Abstract
Two sorbitol-fermenting (SF) Shiga toxin-producing Escherichia coli (STEC) O157:H− strains were isolated from patients with hemolytic-uremic syndrome in the Czech Republic in 1995. Their phenotypic and genotypic characteristics and genomic DNA fingerprints were identical or closely related to those of SF STEC O157:H− strains isolated in Germany in 1988 to 1997. This indicates that the Czech isolates belong to the SF STEC O157 clone which is widespread in Germany. It is the first finding of the clone outside Germany.
Shiga toxin (verocytotoxin)-producing Escherichia coli (STEC) strains of serotype O157:H− (nonmotile) which ferment sorbitol and exhibit β-glucuronidase activity were first recognized in a 1988 outbreak of hemolytic-uremic syndrome (HUS) in Bavaria, Germany (9). Since then, they have been identified as a significant cause of HUS and diarrhea in Germany (6). Based on their phenotypic and genotypic features (1, 6, 12) and closely related pulsed-field gel electrophoresis patterns (10, 12), sorbitol-fermenting (SF) STEC O157:H− strains represent a new clone within E. coli serogroup O157 which shares pathogenic characteristics with non-sorbitol-fermenting (NSF) STEC O157:H7 (10, 12). Here we report the isolation of two SF STEC O157:H− strains in the Czech Republic. The objective of the study was to compare phenotypic and genotypic characteristics and to determine genetic relatedness of the Czech and German SF STEC O157:H− strains to find out whether the Czech isolates belong to the clone which is widespread in Germany.
(This work was presented in part at the 3rd International Symposium and Workshop on Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infections, Baltimore, Md., June 22 to 26, 1997 [abstr. V9/I].)
The Czech SF STEC O157:H− strains were isolated in August and October 1995 from two epidemiologically unrelated patients, aged 17 and 19 months, who were admitted to the University Hospital Motol, Prague, Czech Republic, for HUS preceded by bloody diarrhea. Although no NSF colonies were found in the patients’ stool cultures on sorbitol MacConkey agar (SMAC), both patients had evidence of E. coli O157 infection. This was based on the presence of E. coli O157 antigen in their stools as detected by the E. coli O157 Antigen Detection enzyme-linked immunosorbent assay kit (LMD Laboratories, Carlsbad, Calif.) (15) and on significantly elevated titers of anti-O157 lipopolysaccharide antibodies in their sera (1:10,240 and 1:20,480) as detected by the indirect hemagglutination assay (3, 4). Slide agglutination of SF colonies with anti-O157 antiserum (ITEST, Hradec Králové, Czech Republic) and subsequent biochemical identification of such colonies revealed SF E. coli O157 strains in stool cultures of both patients. Serotyping by standard procedures (5) identified serotype O157:H−. The vehicle of infection was not determined for either patient.
Both E. coli O157:H− isolates were tested for fermentation of d-sorbitol and β-d-glucuronidase activity by tube tests (1), assayed for Shiga toxin 1 (Stx1), Stx2, and Stx2c production by the Vero cell neutralization tests (8, 13), and examined for enterohemorrhagic E. coli hemolysin (EHEC Hly) on enterohemolysin agar (2). Phage patterns were determined (14) and compared with those of German SF STEC O157 strains. The presence of stx1, stx2, stx2c, eaeA, and EHEC hly genes was tested for by PCR procedures (10, 16, 17). Clonal relatedness of the isolates with German SF STEC O157 strains was determined by genomic DNA fingerprinting performed by randomly amplified polymorphic DNA PCR (RAPD PCR) with primer 1247 (7). The RAPD PCR profiles were visualized under UV light and photographed. A digital image of the gel was used to further analyze the profiles by the GelCompar software package (Applied Maths, Kortrijk, Belgium). Calculation of the similarity matrix was done by the Pearson product-moment correlation coefficient method (18). Hierarchic clustering was achieved by using the unweighted-pair-group method with the arithmetic averages clustering algorithm (18).
As shown in Table 1, the Czech SF STEC O157:H− isolates had identical phenotypic and genotypic characteristics which were at the same time identical with those of 24 German SF STEC O157:H− strains isolated in 1988 to 1996. The only exception was the absence of the stx2 gene in isolate 230/95, which lost Stx2 production within 1 month after isolation, before it was genotyped. A new phage type designation (phage type 88) was assigned to the Czech and German SF STEC O157:H− isolates which shared a phage pattern that did not correspond to any of the previously recognized E. coli O157 phage types (14).
