Breast Cancer. Inorg. Chem. 2021, 60, 17593–17607. 10.1021/acs.inorgchem.1c02193. [DOI] [PubMed] [Google Scholar]
- a He J.; Duan Q.; Ran c.; Fu T.; Liu Y.; Tan W. Recent progress of aptamer–drug conjugates in cancer therapy. Acta Pharm. Sin. B 2023, 13, 1358–1370. 10.1016/j.apsb.2023.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang X.; Zhang H.; Chen X. Drug resistance and combating drug resistance in cancer.. Cancer Drug Resist. 2019, 2, 141–160. 10.20517/cdr.2019.10. [DOI] [PMC free article] [PubMed] [Google Scholar]; c He J.; Duan Q.; Ran C.; Fu T.; Liu Y.; Tan W. Recent progress of aptamer–drug conjugates in cancer therapy. Acta Pharm. Sin. B 2023, 13, 1358–1370. 10.1016/j.apsb.2023.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fan R.; Tao X.; Zhai X.; Zhu Y.; Li Y.; Chen Y.; Dong D.; Yang S.; Lv L. Application of aptamer-drug delivery system in the therapy of breast cancer. Biomed. Pharmacotherap. 2023, 161, 114444 10.1016/j.biopha.2023.114444. [DOI] [PubMed] [Google Scholar]; b Lakhin A. V.; Tarantul V. Z.; Gening L. V. Aptamers: Problems, solutions and prospects. Acta Naturae 2013, 5, 34–43. 10.32607/20758251-2013-5-4-34-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kuai H.; Zhao Z.; Mo L.; Liu H.; Hu X.; Fu T.; Zhang X.; Tan W. Circular Bivalent Aptamers Enable in vivo Stability and Recognition. J. Am. Chem. Soc. 2017, 139, 9128–9131. 10.1021/jacs.7b04547. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Amero P.; Lokesh G. L. R.; Chaudhari R. R.; Cardenas-Zuniga R.; Schubert T.; Attia Y. M.; Montalvo-Gonzalez E.; Elsayed A. M.; Ivan C.; Wang Z.; Cristini V.; Franciscis V. d.; Zhang S.; Volk D. E.; Mitra R.; Rodriguez-Aguayo C.; Sood A. K.; Lopez-Berestein G. Conversion of RNA Aptamer into Modified DNA Aptamers Provides for Prolonged Stability and Enhanced Antitumor Activity. J. Am. Chem. Soc. 2021, 143, 7655–7670. 10.1021/jacs.9b10460. [DOI] [PubMed] [Google Scholar]; c Hausmann J.; Kamtekar S.; Christodoulou E.; Day J. E.; Wu T.; Fulkerson Z.; Albers H. M.; van Meeteren L. A.; Houben A. J.; van Zeijl L.; Jansen S.; Andries M.; Hall T.; Pegg L. E.; Benson T. E.; Kasiem M.; Harlos K.; Kooi C. W. V.; Smyth S. S.; Ovaa H.; Bollen M.; Morris A. J.; Moolenaar W. H.; Perrakis A. Structural basis of substrate discrimination and integrin binding by autotaxin. Nat. Struct. Mol. Biol. 2011, 18, 198–204. 10.1038/nsmb.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Kawaguchi M.; Okabe T.; Okudaira S.; Nishimasu H.; Ishitani R.; Kojima H.; Nureki O.; Aoki J.; Nagano T. Screening and X-ray crystal structure-based optimization of autotaxin (ENPP2) inhibitors, using a newly developed fluorescence probe. ACS Chem. Biol. 2013, 8, 1713–1721. 10.1021/cb400150c. [DOI] [PubMed] [Google Scholar]
- a Bakshi S.; Sanz Garcia R.; Van der Weken H.; Tharad A.; Pandey S.; Juarez P.; Virdi V.; Devriendt B.; Cox E.; Depicker A. Evaluating single-domain antibodies as carriers for targeted vaccine delivery to the small intestinal epithelium. J. Controlled Release 2020, 321, 416–429. 10.1016/j.jconrel.2020.01.033. [DOI] [PubMed] [Google Scholar]; b Liu X.; Liu C.; Zheng X.; Chen S.; Pang X.; Xiang X.; Tang J.; Ren R.; Chen Y.; You M.; Wang X.; Chen X.; Luo W.; Liu G.; Xia N. Vesicular Antibodies: A Bioactive Multifunctional Combination Platform for Targeted Therapeutic Delivery and Cancer Immunotherapy. Adv. Mater. 2019, 31, 1808294 10.1002/adma.201808294. [DOI] [PubMed] [Google Scholar]