TABLE 1.
Phenotypic and genotypic characteristics of Czech SF STEC O157:H− isolates compared with those of German SF STEC O157:H− strains
| SF STEC O157:H− isolatea | Sorbitol fermentation/β-glucuronidase activityb | Result forc:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stx phenotype
|
Stx genotype
|
EHEC Hly production | EHEC hly gene | eaeA gene | ||||||
| Stx1 | Stx2 | Stx2c | stx1 | stx2 | stx2c | |||||
| Czech | ||||||||||
| 221/95 | +/+ | − | + | − | − | + | − | − | + | + |
| 230/95 | +/+ | − | +d | − | − | −d | − | − | + | + |
| Germane | +/+ | − | + | − | − | + | − | − | + | + |
All isolates were phage type 88, a new phage type in E. coli serogroup O157.
+, positive after 24 h.
+, positive; −, negative.
The isolate lost Stx2 production before genotyping.
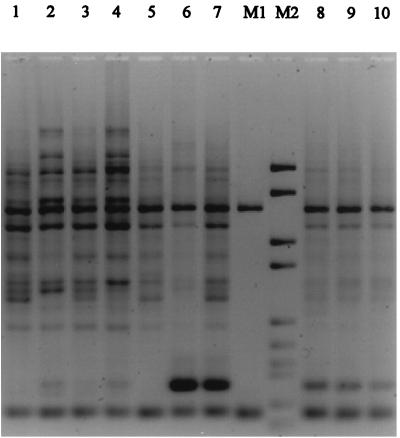
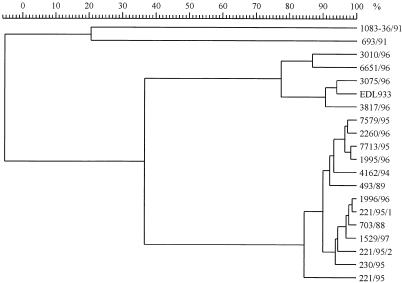
RAPD PCR fingerprinting of genomic DNA (for a list and characteristics of the strains tested, see Table 2) showed that the Czech and German SF STEC O157:H− isolates had identical or closely related profiles that markedly differed from those of NSF STEC O157:H7/H− (Fig. 1). Analysis of the RAPD PCR profiles by the Pearson product-moment correlation method and by the unweighted-pair-group method with arithmetic averages clustering clearly distinguished three clusters of strains (Fig. 2). Nine SF STEC O157:H− strains from Germany and the Czech SF STEC O157:H− isolates gave a cluster with a nearly congruent pattern, thus showing high relatedness. The second cluster contained all NSF STEC O157:H7/H− strains; they were also closely related to each other but could be clearly distinguished from the SF STEC O157:H− strains. The third group consisted of two Stx-negative strains of serotypes O157:H19 and O157:H45; these strains were not related to either NSF or SF STEC O157:H7/H− (Fig. 2). Taken together with the other phenotypic and genotypic results, it can be concluded that the Czech SF STEC O157:H− strains belong to the clone which is resident in Germany.
TABLE 2.
List and characteristics of SF and NSF E. coli O157 strains analyzed by RAPD PCR fingerprinting
| Strain | Serotype | Sorbitol fermen- tationa | Stx phenotype | Disease; originb (referencec) |
|---|---|---|---|---|
| 1083-36/91 | O157:H45 | + | Stx negative | ID; G |
| 693/91 | O157:H19 | + | Stx negative | ID; G |
| 3010/96 | O157:H7 | − | Stx2 | D; G |
| 6651/96 | O157:H7 | − | Stx2 | HUS; G |
| 3075/96 | O157:H7 | − | Stx2c | D; G |
| EDL933 | O157:H7 | − | Stx1+Stx2 | CDC (19) |
| 3817/96 | O157:H− | − | Stx1+Stx2 | HUS; G |
| 7579/95 | O157:H− | + | Stx2 | HUS; G |
| 2260/96 | O157:H− | + | Stx2 | HUS; G |
| 7713/95 | O157:H− | + | Stx2 | HUS; G |
| 1995/96 | O157:H− | + | Stx2 | HUS; G |
| 4162/94 | O157:H− | + | Stx2 | D; G |
| 493/89 | O157:H− | + | Stx2 | HUS; G (10) |
| 1996/96 | O157:H− | + | Stx2 | D; G |
| 221/95/1d | O157:H− | + | Stx2 | HUS; CR |
| 703/88 | O157:H− | + | Stx2 | HUS; G (1) |
| 1529/97 | O157:H− | + | Stx2 | HUS; G |
| 221/95/2d | O157:H− | + | Stx2 | HUS; CR |
| 230/95 | O157:H− | + | —e | HUS; CR |
| 221/95d | O157:H− | + | Stx2 | HUS; CR |
+, positive after 24 h; −, negative after 24 h.
ID, infantile diarrhea; D, diarrhea; CDC, Centers for Disease Control and Prevention, Atlanta, Ga.; CR, Czech Republic; G, Germany (nine German SF STEC O157:H− strains are representative isolates from 1988 to 1997).
Strains for which no references are given are from this study.
Sequential isolates from the same patient obtained on days 1 (221/95), 2 (221/95/1), and 4 (221/95/2) after admission.
The isolate lost Stx2 production before RAPD PCR fingerprinting.
FIG. 1.
Agarose gel showing RAPD PCR fingerprints of representative SF and NSF STEC O157 strains obtained with primer 1247. NSF E. coli O157:H7/H− strains 3075/96 (lane 1), 3010/96 (lane 2), EDL933 (lane 3), 6651/96 (lane 4), and 3817/96 (lane 5) are depicted. The SF E. coli O157:H− strains were Czech isolates 221/95 (lane 6) and 230/95 (lane 7) and German isolates 1529/97 (lane 8), 7713/95 (lane 9), and 1995/96 (lane 10). The molecular weight marker (M2) was DNA marker VI (Boehringer Gmbh); the molecular sizes of the fragments are (in base pairs) 2,176, 1,766, 1,230, 1,033, 653, 517, 453, 394, 298, 234, and 220. The internal standard (M1) consisted of a 1,600-bp and a 244-bp PCR product. In addition, internal standards were included in each lane.
FIG. 2.
Dendrogram derived from RAPD PCR data for Czech and German SF STEC O157:H− strains, NSF STEC O157:H7/H− strains, and SF Stx-negative E. coli O157:H19/H45 strains with the GelCompar software package. The characteristics of the strains are given in Table 2. The similarity scale is shown above the dendrogram (a similarity index of ≥80% indicates clonal relatedness).
This is the first report that SF STEC O157:H− strains belonging to the German clone can be a cause of HUS outside Germany. Although the vehicle of infection was not identified, the fact that none of the Czech patients had histories of travelling in Germany or consumption of foods imported from Germany makes domestic origin of infection very likely. Our findings thus suggest that the SF STEC O157 clone has begun to spread from Germany and that these strains can emerge as a public health problem in other countries. This has important diagnostic implications, emphasizing the need for diagnostic procedures which allow detection of infection with both NSF and SF E. coli O157 strains. In our study, combination of stool culture on SMAC with E. coli O157 stool enzyme-linked immunosorbent assay and anti-O157 serology enabled us to detect E. coli O157 infection despite the absence of NSF colonies on SMAC, thus aiming our diagnostic efforts towards searching for SF E. coli O157 strains. The procedures which have been successfully used to detect SF STEC O157 strains in German studies have included genetic methods (6, 9, 11) and the technique of immunomagnetic separation followed by plating magnetic particles with attached O157 bacteria on SMAC (11). Here, it should be remembered that SF STEC O157 strains, in contrast to NSF STEC O157:H7, do not grow on a selective cefixime-tellurite SMAC (11), since they do not tolerate high tellurite concentrations (12). Although the SF STEC O157:H− strains possess EHEC hly genes, no enterohemolytic phenotype could be observed (Table 1). This finding has consequences for the detection of such STEC in stool samples. While enterohemolysin agar plates have been successfully used for detecting EHEC Hly-producing NSF STEC O157:H7 (2), this method fails to detect nonhemolytic SF STEC O157:H− as characterized in this study. Consistent use of appropriate diagnostic methods for clinical and epidemiological studies is necessary to further evaluate significance of SF STEC O157 strains in human disease, to identify their reservoirs, and, based on that, to implement effective prevention of human disease.
Acknowledgments
This study was supported by grant IGA 2063-3 from the Ministry of Health of the Czech Republic and by grant BMH4-CT96-0970 from the European Communities.