- a Li L.; Xu S.; Yan H.; Li X.; Yazd H. S.; Li X.; Huang T.; Cui C.; Jiang J.; Tan W. Nucleic Acid Aptamers for Molecular Diagnostics and Therapeutics: Advances and Perspectives. Angew. Chem. 2021, 60, 2221–2231. 10.1002/anie.202003563. [DOI] [PubMed] [Google Scholar]; b Fu Z.; Xiang J. Aptamer-Functionalized Nanoparticles in Targeted Delivery and Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9123. 10.3390/ijms21239123. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Liu M.; Wang L.; Lo Y.; Shiu S. C.; Kinghorn A. B.; Tanner J. A. Aptamer-Enabled Nanomaterials for Therapeutics, Drug Targeting and Imaging. Cells 2022, 11, 159. 10.3390/cells11010159. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ni S.; Yao H.; Wang L.; Lu J.; Jiang F.; Lu A.; Zhang G. Chemical Modifications of Nucleic Acid Aptamers for Therapeutic Purposes. Int. J. Mol. Sci. 2017, 18, 1683. 10.3390/ijms18081683. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Kim M.; Kim D.-M.; Kim K.-S.; Jung W.; Kim D.-U. Applications of Cancer Cell-Specific Aptamers in Targeted Delivery of Anticancer Therapeutic Agents. Molecules 2018, 23, 830. 10.3390/molecules23040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Odeh F.; Nsairat H.; Alshaer W.; Ismail M.-A.; Esawi E.; Qaqish B.; Bawab A. A.; Ismail S.-I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2020, 25, 3. 10.3390/molecules25010003. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Presolski S.-I.; Hong V.-P.; Finn M.-G. Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Curr. Protoc. Chem. Biol. 2011, 3, 153–162. 10.1002/9780470559277.ch110148. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Stephanopoulos N.; Tong G.-J.; Hsiao S.-C.; Francis M.-B. Dual-surface modified virus capsids for targeted delivery of photodynamic agents to cancer cells. ACS Nano 2010, 4, 6014–6020. 10.1021/nn1014769. [DOI] [PubMed] [Google Scholar]; d Ninomiya K.; Yamashita T.; Kawabata S.; Shimizu N. Targeted and ultrasound-triggered drug delivery using liposomes co-modified with cancer cell-targeting aptamers and a thermosensitive polymer. Ultrason. Sonochem. 2014, 21, 1482–1488. 10.1016/j.ultsonch.2013.12.023. [DOI] [PubMed] [Google Scholar]; e Danesh N. M.; Lavaee P.; Ramezani M.; Abnous K.; Taghdisi S. M. Targeted and controlled release delivery of daunorubicin to T-cell acute lymphoblastic leukemia by aptamer-modified gold nanoparticles. Int. J. Pharm. 2015, 489, 311–317. 10.1016/j.ijpharm.2015.04.072. [DOI] [PubMed] [Google Scholar]
- a Keefe A. D.; Pai S.; Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discovery 2010, 9, 537–550. 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhang N.; Chen Z.; Liu D.; Jiang H.; Zhang Z. K.; Lu A.; Zhang B. T.; Yu Y.; Zhang G. Structural Biology for the Molecular Insight between Aptamers and Target Proteins. Int. J. Mol. Sci. 2021, 22, 4093. 10.3390/ijms22084093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Bruno J. G. Potential Inherent Stimulation of the Innate Immune System by Nucleic Acid Aptamers and Possible Corrective Approaches. Pharmaceuticals (Basel) 2018, 11, 62. 10.3390/ph11030062. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Camorani S.; d’Argenio A.; Agnello L.; Nilo R.; Zannetti A.; Ibarra L. E.; Fedele M.; Cerchia L. Optimization of Short RNA Aptamers for TNBC Cell Targeting. Int. J. Mol. Sci. 2022, 23, 3511. 10.3390/ijms23073511. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Endoh T.; Ohyama T.; Sugimoto N. RNA-Capturing Microsphere Particles (R-CAMPs) for Optimization of Functional Aptamers. Small 2019, 15, e1805062 10.1002/smll.201805062. [DOI] [PubMed] [Google Scholar]; d Espah Borujeni A.; Mishler D. M.; Wang J.; Huso W.; Salis H. M. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2016, 44, 1–13. 10.1093/nar/gkv1289. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Wang G.; Wang Y.; Chen L.; Choo J. Nanomaterial-assisted aptamers for optical sensing. Biosens. Bioelectron. 2010, 25, 1859–68. 10.1016/j.bios.2009.11.012. [DOI] [PubMed] [Google Scholar]; f Roberts T. C.; Langer R.; Wood M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discovery 2020, 19, 673–694. 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Liu P.; Ga L.; Aodeng G.; Wang Y.; Ai J. Aptamer-drug conjugates: New probes for imaging and targeted therapy. Biosens. Bioelectron.: X 2022, 10, 100126 10.1016/j.biosx.2022.100126. [DOI] [Google Scholar]; h Kang M.-S.; Kong T. W. S.; Khoo J. Y. X.; Loh T.-P. Recent developments in chemical conjugation strategies targeting native amino acids in proteins and their applications in antibody-drug conjugates. Chem. Sci. 2021, 12, 13613–13647. 10.1039/D1SC02973H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Huang Y.-F.; Shangguan D.; Liu H.; Phillips J. A.; Zhang X.; Chen Y.; Tan W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem 2009, 10, 862–868. 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang R.; Zhu G.; Mei L.; Xie Y.; Ma H.; Ye M.; Qing F.-L.; Tan W. Automated modular synthesis of aptamer-drug conjugates for targeted drug delivery. J. Am. Chem. Soc. 2014, 136, 2731–2734. 10.1021/ja4117395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Zhou Z.; Liu S.; Zhang Y.; Yang X.; Ma Y.; Guan Z.; Wu Y.; Zhang L.; Yang Z. Reductive nanocomplex encapsulation of cRGD-siRNA conjugates for enhanced targeting to cancer cells. Int. J. Nanomed. 2017, 12, 7255–7272. 10.2147/IJN.S136726. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yang Q.; Deng Z.; Wang D.; He J.; Zhang D.; Tan Y.; Peng T.; Wang X. Q.; Tan W. Conjugating Aptamer and Mitomycin C with Reductant-Responsive Linker Leading to Synergistically Enhanced Anticancer Effect. J. Am. Chem. Soc. 2020, 142, 2532–2540. 10.1021/jacs.9b12409. [DOI] [PubMed] [Google Scholar]; c Li Y.; Peng Y.; Tan Y.; Xuan W.; Fu T.; Wang X. Q.; Tan W. A new paradigm for artesunate anticancer function: considerably enhancing the cytotoxicity via conjugating artesunate with aptamer. Signal Transduct. Target Ther. 2021, 6, 327. 10.1038/s41392-021-00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Li F.; Lu J.; Liu J.; Liang C.; Wang M.; Wang L.; Li D.; Yao H.; Zhang Q.; Wen J.; Zhang Z. K.; Li J.; Lv Q.; He X.; Guo B.; Guan D.; Yu Y.; Dang L.; Wu X.; Li Y.; Chen G.; Jiang F.; Sun S.; Zhang B. T.; Lu A.; Zhang G. A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat. Commun. 2017, 8, 1390. 10.1038/s41467-017-01565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusuluri A.; Krishnan V.; Lensch V.; Sarode A.; Bunyan W.; Vogus D. R.; Menegatti S.; Soh H. T.; Mitragotri S. Treating Tumors at Low Drug Doses Using an Aptamer-Peptide Synergistic Drug Conjugate. Angew. Chem., Int. Ed. Engl. 2019, 58, 1437–1441. 10.1002/anie.201812650. [DOI] [PubMed] [Google Scholar]