We thank T. Cheasty, Central Public Health Laboratory, London, United Kingdom, for confirming the serotype, Stx genotype, and phage type of Czech E. coli O157:H− isolates. The excellent technical assistance of B. Plaschke and A. Reischelová is greatly appreciated.
REFERENCES
- 1.Aleksić S, Karch H, Bockemühl J. A biotyping scheme for Shiga-like (Vero) toxin-producing Escherichia coli O157 and a list of serological cross-reactions between O157 and other Gram-negative bacteria. Zentralbl Bakteriol. 1992;276:221–230. doi: 10.1016/s0934-8840(11)80009-5. [DOI] [PubMed] [Google Scholar]
- 2.Beutin L, Montenegro M A, Orskov I, Orskov F, Prada J, Zimmermann S, Stephan R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J Clin Microbiol. 1989;27:2559–2564. doi: 10.1128/jcm.27.11.2559-2564.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bielaszewska M, Janda J, Bláhová K, Feber J, Potuz̆ník V, Souc̆ková A. Verocytotoxin-producing Escherichia coli in children with hemolytic uremic syndrome in the Czech Republic. Clin Nephrol. 1996;46:42–44. [PubMed] [Google Scholar]
- 4.Bitzan M, Karch H. Indirect hemagglutination assay for diagnosis of Escherichia coli O157 infection in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1174–1178. doi: 10.1128/jcm.30.5.1174-1178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards P R, Ewing W H. Identification of Enterobacteriaceae. 3rd ed. Minneapolis, Minn: Burgess Publishing; 1972. pp. 67–101. [Google Scholar]
- 6.Gunzer F, Böhm H, Rüssmann H, Bitzan M, Aleksić S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuwelink A E, van de Kar N C A J, Meis J F G M, Monnens L A H, Melchers W J G. Characterization of verocytotoxin-producing Escherichia coli O157 isolates from patients with haemolytic uraemic syndrome in Western Europe. Epidemiol Infect. 1995;115:1–14. doi: 10.1017/s0950268800058064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hii J H, Gyles C, Morooka T, Karmali M A, Clarke R, DeGrandis S, Brunton J L. Development of verotoxin 2- and verotoxin 2 variant (VT2v)-specific oligonucleotide probes on the basis of the nucleotide sequence of the B cistron of VT2v from Escherichia coli E32511 and B2F1. J Clin Microbiol. 1991;29:2704–2709. doi: 10.1128/jcm.29.12.2704-2709.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karch H, Wiss R, Gloning H, Emmrich P, Aleksić S, Bockemühl J. Hämolytisch-urämisches Syndrom bei Kleinkindern durch Verotoxin-produzierende Escherichia coli. Dtsch Med Wochenschr. 1990;115:485–495. doi: 10.1055/s-2008-1065036. [DOI] [PubMed] [Google Scholar]
- 10.Karch H, Böhm H, Schmidt H, Gunzer F, Aleksic S, Heesemann J. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J Clin Microbiol. 1993;31:1200–1205. doi: 10.1128/jcm.31.5.1200-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karch H, Janetzki-Mittmann C, Aleksić S, Datz M. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods, and direct culture. J Clin Microbiol. 1996;34:516–519. doi: 10.1128/jcm.34.3.516-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karch H, Schmidt H, Swarzkopf A, Böhler O, Aleksić S, Rowe B. Molecular characterization and epidemiology of sorbitol-fermenting Escherichia coli O157:H−. Fortschr Med. 1997;115:48. . (Abstract.) [Google Scholar]
- 13.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 14.Khakhria R, Duck D, Lior H. Extended phage-typing scheme for Escherichia coli O157:H7. Epidemiol Infect. 1990;105:511–520. doi: 10.1017/s0950268800048135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park C H, Vandel N M, Hixon D L. Rapid immunoassay for detection of Escherichia coli O157 directly from stool specimens. J Clin Microbiol. 1996;34:988–990. doi: 10.1128/jcm.34.4.988-990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rüsmann H, Schmidt H, Heesemann J, Caprioli A, Karch H. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J Med Microbiol. 1994;40:338–343. doi: 10.1099/00222615-40-5-338. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sneath P H, Sokal R R. Numerical taxonomy. The principles and practice of numerical classification. San Francisco, Calif: W. H. Freeman and Co.; 1973. [Google Scholar]
- 19.Strockbine N A, Marques L R M, Newland J W, Smith H W, Holmes R K, O’Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biological activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]