- Zhu G.; Zheng J.; Song E.; Tan W.; et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics.. Proc. Natl. Acad. Sci. U S A 2013, 110, 7998–8003. 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wen J.; Tao W.; Hao S.; Iyer S. P.; Zu Y. A unique aptamer-drug conjugate for targeted therapy of multiple myeloma. Leukemia 2016, 30, 987–91. 10.1038/leu.2015.216. [DOI] [PubMed] [Google Scholar]; b Zeng Z.; Qi J.; Wan Q.; Zu Y. Aptamers with Self-Loading Drug Payload and pH-Controlled Drug Release for Targeted Chemotherapy. Pharmaceutics 2021, 13, 1221. 10.3390/pharmaceutics13081221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S.; Sun W.; Fu T.; Liu X.; Chen P.; Qiu L.; Qu F.; Tan W. Aptamer-Based Targeted Delivery of Functional Nucleic Acids. J. Am. Chem. Soc. 2023, 145, 7677–7691. 10.1021/jacs.3c00841. [DOI] [PubMed] [Google Scholar]
- a Sargazi S.; Arshad R.; Ghamari R.; Rahdar A.; Bakhshi A.; Karkan S. F.; Ajalli N.; Bilal M.; Diez-Pascual A. M. siRNA-based nanotherapeutics as emerging modalities for immune-mediated diseases: A preliminary review. Cell Biol. Int. 2022, 46, 1320–1344. 10.1002/cbin.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Chen C.; Jing N.; Wang Z.; Zhang Y.; Chen W.; Tang X. Multimerized self-assembled caged two-in-one siRNA nanoparticles for photomodulation of RNAi induced gene silencing. Chem. Sci. 2020, 11, 12289–12297. 10.1039/D0SC03562A. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Huang F.; Lin M.; Duan R.; Lou X.; Xia F.; Willner I. Photoactivated Specific mRNA Detection in Single Living Cells by Coupling ″Signal-on″ Fluorescence and ″Signal-off″ Electrochemical Signals. Nano Lett. 2018, 18, 5116–5123. 10.1021/acs.nanolett.8b02004. [DOI] [PubMed] [Google Scholar]
- a Esposito C.-L.; Catuogno S.; de Franciscis V. Aptamer-mediated selective delivery of short RNA therapeutics in cancer cells. J. RNAi Gene Silencing 2014, 10, 500–506. [PMC free article] [PubMed] [Google Scholar]; b Garrido G.; Schrand B.; Rabasa A.; Levay A.; D’Eramo F.; Berezhnoy A.; Modi S.; Gefen T.; Marijt K.; Doorduijn E.; Dudeja V.; van Hall T.; Gilboa E. Tumor-targeted silencing of the peptide transporter TAP induces potent antitumor immunity. Nat. Commun. 2019, 10, 3773. 10.1038/s41467-019-11728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Shaban S. M.; Kim D.-H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. 10.3390/s21030979. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhao R.; Zhao L.; Feng H.; Chen X.; Zhang H.; Bai Y.; Feng F.; Shuang S. A label-free fluorescent aptasensor based on HCR and G-quadruplex DNAzymes for the detection of prostate-specific antigen. Analyst 2021, 146, 1340–1345. 10.1039/D0AN02188A. [DOI] [PubMed] [Google Scholar]; c Ma C.; Liu H.; Tian T.; Song X.; Yu J.; Yan M. A. A simple and rapid detection assay for peptides based on the specific recognition of aptamer and signal amplification of hybridization chain reaction. Biosens Bioelectron. 2016, 83, 15–18. 10.1016/j.bios.2016.04.030. [DOI] [PubMed] [Google Scholar]; d Zhao X.; Dai X.; Zhao S.; Cui X.; Gong T.; Song Z.; Meng H.; Zhang X.; Yu B. Aptamer-based fluorescent sensors for the detection of cancer biomarkers. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 247, 119038 10.1016/j.saa.2020.119038. [DOI] [PubMed] [Google Scholar]
- a Choi E. J.; Drago N. P.; Humphrey N. J.; Van Houten J.; Ahn J.; Lee J.; Kim I.-D.; Ogata A. F.; Penner R. M. Electrodeposition-enabled, electrically-transduced sensors and biosensors. Mater. Today 2023, 62, 129–150. 10.1016/j.mattod.2022.11.021. [DOI] [Google Scholar]; b Odeh F.; Nsairat H.; Alshaer W.; Ismail M. A.; Esawi E.; Qaqish B.; Bawab A. A.; Ismail S. I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2020, 25, 3. 10.3390/molecules25010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Neupane D.; Stine K. J. Electrochemical Sandwich Assays for Biomarkers Incorporating Aptamers, Antibodies and Nanomaterials for Detection of Specific Protein Biomarkers. Appl. Sci. 2021, 11, 7087 10.3390/app11157087. [DOI] [Google Scholar]; b Hashkavayi A. B.; Raoof J. B.; Ojani R.; Kavoosian S. Ultrasensitive electrochemical aptasensor based on sandwich architecture for selective label-free detection of colorectal cancer (CT26) cells. Biosens. Bioelectron. 2017, 92, 630–637. 10.1016/j.bios.2016.10.042. [DOI] [PubMed] [Google Scholar]
- Napolitano E.; Riccardi C.; Gaglione R.; Arciello A.; Pirota V.; Triveri A.; Doria F.; Musumeci D.; Montesarchio D. Selective light-up of dimeric G-quadruplex forming aptamers for efficient VEGF(165) detection. Int. J. Biol. Macromol. 2023, 224, 344–357. 10.1016/j.ijbiomac.2022.10.128. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Gao P.; Pan W.; Shi M.; Liu S.; Li N.; Tang B. Polyvalent spherical aptamer engineered macrophages: X-ray-actuated phenotypic transformation for tumor immunotherapy. Chem. Sci. 2021, 12, 13817–13824. 10.1039/D1SC03997K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota K.; Dhakal S. FRET-Based Aptasensor for the Selective and Sensitive Detection of Lysozyme. Sensors (Basel) 2020, 20, 914. 10.3390/s20030914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X.; Yao F.; An Y.; Li X.; Yang X.-d. Novel Nanotherapeutics for Cancer Immunotherapy by PD-L1-Aptamer-Functionalized and Fexofenadine-Loaded Albumin Nanoparticles. Molecules 2023, 28, 2556. 10.3390/molecules28062556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Gao P.; Pan W.; Shi M.; Liu S.; Li N.; Tang B. Polyvalent spherical aptamer engineered macrophages: X-ray-actuated phenotypic transformation for tumor immunotherapy. Chem. Sci. 2021, 12, 13817–13824. 10.1039/D1SC03997K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Lai X.; Yao F.; An Y.; Li X.; Yang X.-D. Novel Nanotherapeutics for Cancer Immunotherapy by PD-L1-Aptamer-Functionalized and Fexofenadine-Loaded Albumin Nanoparticles. Molecules 2023, 28, 2556. 10.3390/molecules28062556. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Benenson Y.; Gil B.; Ben-Dor U.; Adar R.; Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature 2004, 429, 423–429. 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- García Melián M. F.; Moreno M.; Cerecetto H.; Calzada V. Aptamer-Based Immunotheranostic Strategies. Cancer Biother. Radiopharm. 2023, 38, 246–255. 10.1089/cbr.2022.0064. [DOI] [PubMed] [Google Scholar]
- a Agnello L.; d’Argenio A.; Nilo R.; Fedele M.; Camorani S.; Cerchia L. Aptamer-Based Strategies to Boost Immunotherapy in TNBC. Cancers (Basel) 2023, 15, 2010. 10.3390/cancers15072010. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhang J.; Li W.; Qi Y.; Wang G.; Li L.; Jin Z.; Tian J.; Du Y. PD-L1 Aptamer-Functionalized Metal-Organic Framework Nanoparticles for Robust Photo-Immunotherapy against Cancer with Enhanced Safety. Angew. Chem., Int. Ed. Engl. 2023, 62, e202214750 10.1002/anie.202214750. [DOI] [PubMed] [Google Scholar]
- a Sun H.; Zu Y. Aptamers and their applications in nanomedicine. Small 2015, 11, 2352–2364. 10.1002/smll.201403073. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Li X.; Li X.; Cheng X.; Bian X.; Shen B.; Ding X.; Ding S. Single-Step and Highly Sensitive Imaging of Exosomal PD-L1 through Aptamer-Activated Cascade Primer Exchange Reaction-Generated Branched DNA Nanostructures. ACS Sens. 2022, 7, 3571–3579. 10.1021/acssensors.2c01614. [DOI] [PubMed] [Google Scholar]